The MYB protein DUO1 is a key determinant of sperm cell development in Arabidopsis. This study identifies a diverse range of downstream genes regulated by DUO1 and provides molecular insight into the regulatory networks associated with the differentiation of precursor germ cells into functional sperm cells.
Abstract
The male germline in flowering plants arises through asymmetric division of a haploid microspore. The resulting germ cell undergoes mitotic division and specialization to produce the two sperm cells required for double fertilization. The male germline-specific R2R3 MYB transcription factor DUO1 POLLEN1 (DUO1) plays an essential role in sperm cell specification by activating a germline-specific differentiation program. Here, we show that ectopic expression of DUO1 upregulates a significant number (~63) of germline-specific or enriched genes, including those required for fertilization. We validated 14 previously unknown DUO1 target genes by demonstrating DUO1-dependent promoter activity in the male germline. DUO1 is shown to directly regulate its target promoters through binding to canonical MYB sites, suggesting that the DUO1 target genes validated thus far are likely to be direct targets. This work advances knowledge of the DUO1 regulon that encompasses genes with a range of cellular functions, including transcription, protein fate, signaling, and transport. Thus, the DUO1 regulon has a major role in shaping the germline transcriptome and functions to commit progenitor germ cells to sperm cell differentiation.
INTRODUCTION
In flowering plants, the process of male gametogenesis takes place within specialized male reproductive organs, the stamens, where meiosis occurs to produce haploid unicellular microspores. Each microspore undergoes a highly asymmetric division to form a large vegetative cell encapsulating a small generative or germ cell. The germ cell then undergoes a second round of mitosis to produce two sperm cells. Upon successful pollination, the vegetative cell grows a pollen tube through the female stylar tissue to deliver the sperm cells to the embryo sac. While one sperm cell fertilizes the egg cell to give rise to the embryo, the other fertilizes the central cell to form the endosperm. The production of fully differentiated sperm cells is thus critical for double fertilization and has major implications for crop fertility and seed production.
Although there have been significant advances in understanding regulatory gene cascades in the sporophytic tissues that nurture male gametophyte development, relatively little is known about the scale and complexity of gametophytic regulatory networks (Wilson and Zhang, 2009). In the male gametophyte, a pioneering study established a late pollen regulatory network governed by five pollen-specific MIKC* MADS box proteins that is important in the vegetative cell for pollen maturation (Verelst et al., 2007a, 2007b). A corresponding example in the female gametophyte is a regulatory network modulated by the R2R3-type MYB transcription factor MYB98, which regulates a battery of synergid cell-expressed genes that are required for pollen tube guidance and formation of the filliform apparatus (Punwani et al., 2007). In terms of the gametes, transcriptional regulation is likely to be an important aspect in germline development as plant male germ cells have a distinct and diverse transcriptome (Engel et al., 2003; Okada et al., 2006; Borges et al., 2008). Despite this, there are currently no well-characterized regulatory networks described in either the male or female plant germline.
DUO POLLEN1 (DUO1) was the first male germline-specific transcription factor to be identified in plants, and mutation of DUO1 results in a single mutant germ cell that is unable to undergo fertilization (Durbarry et al., 2005; Rotman et al., 2005). We have subsequently shown that DUO1 influences sperm cell specification by regulating three male germline genes and integrating their expression with germ cell cycle progression through the G2/M phase-specific accumulation of CYCB1;1 (Brownfield et al., 2009a). The three genes known to be regulated by DUO1 are MGH3 (HTR10), which encodes a male germline-specific histone H3.3 variant (Okada et al., 2005; Ingouff et al., 2007), GEX2, which encodes a membrane-associated protein (Engel et al., 2005), and GCS1/HAP2, encoding an ancestral membrane-associated protein required for gamete fusion (Mori et al., 2006; von Besser et al., 2006; Hirai et al., 2008; Liu et al., 2008; Steele and Dana, 2009; Wong and Johnson, 2010). DUO3 is another regulatory protein that, like DUO1, is required to coordinate germ cell division with gamete specification (Brownfield et al., 2009b). DUO3 shares regulatory targets in common with DUO1 since it is required for the expression of GEX2 and GCS1/HAP2, although the precise reason for failed division in duo3 germ cells remains unknown as CYCB1;1 accumulation is unaffected (Brownfield et al., 2009b).
Detailed analysis of DUO1 and the discovery of novel target genes present a timely opportunity to uncover the scale and organization of a germline transcriptional network that shapes aspects of the sperm cell transcriptome and influences the differentiation of the male gametes. We recently reported that ectopic expression of DUO1 in seedlings results in the detection of known DUO1 target transcripts MGH3, GEX2, and GCS1 (Brownfield et al., 2009a). Here, we exploit this inducible system to screen for novel DUO1 target genes. We go on to describe the validation of 14 of these target genes by demonstrating DUO1-dependent promoter activity in the male germline and transactivation in transient luciferase assays. We analyze the expression profiles of several DUO1 target genes during pollen development and identify two promoters with sperm cell–specific activity. Furthermore, we describe a series of experiments that provide evidence that transactivation of DUO1 target genes involves binding by the DUO1 MYB domain to conserved sequences in target gene promoters and show that at least one of these downstream genes is a direct DUO1 target. Collectively, these data provide valuable insight into the downstream genes involved in the DUO1 male germline regulatory network and significantly expand on the current models of male gamete differentiation.
RESULTS
Novel DUO1 Targets Identified by Ectopic Expression of DUO1
We previously reported transgenic lines that ectopically express DUO1 in an estradiol-inducible manner and showed that ectopic induction of DUO1 in seedlings results in the expression of three known targets of DUO1: MGH3, GEX2, and GCS1 (Brownfield et al., 2009a). This ectopic system utilizes mDUO1, a version of the DUO1 cDNA that is resistant to miR159 (Palatnik et al., 2007). We exploited this inducible system to explore the scale of the DUO1 regulon by screening for novel DUO1 target genes. We performed comparative microarray analysis with RNA isolated from transgenic seedlings that were transferred to plates with or without inducer for 6, 12, or 24 h. RNA was isolated from three biological replicates for uninduced and induced seedlings at each time point and used to interrogate Affymetrix Arabidopsis thaliana ATH1 genome arrays.
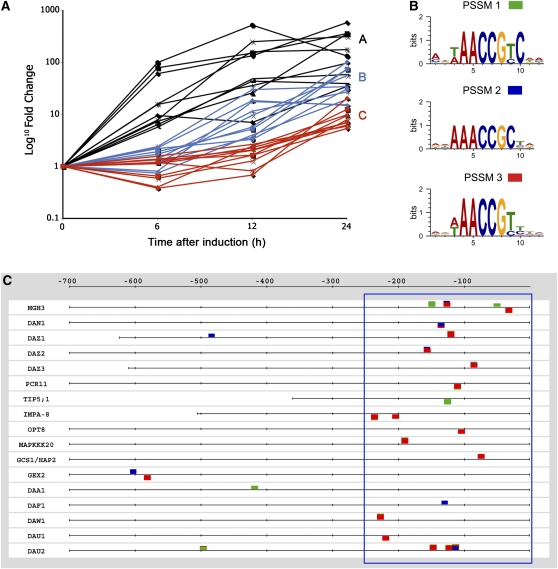
To identify putative DUO1 target genes, we applied several filters to the data from the 24-h induction. Our first filter was to only select genes that had a 3-fold change between the mean expression values of the uninduced and induced replicates and a statistically significant difference (Student’s t test, P value < 0.05). Candidate targets were also limited to those with a present call in at least two of the three induced samples. These criteria yielded a total of 124 putative target genes (see Supplemental Data Set 1 online). Given that DUO1 is specifically expressed in the male germline (Rotman et al., 2005), its target genes should also be expressed in sperm cells. Hence, our second filter was to consider only those genes with a present call in sperm cell transcriptomic data (Borges et al., 2008). Applying this criterion narrowed down the final list of candidate DUO1 targets to 63 genes, including the known DUO1 targets MGH3 and GEX2 (see Supplemental Data Set 2 online). Herein, we will refer to our final list of candidate genes as DUO1-activated target (DAT) genes. The putative DAT genes were divided into three groups based on their responses in the time course experiments (Figure 1A; see Supplemental Data Set 2 online). Group A consists of 24 genes that show a 3-fold increase between uninduced and induced samples at all time points (6, 12, and 24 h). Group B consists of 19 genes that have a 3-fold increase at 12 and 24 h, while Group C contains 20 genes that show a 3-fold increase only after 24 h of induction.
Figure 1.
MYB Binding Sites Are Overrepresented in DUO1 Target Gene Promoters.
(A) DUO1 candidate targets divided into three groups based on their response time to ectopic DUO1 expression. Group A genes (black) have a 3-fold increase after 6 h of DUO1 induction, Group B (blue) after 12 h, and Group C (red) only after 24 h of induction.
(B) Bioinformatic analysis of the promoters of candidate DAT genes identified three overrepresented MYB binding motifs. The consensus sequence of the motifs is represented as a PSSM logo.
(C) A feature map encompassing positional information of the overrepresented MYB binding motifs in the 17 validated DAT gene promoters. The colored boxes indicate the position of each MYB binding site with each color corresponding to the PSSM logos in (B).
To gain a broad perspective of the expression of the DAT genes, we referred to publicly available microarray data. The vast majority of rapidly responding genes in Group A, as well as several of the later responding genes in Group B and C, appear not be expressed in sporophytic tissues as indicated by the MAS5.0-assigned absent calls in the array data (see Supplemental Data Set 2 online). To further illustrate tissue specificity, we assessed whether DUO1-induced genes have sperm cell–enriched expression according to the transcriptomic analysis of Borges et al. (2008). Of the 63 DAT genes, nearly 80% show sperm cell–enriched expression (see Supplemental Data Set 2 online). Interestingly, the earlier responding genes in Group A contain the most sperm cell–enriched members (Groups A, B, and C consist of 87.5, 78.9, and 65.0% sperm cell–enriched genes, respectively). The number of sperm cell–enriched genes in our list of putative targets is significantly increased compared with the number occurring by chance in random lists of 63 genes called present in sperm (two-tailed χ2 test: χ2 = 6.03, P < 0.05). These findings are consistent with the male germline-specific expression of DUO1 (Rotman et al., 2005) and show it induces transcription of predominantly male germline-enriched genes.
MYB Binding Sites Are Highly Overrepresented in Candidate DUO1 Target Gene Promoters
If the candidate DAT genes are direct targets of DUO1, then their upstream regulatory regions are expected to contain common DNA motifs bound by DUO1. To explore this possibility, the motif analysis suite Regulatory Sequence Analysis Tools (van Helden, 2003; Thomas-Chollier et al., 2008) was used to identify any overrepresented motifs in the promoter regions of our candidate DAT genes. Three highly overrepresented motifs (motif 1, mwwAACCGTCwa; motif 2, awAAACCGCta; and motif 3, adwAACCGTywh) were identified that each have a typical MYB core containing the nucleotides AAC and are presented as a position-specific scoring matrix (PSSM) in Figure 1B. Each PSSM has high information content (9.7, 8.2, and 8.6, respectively), showing a high sequence conservation within the PSSM. A map of the distribution of these MYB binding sites (MBSs) among our DAT gene promoters reveals a strong bias, with the majority lying within the proximal 250 bp upstream of the start codon (Figure 1C for validated candidates, see below; see Supplemental Figure 1 online for remaining candidates). These overrepresented MBSs are likely to be important for DUO1-dependent transcription of target genes and suggest that DUO1 may activate some DAT genes directly. Consistent with this, the earliest responding genes possess on average more MBSs in their promoters than later responding genes, with Groups A, B, and C containing 1.2, 0.5, and 0.3 MBSs, respectively (see Supplemental Data Set 3 online). Furthermore, most of the genes in Group A (19 of 24) have at least one MBS within −250 bp of ATG compared with Groups B (8 of 19) and C (6 of 20). Thus, the rapid response of Group A genes may relate to the increased number of proximal MBSs and direct activation by DUO1.
Candidate DAT Genes Exhibit DUO1-Dependent Expression in the Male Germline
To validate candidate DAT genes and explore the specificity of their expression, we generated stable transgenic H2B-green fluorescent protein (GFP) marker lines driven by a selection of DAT gene promoters and monitored their expression in the duo1 background (Figure 2). A total of 19 candidate target gene promoters were tested, nine genes from Group A, five from Group B, four from Group C, and an additional gene (At5g02390) that failed the second selection filter (see Supplemental Data Set 2 online). The stringency of our selection criteria is likely to exclude native targets that have a low and/or variable response to DUO1. For example, GCS1 is a known regulatory target of DUO1 (Brownfield et al., 2009a) but is not in our final list because its response was variable between uninduced and induced samples (see Supplemental Data Set 2 online). Like GCS1, At5g02390 is called present in sperm cells (Borges et al., 2008) and its induction was >3-fold, but this induction was unreliable (see Supplemental Data Set 2 online). Thus, we included At5g02390 in our analysis to confirm that we may have missed target genes due to our strict criteria. For those target genes we validated (see below) without an official gene symbol, we have adapted the DAT nomenclature and named the genes according to their predicted function (Table 1). For example, we termed the protein encoded by At3g04620 DAN1 because it is a DUO1-activated nucleic acid binding protein.
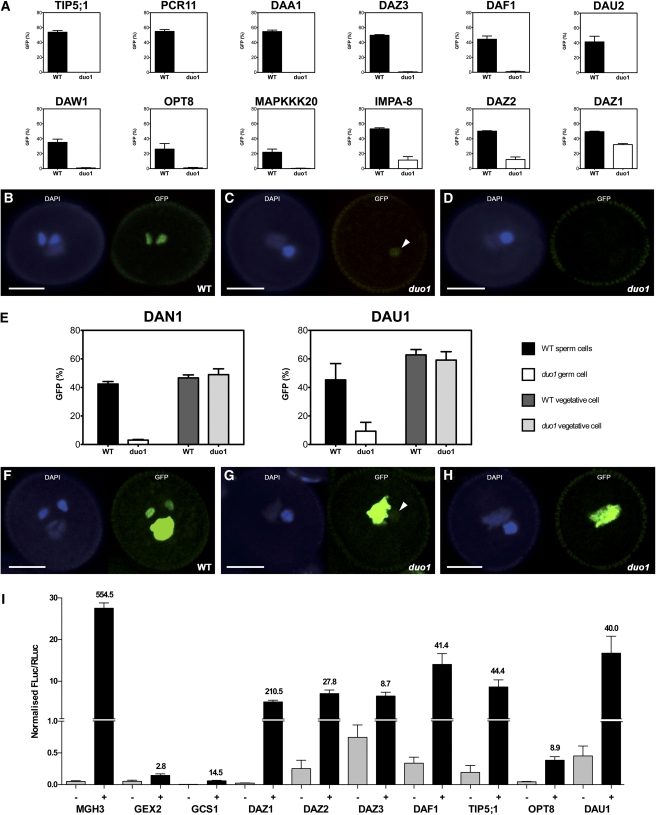
Figure 2.
A Selection of Induced Target Genes Exhibit DUO1-Dependent Expression in the Male Germline.
(A) Promoter activity of male germline-specific DAT genes. GFP expression was scored in single insert hemizygous lines in the duo1-1/+ background. Wild-type (WT) pollen grains (black) were distinguished from duo1 pollen grains (white) by germ cell number. Each bar represents the mean of at least three independent lines and error bars represent the se. All 12 promoters show significantly reduced activity in duo1 germ cells compared with wild-type sperm cells (two-tailed χ2 test: P < 0.0001).
(B) to (D) Examples of male germline-specific DAT gene promoter activity in wild-type and duo1 pollen using CLSM. Each panel shows a representative pollen grain under DAPI fluorescence (left) and GFP fluorescence (right). Sperm cell nuclei in wild-type pollen grains show a GFP signal (B), while duo1 germ cell nuclei show a residual level (C) or no detectable GFP signal (D). Arrowheads indicate a residual level of GFP signal in duo1 germ cells. Bars = 10 μm.
(E) Promoter activity of DAT genes expressed in the vegetative and sperm cells. GFP expression was scored in single insert hemizygous lines in the duo1-1/+ background for wild-type sperm cells (black), wild-type vegetative nuclei (dark gray), duo1 germ cells (white), and duo1 vegetative nuclei (light gray). Each bar chart represents the mean of at least three independent lines, and error bars show the se. The two promoters show significantly reduced activity in duo1 germ cells compared with wild-type sperm cells (two-tailed χ2 test: P < 0.0001), but there is no significant difference between activity in the vegetative cells.
(F) to (H) Examples of non-male germline-specific DAT gene promoter activity in wild-type and duo1 pollen grains using CLSM. Each panel shows a representative pollen grain under DAPI fluorescence (left) and GFP fluorescence (right). Both sperm cell and vegetative cell nuclei in wild-type pollen grains show a GFP signal (F), while duo1 germ cell nuclei either show a residual level (G) or no detectable GFP signal (H), and the GFP signal of vegetative cell nuclei is unaffected in duo1 pollen grains ([G] and [H]). Arrowheads indicate a residual level of GFP signal in duo1 germ cells. Bars = 10 μm.
(I) DUO1-dependent transactivation of validated DAT promoters in tobacco leaves. The relative luciferase activity (FLuc/RLuc) of each target promoter (ProDAT:LUC) alone (−; light gray) and upon coinfiltration with Pro35S:mDUO1 (+; dark gray) is shown with the fold change indicated above the (+) column. Each bar represents the mean of at least four independent infiltrations, and error bars show the se. A split y axis is presented in order to illustrate lower level activity.
[See online article for color version of this figure.]
Table 1.
Summary of Validated DUO1 Target Genes
| Promoter Activity |
|||||||
| Groupa | AGI | Gene | Name | GO | Protein | SC | VC |
| A | At1g19890 | MGH3/HTR10b | MALE GAMETE-SPECIFIC HISTONE H3 | Chromatin | Histone H3 | X | |
| At2g17180 | DAZ1 | DUO1-ACTIVATED ZINC FINGER1 | Transcription | C2H2-type zinc finger protein | X | ||
| At3g04620 | DAN1 | DUO1-ACTIVATED NUCLEIC ACID BINDING PROTEIN1 | DNA/RNA | Nucleic acid binding protein | X | X | |
| At3g47440 | TIP5;1 | TONOPLAST INTRINSIC PROTEIN 5;1 | Transport | Tonoplast intrinsic protein | X | ||
| At3g62230 | DAF1 | DUO1-ACTIVATED F-BOX1 | Protein fate | F-box protein | X | ||
| At4g35280 | DAZ2 | DUO1-ACTIVATED ZINC FINGER2 | Transcription | C2H2-type zinc finger protein | X | ||
| At5g39650 | DAU2 | DUO1-ACTIVATED UNKNOWN2 | Unknown | DUF679 protein | X | ||
| At5g49150 | GEX2b | GAMETE EXPRESSED2 | Signaling | Membrane protein | X | ||
| At5g52000 | IMPa-8 | IMPORTIN α-8 | Transport | Importin α | X | ||
| At5g53520 | OPT8 | OLIGOPEPTIDE TRANSPORTER8 | Transport | Oligopeptide transporter | X | ||
| B | At1g64110 | DAA1 | DUO1-ACTIVATED ATPASE1 | Protein remodeling | AAA+ type ATPase | X | |
| At1g68610 | PCR11 | PLANT CADMIUM RESISTANCE 11 | Transport | Zinc transporter | X | ||
| C | At3g50310 | MAPKKK20 | MITOGEN ACTIVATED PROTEIN KINASE KINASE KINASE 20 | Signaling | Protein kinase | X | |
| At4g35560 | DAW1 | DUO1-ACTIVATED WD40 1 | Protein fate | WD40 protein | X | ||
| At4g35700 | DAZ3 | DUO1-ACTIVATED ZINC FINGER3 | Transcription | C2H2-type zinc finger protein | X | ||
| FSC | At4g11720 | GCS1/HAP2b | GENERATIVE CELL SPECIFIC1/HAPLESS 2 | Signaling | Membrane protein | X | |
| At5g02390 | DAU1 | DUO1-ACTIVATED UNKNOWN1 | Unknown | DUF3741 protein | X | X | |
AGI, Arabidopsis Genome Initiative.
Groups defined according to response time (≥3-fold induction) in seedlings ectopically expressing DUO1 (A = 6 h, B = 12 h, and C = 24 h). FSC indicates genes that failed the selection criteria.
Genes previously validated by Brownfield et al. (2009a).
Promoter activity determined in ProDAT:H2B-GFP marker lines by GFP signal in sperm cells (SC) and vegetative cell (VC) of mature pollen.
We first confirmed expression of each candidate promoter in sperm cells by monitoring the nuclear-localized H2B-GFP reporter in mature pollen from several independent T1 lines. Of the 19 genes tested, three (At3g60780, At1g73510, and At5g08240) showed no detectable expression in pollen, while one (At5g45840) showed only vegetative cell expression (see Supplemental Data Set 2 online), even although transcripts of these four genes have been detected in Arabidopsis sperm cells (Borges et al., 2008). This suggests that the promoter activity of these genes is likely to be below the detection threshold of the H2B-GFP reporter or that sequences downstream of these promoters could be important for expression in sperm cells. These four genes were thus excluded from further analysis. Of the remaining 15 genes, 13 showed sperm cell–specific expression in mature pollen (Figure 2B; see Supplemental Data Set 2 online), while the remaining two (DAN1 and DAU1) showed fluorescence in both the sperm cell and vegetative cell (Figure 2F; see Supplemental Data Set 2 online). Having established that these 15 promoters were expressed in the male germline, we analyzed their expression in duo1 germ cells.
Heterozygous duo1 plants produce 50% wild-type pollen with two sperm cells and 50% duo1 pollen with a single germ cell (Durbarry et al., 2005), enabling pollen genotype to be determined by germ cell number. Most lines displayed GFP fluorescence in >90% of wild-type sperm cells (Figures 2A and 2B). However, several lines (DAF1, DAN1, DAU2, DAW1, OPT8, and MAPKKK20) had a reduced percentage of wild-type sperm cells with GFP fluorescence, which is likely due to a low GFP signal resulting in a lack of detection in some sperm cells (Figures 2A and 2E). For the 15 promoters analyzed, all but one (At5g60250) showed a statistically significant reduction in the proportion of germ cells with GFP fluorescence in duo1 compared with wild-type pollen (Figures 2A and 2E). In duo1 germ cells little (Figures 2C and 2G) or no (Figures 2D and 2H) GFP fluorescence was detected. For those promoters that also displayed vegetative cell activity (DAN1 and DAU1), the GFP fluorescence was unaffected in the vegetative cell of duo1 pollen (Figures 2E to 2H), with no significant difference between the percentage of GFP-positive vegetative nuclei in wild-type and duo1 pollen. These results demonstrate that DUO1 is necessary for the activation of 14 promoters in the male germline and confirms them as DUO1 target genes (Table 1).
To complement the DUO1-dependent activity of DAT promoters observed in the male germline, transient luciferase assays were conducted in tobacco (Nicotiana tabacum) leaves to determine whether DUO1 is sufficient for the transactivation of target gene promoters in a heterologous system. Reporter vectors were generated for 10 DAT genes (MGH3, GEX2, GCS1, DAZ1, DAZ2, DAZ3, DAF1, ATOPT8, TIP5;1, and DAU1) by fusion of the firefly luciferase open reading frame with each target gene promoter (ProDAT:LUC). Infiltration of leaves with Agrobacterium tumefaciens strains harboring the reporters alone resulted in a low background level of relative luciferase activity that is likely attributed to nonspecific basal transcription (Figure 2I). By contrast, coinfiltration of each ProDAT:LUC reporter strain with a Pro35S:mDUO1 effector strain resulted in an increase in activity that ranged from nearly 3-fold to >500-fold (Figure 2I). This result further demonstrates that DUO1 is sufficient for the transactivation of these target gene promoters in a heterologous system, which implies that their activation is more likely to be direct rather than through intermediate regulators.
Developmental Expression Profiles of DUO1 Target Genes
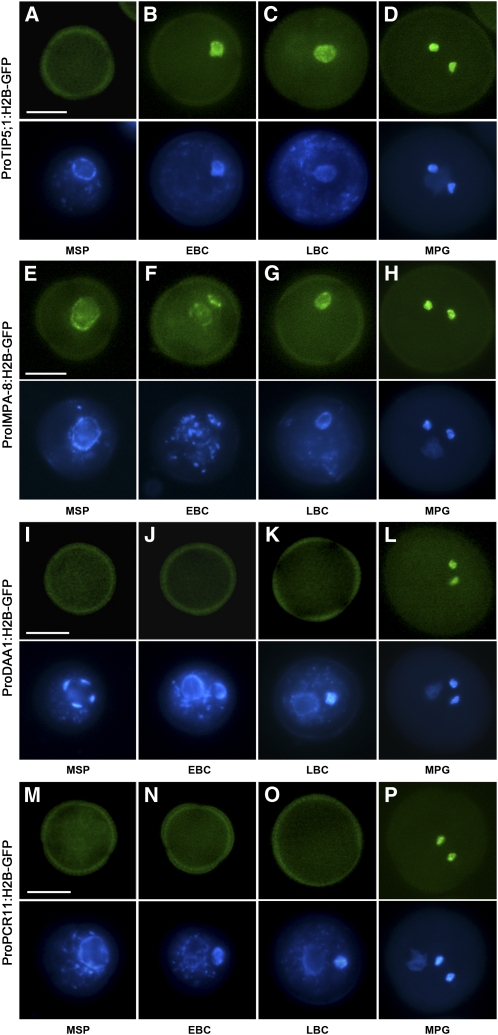
Previous analysis of DUO1 target genes has shown that the DUO1 target genes MGH3, GEX2, and GCS1 are expressed throughout male germline development soon after asymmetric microspore division through to mature pollen (Brownfield et al., 2009a). To determine if other DUO1 target genes show similar profiles, we examined the developmental expression of a subset of DAT genes, namely, TIP5;1, IMPa-8, DAA1, PCR11, and DAN1. The activity of the TIP5;1 promoter was typical of the other three genes we have previously described (Brownfield et al., 2009a), with GFP fluorescence absent from the microspore (Figure 3A) and appearing specifically in the germ cell soon after its inception and persisting in sperm cells of mature pollen (Figures 3B to 3D).
Figure 3.
Promoter Activity of DUO1 Target Genes during Male Gametogenesis
Expression of ProTIP5;1:H2B-GFP ([A] to [D]), ProIMPa-8:H2B-GFP ([E] to [H]), ProDAA1:H2B-GFP ([I] to [L]), and ProPCR11:H2B-GFP ([M] to [P]) during wild-type pollen development as observed with fluorescence microscopy. Representative pollen grains show GFP fluorescence (top, green) and DAPI fluorescence (bottom, blue) at microspore (MSP), early bicellular (EBC), late bicellular (LBC), and mature pollen (MPG) stages. Bars = 10 μm.
In contrast with TIP5;1, IMPa-8 promoter activity was clearly detectable at the microspore stage (Figure 3E). While the fluorescence signal in the vegetative cell appears to decline following microspore division (Figure 3F), it continues to accumulate in the incipient germ cell and persists in sperm cells (Figures 3F to 3H). This is consistent with DUO1-independent IMPa-8 expression in the microspore followed by DUO1-dependent expression in the germline, with a declining vegetative cell signal due to turnover of inherited H2B-GFP protein. Unlike IMPa-8, no fluorescence signal is detected at the microspore stage in DAN1 marker lines (see Supplemental Figure 2A online), with promoter activity first detected after microspore division in both the vegetative cell and the incipient germ cell (see Supplemental Figure 2B online). While DUO1-independent DAN1 promoter activity appears to peak midway through pollen development in the vegetative cell (see Supplemental Figure 2C online), the DUO1-dependent activity in the germline increases progressively and persists in mature pollen (see Supplemental Figures 1C to 1F online).
Interestingly, the promoter activity of the two target genes DAA1 and PCR11 appears to be sperm cell specific. A GFP fluorescence signal in these marker lines was detected only after germ cell division but not earlier in development (Figures 3I to 3L for DAA1 and Figures 3M to 3P for PCR11). These genes both belong to Group B, which respond to DUO1 expression only after 12 h (see Supplemental Data Set 2 online), a delay that also appears to be evident during male germline development. Collectively, these observations highlight the diverse expression patterns and complex regulation of male germline-expressed DUO1 target genes during pollen development.
DAT Genes Belong to Several Functional Classes
To determine the range of functions potentially regulated by DUO1, we undertook gene ontology (GO) analysis on the list of 65 candidate DAT genes. Although not part of our final list, GCS1 and DAN1 were also included since they are both validated DAT genes (Brownfield et al., 2009a; Figure 2E). Using the Classification SuperViewer tool at the Bio-Array Resource (Provart and Zhu, 2003), the 65 genes were functionally classified according to the three main GO categories (biological process, molecular function, and cellular component) and the level of representation compared with all Arabidopsis genes (see Supplemental Figure 2 online).
In terms of molecular function, transporter activity (GO:0005215), hydrolase activity (GO:0016787), nucleic acid binding (GO:0003676), and other binding (GO:0005488) are significantly overrepresented in our list of DAT genes (see Supplemental Figure 3 online). Genes associated with the endomembrane system are also highly overrepresented, with the endoplasmic reticulum (GO:0005783) most notably being a significantly overrepresented GO term (see Supplemental Figure 3 online). In parallel, we also used the DAVID Gene Functional Classification tool (Huang et al., 2008) to group this list of DAT genes into functionally related classes based on significant co-occurrences of GO annotation terms (see Supplemental Data Set 4 online). The enrichment scores for the condensed groups were calculated by comparing the 65 DAT genes with all genes called present in Arabidopsis sperm cells (Borges et al., 2008). The low enrichment scores indicate that no functional classes are highly enriched compared with the sperm cell transcriptome, suggesting that DUO1 has a wide role in sperm cell specification rather than regulating defined pathways. Consistent with the idea that DUO1 regulates genes important during male germline development, genes involved in intracellular transport are enriched in both the sperm cell transcriptome (Borges et al., 2008) and candidate DAT genes.
While not showing a significant overrepresentation in comparison to the entire Arabidopsis genome, other functional classes enriched in the sperm cell transcriptome are also present in our list of DAT genes and include ubiquitin-degradation pathways, ATPase activity, and G-protein signal transduction (see Supplemental Data Set 2 online). Although transcription factor activity appears to be underrepresented in our DAT gene list compared with the Arabidopsis genome (see Supplemental Figure 2 online), four putative transcription factors were induced, and three that belong to the C2H2-type zinc finger family of proteins (DAZ1, DAZ2, and DAZ3) have been validated as DUO1-regulated target genes (Figure 2A).
MYB Binding Sites Are Critical for Transactivation of the MGH3 Promoter
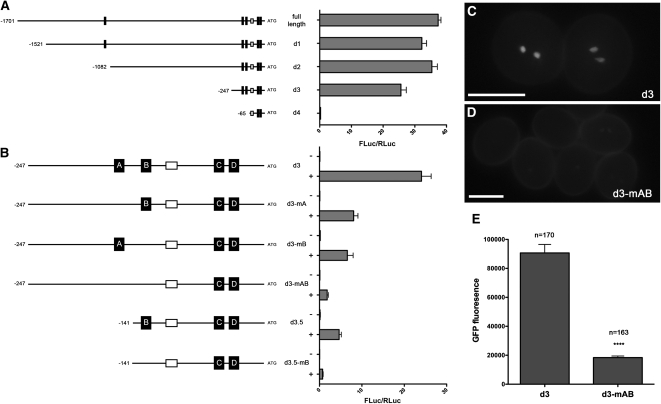
Having validated several DAT genes and identified overrepresented MBS motifs in DAT promoter regions, we investigated the mechanism through which DUO1 regulates its targets. As a model target promoter, we chose the MGH3 promoter since it is strongly activated by DUO1 (Figure 2I; see Supplemental Data Set 2 online) and contains five MBSs, four of which surround a canonical TATA box within the −250-bp region upstream of the ATG start codon (Figure 4A). A 5′ deletion series was analyzed using transient luciferase assays to narrow down regions of the MGH3 promoter required for DUO1-dependent transactivation (Figure 4A). Deletions involving the removal of sequence up to −1082 bp, including the MBS furthest upstream of ATG, showed a small reduction in relative luciferase activity (rLUC) compared with the full-length promoter. A clear decrease in rLUC was observed in deletion 3 (d3), where removal of a 1454-bp sequence reduced activity by ~30% (Figure 4A). The remaining 247 bp of promoter fragment has 69% the activity of the full-length MGH3 promoter and contains four MBSs surrounding the TATA box (Figures 4A and 4B). Further removal of the region harboring the two MBSs upstream of the TATA box (deletion 4) all but abolished rLUC (Figure 4A).
Figure 4.
MYB Binding Sites Are Critical for DUO1-Dependent Transactivation of the MGH3 Promoter.
(A) Relative luciferase activity of 5′ deletions in the MGH3 promoter. Left shows the promoter sequences highlighting MYB sites (black boxes) and the canonical TATA box (white box). Right shows the mean relative luciferase activity (FLuc/RLuc) for at least four independent infiltrations, and error bars show the se.
(B) The significance of MYB sites for DUO1-dependent transactivation of the MGH3 promoter. Left shows the promoter fragments with targeted mutations and/or deletions in context of d3 of the MGH3 promoter with MYB sites named A to D. Right shows the mean FLuc/RLuc for at least four independent infiltrations, and error bars show the se.
(C) Example of ProMGH3d3:H2B-GFP activity in mature pollen. Bar = 15 μm.
(D) Example of ProMGH3d3_mAB:H2B-GFP activity in mature pollen. A clear decrease in GFP signal was observed in several independent lines. Error bars represent the se. Bar = 15 μm.
(E) The decrease in GFP signal observed in ProMGH3d3_mAB:H2B-GFP lines. GFP fluorescence represents the mean total pixel intensity corrected for background in (n) sperm cells analyzed. Asterisks indicate statistically significant differences determined using a two-tailed Mann-Whitney U test (P < 0.0001, U = 2768).
This result indicates that these proximal MBSs are likely to play a major role in DUO1-dependent transactivation of the MGH3 promoter. This hypothesis was tested in context of d3 by generating further deletions and promoter fragments in which MBSs in this promoter fragment (termed A and B) were specifically ablated by site-directed mutagenesis (Figure 4B). Mutagenesis of MBS A (d3-mA) or B (d3-mB) reduced the rLUC by 67 and 73% compared with d3, respectively (Figure 4B). A fragment mutating both these MBSs (d3-mAB) reduced the activity even further, demonstrating that both MBSs act synergistically to account for 93% of the rLUC of d3 (Figure 4B). This residual activity is most likely due to the two remaining MBSs (C and D) lying just upstream of the ATG start codon (Figure 4B). A deletion fragment prior to MBS B (d3.5) reduced the rLUC by 81% compared with d3, which is lower than that observed with the d3-mA fragment (Figure 4B), indicating that the sequence context surrounding the MBS is important for transactivation by DUO1. Finally, mutagenesis of MBS B in context of d3.5 almost abolished luciferase activity, showing just 3% activity of the native d3.5 promoter (Figure 4B). These results demonstrate that these MBSs are critical for DUO1-dependent transactivation of the MGH3 promoter.
To determine whether the MBSs analyzed are necessary for expression of MGH3 in the male germline, a two-way comparison of promoter activity in mature pollen was performed between the activity of d3 of the MGH3 promoter (ProMGH3d3), and the same deletion in which MYB sites A and B were both mutagenized (ProMGH3d3_mAB). Both promoters were used to drive expression of an H2B-GFP fusion and their activity was monitored in wild-type plants. The GFP signal in the ProMGH3d3:H2B-GFP lines (Figure 4C) was stronger than that observed in the ProMGH3d3_mAB:H2B-GFP lines (Figure 4D). To assess this difference quantitatively, a pooled pollen sample from six single locus lines was analyzed by microscopy and the GFP fluorescence of sperm cell nuclei quantified. As shown in Figure 4E, mutagenesis of both MBSs in ProMGH3d3_mAB significantly reduced the GFP signal by almost 5-fold (Mann-Whitney U test; U = 2768, P < 0.0001). These data demonstrate that the MBSs present in the MGH3 promoter are necessary for high levels of MGH3 expression in the male germline.
DUO1 Is Able to Bind to a MYB Binding Site in the MGH3 Promoter through Its R2R3 MYB Domain
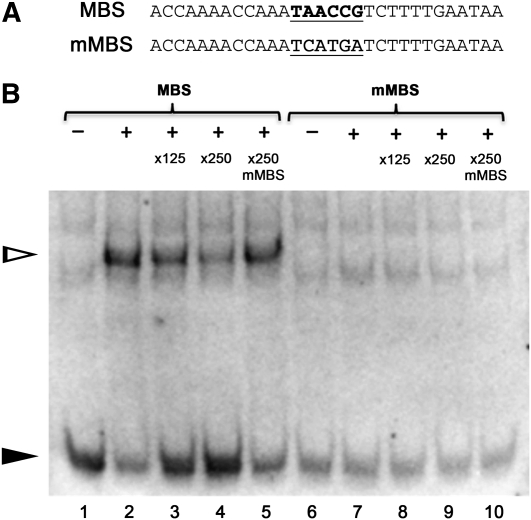
To complement our findings that demonstrate the importance of MBSs for DUO1-dependent transactivation, an electromobility shift assay (EMSA) was performed to determine whether the DUO1 MYB domain is able to bind to these MBSs in vitro. EMSA assays were performed with recombinant DUO1 MYB domain and an oligonucleotide containing MBS-A of the MGH3 promoter (MBS; Figure 5A). DUO1 is able to bind this oligonucleotide in vitro and this binding is competed with excess unlabeled MBS competitor (Figure 5B). As a control, an additional oligonucleotide was assayed in which the MBS was mutagenized (mMBS; Figure 5A). DUO1 is unable to bind to mMBS, and an unlabeled version of this oligonucleotide is unable to compete with binding of DUO1 to the MBS oligonucleotide (Figure 5B). These biochemical data demonstrate that DUO1 is able to bind specifically to a MBS present in the promoter of the target gene MGH3.
Figure 5.
DUO1 Binds in Vitro to a MYB Binding Site in the MGH3 Promoter.
(A) Sequences of the oligos used in the EMSA experiments. MBS is the region flanking MYB site A in the MGH3 promoter (bold and underlined), and mMBS is a mutagenized version in which the MYB site has been ablated (underlined).
(B) EMSA experiments using recombinant DUO1 MYB domain with MBS or mMBS oligos. – and + indicate the absence or presence of the DUO1 MYB domain, respectively, and ×125, ×250, and ×250 indicate competing unlabeled oligos. Addition of DUO1 MYB protein (lane 2 to 5) causes a clear shift (white triangle) accompanied by a reduction in free probes (black triangle), with free probes increasing in the presence of more competing MBS oligos. In lanes 7 to 10, no shift was observed in EMSA experiments using mMBS oligos. Results were confirmed in independent EMSA experiments.
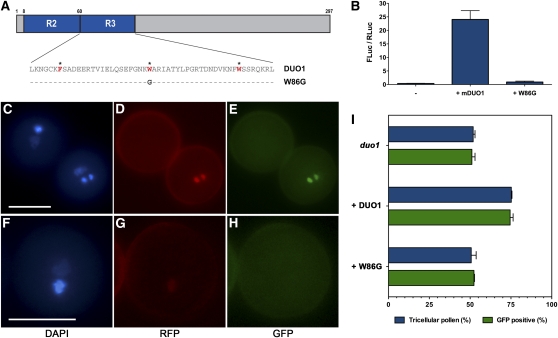
We wanted to confirm that the interaction between DUO1 and the MBS occurs through the DUO1 MYB domain. Thus, we generated a variant cDNA in which the DNA binding capacity of the MYB domain is compromised. Repeated Trp residues in the MYB domain form a hydrophobic scaffold that maintains a helix-turn-helix structure and DNA binding activity (Saikumar et al., 1990), so a variant mDUO1 cDNA was generated by substituting the second Trp of the R3 repeat at position 86 with a Gly residue (mW86G; Figure 6A). To determine whether this mW86G variant was able to function, its ability to transactivate the MGH3 promoter was compared with mDUO1 in transient luciferase assays. To increase sensitivity of the assay, the effector cDNAs had a disrupted miR159 binding site (mDUO1 and mW86G). While coinfiltration with Pro35S:mDUO1 caused a 71.8-fold increase in relative luciferase activity, coinfiltration with Pro35S:mW86G resulted in only 2.7-fold induction (Figure 6B). This low level of luciferase activity demonstrates that the integrity of the DUO1 R2R3 MYB domain is important for wild-type levels of MGH3 promoter transactivation.
Figure 6.
DNA Binding Integrity of the R3R3 MYB Domain Is Critical for DUO1 Function.
(A) Schematic diagram of the DUO1 protein with the R2 and R3 MYB repeats shown in blue. Below is the amino acid sequence of the R3 MYB repeat, with the red and asterisked amino acids representing the hydrophobic scaffold. Below the native sequence (DUO1) is the variant sequence (W86G) showing the mutation that compromises DNA binding capacity.
(B) Transactivation of the MGH3 promoter by mDUO1 and mW86G. Bars show the relative luciferase activity (Fluc/Rluc) of ProMGH3:LUC in tobacco leaves with and without coinfiltration of Pro35S:mDUO1 and Pro35S:mW86G in at least four independent infiltrations, with error bars showing the se.
(C) to (H) Mature pollen from transgenic lines expressing ProDUO1:DUO1-mRFP and ProDUO1:W86G-mRFP in duo1/+ plants homozygous for a ProMGH3:H2B-GFP marker. Each panel from left to right shows the same pollen grains with DAPI, mRFP, and GFP fluorescence as illustrated below the images.
(C) to (E) Top left is an unrescued duo1 pollen grain with a bicellular phenotype (C) and no expression of the MGH3 marker (E). Bottom right is a fully rescued pollen grain, with expression of ProDUO1:DUO1-mRFP (D) restoring tricellularity (C) and MGH3 marker expression (E).
(F) to (H) duo1 pollen grains expressing ProDUO1:W86G-mRFP (G) remain bicellular (F) and do not express the MGH3 marker (H).
(I) Ability of ProDUO1:DUO1-mRFP and ProDUO1:W86G-mRFP to rescue germ cell division (blue column) and MGH3 transactivation (green column) in duo1 pollen. In duo1/+ plants (top columns), ~50% of the pollen is wild type and has two GFP-positive sperm cells. Expression of DUO1-mRFP (middle columns) rescues both germ cell division and activates MGH3 expression in the 50% of duo1 germ cells (25% total pollen) that contain the transgene. Expression of W86G-mRFP (bottom columns) is unable to rescue germ cell division or transactivate the MGH3 marker line. Bars show the mean of at least four hemizygous single locus lines, and error bars represent the se.
[See online article for color version of this figure.]
To determine whether DNA binding capacity was essential for DUO1 function in the male germline, the potential of the W86G variant to complement the duo1 mutation was compared with that of native DUO1. Expression constructs were created in which DUO1 and W86G were fused to monomeric red fluorescent protein (mRFP) and driven by the DUO1 promoter to generate ProDUO1:DUO1-mRFP1 and ProDUO1:W86G-mRFP and stably introduced into duo1-1/+ plants homozygous for a ProMGH3:H2B-GFP marker. As predicted in single locus lines, the percentage of tricellular pollen expressing GFP in independent T1 plants hemizygous for the ProDUO1:DUO1-mRFP transgene (n = 5) was increased to ~75% (Figure 6I), showing rescue of the duo1 phenotype by restored mitotic division and GFP expression in the half of the duo1 pollen that contains the transgene (Figures 6C to 6E). Conversely, in plants hemizygous for a single ProDUO1:W86G-mRFP transgene (n = 5), neither mitotic rescue nor transactivation of the ProMGH3:H2B-GFP marker line was observed (Figure 6I). This lack of complementation is supported by the detection of W6G-mRFP fluorescence in GFP-negative duo1 germ cells in W86G lines (Figures 6F to 6H). These experiments provide robust evidence that DNA binding capacity of the R2R3-MYB domain is essential for the transactivation of DUO1 target genes in the male germline.
DISCUSSION
Here, we pioneered a detailed exploration of the plant germline regulatory network that is modulated by the MYB transcription factor DUO1. Building on our previous observations that DUO1 is required for sperm cell specification, we have shown that DUO1 regulates the expression of a plethora of sperm cell expressed genes by identifying 61 new candidate targets through the ectopic expression of DUO1 in seedlings. Since we employed strict criteria for the selection of candidate target genes, this number is likely to be an underrepresentation. Indeed, GCS1 and DAU1 both failed to meet our selection criteria but were validated as DUO1 target genes. Our strict selection criteria and the demonstration of DUO1 dependence for the majority (13 of 14) of candidate target gene promoters tested in the male germline give us confidence that most candidates represent native DUO1 target genes. This has increased the number of validated DUO1 target genes from three to 17 and has identified nearly 50 additional genes that are likely to be DUO1 targets. This increased knowledge of genes that form part of the DUO1 regulon reveals the functional diversity of pathways under DUO1 regulation.
DUO1 Positively Regulates a Host of Functionally Diverse Target Genes
During male germline development, a diverse array of genes is expressed as shown by the unique transcriptional profile of Arabidopsis sperm cells (Borges et al., 2008). DUO1 plays a critical role in regulating sperm cell production, and as such its downstream target genes are likely to be important for the development of sperm cells as well as for fertilization. Our investigation of DUO1 target genes revealed that the majority are preferentially expressed in the male germline and belong to several functional classes (Table 1).
The apparent overrepresentation of transmembrane transporter proteins in our list of candidate DAT genes (see Supplemental Figure 2 online) suggests that DUO1 is likely to play a significant role in shaping this aspect of the sperm cell transcriptome. Our analysis validated four DUO1-dependent transport-associated genes (Table 1). PCR11 belongs to the PLANT CADMIUM RESISTANCE family of genes, which encodes small plasma membrane proteins involved in the efflux of heavy metals in Arabidopsis (Song et al., 2004, 2010). Another DUO1 target that could have a role in metal transport is OPT8, one of nine oligopeptide transporter (OPT) proteins in Arabidopsis. OPT proteins have been implicated in the transport of diverse substrates (Lubkowitz, 2006), with OPT3 functioning in metal homeostasis and transport during Arabidopsis seed development (Stacey et al., 2008).
TIP5;1 belongs to a subclass of the plant major intrinsic protein family of aquaporins called tonoplast intrinsic proteins (TIPs; Forrest and Bhave, 2007). TIP5;1 is involved in the transport of water and urea (Vander Willigen et al., 2006; Soto et al., 2008) as well as of boron in seedlings (Pang et al., 2010). TIP5;1 localizes to mitochondria, and tip5;1 pollen tubes grow shorter in the absence of exogenous nitrogen (Soto et al., 2010). Interestingly, the TIP5;1 transcript is enriched in sperm cells (Borges et al., 2008), and TIP5;1 promoter activity appears to be sperm cell specific in mature pollen (Figure 3D). This suggests that sperm cell function can influence pollen tube growth and that TIP5;1 could have an important role in nitrogen recycling within sperm cells. Another DUO1 target involved in transport activity is IMPa-8, which belongs to the importin α class of adaptor proteins involved in nuclear protein import (Lange et al., 2007).
Protein metabolism is likely to be an important process during germline differentiation, and as such, genes involved in ubiquitin-mediated proteolysis are highly enriched in the sperm cell transcriptome (Borges et al., 2008) and have an important role in germ cell division (Kim et al., 2008). DUO1 appears to have a role in regulating ubiquitin proteasome-dependent proteolysis since the expression of DAF1, an F-box protein, is under DUO1 regulation. Another target, DAA1, belongs to a large superfamily of ATPases that are involved in diverse processes, including protein unfolding and degradation as well as in DNA recombination, replication, and repair (Snider and Houry, 2008).
The chromatin state of sperm cells, both its highly condensed structure and its epigenetic marks, may be important for successful karyogamy and subsequent development of the zygote and endosperm (Ingouff et al., 2007; Berger et al., 2008). DUO1 is known to regulate genes involved in chromatin structure since the male gamete-specific histone H3 MGH3 (HTR10) is a DUO1 target gene (Brownfield et al., 2009a). Similarly, DAN1 has a putative role in chromatin organization due its similarity with Alba (Bell et al., 2002; Sandman and Reeve, 2005), an archaeal protein that affects the topology of chromosomal DNA in a temperature-dependent manner (Xue et al., 2000).
DUO1 also regulates a number of putative transcription factors. Three of the validated target genes, DAZ1, DAZ2, and DAZ3, all belong to the C2H2-type zinc finger family of transcription factors (Englbrecht et al., 2004) and are among the most highly expressed genes in Arabidopsis sperm cells (Borges et al., 2008). C2H2-type zinc finger transcription factors are known to influence various developmental processes, including leaf/shoot initiation (Takatsuji, 1998, 1999), floral organogenesis (Bowman et al., 1992; Sakai et al., 1995), gametogenesis (Kobayashi et al., 1998), and seed development (Gaiser et al., 1995; Meister et al., 2002). The DUO1-dependent germline expression of three DAZ genes (Figure 2) suggests that DUO1 is responsible for activating subordinate regulons during male germline differentiation.
Cell-to-cell communication between pollen tubes and female stylar tissues is critical during plant reproduction, and such signaling processes appear to be at least partially under DUO1 regulation. DUO1 regulates the expression of GCS1/HAP2 (Brownfield et al., 2009a), an ancestral signaling molecule that is required for efficient pollen tube guidance (von Besser et al., 2006) and fertilization (Mori et al., 2006). Our demonstration that MAPKKK20 is under DUO1 regulation (Figure 2A) suggests that DUO1 also regulates mitogen-activated protein kinase (MAPK) pathways in the germline. Interestingly, MAPKKK20 is phylogenetically related to FERTILIZATION-RELATED KINASE2 from Chaco potato (Solanum chacoense), which is involved in seed and fruit development (Gray-Mitsumune et al., 2006) as well as pollen development and viability (O’Brien et al., 2007).
The DUO1 target gene DAW1 encodes a protein homologous to the lethal giant larvae (Lgl) family of tumor suppressor pathway genes, which play critical roles in cell polarization in a variety of eukaryotic organisms (Baek, 1999; Bilder, 2001; Humbert et al., 2003; Justice and Jan, 2003; Klezovitch et al., 2004). Recent characterization of DAW1 in Arabidopsis extends Lgl conservation to the plant kingdom since DAW1 is able to partially substitute for the function of yeast Lgl homologs (Forsmark, 2009). Furthermore, daw1 mutations result in decreased lateral root growth, consistent with a role for DAW1 in cell and tissue polarity (Forsmark, 2009).
DUO1 Target Genes Are Differentially Expressed during Male Germline Development
Not only does DUO1 appear to regulate proteins with a diverse range of functions, but there also appears to be some variability in the expression pattern of DUO1 target genes. We previously reported that sperm cell specification begins early after inception of the germline, with the expression of target genes mirroring DUO1 expression and beginning shortly after asymmetric division of the microspore (Brownfield et al., 2009a). Our developmental analysis of several DUO1 target genes has verified our previous findings but also identified a novel class of male germline genes that show sperm cell–specific expression (DAA1 in Figures 3I to 3L and PCR11 in Figures 3M to 3P). Interestingly, DAA1 and PCR11 were induced above 3-fold only after 12 h of induction (Group B; see Supplemental Data Set 2 online), mirroring the delayed activation of these promoters in the germline. This delayed response to DUO1 suggests that these promoters could be subject to chromatin modifications and/or derepression only in the presence of DUO1. The identification of sperm cell–specific genes highlights the varied temporal patterns of gene expression operating in the male germline and alludes to a sperm cell–specific differentiation program in which DUO1 plays a regulatory role. The DAA1 and PCR11 promoters offer novel molecular tools for the manipulation of gene expression specifically in Arabidopsis sperm cells.
The use of previously assigned criteria to characterize the sperm cell–expressed genes (Borges et al., 2008) indicates that the majority of DUO1 target genes appear to be enriched in sperm cells. However, in contrast with the male germline-specific expression of DUO1 (Rotman et al., 2005), some DUO1 target genes show expression in sporophytic tissues and/or other cell types of the male gametophyte (see Supplemental Data Set 2 online). We have shown that two DUO1 target gene promoters, DAN1 and DAU1, are active in the vegetative cell, while the IMPa-8 promoter shows microspore activity that subsequently becomes sperm cell specific. This indicates that germline-independent mechanisms also operate to regulate the expression of some DUO1 target genes. Consistent with this, the expression of DAN1 and DAU1 in the vegetative cell is unaffected in duo1 pollen grains (Figure 2E). It is likely that parallel mechanisms involving other male germline regulators also contribute to the activation of DAT genes as five DUO1 target genes (IMPa-8, DAZ1, DAZ2, DAN1, and DAU1) have a residual level of expression in duo1 germ cells (Figures 2A and 2E). Indeed, germline expression of GEX2 and GCS1 has been shown to be dependent on the regulator DUO3 (Brownfield et al., 2009b). However, the low residual expression suggests that DUO1 is the major factor required for the activation of these DAT genes in the male germline.
DUO1 Binds to a MYB Consensus Sequence and Directly Activates MGH3
An important aspect in defining a transcriptional network is to determine whether the relationship between a transcription factor and its target genes is direct or indirect. We identified conserved MYB sites that occur frequently in the promoter region of DAT genes (Figures 1B and 1C; see Supplemental Data Set 3 online). The MYB sequences each contain a typical MYB core AAC, which is bound by several other R2R3-type MYB transcription factors, including C1 (Roth et al., 1991), P (Grotewold et al., 1994), MYB.Ph3 (Solano et al., 1995), GAMYB (Gubler et al., 1995), and MYB98 (Punwani et al., 2007). The majority of MYB binding sites are present in the −250-bp proximal region upstream of ATG and are often adjacent to one another (Figure 1C), suggesting that this architecture may be an important feature of DUO1 target promoters. We have shown that these MYB sequences are required for DUO1-dependent transactivation of the MGH3 promoter (Figure 4). Furthermore, the DUO1 MYB domain is able to bind to one of these proximal MYB binding sites in vitro (Figure 5), strongly suggesting that DUO1 directly activates MGH3 through these motifs.
While the MGH3 gene appears to be a direct target of DUO1, it is not known for certain whether this is the case for the other DAT genes in this regulon. Most of the DAT genes, including the 16 validated DATs, contain at least one MBS motif in their promoter, strongly suggesting that they too are direct DUO1 targets. Thus, DUO1 appears to directly regulate numerous genes involved in male germline development, and it is likely that the timing and magnitude of activation depends upon factors such as the architecture of the promoter region, the number and arrangement of MBS motifs, and interactions with other regulatory proteins. It is worth noting that other candidate genes upregulated in seedlings in response to ectopic DUO1 expression do not contain an MBS motif (see Supplemental Figure 3 online). These genes may be indirectly regulated by DUO1 through intermediate factors that are themselves under DUO1 regulation.
In conclusion, we uncovered an essential germline-specific regulon that specifies major features of male germ cell fate in Arabidopsis. The target genes that form the DUO1 regulon highlight the range of functions required for sperm cell formation as well as the unique characteristics integral to fertilization. In particular, target genes like GCS1/HAP2, which is essential for pollen tube guidance and fertilization, as well as TIP5;1, which has a conditional role in pollen tube growth, demonstrate the importance of the DUO1 regulon in sperm cell development and function. Future work must endeavor to define the regulatory networks operating in the male germline by uncovering the full complement of genes regulated by DUO1 and other key germline determinants like DUO3. The analysis of downstream regulatory proteins, including the zinc finger proteins identified in this work, coupled with comparative analysis in other species like rice (Oryza sativa), will help to establish robust models of the regulatory networks that modulate male gamete differentiation in plants.
METHODS
Plant Material and Transformation
Arabidopsis thaliana plants were grown at 21°C with a 16-h-light and 8-h-dark cycle or with 24 h light (120 to 140 μmol/m2/s) with variable humidity. Marker lines were generated in the +/duo1-1 Nossen-0 background or in wild-type Columbia-0 for ProMGH3d3 and ProMGH3d3_mAB marker lines. Plants were transformed with Agrobacterium tumefaciens (GV3101) using a standard floral dipping method (Clough and Bent, 1998). Transformants were selected on soil with 30 μg/mL BASTA (glufosinate ammonium; DHAI PROCIDA) fed by subirrigation or on Murashige and Skoog agar containing 50 mg/mL kanamycin.
Microarray Analysis
Comparative microarray analysis was performed with RNA isolated from transgenic 12-d-old seedlings harboring pMDC7-mDUO1 and grown in the presence and absence of inducer for 6, 12, and 24 h as described by Brownfield et al. (2009a). RNA was isolated from three biological replicates using an RNeasy Mini Kit (Qiagen) and hybridized to Affymetrix Arabidopsis ATH1 genome arrays at the NASCarrays facility. The raw expression data generated from these 18 experiments are available through the NASCarrays repository. The expression data were normalized using the MAS5.0 algorithm and scaled with a 2% trimmed mean. Data from triplicate experiments were used to calculate a mean expression value for each probe set, and the fold change between the uninduced and induced values determined at each time point.
Vector Construction
Single and multisite Gateway recombination (Invitrogen) was used to generate vectors as described by Brownfield et al. (2009a). The promoter region of candidate genes was amplified from Columbia-0 genomic DNA. The firefly and Renilla luciferase open reading frames were amplified from pRT2ΩLUC (Man-Kim Cheung) and pK82 (Ralph Panstruga), respectively. DUO1 variants were amplified from existing clones (Rotman et al., 2005) or from a miR159-resistant DUO1 cDNA (Palatnik et al., 2007). PCR reactions were performed with high-fidelity Phusion DNA polymerase (Finnzymes) and primers with suitable attachment (attB) adapters (see Supplemental Data Set 5 online; attB adapters in bold). Promoter fragments were cloned into pDONRP4P1R and coding regions into pDONR221 and verified by sequencing. The entry clones generated and those described by Brownfield et al. (2009a) were recombined using Multisite Gateway LR reactions with LR Clonase II Plus (Invitrogen) into the destination vectors pB7m34GW or pK7m24GW,3 (Karimi et al., 2005) to generate ProDAT:H2B-GFP marker constructs and ProDAT:LUC luciferase reporter constructs, respectively. Pro35S:mDUO1 and Pro35S:mW86G effector constructs and the Pro35S:RenLUC control construct were generated by single-site Gateway recombination into pB2GW7 (Karimi et al., 2002) using LR Clonase II.
Microscopy Analysis
Mature pollen was stained with 4',6-diamidino-2-phenylindole (DAPI) as described previously (Park et al., 1998). For developmental analysis, pollen from buds at different stages of development was teased out of staged anthers with a hypodermic needle and mounted directly into DAPI solution or 0.3 M mannitol. Fluorescence and confocal laser scanning microscopy (CLSM) were performed using the methods and equipment described by Brownfield et al. (2009a).
Marker Line Analysis
For analysis of H2B-GFP marker lines, mature pollen shed from ~24 T1 lines were stained with DAPI and analyzed by fluorescence microscopy. Representative duo1/+ lines with a reasonable GFP signal and an apparent single locus line (i.e., having ~50% GFP-positive wild type) were chosen for further analysis. Detailed counts of GFP-positive and GFP-negative sperm and germ cells in the wild type and duo1 portion of the population, respectively, in at least three independent T1 lines for each marker analyzed. For markers also expressed in the vegetative cells, counts of GFP-positive and GFP-negative vegetative cell nuclei in wild-type and duo1 pollen were also performed.
Complementation Analysis
For analysis of complementation lines, mature pollen from ~48 T1 lines was examined by fluorescence microscopy. The T1 lines were initially screened to identify representative duo1/+ lines with a single insertion for the transgene (i.e., having ~50% RFP-positive pollen grains) and a comparative mRFP signal. For all of the lines analyzed, the frequency of duo1 pollen grains was determined by scoring the number of bicellular and tricellular pollen grains by DAPI staining. The ability of the variants to transactivate the marker line was assessed by counting the frequency of pollen grains expressing GFP. The efficiency of rescue of the cell cycle defect was determined by expressing the number of observed rescued tricellular pollen grains as a percentage of the scored population. Similarly, the efficiency of the variants to transactivate the MGH3 marker line was determined by expressing the number of GFP-positive pollen grains as a percentage of pollen grains scored for GFP expression.
Quantification of GFP Fluorescence
For quantification of GFP fluorescence in ProMGH3d3:H2B-GFP and ProMGH3d3_mAB:H2B-GFP marker lines, mature pollen from ~24 T1 lines were examined by fluorescence microscopy to identify six representative single locus lines (i.e., having ~50% GFP-positive pollen grains). A pooled pollen sample from the six representative lines was analyzed using a Nikon ECLIPSE 80i fluorescence microscope using a DS-QiMc cooled CCD camera (Nikon) and Plan Fluor ×60/1.25 NA oil immersion objective. The GFP fluorescence of sperm cell nuclei was quantified in randomly selected pollen grains by image capture under standardized conditions. The exposure time was determined empirically to avoid image saturation and kept constant during image capture of all mutants or trangenic lines analyzed. NIS-Elements BR v3.0 software (Nikon) was used to process the images and determine the total pixel intensity (TPI) of manually defined regions of interest encompassing sperm cell nuclei. This region of interest was duplicated and used to measure the TPI of the cytoplasmic background within the same pollen grain. True fluorescence of sperm cell nuclei was determined by subtracting the cytoplasmic background TPI from the nuclear TPI and a mean TPI calculated for each marker line.
Transient Transformation of Tobacco Leaf
Argrobacterium-mediated transient transformation of Nicotiana tabacum leaf was performed as described by Sparkes et al. (2006) with some modifications. A single colony from a desired Agrobacterium strain was used to inoculate 5 mL of Luria-Bertani medium containing appropriate antibiotics and cultured to saturation overnight at 28°C with vigorous shaking (220 rpm). Cells were harvested from a 1.5-mL aliquot of the overnight culture by centrifugation at 1000g, and the pellet was resuspended in 1 mL of infiltration medium (280 mM d-glucose, 50 mM MES, 2 mM Na3PO4·12H2O, and 0.1 mM acetosyringone). The cells were washed by resuspending in another 1 mL of infiltration medium to remove traces of antibiotic, and the OD600 was measured using a Pye Unicam PU 8650 spectrophotometer (Philips).
Agrobacterium solutions were then combined according to the experiment at an OD600 of 0.1 for reporter and effector vectors and an OD600 of 0.02 for the Renilla luciferase control vector. Four- to six-week-old tobacco plants were grown in greenhouse conditions but placed under a white fluorescent lamp for 1 h before infiltration to ensure fully open stomata. Generally the third and fourth largest leaves from the apical mersitem were chosen for infiltration. Each infiltration was performed four times, on both sides of the midrib region on two separate leaves. The Agrobacterium solutions were taken up in 1-mL syringes and the underside of the leaf prepared by gently rubbing a 0.5-cm2 region to remove the cuticle. The syringe tip was then placed against the rubbed regions and the Agrobacterium solutions gently infiltrated while directly supporting the front of the leaf with a finger. Infiltrated areas were marked and labeled with a permanent black marker pen. Gloves were sprayed with 70% ethanol in between infiltrations to prevent cross-contamination. Plants were placed in a growth chamber under normal growth conditions and left for 2 d. Infiltrated regions were excised and used in dual luciferase assays.
Transient Luciferase Assays
Agrobacterium-mediated transient transformation of tobacco SR1 leaves was performed as described by Sparkes et al. (2006) with modifications. A standard size of Agrobacterium-infiltrated leaf was excised using a cork borer and ground in 500 μL of 1× Passive Lysis Buffer (Promega) until extracts appeared homogenous. Leaf extracts were centrifuged at 4°C at 14,000g to pellet cell debris. The firefly luciferase assay buffer (25 mM glycylglycine, 15 mM KPO4, pH 8.0, 4 mM EGTA, 2 mM ATP, 1 mM DTT, 15 mM MgSO4, 0.1 mM CoA, and 75 μM luciferin with final pH adjusted to 8.0) and Renilla luciferase assay buffer (1.1 M NaCl, 2.2 mM Na2EDTA, 0.22 M KPO4, pH 5.1, 0.44 mg/mL BSA, and 1.43 μM coelenterazine with final pH adjusted to 5.0) were prepared as described by Dyer et al. (2000). Two 25-μL aliquots were separately assayed with 200 μL of each assay buffer using a Clinilumat LB9502 luminometer (Berthold). Relative luciferase activity (FLuc/RLuc) was calculated for each infiltration by dividing firefly luminescence (FLuc) with Renilla luminescence (RLuc).
EMSAs
The MYB domain sequence of DUO1 (amino acids 1 to 122) was inserted via Gateway recombination into pDEST-544 (Addgene plasmid 11519) to create a 6xHis-NusA-DUO11-122 translational fusion. Recombinant proteins were purified using Ni-NTA spin columns (Qiagen) under native conditions and dialyzed against storage buffer (20% glycerol, 0.5 mM DTT, 6 mM MgCl2, 50 mM KCl, 0.1 mg/mL BSA, 10 mM Tric-Cl, and 1 mM EDTA, pH 8.0). Double-stranded oligos (Sigma-Aldrich) were labeled with digoxigenin using a Roche DIG Gel Shift Kit (v2) according to the manufacturer’s instructions. The binding reactions were performed according to Punwani et al. (2007) in binding buffer (10% glycerol, 0.5 mM DTT, 6 mM MgCl2, 50 mM KCl, 0.1 mg/mL BSA, 10 mM Tris-Cl, and 1 mM EDTA, pH 8.0) for 15 min at room temperature. Each 20-μL reaction contained 32 fM of DIG-labeled oligos, an appropriate concentration of unlabeled oligos, 300 ng of recombinant protein, 500 ng poly[d(I-C)], and 500 ng poly[d(A-T)]. Binding reactions were incubated on ice for 5 min and resolved on native PAGE gels in 0.5× TBE buffer. The oligos were transferred onto Genescreen Plus Charged Nylon Membrane (Perkin-Elmer) by electroblotting in 0.5× TBE and cross-linked with a UV Stratalinker (Stratagene). The membrane was developed according to Roche DIG Gel Shift Kit instructions and imaged using an ICCD225 intensified CCD camera (Photek).
Bioinformatic Analyses
Overrepresented motifs were identified in candidate DAT gene promoters using the online suite Regulatory Sequence Analysis Tools using default settings (van Helden, 2003; Thomas-Chollier et al., 2008). The −700-bp intergenic sequence upstream of the start codon of each DAT gene was compared with a background model consisting of the intergenic upstream sequence of all genes in Arabidopsis. PSSM logos were drawn using WebLogo (Crooks et al., 2004). GO analysis was performed with the Classification SuperViewer tool (Provart and Zhu, 2003) and functionally classified according to the three main GO categories: biological process, molecular function, and cellular component. The DAVID gene functional classification tool (Huang et al., 2008) was used to compare DAT genes with all sperm cell–expressed genes (Borges et al., 2008) using the lowest classification stringency setting.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the accession numbers provided in Supplemental Data Sets 1 to 4 online.
Author Contributions
M.B., L.B., and D.T. conceived and designed the experiments. M.B., L.B., H.K., M.L., and A.S. performed the experiments. M.B., L.B., H.K., and D.T. analyzed the data. M.B., L.B., and D.T. wrote the manuscript.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Feature Map of MYB Sites in Candidate DAT Gene Promoters.
Supplemental Figure 2. Developmental Expression of the DAN1 Promoter during Male Gametogenesis.
Supplemental Figure 3. Gene Ontology Analysis of Candidate DAT Genes.
Supplemental Data Set 1. Genes Upregulated in Seedlings in Response to Ectopic mDUO1 Expression.
Supplemental Data Set 2. List of Candidate DAT Genes.
Supplemental Data Set 3. Occurrence of MYB Motifs in Candidate DAT Gene Promoters.
Supplemental Data Set 4. Output from the DAVID Gene Functional Classification Analysis.
Supplemental Data Set 5. Primers Used in This Study.
Acknowledgments
We acknowledge David Honys (Czech Academy of Sciences, Prague, Czech Republic) and Gael Le Trionnaire (University of Leicester, United Kingdom) for the collection and normalization of publicly available microarray data. We thank Man-Kim Cheung (University of the West of England, Bristol, United Kingdom) and Ralph Panstruga (Max-Planck Institute for Plant Breeding Research, Cologne, Germany) for the Firefly and Renilla luciferase cDNA, respectively. We thank June Saddington at the University of Leicester Botanical Gardens for support with plant cultivation. We acknowledge the Biotechnology and Biological Sciences Research Council (BBSRC) for funding this work as well as for supporting M.B. through a BBSRC-funded PhD studentship.
References
- Baek K.H. (1999). The first oncogene in Drosophila melanogaster. Mutat. Res. 436: 131–136 [DOI] [PubMed] [Google Scholar]
- Bell S.D., Botting C.H., Wardleworth B.N., Jackson S.P., White M.F. (2002). The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296: 148–151 [DOI] [PubMed] [Google Scholar]
- Berger F., Hamamura Y., Ingouff M., Higashiyama T. (2008). Double fertilization - caught in the act. Trends Plant Sci. 13: 437–443 [DOI] [PubMed] [Google Scholar]
- Bilder D. (2001). Cell polarity: Squaring the circle. Curr. Biol. 11: R132–R135 [DOI] [PubMed] [Google Scholar]
- Borges F., Gomes G., Gardner R., Moreno N., McCormick S., Feijó J.A., Becker J.D. (2008). Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 148: 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Sakai H., Jack T., Weigel D., Mayer U., Meyerowitz E.M. (1992). SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114: 599–615 [DOI] [PubMed] [Google Scholar]
- Brownfield L., Hafidh S., Borg M., Sidorova A., Mori T., Twell D. (2009a). A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet. 5: e1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield L., Hafidh S., Durbarry A., Khatab H., Sidorova A., Doerner P., Twell D. (2009b). Arabidopsis DUO POLLEN3 is a key regulator of male germline development and embryogenesis. Plant Cell 21: 1940–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbarry A., Vizir I., Twell D. (2005). Male germ line development in Arabidopsis. duo pollen mutants reveal gametophytic regulators of generative cell cycle progression. Plant Physiol. 137: 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer B.W., Ferrer F.A., Klinedinst D.K., Rodriguez R. (2000). A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 282: 158–161 [DOI] [PubMed] [Google Scholar]
- Engel M.L., Chaboud A., Dumas C., McCormick S. (2003). Sperm cells of Zea mays have a complex complement of mRNAs. Plant J. 34: 697–707 [PubMed] [Google Scholar]
- Engel M.L., Holmes-Davis R., McCormick S. (2005). Green sperm. Identification of male gamete promoters in Arabidopsis. Plant Physiol. 138: 2124–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englbrecht C.C., Schoof H., Böhm S. (2004). Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest K.L., Bhave M. (2007). Major intrinsic proteins (MIPs) in plants: A complex gene family with major impacts on plant phenotype. Funct. Integr. Genomics 7: 263–289 [DOI] [PubMed] [Google Scholar]
- Forsmark A. (2009). Functional Characterisation of the Yeast Tumour Suppressor Homologue Sro7p. Master’s thesis (Gothenburg, Sweden: University of Gothenburg; ). [Google Scholar]
- Gaiser J.C., Robinson-Beers K., Gasser C.S. (1995). The Arabidopsis SUPERMAN gene mediates asymmetric growth of the outer integument of ovules. Plant Cell 7: 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Mitsumune M., O’Brien M., Bertrand C., Tebbji F., Nantel A., Matton D.P. (2006). Loss of ovule identity induced by overexpression of the fertilization-related kinase 2 (ScFRK2), a MAPKKK from Solanum chacoense. J. Exp. Bot. 57: 4171–4187 [DOI] [PubMed] [Google Scholar]
- Grotewold E., Drummond B.J., Bowen B., Peterson T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553 [DOI] [PubMed] [Google Scholar]
- Gubler F., Kalla R., Roberts J.K., Jacobsen J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 7: 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M., Arai M., Mori T., Miyagishima S.Y., Kawai S., Kita K., Kuroiwa T., Terenius O., Matsuoka H. (2008). Male fertility of malaria parasites is determined by GCS1, a plant-type reproduction factor. Curr. Biol. 18: 607–613 [DOI] [PubMed] [Google Scholar]
- Huang B.T.S., Sherman B.T., Lempicki R.A. (2008). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Humbert P., Russell S., Richardson H. (2003). Dlg, Scribble and Lgl in cell polarity, cell proliferation and cancer. Bioessays 25: 542–553 [DOI] [PubMed] [Google Scholar]
- Ingouff M., Hamamura Y., Gourgues M., Higashiyama T., Berger F. (2007). Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr. Biol. 17: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Justice N.J., Jan Y.N. (2003). A lethal giant kinase in cell polarity. Nat. Cell Biol. 5: 273–274 [DOI] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim H.J., Oh S.A., Brownfield L., Hong S.H., Ryu H., Hwang I., Twell D., Nam H.G. (2008). Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature 455: 1134–1137 [DOI] [PubMed] [Google Scholar]
- Klezovitch O., Fernandez T.E., Tapscott S.J., Vasioukhin V. (2004). Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 18: 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A., Sakamoto A., Kubo K., Rybka Z., Kanno Y., Takatsuji H. (1998). Seven zinc-finger transcription factors are expressed sequentially during the development of anthers in petunia. Plant J. 13: 571–576 [DOI] [PubMed] [Google Scholar]
- Lange A., Mills R.E., Lange C.J., Stewart M., Devine S.E., Corbett A.H. (2007). Classical nuclear localization signals: Definition, function, and interaction with importin α. J. Biol. Chem. 282: 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Tewari R., Ning J., Blagborough A.M., Garbom S., Pei J., Grishin N.V., Steele R.E., Sinden R.E., Snell W.J., Billker O. (2008). The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 22: 1051–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubkowitz M. (2006). The OPT family functions in long-distance peptide and metal transport in plants. Genet. Eng. (N. Y.) 27: 35–55 [DOI] [PubMed] [Google Scholar]
- Meister R.J., Kotow L.M., Gasser C.S. (2002). SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development 129: 4281–4289 [DOI] [PubMed] [Google Scholar]
- Mori T., Kuroiwa H., Higashiyama T., Kuroiwa T. (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8: 64–71 [DOI] [PubMed] [Google Scholar]
- O’Brien M., Gray-Mitsumune M., Kapfer C., Bertrand C., Matton D.P. (2007). The ScFRK2 MAP kinase kinase kinase from Solanum chacoense affects pollen development and viability. Planta 225: 1221–1231 [DOI] [PubMed] [Google Scholar]
- Okada T., Bhalla P.L., Singh M.B. (2005). Transcriptional activity of male gamete-specific histone gcH3 promoter in sperm cells of Lilium longiflorum. Plant Cell Physiol. 46: 797–802 [DOI] [PubMed] [Google Scholar]
- Okada T., Bhalla P.L., Singh M.B. (2006). Expressed sequence tag analysis of Lilium longiflorum generative cells. Plant Cell Physiol. 47: 698–705 [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Wollmann H., Schommer C., Schwab R., Boisbouvier J., Rodriguez R., Warthmann N., Allen E., Dezulian T., Huson D., Carrington J.C., Weigel D. (2007). Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 13: 115–125 [DOI] [PubMed] [Google Scholar]
- Pang Y., Li L., Ren F., Lu P., Wei P., Cai J., Xin L., Zhang J., Chen J., Wang X. (2010). Overexpression of the tonoplast aquaporin AtTIP5; 1 conferred tolerance to boron toxicity in Arabidopsis. J. Genet. Genomics 37: 389–397 [DOI] [PubMed] [Google Scholar]
- Park S.K., Howden R., Twell D. (1998). The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125: 3789–3799 [DOI] [PubMed] [Google Scholar]
- Provart N., Zhu T. (2003). A browser-based functional classification SuperViewer for Arabidopsis genomics. Mol. Biol. 2003: 271–272 [Google Scholar]
- Punwani J.A., Rabiger D.S., Drews G.N. (2007). MYB98 positively regulates a battery of synergid-expressed genes encoding filiform apparatus localized proteins. Plant Cell 19: 2557–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth B.A., Goff S.A., Klein T.M., Fromm M.E. (1991). C1- and R-dependent expression of the maize Bz1 gene requires sequences with homology to mammalian myb and myc binding sites. Plant Cell 3: 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman N., Durbarry A., Wardle A., Yang W.C., Chaboud A., Faure J.E., Berger F., Twell D. (2005). A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15: 244–248 [DOI] [PubMed] [Google Scholar]
- Saikumar P., Murali R., Reddy E.P. (1990). Role of tryptophan repeats and flanking amino acids in Myb-DNA interactions. Proc. Natl. Acad. Sci. USA 87: 8452–8456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Medrano L.J., Meyerowitz E.M. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378: 199–203 [DOI] [PubMed] [Google Scholar]
- Sandman K., Reeve J.N. (2005). Archaeal chromatin proteins: Different structures but common function? Curr. Opin. Microbiol. 8: 656–661 [DOI] [PubMed] [Google Scholar]
- Snider J., Houry W.A. (2008). AAA+ proteins: Diversity in function, similarity in structure. Biochem. Soc. Trans. 36: 72–77 [DOI] [PubMed] [Google Scholar]
- Solano R., Nieto C., Avila J., Cañas L., Diaz I., Paz-Ares J. (1995). Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 14: 1773–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.Y., Choi K.S., Kim D.Y., Geisler M., Park J., Vincenzetti V., Schellenberg M., Kim S.H., Lim Y.P., Noh E.W. (2010). Arabidopsis PCR2 is a zinc exporter involved in both zinc extrusion and long-distance zinc transport. Plant Cell 22: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.Y., Martinoia E., Lee J., Kim D., Kim D.Y., Vogt E., Shim D., Choi K.S., Hwang I., Lee Y. (2004). A novel family of cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol. 135: 1027–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto G., Alleva K., Mazzella M.A., Amodeo G., Muschietti J.P. (2008). AtTIP1;3 and AtTIP5;1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 582: 4077–4082 [DOI] [PubMed] [Google Scholar]
- Soto G., Fox R., Ayub N., Alleva K., Guaimas F., Erijman E.J., Mazzella A., Amodeo G., Muschietti J. (2010). TIP5;1 is an aquaporin specifically targeted to pollen mitochondria and is probably involved in nitrogen remobilization in Arabidopsis thaliana. Plant J. 64: 1038–1047 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Stacey M.G., Patel A., McClain W.E., Mathieu M., Remley M., Rogers E.E., Gassmann W., Blevins D.G., Stacey G. (2008). The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol. 146: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele R.E., Dana C.E. (2009). Evolutionary history of the HAP2/GCS1 gene and sexual reproduction in metazoans. PLoS ONE 4: e7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji H. (1998). Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. 54: 582–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji H. (1999). Zinc-finger proteins: The classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39: 1073–1078 [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M., Sand O., Turatsinze J.V., Janky R., Defrance M., Vervisch E., Brohée S., van Helden J. (2008). RSAT: Regulatory sequence analysis tools. Nucleic Acids Res. 36(Web Server issue): W119–W127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J. (2003). Regulatory sequence analysis tools. Nucleic Acids Res. 31: 3593–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Willigen C., Postaire O., Tournaire-Roux C., Boursiac Y., Maurel C. (2006). Expression and inhibition of aquaporins in germinating Arabidopsis seeds. Plant Cell Physiol. 47: 1241–1250 [DOI] [PubMed] [Google Scholar]
- Verelst W., Saedler H., Münster T. (2007a). MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol. 143: 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verelst W., Twell D., de Folter S., Immink R., Saedler H., Münster T. (2007b). MADS-complexes regulate transcriptome dynamics during pollen maturation. Genome Biol. 8: R249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Besser K., Frank A.C., Johnson M.A., Preuss D. (2006). Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133: 4761–4769 [DOI] [PubMed] [Google Scholar]
- Wilson Z.A., Zhang D.B. (2009). From Arabidopsis to rice: Pathways in pollen development. J. Exp. Bot. 60: 1479–1492 [DOI] [PubMed] [Google Scholar]
- Wong J.L., Johnson M.A. (2010). Is HAP2-GCS1 an ancestral gamete fusogen? Trends Cell Biol. 20: 134–141 [DOI] [PubMed] [Google Scholar]
- Xue H., Guo R., Wen Y., Liu D., Huang L. (2000). An abundant DNA binding protein from the hyperthermophilic archaeon Sulfolobus shibatae affects DNA supercoiling in a temperature-dependent fashion. J. Bacteriol. 182: 3929–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]