This study characterizes the vanishing tassel2 (vt2) mutant of maize, which has reduced levels of auxin and severe defects in vegetative and reproductive development. It finds that vt2 encodes a co-ortholog of TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1, which functions in auxin biosynthesis, and provides evidence that vt2 and spi1, a YUCCA-like gene, may act in the same auxin biosynthetic pathway.
Abstract
Auxin plays a fundamental role in organogenesis in plants. Multiple pathways for auxin biosynthesis have been proposed, but none of the predicted pathways are completely understood. Here, we report the positional cloning and characterization of the vanishing tassel2 (vt2) gene of maize (Zea mays). Phylogenetic analyses indicate that vt2 is a co-ortholog of TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1), which converts Trp to indole-3-pyruvic acid in one of four hypothesized Trp-dependent auxin biosynthesis pathways. Unlike single mutations in TAA1, which cause subtle morphological phenotypes in Arabidopsis thaliana, vt2 mutants have dramatic effects on vegetative and reproductive development. vt2 mutants share many similarities with sparse inflorescence1 (spi1) mutants in maize. spi1 is proposed to encode an enzyme in the tryptamine pathway for Trp-dependent auxin biosynthesis, although this biochemical activity has recently been questioned. Surprisingly, spi1 vt2 double mutants had only a slightly more severe phenotype than vt2 single mutants. Furthermore, both spi1 and vt2 single mutants exhibited a reduction in free auxin levels, but the spi1 vt2 double mutants did not have a further reduction compared with vt2 single mutants. Therefore, both spi1 and vt2 function in auxin biosynthesis in maize, possibly in the same pathway rather than independently as previously proposed.
INTRODUCTION
Auxin has been shown to play a critical role in all stages of plant development. Through its functions in cell division and cell expansion, auxin is required for the initiation of lateral roots, vascular strands, leaves, flowers, and floral organs (Benjamins and Scheres, 2008). Evidence from genetics, molecular biology, and modeling has shown that auxin transport is crucial for providing the source of auxin required for organogenesis (Petrásek and Friml, 2009). More recently, the importance of auxin biosynthesis in providing a localized source of auxin for organogenesis has been appreciated (Chandler, 2009).
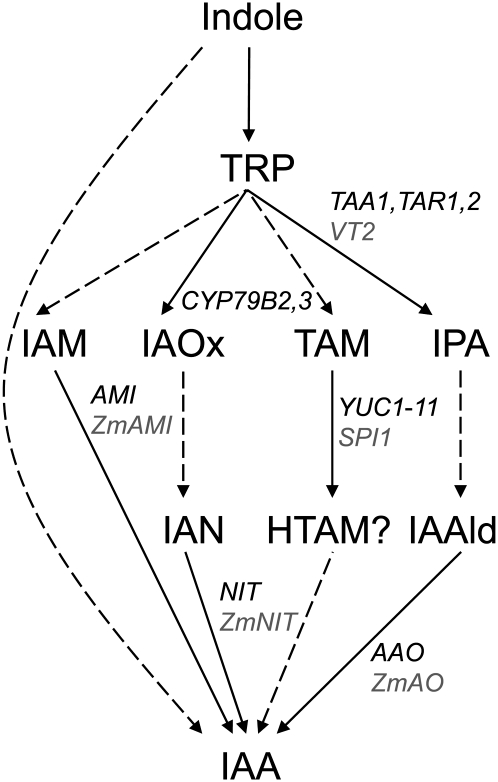
In plants, there are hypothesized to be four Trp-dependent and one Trp-independent pathway for the biosynthesis of auxin in its most common form, indole-3-acetic acid (IAA) (Figure 1) (Bartel, 1997; Woodward and Bartel, 2005; Pollmann et al., 2006; Kriechbaumer et al., 2008; Sugawara et al., 2009; McSteen, 2010). Each pathway is named after an intermediate that it is proposed to use, but few genes encoding enzymes in each pathway have been identified. Therefore, none of the proposed pathways have been fully elucidated with respect to all the intermediates formed nor is it clear how the pathways might intersect with each other. (1) The IAM pathway. The conversion of Trp to IAA through an indole-3-acetamide (IAM) intermediate has been demonstrated in Arabidopsis thaliana (Pollmann et al., 2009). Genes encoding enzymes that catalyze the conversion of Trp to IAM are unknown, but AMIDASE1 (AMI1), which converts IAM to IAA, has been identified in Arabidopsis (Pollmann et al., 2003). A maize (Zea mays) sequence with similarity to AMI1 has recently been reported, but function has not yet been demonstrated (Lehmann et al., 2010). (2) The IAOx pathway. Genes encoding the cytochrome P450 enzymes, CYP79B2/CYP79B3, which convert Trp to indole-3-acetaldoxime (IAOx) have been identified in Arabidopsis but are not present outside of the Brassicaceae (Zhao et al., 2002; Sugawara et al., 2009). IAOx is converted to indole-3-acetonitrile by unknown means, and indole-3-acetonitrile is converted to IAA by nitrilases. Genes encoding nitrilases have been identified from both maize and Arabidopsis (Bartling et al., 1992; Park et al., 2003; Kriechbaumer et al., 2007). However, the existence of this pathway in maize has been questioned due to the absence of genes co-orthologous to CYP79B2/3 genes and the lack of detectable IAOx levels (Sugawara et al., 2009). (3) The tryptamine (TAM) pathway. The enzymes converting Trp to TAM are not known, but the conversion of TAM to N-hydroxyl tryptamine is reported to be catalyzed in vitro by the YUCCA (YUC) genes of Arabidopsis, which play important roles in various aspects of development (Zhao et al., 2001; Cheng et al., 2006, 2007a). However, the biochemical function of YUC and, in particular, the role of N-hydroxyl tryptamine have been called into question recently, indicating that further in vivo research is needed (Tivendale et al., 2010; Nonhebel et al., 2011). In maize, the sparse inflorescence1 (spi1) gene, a monocot-specific member of the YUC gene family, shows that this pathway is also important for maize inflorescence development (Gallavotti et al., 2008b). Another member of the gene family (Zm-YUC1, which is more similar to At-YUC10 and At-YUC11) is specifically expressed in endosperm, illustrating the tissue-specific regulation of the gene family (LeClere et al., 2010). (4) The IPA pathway. Trp is converted to indole-3-pyruvic acid (IPA) by TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) and related proteins, TAR1 and TAR2 (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009). It is not known how IPA is converted to indole-3-acetaldehyde (IAAld), but the conversion of IAAld to IAA is catalyzed by aldehyde oxidases, which have been identified in both maize and Arabidopsis (Sekimoto et al., 1997, 1998). Here, we identify vanishing tassel2 (vt2), a maize co-ortholog of TAA1/TAR1/TAR2.
Figure 1.
Proposed Auxin Biosynthesis Pathways in Arabidopsis and Maize.
One Trp-independent and four Trp-dependent pathways have been proposed. Dotted lines indicate steps that are inferred. Solid lines indicate steps for which genes encoding enzymes catalyzing the reaction have been identified from either Arabidopsis (black) or maize (gray). IAN, indole-3-acetonitrile; HTAM, N-hydroxyl tryptamine. Adapted from Bartel (1997), Woodward and Bartel (2005), Kriechbaumer et al. (2006), and Sugawara et al. (2009).
taa1 mutants were identified in three different genetic screens in Arabidopsis, as indicated by the different phenotypes and nomenclature of respective mutants: insensitivity to ethylene-induced root shortening, weak ethylene insensitive8 (Stepanova et al., 2008); insensitivity to shade-induced hypocotyl elongation, shade avoidance3 (Tao et al., 2008); and insensitivity to naphthylphthalamic acid–induced root shortening, transport inhibitor response2 (Yamada et al., 2009). Unlike the mild phenotype of taa1 single mutants, double mutants with the paralogous gene tar2 show more severe defects, producing dwarf, bushy plants with agravitropic roots, reduced vasculature, and sterile flowers (Stepanova et al., 2008). taa1 tar1 tar2 triple mutants lack roots and are seedling lethal, similar to the phenotype of plants containing quadruple knockouts of the YUC gene family (Cheng et al., 2007a; Stepanova et al., 2008). Therefore, both the TAA1 and YUC gene families exhibit genetic redundancy and function in auxin biosynthesis, which raises the question of why neither pathway can compensate for the other.
Developmental defects are also seen in mutants with altered auxin transport. For example, mutations in the auxin efflux carrier PINFORMED1 (PIN1) and the PINOID (PID) kinase, which regulates PIN1 subcellular localization, produce an inflorescence with no flowers known as a pin inflorescence (Bennett et al., 1995; Gälweiler et al., 1998; Christensen et al., 2000; Friml et al., 2004; Kleine-Vehn et al., 2009; Zhang et al., 2010). Mutations in the PID co-ortholog in maize, barren inflorescence2 (bif2), result in an equivalent phenotype called a barren inflorescence (bif) phenotype: no branches and few spikelets (small branches that bear the florets) are produced in the male inflorescence (the tassel), and few kernels are produced in the female inflorescence (the ear) (McSteen and Hake, 2001). Fewer flowers are also seen in yuc1 yuc2 double mutants in Arabidopsis and spi1 single mutants in maize (Cheng et al., 2006; Gallavotti et al., 2008b), indicating that both auxin transport and auxin biosynthesis are required for the initiation of flowers. The importance of these two processes in development is further illustrated by the synergistic interactions observed between auxin biosynthesis and transport mutants. For example, yuc1 yuc4 pin1 triple mutants do not produce leaves, and spi1 bif2 double mutants have dramatically reduced leaf number, indicating that both auxin biosynthesis and transport are required for leaf initiation in addition to flower initiation (Cheng et al., 2007a; Gallavotti et al., 2008b).
Here, we report on the identification of the vt2 mutant of maize, which exhibits a severe barren inflorescence phenotype with no branches or spikelets, as well as a semidwarf vegetative phenotype due to the production of fewer leaves. Positional cloning and phylogenetic analysis indicate that vt2 encodes a co-ortholog of the TAA1/TAR1/TAR2 genes of Arabidopsis, which function in the IPA pathway for auxin biosynthesis (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009). The dramatic phenotype of vt2 loss-of-function mutants indicates that the IPA pathway plays a critical role in maize vegetative and reproductive development. Furthermore, due to the reduced redundancy of the vt2 and spi1 genes in maize, we were able to test the relative contributions of the IPA and TAM pathways to auxin biosynthesis.
RESULTS
vt2 Functions in Vegetative Development
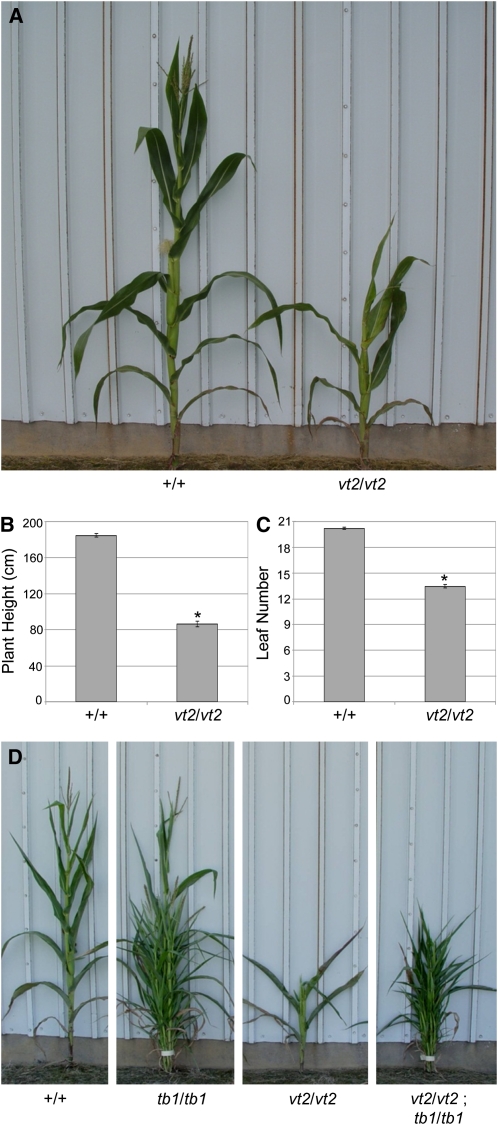
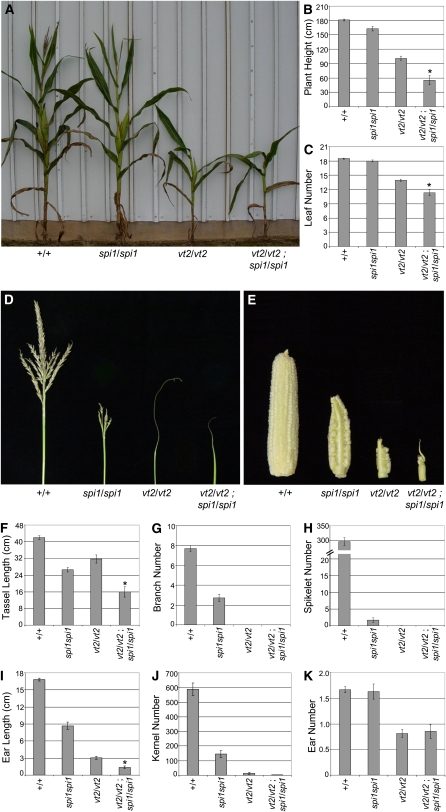
At maturity, vt2 mutants were visibly shorter than wild-type siblings (Figure 2A). As expected, quantification revealed a statistically significant reduction in plant height in vt2 mutants compared with the wild-type (Figure 2B). To determine if this decrease in plant height was caused by a reduction in the number of leaves produced, leaf number was counted. Wild-type maize plants produced an average of 20.2 (±0.1 se) leaves at maturity, whereas vt2 mutants produced ~13.5 (±0.18 se) leaves (Figure 2C). Therefore, the reduction in plant height in vt2 is due in part to a reduction in leaf number, although reduced tassel length also contributes to the reduced plant height (see below).
Figure 2.
Mature Vegetative Phenotype Analysis of vt2 Mutants.
(A) vt2 mutants are much shorter than their wild-type siblings.
(B) Quantification of plant height.
(C) Quantification of leaf number.
(D) Mature vegetative phenotypes of individuals from a segregating vt2 tb1 double mutant family show that vt2/vt2 tb1/tb1 double mutants produce many tillers like tb1/tb1 single mutants. Asterisk indicates significant reduction from normal siblings at P < 0.05; error bars represent the se; n = 37 vt2 and 113 normal.
[See online article for color version of this figure.]
The decrease in leaf number in vt2 mutants could be due to the production of fewer juvenile leaves or fewer adult leaves. To determine which leaves were missing in vt2 mutants, the juvenile-to-adult transition was analyzed through visual inspection of leaf waxes. Due to the production of epicuticular waxes, the surface of juvenile maize leaves appears dull while adult leaves appear glossy, and transitional leaves (with a glossy appearance at the tip and a matte appearance at the base and margins) are produced at the juvenile-to-adult transition (Kerstetter and Poethig, 1998). In wild-type siblings, the juvenile-to-adult transition began at leaf six when transitional leaves were produced, and the transition continued through leaf eight after which adult glossy leaves were produced (see Supplemental Table 1 online). vt2 mutants showed no substantial difference in the transition point from juvenile to transitional leaves or from transitional leaves to adult leaves (see Supplemental Table 1 online). As there is no difference in the timing of the juvenile-to-adult transition in vt2 mutants, this indicates that the later-formed adult leaves are those that are missing in vt2 mutants.
vt2 Functions in Inflorescence Development
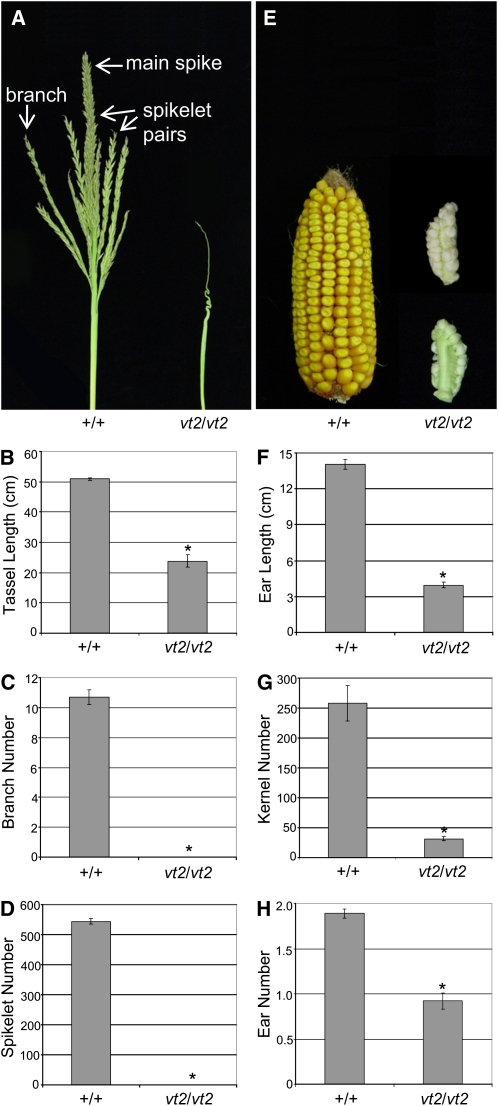
In maize, the tassel inflorescence normally produces a main spike with several long lateral branches extending near the base (Figure 3A) (McSteen et al., 2000). Short branches known as spikelet pairs house the florets and cover both the main spike and long branches (Figure 3A). vt2 mutant tassels were smaller at maturity and exhibited a severely barren phenotype compared with wild-type siblings, including a complete lack of lateral branches and functional spikelets (Figure 3A). Quantitative analysis of tassel length (Figure 3B), branch number (Figure 3C), and spikelet number (Figure 3D) confirmed a significant reduction in vt2 mutants compared with the wild type.
Figure 3.
Mature Inflorescence Phenotype Analysis of vt2 Mutants.
(A) Wild-type tassels normally produce multiple lateral branches. The branches and the main spike are covered in pairs of spikelets. vt2 mutant tassels produce no lateral branches or functional spikelets.
(B) Quantification of tassel length.
(C) Quantification of tassel branch number.
(D) Quantification of spikelet number in the tassel.
(E) Wild-type ears normally produce hundreds of kernels in regular rows from the base to the tip, whereas vt2 mutant ears are smaller in size, produce very few kernels, and typically have barren patches on one or both sides of the ear (both sides of the same vt2 ear are shown on the right).
(F) Quantification of ear length.
(G) Quantification of kernel number.
(H) Quantification of visible ear shoot number. Asterisk indicates significant reduction compared with normal siblings at P < 0.05; error bars represent the se; n = 10 vt2 and 10 normal.
[See online article for color version of this figure.]
Reduction in tassel length in other barren inflorescence mutants has been shown to be caused by reduced cell size (Barazesh et al., 2009). To test whether the reduction in tassel length in vt2 was caused by a reduction in cell size, the length of cells in the epidermis of the mature tassel was measured. vt2 mutants exhibited a 63% reduction in epidermal cell length compared with normal siblings (see Supplemental Table 2 online). Therefore, the reduced tassel length in vt2 mutants is likely due to reduced cell elongation.
In addition to male inflorescence defects, vt2 mutants also showed severe defects in the female inflorescence. vt2 mutant ears showed obvious defects in length and kernel number with a barren patch devoid of kernels often extending along the adaxial side of the ear (Figure 3E). Quantification of traits at maturity revealed a statistically significant reduction in both ear length (Figure 3F) and kernel number (Figure 3G) in vt2 mutants compared with the wild type. In addition, segregating families were scored to determine if the number of visible ear shoots produced by vt2 mutants was altered compared with wild-type siblings, and a statistically significant reduction in vt2 ear shoot number was detected (Figure 3H). These data show that vt2 ear inflorescences exhibit similar defects to those observed in tassel inflorescences and together indicate that vt2 plays an important role in inflorescence development. As all of the reproductive structures missing arise from axillary meristems (ear shoots, tassel branches, spikelets, and kernels), it appears that vt2 mutants are defective in axillary meristem formation similar to other maize mutants with defects in auxin transport or biosynthesis (McSteen and Hake, 2001; Barazesh and McSteen, 2008; Gallavotti et al., 2008b).
vt2 Functions in Axillary Meristem Formation during Inflorescence Development
To determine whether the absence of branches and spikelets in vt2 inflorescences was caused by altered axillary meristem formation, we observed tassel inflorescences at early stages of development using scanning electron microscopy. Early in development, wild-type tassels produce branch meristems (BMs) at the base of the inflorescence and spikelet pair meristems (SPMs) in regular rows on the flanks of both the branches and main spike (Figure 4A, arrows) (Cheng et al., 1983). By contrast, field-grown vt2 mutant tassels showed a complete lack of formation of both BMs and SPMs early in development (Figure 4B). Later in development, a few SPMs formed sporadically near the tip of the inflorescence (Figure 4C). Scanning electron microscopy was also performed on developing ear inflorescences, and similar defects in axillary meristem formation were observed (see Supplemental Figure 1 online).
Figure 4.
Scanning Electron Micrographs of Developing Inflorescences.
(A) Wild-type field-grown tassel at 3-mm stage exhibiting BMs at the base and SPMs covering the branches and main spike.
(B) vt2 field-grown tassel at 3-mm stage exhibiting complete lack of BM and SPM initiation.
(C) vt2 field-grown tassel at 4- to 5-mm stage producing a few SPMs near the tip.
(D) vt2 greenhouse-grown tassel at 4- to 5-mm stage displaying a weak mutant phenotype, with several BMs at the base and many SPM on the branches and main spike.
(E) Wild-type greenhouse-grown tassel later in development at 6- to 7-mm stage exhibiting production of paired SMs in regular rows on all branches and the main spike.
(F) vt2 greenhouse-grown tassel later in development at 6- to 7-mm stage showing production of paired SMs on the branches and paired or single SMs on the main spike.
(G) vt2 mutant tassel at 6- to 7-mm stage grown at cooler greenhouse temperatures compared with typical greenhouse conditions for maize. Mutants grown in these conditions display intermediate phenotypes between field-grown and typical greenhouse-grown tassels, with no BMs and few SPMs, which give rise to paired or single SM.
(H) Close-up of 4- to 5-mm vt2 tassel grown at cooler greenhouse temperatures, displaying two files of single SMs produced along the main spike.
(I) Close-up of 6- to 7-mm vt2 tassel (from [G]) grown at cooler greenhouse temperatures, displaying both paired and single SMs along the main spike.
PS, paired spikelet; SS, single spikelet. Bars = 250 μm.
Although field-grown vt2 mutant tassels never produced functional spikelets, we observed that temperature conditions during development greatly impacted the severity of the phenotype. In vt2 mutant tassels grown under typical warm maize greenhouse growing conditions, we observed a very weak phenotype with the production of several BMs and many irregularly placed SPMs (Figure 4D). To explore the role of temperature in vt2 inflorescence development further, we grew plants under normal warm and cooler greenhouse growing conditions until later in development. In wild-type greenhouse-grown plants, SPMs on the branches and main spike gave rise to two spikelet meristems (SMs) (Figures 4A and 4E). vt2 mutants grown in warm greenhouse conditions produced both single and paired SMs (Figure 4F), which is characteristic of barren inflorescence mutants in maize (McSteen and Hake, 2001; Barazesh and McSteen, 2008). However, vt2 mutants grown in cooler greenhouse conditions had a more severe phenotype in which BMs were typically not produced, and the SPMs that were produced often gave rise to single SMs (Figures 4G to 4I). Hence, in cooler greenhouse conditions, the vt2 mutant phenotype was more severe than that observed under warmer greenhouse conditions, although the most severe phenotypes were seen in field-grown plants that were exposed to very cold minimum nighttime temperatures (see Methods).
Therefore, vt2 mutants produce few branches and spikelets due to defects in BM, SPM, and SM formation. Furthermore, the phenotype is temperature dependent. In addition to vt2-ref, this temperature dependence was also observed in the vt2-RM123 and vt2-DB1845 alleles; hence, this phenomenon is not due to vt2-ref being a temperature-sensitive allele.
vt2 Does Not Function in Axillary Meristem Formation during Vegetative Development
As vt2 played an important role in axillary meristems during inflorescence development, we also tested the role of vt2 in axillary meristems during vegetative development by constructing double mutants with teosinte branched1 (tb1). The tb1 gene functions to suppress the outgrowth of branches (tillers) from vegetative axillary meristems located in the axil of each leaf node (Doebley et al., 1997; Hubbard et al., 2002). Loss of function of tb1 allows the outgrowth of vegetative axillary meristems, resulting in mutants that produce many tillers (Figure 2D). vt2 tb1 double mutants had an additive phenotype, producing short plants with many tillers topped by vt2 tassels (Figure 2D). Quantification of tiller number on the main stem indicated no statistical difference between the number of tillers in tb1 (9.65 ± 0.69) compared with vt2 tb1 (9.78 ± 1.29, P value = 0.922). This additive genetic interaction indicates that vt2 does not play a role in axillary meristem formation during vegetative development. Furthermore, the production of a similar number of tillers in tb1 and vt2 tb1 provides further support that later arising leaves are missing in vt2 mutants, as tb1 mutants do not produce tillers from the uppermost nodes.
Positional Cloning of vt2
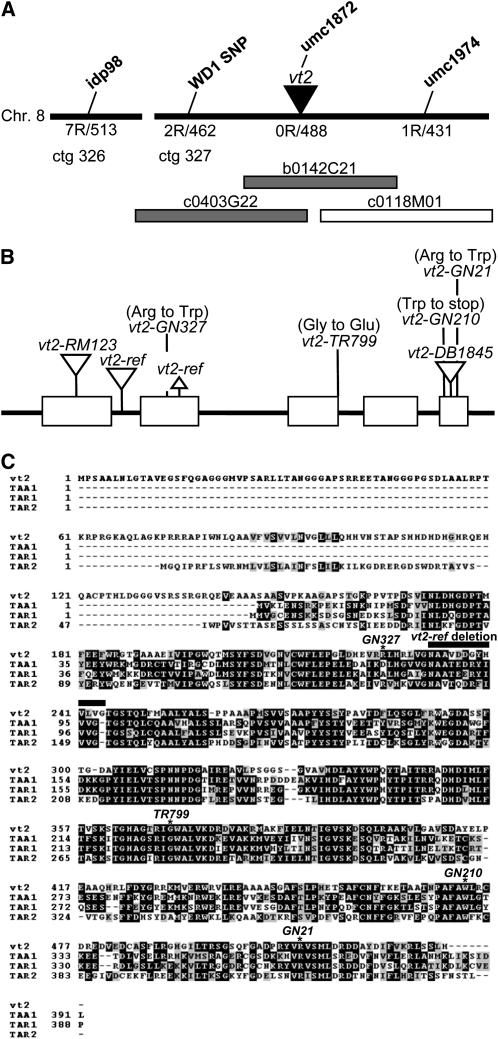
vt2 was proposed to be allelic to the semidominant Bif1 mutant (Smith and Hake, 1993); however, allelism tests revealed that the two genes were not allelic, but closely linked on chromosome 8L. To define the map position of vt2 further, two F2 mapping populations, vt2-ref-B73 × Mo17 and vt2-TR799-A619 × B73, were constructed. Using simple sequence repeat and insertion-deletion polymorphism markers, vt2 was fine-mapped to within two BAC contigs in bin 8.02. Marker idp98 was identified as the closest public flanking marker available on the north side of vt2 (1.36 centimorgans [cM], contig 326), and umc1974 was identified as the closest public marker on the south side of vt2 (0.12 cM, contig 327) (Figure 5A). Between these two flanking markers, a third public marker, umc1872, was found to be polymorphic but did not detect any recombinants, indicating that it was tightly linked to the mutant (0 cM, contig 327). Using single nucleotide polymorphism (SNP) markers identified in neighboring genes in the region, the number of recombinants on the north (idp98) side was narrowed down (WD1 SNP, 0.22 cM), placing vt2 on contig 327 within a region containing three overlapping BAC clones (Figure 5A). A candidate gene search in the region revealed a Trp aminotransferase-like gene (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009) located on the overlapping portion of two of the three BAC clones (Figure 5A).
Figure 5.
Cloning and Sequence Analysis of vt2.
(A) Diagram representing the vt2 region in maize after mapping with public and SNP markers (not to scale). The number of recombinant chromosomes (R) out of the total number of chromosomes is displayed below each marker. Maize BAC clones within this region are represented by rectangles, with the shaded rectangles indicating the overlapping clones on which vt2 was identified.
(B) Schematic of the vt2 gene structure including the position and mutations of seven alleles. Exons are represented by boxes; insertions and deletions are represented by downward and upward triangles, respectively.
(C) Sequence alignment of the predicted vt2 protein and three Arabidopsis Trp aminotransferases. vt2 shows the highest similarity to TAR2. Asterisks indicate the position of mutations in each EMS allele. Bar line indicates the position of the vt2-ref deletion.
To test if the Trp aminotransferase-like candidate gene was vt2, overlapping gene-specific PCR primers (see Supplemental Table 3 online) were designed to amplify and sequence the gene from all vt2 alleles. Point mutations that were not present in the progenitor backgrounds were identified in the coding regions of each of four ethyl methanesulfonate (EMS)-induced alleles, three of which caused a single amino acid substitution and one of which caused a premature stop codon in the predicted protein (Figures 5B and 5C). PCR amplification of the vt2-DB1845 allele using primers near the 3′ end of the gene revealed an insertion of ~300 bp in the 5th exon (Figure 5B). The Mutator (Mu) transposon-induced alleles, vt2-ref and vt2-RM123, were screened with a conserved Mu terminal inverted repeat primer (Mu3456) and gene-specific primers to identify potential Mu insertions. Sequencing of the PCR products revealed a Mu1 insertion in the first exon of vt2-RM123 and in the first intron of vt2-ref (Figure 5B). The vt2-ref allele was also determined to have a 12–amino acid deletion in the second exon (Figures 5B and 5C). These data indicate significant sequence changes in seven independent alleles and confirm that the vt2 gene encodes a Trp aminotransferase.
vt2 Is Co-Orthologous to Trp Aminotransferases from Arabidopsis
Sequence analysis of vt2 revealed highest similarity to the Trp aminotransferase gene of Arabidopsis, TAA1, and two Trp aminotransferase-related genes, TAR1 and TAR2 (Figure 5C) (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009). These genes have been categorized in the superfamily of pyridoxal-5′-phosphate–dependent enzymes, which differ from typical alliinases like TAR3 and TAR4 by their lack of an epidermal growth factor domain (Stepanova et al., 2008; Tao et al., 2008). The similarity between VT2 and the founding member of the alliinase family, garlic (Allium sativum) alliinase, extends throughout most of the coding region of VT2 containing the alliinase C-terminal domain (35% amino acid identity). Sequence alignment in this region indicated that VT2 shares 51% amino acid identity with TAA1, 51% identity with TAR1, and 56% identity with TAR2. The predicted amino acid sequence of VT2 is 90 amino acids longer than the longest Arabidopsis gene TAR2 due to an extended N terminus. Sequence alignment also indicated that the vt2 EMS-induced alleles and the vt2-ref deletion caused mutations in regions conserved with garlic alliinase and the TAA1/TAR genes from Arabidopsis. Notably, the vt2-GN21 allele had an Arg to Trp amino acid substitution at one of the known enzyme active sites (Kuettner et al., 2002; Tao et al., 2008). As all alleles have a similar phenotype to that of vt2-GN21, all alleles are assumed to be null.
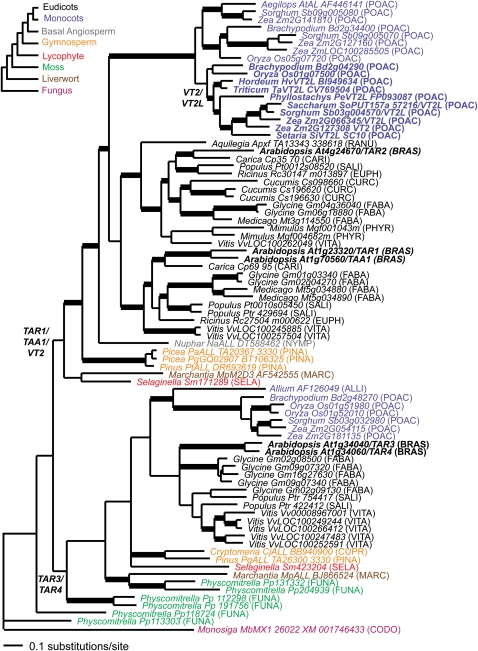
Bayesian phylogenetic analysis of 82 land plant and fungal alliinases recovered two major clades of Trp aminotransferases that are estimated products of a duplication event prior to the origin of extant land plants ~450 million years ago (Sanderson, 2003) (Figure 6). The first clade comprised liverwort (Marchantia), moss (Physcomitrella), gymnosperm (Pinus and Picea), and flowering plant sequences, including Arabidopsis TAA1/TAR1/TAR2 and maize vt2. Although deeper branches in this clade were not strongly supported (clade credibility [CC] ≥ 0.95), the seed plant, flowering plant, and monocot subclades were all strongly supported (1.00 CC). The second clade also comprised liverwort, moss, gymnosperm, and flowering plant sequences (including Arabidopsis TAR3 and TAR4), but only the monocot subclade was strongly supported.
Figure 6.
Phylogenetic Analysis.
Bayesian consensus phylogram of 82 vt2-like alliinases using the GTR model of evolution with some invariant sites and Γ distributed rates (GTR + I + Γ). Bold branches are supported by clade credibility ≥ 0.95. Color coding depicts different taxa. The cartoon depicts the accepted relationships among sampled taxa. Family abbreviations are included in parentheses (ALLI, Alliaceae; BRAS, Brassicaceae; CARI, Caricaceae; CODO, Codonosigidae; CUPR, Cupressaceae; CURC, Curcubitaceae; EUPH, Euphorbiaceae; FABA, Fabaceae; FUNA, Funariaceae; MARC, Marchantiaceae; NYMP, Nymphaeaceae; PHYR, Phyrmaceae; PINA, Pinaceae; POAC, Poaceae; RANU, Ranunculaceae; SALI, Salicaceae; SELA, Selaginaceae; SOLA, Solanaceae; and VITA, Vitaceae). The alignment used for this analysis is available as Supplemental Data Set 1 online.
vt2 was placed in a well-supported clade (1.00 CC) composed of grass species from both the BEP (Brachypodium, Hordeum, Oryza, Phyllostachys, and Triticum) and PACCMAD (Setaria, Sorghum, Saccharum, and Zea) clades, suggesting an origin for the vt2/vt2-like clade at least near the base of the grass family and possibly deeper within monocots (Figure 6). vt2 was sister to a clade of andropogonoid sequences comprising maize Zm2G066345 (vt2-like), Sorghum Sb03g004570, and Saccharum SoPUT157a-57216, and this clade was in turn sister to a vt2-like sequence from Setaria italica, suggesting that the maize vt2 and andropogonoid vt2-like sequences are products of a duplication event near the base of the andropogonoid clade. In agreement with this interpretation, filtering the 15,000 optimal Bayesian trees with a constraint tree where the maize vt2 and vt2-like (Zm2G066345) are sister taxa yielded only 73 trees or a probability of only 0.49% (73/15000) that the maize vt2 and vt2-like sequences are products of the maize tetraploidy event.
The placement of vt2 in a grass (likely monocot)-specific clade that is sister to a eudicot clade containing TAA1, TAR1, and TAR2 from Arabidopsis indicates that vt2 and vt2-like genes from maize are co-orthologous to TAA1, TAR1, and TAR2 from Arabidopsis.
vt2 Is Expressed in the Epidermis and Vasculature
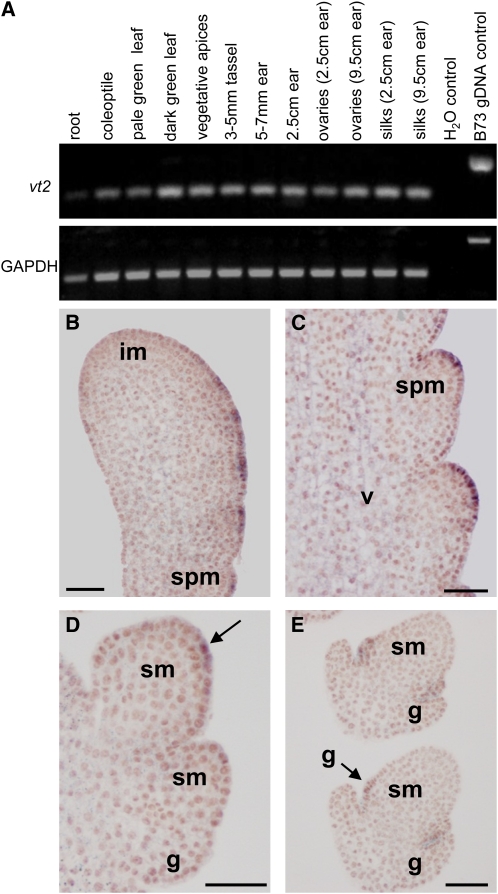
Sequence alignment of vt2 with its closest homologs in maize allowed the design of gene-specific primers. RT-PCR confirmed the intron-exon boundaries predicted in vt2 and revealed that vt2 was broadly expressed, with expression detected in all tissues tested (Figure 7A). RNA in situ hybridization with an antisense probe to vt2 was performed on immature tassel and ear inflorescences to determine the cell-type specificity of expression. Similar to TAA1 (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009), vt2 expression was detected in the epidermis of axillary meristems and in the vasculature in the inflorescence stem (Figures 7B to 7E). vt2 expression was first detected in the epidermis on the flanks of the inflorescence meristem prior to SPM initiation (Figure 7B). As the SPM initiated, vt2 was expressed in the epidermis in the most apical dome of the SPM (Figure 7C). As the SPM produced two SM, vt2 was expressed in the epidermis of both SM. As meristems gave rise to lateral organs, vt2 was again expressed in the epidermis as organs initiated (Figure 7E). We did not detect any signal using sense probes on similarly staged and treated tissues (see Supplemental Figure 2 online). Therefore, vt2, like TAA1in Arabidopsis, is expressed in epidermal and vascular tissue in the inflorescence (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009). In contrast, another gene family member, Zm-TAR1 (Zm2g127160 in Figure 6), was recently reported to be specifically expressed in the endosperm in maize (Chourey et al., 2010).
Figure 7.
vt2 Is Expressed in the Epidermis and Vasculature.
(A) Qualitative RT-PCR shows that vt2 is expressed in all tissues tested.
(B) to (E) RNA in situ hybridization on immature B73 tassels using pooled antisense probes from the 5′ and 3′ ends of vt2. im, inflorescence meristem; g, glume; v, vasculature; GAPDH, glyceraldehyde 3-phosphate dehydrogenase control. Bars = 50 μm.
(B) Apical inflorescence meristem (from the top of a branch) showing vt2 expression in the epidermis along the flanks of the inflorescence.
(C) Longitudinal section showing vt2 expression in the epidermis of SPM. Weak signal is also detected in the vasculature within the inflorescence.
(D) Transverse section showing two SM. The arrow indicates vt2 expression in the epidermis of the SM.
(E) Longitudinal section showing SM in the process of producing the outer and inner glumes. The arrow indicates vt2 expression in the epidermis of the inner glume.
[See online article for color version of this figure.]
vt2 spi1 Double Mutants Have a Slightly More Severe Phenotype Than vt2 Single Mutants
In Arabidopsis, the knockout of multiple YUC or TAA genes is required to produce a severe morphological phenotype (Cheng et al., 2006, 2007a; Stepanova et al., 2008). Therefore, the effect of eliminating these two proposed pathways of Trp-dependent auxin biosynthesis has not yet been examined. As a single mutant knockout of either spi1 or vt2 causes a significant phenotype on its own, we constructed double mutants to determine if these two genes have overlapping functions.
spi1 mutants are slightly shorter and produce one or two fewer leaves than the wild type, hence the overall vegetative phenotype is not nearly as severe as that of vt2 mutants (Figures 8A to 8C) (Gallavotti et al., 2008b). vt2 spi1 double mutants revealed a vegetative phenotype very similar to that of vt2 alone (Figure 8A). However, quantitative analysis revealed a small but statistically significant reduction in plant height (Figure 8B) and leaf number (Figure 8C) in double mutants compared with vt2 alone.
Figure 8.
vt2 spi1 Double Mutant Analysis.
(A) vt2 spi1 plants resemble vt2 single mutants except with a reduction in height.
(B) Quantification of plant height.
(C) Quantification of leaf number.
(D) vt2 spi1 tassels resemble vt2 single mutants except with a reduction in length.
(E) vt2 spi1 ears exhibit reduced length and kernel number.
(F) Quantification of tassel length.
(G) Quantification of tassel branch number.
(H) Quantification of tassel spikelet number.
(I) Quantification of ear length.
(J) Quantification of kernel number.
(K) Quantification of visible ear shoot number. Asterisk indicates significant reduction at P < 0.05 compared with either single mutant alone; error bars represent the se; n = 55 normal, 17 spi1, 27 vt2, 7 vt2 spi1 for (B), (C), and (K); n = 10 normal, spi1, and vt2 and 7 vt2 spi1 for (F) to (H); n = 5 each of normal, spi1, vt2, and vt2 spi1 for (I) and (J).
[See online article for color version of this figure.]
The tassel inflorescence phenotype of spi1 mutants is less severe than that of vt2 mutants, with nonetheless a strong reduction in branch and functional spikelet number compared with normal (Figure 8D) (Gallavotti et al., 2008b). vt2 spi1 double mutant tassels resembled those of vt2 single mutants (Figure 8D), with the complete absence of branches (Figure 8G) or spikelets (Figure 8H). However, tassel length was found to be statistically significantly reduced compared with either single mutant (Figure 8F). In the female inflorescence, spi1 mutant ears resemble vt2 ears but are typically much less severely affected (Figure 8E). By contrast, vt2 spi1 double mutant ears revealed a phenotype that was slightly more severe than that of vt2 (Figure 8E). Quantification revealed a small but statistically significant reduction of ear length (Figure 8I) and kernel number (Figure 8J) in double mutants compared with vt2. Since double mutants could produce some ears, we also tested whether they produced an altered number of visible ear shoots. Quantification revealed that vt2 spi1 plants produced an equivalent number of ear shoots as vt2 plants, whereas spi1 plants produced a similar number as wild-type plants (Figure 8K).
Although the vegetative and reproductive phenotypes initially appeared to indicate that vt2 spi1 was similar to vt2, quantification revealed that vt2 spi1 double mutants had a slightly more severe phenotype than vt2 with respect to vegetative phenotypes and inflorescence length.
Auxin Levels in Single and Double Mutants
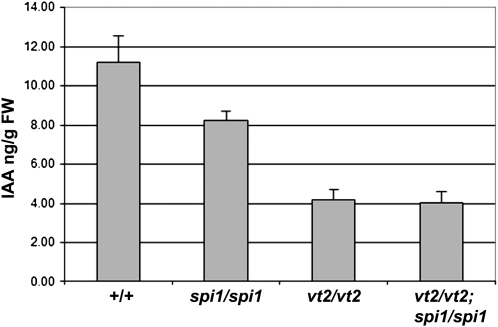
To investigate further the functions of spi1 and vt2 in the biosynthesis of auxin, free IAA levels were measured (Barkawi et al., 2010). These experiments were performed in developing leaves due to the difficulties of normalizing auxin levels in developing inflorescence meristems with such dramatic morphological defects (Skirpan et al., 2009). Normal leaves had ~10 to 11 ng/g fresh weight of IAA, while spi1 and vt2 mutants had an average 82 and 34% of normal IAA levels, respectively (Figure 9). Surprisingly, the reduction in IAA in spi1 vt2 double mutants was not statistically significantly different from the levels detected in vt2 single mutants (P value = 0.845). These results provide experimental support for the hypothesis that vt2 and spi1 may actually function in the same pathway for auxin biosynthesis, at least in leaf tissue.
Figure 9.
Measurement of Free IAA Levels in Normal, spi1, vt2, and spi1 vt2 Mutants.
Free IAA levels were measured in developing leaves of 2-week-old seedlings. Error bars represent the se. n = 5 each of normal, spi1, vt2, and vt2 spi1. One replicate of three is shown. FW, fresh weight.
vt2 and bif2 Exhibit a Synergistic Interaction
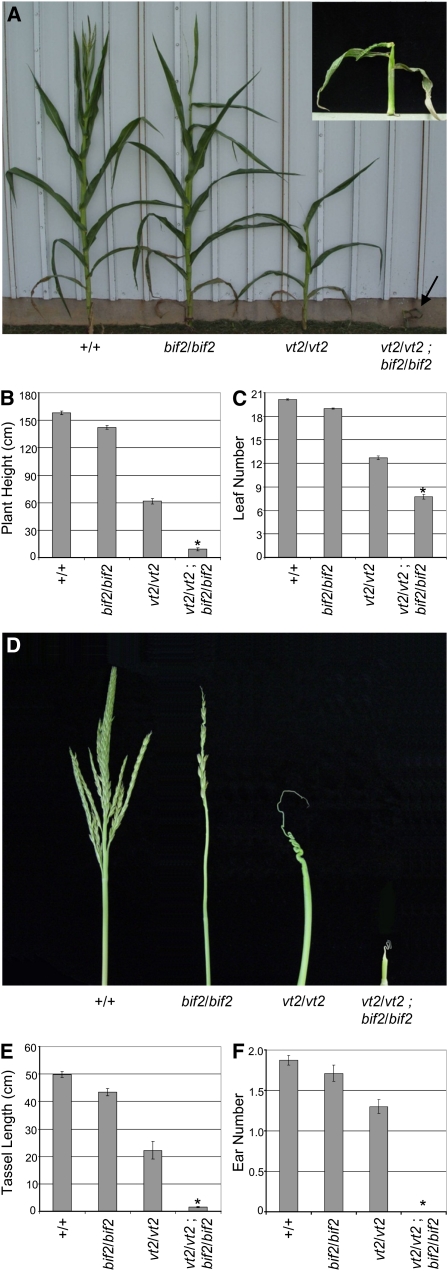
The effects of eliminating both a gene in auxin biosynthesis and a gene in auxin transport have been examined in maize through double mutant combinations with spi1 and bif2 (Gallavotti et al., 2008b). The results of those studies revealed a synergistic interaction that produced double mutant plants with a very similar vegetative phenotype to that of vt2 mutants (Gallavotti et al., 2008b). To test the interaction of vt2 and bif2, we constructed double mutants.
In the vegetative phase of growth, bif2 mutants have a slight reduction in plant height and leaf number compared with wild-type plants (Figures 10A to 10C) (McSteen et al., 2007). By contrast, the vt2 bif2 double mutant vegetative phenotype was extremely severe (Figure 10A, inset). Double mutants exhibited a significant reduction in plant height to ~10% of normal (Figure 10B) and significantly fewer leaves with only about seven total leaves produced (Figure 10C). In fact, the phenotype was so severe that double mutants died weeks before siblings flowered.
Figure 10.
vt2 bif2 Double Mutant Analysis.
(A) vt2 bif2 plants (arrow and inset) exhibit a drastic reduction in vegetative growth compared with vt2 or bif2 single mutants.
(B) Quantification of plant height.
(C) Quantification of leaf number.
(D) vt2 bif2 tassels are severely underdeveloped compared with vt2 or bif2 single mutants.
(E) Quantification of tassel length.
(F) Quantification of ear number. Asterisk indicates significant reduction at P < 0.05 compared with either single mutant alone; error bars represent the se; n = 31 normal, 28 bif2, 18 vt2, and 6 vt2 bif2 for (B), (C), and (F); n = 10 normal, bif2, and vt2 and 8 vt2 bif2 for (E).
[See online article for color version of this figure.]
bif2 single mutants typically produce tassels with a reduced number of branches, spikelets, florets, and floral organs (Figure 10D) (McSteen and Hake, 2001). vt2 bif2 double mutants produced completely barren tassels similar to vt2 single mutants, although they also had an extreme reduction in size (Figure 10D). Quantification of tassel length confirmed a significant reduction in vt2 bif2 mutants compared with either single mutant alone (Figure 10E). In addition, vt2 bif2 mutants never produced visible ear shoots, which is a more severe phenotype than either single mutant (Figure 10F).
Both vegetative and reproductive data for vt2 bif2 double mutants show a phenotype that is significantly more severe than either single mutant alone. This can be interpreted as a synergistic interaction, indicating that vt2 and bif2 have overlapping functions in vegetative and reproductive development in maize.
vt2 Is Epistatic to ba1
To investigate the interaction of vt2 with another barren inflorescence mutant that does not have defects in auxin transport or biosynthesis, we constructed double mutants with barren stalk1 (ba1), which has defects in the production of all axillary meristems (Ritter et al., 2002; Gallavotti et al., 2008a). ba1 mutants do not show an obvious vegetative phenotype except for a reduction in height (see Supplemental Figures 3A and 3B online), which is due to a decrease in tassel length (see Supplemental Figure 3E online), rather than leaf number (see Supplemental Figure 3C online). The vt2 ba1 double mutant vegetative phenotype clearly resembled that of vt2 single mutants (see Supplemental Figure 3A online). Quantification revealed that there was no statistically significant reduction in plant height (see Supplemental Figure 3B online) or leaf number (see Supplemental Figure 3C online) in vt2 ba1 double mutants compared with vt2 alone.
ba1 mutants produce tassel inflorescences similar to those of vt2 mutants, exhibiting a complete lack of branches and spikelets (see Supplemental Figure 3D online). However, unlike vt2 mutants, ba1 tassels produce suppressed bract primordia (visible as bumps) in regular rows along the rachis of the tassel (see Supplemental Figure 3D online, arrow) (Ritter et al., 2002). These bumps indicate pools of auxin that are produced and transported normally to the inflorescence but cannot be used to produce spikelets due to the absence of ba1 gene function (Gallavotti et al., 2008a). vt2 ba1 double mutants produced tassels that resembled vt2 single mutants, with no evidence of bract primordia that are normally observed in ba1 single mutants (see Supplemental Figure 3D online). Tassel length of vt2 ba1 double mutants was not significantly different than either single mutant alone (see Supplemental Figure 3E online). Finally, ba1 mutants never produce ears since they lack the ability to initiate ear axillary meristems, and vt2 ba1 double mutants similarly never produced an ear shoot (see Supplemental Figure 3F online).
These results illustrate that vt2 is completely epistatic to ba1 during both vegetative and tassel inflorescence development. As ba1 mutants do not produce ear shoots, ba1 is epistatic to vt2 during the production of the ear. These results support the idea that vt2 is functioning upstream in auxin production, while ba1 is functioning downstream in the production of axillary meristems.
DISCUSSION
Here, we show that the vt2 plays a significant role in axillary meristem formation during inflorescence development in maize. During vegetative development, vt2 does not play a role in axillary meristem formation but functions in leaf initiation during the adult phase of growth. vt2 encodes an enzyme with similarity to TAA1, which has been demonstrated to convert Trp to IPA in Arabidopsis (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009). Phylogenetic analysis indicates that vt2 is co-orthologous to TAA1, TAR1, and TAR2 in Arabidopsis. Our results suggest that the IPA pathway for Trp-dependent auxin biosynthesis contributes significantly to vegetative and reproductive development in maize.
Auxin Biosynthesis Pathways in Maize and Arabidopsis
The phenotype of vt2 mutants shares many similarities with the phenotype of spi1 mutants, which are defective in the proposed TAM pathway for Trp-dependent auxin biosynthesis in maize (Gallavotti et al., 2008b). Both mutants have defects in axillary meristem formation during reproductive development, shorter inflorescences due to defects in cell elongation, and shorter plant height due to defects in leaf initiation during vegetative development (Gallavotti et al., 2008b; Barazesh et al., 2009). The vt2 mutant, however, has more severe defects than does spi1, although both are presumed to be null alleles. The similarity in phenotype is not due to effects on gene expression as real-time RT-PCR experiments indicate that spi1 expression is not reduced in vt2 mutants nor is vt2 expression reduced in spi1 mutants (see Supplemental Figure 4 online). Both spi1 and vt2 mutants have reduced free auxin levels, indicating that both gene products contribute to the production of auxin in the plant. In Arabidopsis, the similarity of phenotype of yuc and taa multiple mutant combinations has been used as an argument to suggest that these two genes act in the same pathway for the production of auxin rather than being separate independent pathways (Strader and Bartel, 2008).
In Arabidopsis, multiple knockouts of the TAA genes or the YUC genes are required to produce a severe phenotype, whereas in maize, single knockouts of either vt2 or spi1 have a dramatic effect on development (Cheng et al., 2006, 2007a; Gallavotti et al., 2008b; Stepanova et al., 2008). The availability of single gene knockouts with dramatic phenotypes in maize enabled us to test the relative contribution of the vt2 and spi1 genes to development through the construction of double mutants. The results show that vt2 spi1 double mutants have a slightly more severe phenotype than the vt2 single mutant does. This interaction could be interpreted as additive, although synergism, epistasis, and additivity can be difficult to distinguish when mutants have similar phenotypes. When both mutants are null, an additive interaction is interpreted to indicate that the genes function in different pathways. However, when mutants are not null, a more severe phenotype in the double mutant is interpreted to indicate that the genes function in the same pathway (Martienssen and Irish, 1999). The presence of other spi1 and vt2 gene family members expressed during vegetative and reproductive development (K.A. Phillips and P. McSteen, unpublished data) suggests that the function of the gene family has not been completely knocked out in the single mutants. Therefore, the more severe phenotype in the vt2 spi1 double mutant could also be interpreted to imply that the spi1 and vt2 genes function in the same pathway. Although the genetic interaction could be interpreted to support either hypothesis, the observation that the double mutant does not have a statistically significant reduction in auxin levels compared with the vt2 single mutant provides some support for the hypothesis that the two genes may act in the same pathway. However, the alternative hypothesis that they act in different pathways has not been disproven by these results as auxin levels rather than auxin biosynthetic rates were measured. Therefore, further analysis of the in vivo functions of these enzymes is essential.
The hypothesis of multiple auxin biosynthetic pathways also raises the question of why the different pathways do not compensate for each other in either maize or Arabidopsis (Tao et al., 2008). This lack of compensation has been used as an argument to support the hypothesis that the YUC and TAA genes may act in the same pathway (Strader and Bartel, 2008). In fact, in Arabidopsis, upregulation of the IAOx pathway has been shown to compensate for defects in the IPA pathway (Stepanova et al., 2008), and expression of the bacterial iaaM gene, which catalyzes the conversion of Trp to IAM, can rescue the yuc1 yuc4 double mutant in Arabidopsis (Cheng et al., 2006). Therefore, when misregulated, other auxin biosynthetic pathways can compensate for deficiencies in the TAM or IPA pathways in Arabidopsis. This indicates that lack of compensation may be due to differences in expression pattern or availability of intermediates in different cell types. spi1 and vt2 are both expressed as axillary meristems and lateral organs initiate in the inflorescence, although spi1 expression extends several cell layers instead of being restricted to the epidermis as for vt2 (Gallavotti et al., 2008b); therefore, lack of compensation by the two proposed pathways in maize cannot wholly be explained by differences in expression pattern. The identification of spi1 and vt2 gene family members specifically expressed in the endosperm of maize suggests that these enzyme activities may also overlap during endosperm development (Chourey et al., 2010; LeClere et al., 2010). Moreover, the recent findings questioning the biochemical function of YUC indicate that much remains to be learned about how these pathways function (Tivendale et al., 2010; Nonhebel et al., 2011). Much also remains to be learned about the differences in auxin biosynthesis pathways in different plant species and in different tissues (Quittenden et al., 2009; Ishii et al., 2010; Tivendale et al., 2010).
In contrast with the similarity in phenotype seen between vt2 and spi1 mutants, vt2 mutants have few similarities with the phenotypes of taa1, tar1, and tar2 single, double, and triple mutants in Arabidopsis (Stepanova et al., 2008). Some of these differences may be superficial due to the fact that, for example, leaf and flower number have not been quantified in taa1 tar double mutants, and vt2 mutants have similarly not been tested for insensitivity to ethylene, shade, or naphthylphthalamic acid. One clear difference between maize and Arabidopsis is that the Arabidopsis taa1 tar2 double mutants have very significant defects in apical dominance, exhibiting a bushy phenotype presumably due to outgrowth of secondary branches. By contrast, bushiness is not a characteristic of the maize vt2 mutants. In fact, testing the interaction of vt2 with tb1 showed that vt2 did not appear to play a role in suppression or promotion of axillary branch outgrowth. These apparent differences between vt2 and taa1 mutants could be due to roles of additional gene family members that have not yet been fully addressed in either maize or Arabidopsis.
The Role of Temperature in Auxin Biosynthesis
Another point of contrast between vt2 and taa1 mutants is that both exhibit temperature dependence but they have opposite responses to high temperature. Maize vt2 mutants have a weaker inflorescence phenotype at high temperature, whereas some defects in Arabidopsis taa1 mutants can be detected only at high temperature (Yamada et al., 2009). It has been shown in Arabidopsis that free auxin levels in hypocotyls increase at higher temperature (Gray et al., 1998), and it now appears that multiple auxin biosynthetic pathways may be contributing to these increased levels. The TAA1 gene is temperature induced, and temperature-induced hypocotyl elongation is abolished in taa1 mutants, indicating that the IPA pathway is induced by high temperature in Arabidopsis (Yamada et al., 2009). Furthermore, in the IAOx pathway, Arabidopsis cyp79b2 cyp79b3 double mutants were shown to have reduced auxin levels at higher temperature compared with the wild type, indicating that the IAOx pathway also contributes to increased auxin levels at higher temperatures (Zhao et al., 2002). As the IAOx pathway is not supposed to be critical outside the Brassicaceae (Sugawara et al., 2009), one possible explanation for the weaker phenotype of maize vt2 mutants at higher temperature is that closely related vt2-like genes may be capable of providing increased activity at high temperatures. Alternately, spi1 may contribute to auxin biosynthesis at higher temperature, as the spi1 mutant does not have a weaker phenotype at higher temperature. However, YUC gene expression has been shown to be reduced rather than increased in anthers of barley (Hordeum vulgare) and Arabidopsis at high temperature (Sakata et al., 2010). Therefore, future work is required to address the role of all gene family members and their relative contributions to auxin biosynthesis at different temperatures, in different tissues, and in different species.
Synergistic Interaction of Auxin Biosynthesis and Auxin Transport
In contrast with the slightly enhanced phenotype observed in vt2 spi1 double mutants, synergistic interactions are seen when both auxin biosynthesis and auxin transport are reduced. This was demonstrated in spi1 bif2 double mutants, as disruption of each single gene has mild effects on vegetative development, but dramatic effects are observed in the double mutant combination (Gallavotti et al., 2008b). vt2 bif2 double mutants show even more dramatic effects on plant height and leaf number, causing the plants to complete development and senesce weeks earlier than normal. These synergistic interactions indicate that both auxin transport and biosynthesis play overlapping roles in normal development. Synergistic interactions between auxin biosynthesis and transport have previously been reported in Arabidopsis, indicating that this phenomenon is widespread (Weijers et al., 2005; Cheng et al., 2007a, 2007b). Furthermore, it has been shown in Arabidopsis that auxin synthesized by TAA1 in the leaves has to be transported to the stem to exert its effects (Tao et al., 2008). Therefore, both auxin transport and auxin biosynthesis play critical roles in vegetative and reproductive development.
METHODS
Origin of vt2 Alleles
The vt2-ref allele originated by Mu transposon mutagenesis (Smith and Hake, 1993). vt2-TR799, vt2-GN21, vt2-GN210, and vt2-GN327 were obtained from the Maize Inflorescence Project and arose via EMS mutagenesis in defined genetic backgrounds (http://www.maizegdb.org/ems-phenotype.php). vt2-RM123 was obtained from the RescueMu population (http://www.maizegdb.org/rescuemu-phenotype.php). vt2-DB1845 arose by spontaneous mutation in the maize (Zea mays) B73 genetic background (David Braun, Penn State University).
Mature Phenotype Analysis
All mature phenotype data were obtained using the vt2-ref allele backcrossed into the B73 background six times and compared with normal siblings from the same family. Segregating families were planted in two separate field plantings during the summer in Rocksprings, PA, grown to maturity (10 to 12 weeks), and scored for phenotype. Data presented are representative of one field season. Plant height was obtained by measuring from the ground to the tip of the tassel, and ear number was scored by counting all visible ears on each plant. Leaf number was quantified by marking every 5th leaf of developing field-grown plants beginning at 4 weeks until full maturity. Tassel length was calculated by measuring from the tip of the tassel to the base of the flag leaf node, and branch number was obtained by counting all visible lateral branches. Spikelet number was obtained prior to anthesis by removing and counting all spikelets from the branches and main spike. Kernel number was estimated by counting all spikelets on mature open-pollinated ears, and ear length was obtained by measuring these ears from the base to the tip.
For leaf juvenile-to-adult transition analysis, segregating families were greenhouse-grown for 9 weeks and genotyped for vt2 using a TaqMan SNP assay (described below). Leaf number was counted as described above. As leaves fully emerged from the whorl, the presence or absence of epicuticular waxes on the blade was evaluated for leaves 1 to 10.
For double mutant analysis, segregating families were planted twice separated by a few weeks and grown to maturity (10 to 12 weeks) in Rocksprings, PA in the summer of 2007 and 2008 (2008 and 2009 for vt2 spi1). Similar results were also seen in Columbia, MO in the summer of 2010. Data shown are representative of one planting from the 2008 field season except for the ear data from vt2 spi1 double mutants, which are from the 2009 field season. vt2 tb1 double mutant families were generated using the tb1-ref allele (Doebley et al., 1997) in the B73 background. Visible tillers were counted at maturity as those that were derived directly from one of the nodes on the main stalk. vt2 spi double mutant families were generated using the spi1-ref allele (Gallavotti et al., 2008b) in the B73 background. All individuals were genotyped for vt2 using the SNP TaqMan protocol and for spi1 as previously described (Gallavotti et al., 2008b). vt2 bif2 double mutant families were generated using the bif2-77 allele (McSteen et al., 2007) in the B73 background. All individuals were genotyped for vt2 using the SNP TaqMan protocol and for bif2 as previously described (Skirpan et al., 2008). Data for all vt2 bif2 double mutants were obtained about 7 weeks after germination due to the drastically reduced lifespan of the plants. Photos display younger vt2 bif2 mutants next to siblings from a planting 2 weeks earlier to represent all individuals at maturity. vt2 ba1 double mutant families were generated using the ba1-ref allele (Ritter et al., 2002) in the B73 background.
For statistical analysis, results were analyzed using Microsoft Excel 2003. Bar graphs were produced using the mean of each data set, and error bars are the se. Data were considered statistically significant at P value < 0.05 using the Student’s two-tailed t test in Microsoft Excel.
TaqMan vt2 Genotyping Assay
The Panzea database was used to identify SNPs in the vt2 region after preliminary fine-mapping (www.panzea.org). A SNP located on contig 327 was identified to be polymorphic between vt2 mutants (CGA) and the B73 background (CAA). Primers flanking the SNP were designed based on the Panzea sequence, and the SNP was confirmed to be linked to the vt2 mutant background by DNA sequencing. A TaqMan SNP genotyping assay (primers in Supplemental Table 3 online) was then designed using the Custom TaqMan SNP Genotyping Assays design program, File Builder v3.1 (Applied Biosystems). TaqMan assays were performed by the Penn State Huck Institutes Genomics Core Facility using an ABI 7300 sequence detection system.
Measurement of Cell Size
For measurement of cell size, nail polish impressions were obtained from the basal region of five wild-type and vt2 field-grown tassels at maturity as previously described (Barazesh et al., 2009). Slides were analyzed at ×20 magnification using a Nikon Eclipse 80i upright microscope on the bright-field setting and a Nikon DM1200F camera. Approximately 15 to 30 cells were measured per biological replicate depending on cell size. The average lengths of each replicate were used to obtain the final mean presented for each genotype.
Scanning Electron Microscopy
Families segregating for vt2-ref in the B73 genetic background were planted and genotyped using the TaqMan assay. Ears were collected from field-grown plants after ~8 weeks, while tassels were collected from both field- and greenhouse-grown plants after ~5 weeks. Field-grown plants were exposed to an average daily minimum temperature of 12.9°C and an average daily maximum temperature of 23.7°C according to weather history data available for Pennsylvania Furnace, PA from May 19, 2009 through July 10, 2009 (www.almanac.com). Greenhouse temperatures in the maize growth rooms were held at a minimum temperature of 26.7°C for both day and night, with daytime temperatures typically increasing an additional 5 to 10°C. Cooler greenhouse rooms were held at a minimum temperature of 20°C and a maximum temperature of 26.7°C each day. Fixation and scanning electron microscopy of samples were performed as previously described (Wu and McSteen, 2007).
Phylogenetic Analyses
vt2-like alliinase sequences were retrieved using BLAST searches at CoGe (http://synteny.cnr.berkeley.edu/CoGe), the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), Phytozome (http://www.phytozome.org), PlantGDB (http://www.plantgdb.org), and The Institute for Genomic Research (http://blast.jcvi.org/euk-blast/plantta_blast.cgi). The resulting nucleotide sequences were then aligned using the conceptual amino acid translation using MacClade (Maddison and Maddison, 2003) and MUSCLE (Edgar, 2004) before being manually adjusted. Hypervariable regions of the alignment between nucleotide positions 1 to 567 and 2197 to 3027 were considered too divergent for reliable alignment and were excluded from subsequent analyses (see Supplemental Data Set 1 online for the alignment). Phylogenetic relationships were estimated with MrBayes 3.1 (Ronquist and Huelsenbeck, 2003) on the Grethor parallel processing cluster at the University of Missouri–St. Louis using the General Time Reversible (GTR) model of evolution with invariant sites and gamma distributed rates (GTR + I + Γ), with the nucleotide data set partitioned according to codon position. Bayesian phylogenetic analysis consisted of two separate runs of four chains of 10 million generations with trees sampled every 1000 generations and the first 25% (2500 trees) of suboptimal trees removed as burnin. Trees were rooted using a fungal Asp aminotransferase (Monosiga XM_001746433) as an outgroup. Constraint trees were generated in MacClade, and the sets of optimal trees from the Bayesian analyses were filtered using PAUP 3.1 (Swofford, 2000).
Expression Analysis
cDNA was generated from total RNA for each tissue sample using the High Capacity cDNA reverse transcription kit (Applied Biosystems). Three μL of cDNA was amplified by PCR using the vt2 gene-specific primers ex34-F and ex34-R (see Supplemental Table 3 online) for 40 cycles. GAPDH primers L4 and R4 (see Supplemental Table 3 online) were used as controls and amplified 1 μL of cDNA for 35 cycles. RNA in situ hybridization was performed as described (Jackson et al., 1994). The 5′ antisense probe construct was amplified using primers 5′probe-F and 5′probe-R (see Supplemental Table 3 online) and cloned into the pGemTEasy vector (Promega). The 3′ antisense probe construct was amplified using primers 3′probe-F and 3′probe- R (see Supplemental Table 3 online) and cloned into the pGemTEasy vector. DNA template was amplified by PCR using M13 forward and reverse primers and DIG-labeled antisense RNA probe generated with SP6 RNA polymerase. One microliter of both 5′ and 3′ probe was pooled prior to hybridization on slides. Sense probes were generated from the same constructs using T7 RNA polymerase. Immature greenhouse-grown B73 tassels were fixed and embedded in paraffin wax as previously described except that fixation was for ~2 h (Wu and McSteen, 2007). Real-time RT-PCR was performed as described (Barazesh and McSteen, 2008) using Taqman assays (probes and primers listed in Supplemental Table 3 online) and total RNA extracted from immature leaf tissue.
Quantification of Free IAA
Families segregating for both spi1 and vt2 in the B73 genetic background were grown for 2 weeks and genotyped as described above. Samples of leaf tissue were collected from the fifth leaf that was just emerging from the whorl. Leaf tissue (50 to 80 mg) was weighed, frozen in liquid nitrogen, and stored at −80°C. For each sample, 150 μL of homogenization buffer (35% of 0.2 M imidazole and 65% isopropanol, pH 7) containing 3 or 4 ng of [13C6]IAA was added before homogenization. The level of free IAA was analyzed by solid phase extraction followed by selected ion monitoring–gas chromatography–mass spectrometry as previously described (Barkawi et al., 2010).
Accession Numbers
vt2 genomic and cDNA nucleotide sequence data reported are available in the Third Party Annotation Section of the GenBank/EMBL/DDBJ databases under the accession number BK007972 (TPA). The accession numbers for sequences used in the phylogenetic analysis in Figure 6 can be found in Supplemental Table 4 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Scanning Electron Microscopy Analysis of vt2 Ear Inflorescences.
Supplemental Figure 2. RNA in Situ Hybridization with vt2 Sense Probes.
Supplemental Figure 3. vt2 ba1 Double Mutant Analysis.
Supplemental Figure 4. Real-Time Expression Analysis.
Supplemental Table 1. vt2 Juvenile-to-Adult Leaf Transition Is Not Significantly Different from Normal Siblings.
Supplemental Table 2. vt2 Mutant Tassels Exhibit Reduced Cell Length Compared with Normal Siblings.
Supplemental Table 3. Primers Used for vt2 Mapping, Cloning, Genotyping, and Expression Analyses.
Supplemental Table 4. Accession Numbers for Sequences Used in Phylogenetic Analysis in Figure 6.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis in Figure 6.
Acknowledgments
We thank Frank Baker for advice and assistance with scoring juvenile versus adult leaves. We thank W. Scott Harkom, Tony Omeis, and Tom Slewinski for plant care in the greenhouse and the field. We thank Missy Hazen and Ruth Haldeman of the Pennsylvania State University Huck Institutes Electron Microscopy facility for assistance with scanning electron microscopy and Deb Grove and Ashley Price of the Huck Institutes Genomics core facility for assistance with DNA sequencing, TaqMan genotyping, and real-time RT-PCR assays. We thank Jessica Levy and Matthew Phillips for assistance in the field and Peng Yu for assistance with auxin measurements. We also thank the University of Missouri–St. Louis for access to their Grethor parallel-processing cluster and Elizabeth Kellogg (University of Missouri–St. Louis), Jeff Bennetzen and Ryan Percifield (University of Georgia), and the Joint Genome Institute for providing the Setaria italica vt2 genomic sequence. This research was supported by USDA Grant 2007-03036 to P.M., National Science Foundation Grant IOS-0820729 to P.M. and S.M., and National Science Foundation Grants MCB0725149 and IOS-PGRP-0923960 and the Gordon and Margaret Bailey Endowment for Environmental Horticulture to J.D.C.
References
- Barazesh S., McSteen P. (2008). Barren inflorescence1 functions in organogenesis during vegetative and inflorescence development in maize. Genetics 179: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazesh S., Nowbakht C., McSteen P. (2009). sparse inflorescence1, barren inflorescence1 and barren stalk1 promote cell elongation in maize inflorescence development. Genetics 182: 403–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkawi L.S., Tam Y.Y., Tillman J.A., Normanly J., Cohen J.D. (2010). A high-throughput method for the quantitative analysis of auxins. Nat. Protoc. 5: 1609–1618 [DOI] [PubMed] [Google Scholar]
- Bartel B. (1997). Auxin biosynthesis. Annu. Rev. Plant Physiol. 48: 49–64 [DOI] [PubMed] [Google Scholar]
- Bartling D., Seedorf M., Mithöfer A., Weiler E.W. (1992). Cloning and expression of an Arabidopsis nitrilase which can convert indole-3-acetonitrile to the plant hormone, indole-3-acetic acid. Eur. J. Biochem. 205: 417–424 [DOI] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Bennett S.R.M., Alvarez J., Bossinger G., Smyth D.R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8: 505–520 [Google Scholar]
- Chandler J.W. (2009). Local auxin production: A small contribution to a big field. Bioessays 31: 60–70 [DOI] [PubMed] [Google Scholar]
- Cheng P.C., Greyson R.I., Walden D.B. (1983). Organ initiation and the development of unisexual flowers in the tassel and ear of Zea mays. Am. J. Bot. 70: 450–462 [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2007a). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Qin G., Dai X., Zhao Y. (2007b). NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 18825–18829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.F., Dai X.H., Zhao Y.D. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey P.S., Li Q.-B., Kumar D. (2010). Sugar-hormone cross-talk in seed development: two redundant pathways of IAA biosynthesis are regulated differentially in the invertase-deficient miniature1 (mn1) seed mutant in maize. Mol. Plant 3: 1026–1036 [DOI] [PubMed] [Google Scholar]
- Christensen S.K., Dagenais N., Chory J., Weigel D. (2000). Regulation of auxin response by the protein kinase PINOID. Cell 100: 469–478 [DOI] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Gallavotti A., Barazesh S., Malcomber S., Hall D., Jackson D., Schmidt R.J., McSteen P. (2008b). sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA 105: 15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Yang Y., Schmidt R.J., Jackson D. (2008a). The relationship between auxin transport and maize branching. Plant Physiol. 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gälweiler L., Guan C.H., Müller A., Wisman E., Mendgen K., Yephremov A., Palme K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230 [DOI] [PubMed] [Google Scholar]
- Gray W.M., Ostin A., Sandberg G., Romano C.P., Estelle M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard L., McSteen P., Doebley J., Hake S. (2002). Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162: 1927–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T., Soeno K., Asami T., Fujioka S., Shimada Y. (2010). Arabidopsis seedlings over-accumulated indole-3-acetic acid in response to aminooxyacetic acid. Biosci. Biotechnol. Biochem. 74: 2345–2347 [DOI] [PubMed] [Google Scholar]
- Jackson D., Veit B., Hake S. (1994). Expression of maize knotted1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Kerstetter R.A., Poethig R.S. (1998). The specification of leaf identity during shoot development. Annu. Rev. Cell Dev. Biol. 14: 373–398 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Huang F., Naramoto S., Zhang J., Michniewicz M., Offringa R., Friml J. (2009). PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21: 3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V., Park W.J., Gierl A., Glawischnig E. (2006). Auxin biosynthesis in maize. Plant Biol (Stuttg) 8: 334–339 [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V., Park W.J., Piotrowski M., Meeley R.B., Gierl A., Glawischnig E. (2007). Maize nitrilases have a dual role in auxin homeostasis and beta-cyanoalanine hydrolysis. J. Exp. Bot. 58: 4225–4233 [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V., Weigang L., Fiesselmann A., Letzel T., Frey M., Gierl A., Glawischnig E. (2008). Characterisation of the tryptophan synthase alpha subunit in maize. BMC Plant Biol. 8: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuettner E.B., Hilgenfeld R., Weiss M.S. (2002). The active principle of garlic at atomic resolution. J. Biol. Chem. 277: 46402–46407 [DOI] [PubMed] [Google Scholar]
- LeClere S., Schmelz E.A., Chourey P.S. (2010). Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol. 153: 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T., Hoffmann M., Hentrich M., Pollmann S. (2010). Indole-3-acetamide-dependent auxin biosynthesis: a widely distributed way of indole-3-acetic acid production? Eur. J. Cell Biol. 89: 895–905 [DOI] [PubMed] [Google Scholar]
- Maddison D.R., Maddison W.P. (2003). MacClade: Analysis of Phylogeny and Character Evolution. (Sunderland, MA: Sinauer Associates; ). [DOI] [PubMed] [Google Scholar]
- Martienssen R., Irish V. (1999). Copying out our ABCs: The role of gene redundancy in interpreting genetic hierarchies. Trends Genet. 15: 435–437 [DOI] [PubMed] [Google Scholar]
- McSteen P. (2010). Auxin and monocot development. Cold Spring Harb. Perspect. Biol. 2: a001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P., Hake S. (2001). barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128: 2881–2891 [DOI] [PubMed] [Google Scholar]
- McSteen P., Laudencia-Chingcuanco D., Colasanti J. (2000). A floret by any other name: control of meristem identity in maize. Trends Plant Sci. 5: 61–66 [DOI] [PubMed] [Google Scholar]
- McSteen P., Malcomber S., Skirpan A., Lunde C., Wu X., Kellogg E., Hake S. (2007). barren inflorescence2 Encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol. 144: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonhebel H., Yuan Y., Al-Amier H., Pieck M., Akor E., Ahamed A., Cohen J.D., Celenza J.L., Normanly J. (2011). Redirection of tryptophan metabolism in tobacco by ectopic expression of an Arabidopsis indolic glucosinolate biosynthetic gene. Phytochemistry 72: 37–48 [DOI] [PubMed] [Google Scholar]
- Park W.J., Kriechbaumer V., Möller A., Piotrowski M., Meeley R.B., Gierl A., Glawischnig E. (2003). The nitrilase ZmNIT2 converts indole-3-acetonitrile to indole-3-acetic acid. Plant Physiol. 133: 794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrásek J., Friml J. (2009). Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Pollmann S., Düchting P., Weiler E.W. (2009). Tryptophan-dependent indole-3-acetic acid biosynthesis by ‘IAA-synthase’ proceeds via indole-3-acetamide. Phytochemistry 70: 523–531 [DOI] [PubMed] [Google Scholar]
- Pollmann S., Müller A., Weiler E.W. (2006). Many roads lead to “auxin”: Of nitrilases, synthases, and amidases. Plant Biol (Stuttg) 8: 326–333 [DOI] [PubMed] [Google Scholar]
- Pollmann S., Neu D., Weiler E.W. (2003). Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry 62: 293–300 [DOI] [PubMed] [Google Scholar]
- Quittenden L.J., Davies N.W., Smith J.A., Molesworth P.P., Tivendale N.D., Ross J.J. (2009). Auxin biosynthesis in pea: characterization of the tryptamine pathway. Plant Physiol. 151: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter M.K., Padilla C.M., Schmidt R.J. (2002). The maize mutant barren stalk1 is defective in axillary meristem development. Am. J. Bot. 89: 203–210 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Sakata T., Oshino T., Miura S., Tomabechi M., Tsunaga Y., Higashitani N., Miyazawa Y., Takahashi H., Watanabe M., Higashitani A. (2010). Auxins reverse plant male sterility caused by high temperatures. Proc. Natl. Acad. Sci. USA 107: 8569–8574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson M.J. (2003). Molecular data from 27 proteins do not support a precambian origin of land plants. Am. J. Bot. 90: 954–956 [DOI] [PubMed] [Google Scholar]
- Sekimoto H., Seo M., Dohmae N., Takio K., Kamiya Y., Koshiba T. (1997). Cloning and molecular characterization of plant aldehyde oxidase. J. Biol. Chem. 272: 15280–15285 [DOI] [PubMed] [Google Scholar]
- Sekimoto H., Seo M., Kawakami N., Komano T., Desloire S., Liotenberg S., Marion-Poll A., Caboche M., Kamiya Y., Koshiba T. (1998). Molecular cloning and characterization of aldehyde oxidases in Arabidopsis thaliana. Plant Cell Physiol. 39: 433–442 [DOI] [PubMed] [Google Scholar]
- Skirpan A., Culler A.H., Gallavotti A., Jackson D., Cohen J.D., McSteen P. (2009). BARREN INFLORESCENCE2 interaction with ZmPIN1a suggests a role in auxin transport during maize inflorescence development. Plant Cell Physiol. 50: 652–657 [DOI] [PubMed] [Google Scholar]
- Skirpan A., Wu X., McSteen P. (2008). Genetic and physical interaction suggest that BARREN STALK 1 is a target of BARREN INFLORESCENCE2 in maize inflorescence development. Plant J. 55: 787–797 [DOI] [PubMed] [Google Scholar]
- Smith L.G., Hake S. (1993). A new mutation affecting tassel and ear morphology. Maize Newsletter 67: 2–3 [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Strader L.C., Bartel B. (2008). A new path to auxin. Nat. Chem. Biol. 4: 337–339 [DOI] [PubMed] [Google Scholar]
- Sugawara S., Hishiyama S., Jikumaru Y., Hanada A., Nishimura T., Koshiba T., Zhao Y., Kamiya Y., Kasahara H. (2009). Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 5430–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. (2000). PAUP*: Phylogenetic Analysis Using Parsimony. (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Tao Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivendale N.D., Davies N.W., Molesworth P.P., Davidson S.E., Smith J.A., Lowe E.K., Reid J.B., Ross J.J. (2010). Reassessing the role of N-hydroxytryptamine in auxin biosynthesis. Plant Physiol. 154: 1957–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Sauer M., Meurette O., Friml J., Ljung K., Sandberg G., Hooykaas P., Offringa R. (2005). Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell 17: 2517–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward A.W., Bartel B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., McSteen P. (2007). The role of auxin transport during inflorescence development in maize, Zea mays (Poaceae). Am. J. Bot. 11: 1745–1755 [DOI] [PubMed] [Google Scholar]
- Yamada M., Greenham K., Prigge M.J., Jensen P.J., Estelle M. (2009). The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 151: 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nodzynski T., Pencík A., Rolcík J., Friml J. (2010). PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl. Acad. Sci. USA 107: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.D., Christensen S.K., Fankhauser C., Cashman J.R., Cohen J.D., Weigel D., Chory J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zhao Y.D., Hull A.K., Gupta N.R., Goss K.A., Alonso J., Ecker J.R., Normanly J., Chory J., Celenza J.L. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16: 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]