This work shows that Hsp90 and Hsp70 chaperones physically interact with tomato heat shock transcription factors Hsfs A1, A2, and B1, but their influence on Hsf function is realized by distinct and factor-specific mechanisms, which control either activity or abundance of the individual transcription factors and contribute in concert to the regulation of heat stress response.
Abstract
Heat stress transcription factors (Hsfs) regulate gene expression in response to environmental stress. The Hsf network in plants is controlled at the transcriptional level by cooperation of distinct Hsf members and by interaction with chaperones. We found two general mechanisms of Hsf regulation by chaperones while analyzing the three major Hsfs, A1, A2, and B1, in tomato (Solanum lycopersicum). First, Hsp70 and Hsp90 regulate Hsf function by direct interactions. Hsp70 represses the activity of HsfA1, including its DNA binding, and the coactivator function of HsfB1 in the complex with HsfA2, while the DNA binding activity of HsfB1 is stimulated by Hsp90. Second, Hsp90 affects the abundance of HsfA2 and HsfB1 by modulating hsfA2 transcript degradation involved in regulation of the timing of HsfA2 synthesis. By contrast, HsfB1 binding to Hsp90 and to DNA are prerequisites for targeting this Hsf for proteasomal degradation, which also depends on a sequence element in its carboxyl-terminal domain. Thus, HsfB1 represents an Hsp90 client protein that, by interacting with the chaperone, is targeted for, rather than protected from, degradation. Based on these findings, we propose a versatile regulatory regime involving Hsp90, Hsp70, and the three Hsfs in the control of heat stress response.
INTRODUCTION
Exposure to elevated temperatures leads to activation of the cellular heat stress response (HSR), which is conserved throughout all kingdoms of life. The HSR causes the enhanced expression of heat stress (hs) genes, multigene families encoding molecular chaperones (Boston et al., 1996; Bösl et al., 2006; Bukau et al., 2006; Nakamoto and Vígh, 2007; Morimoto, 2008; Hartl and Hayer-Hartl, 2009; Richter et al., 2010). Among these, Hsp70 is one of the most abundant hs proteins in eukaryotic cells. Hsp70s bind in an ATP-dependent manner to hydrophobic patches of partially unfolded proteins and prevent protein aggregation (Mayer and Bukau, 2005). While Hsp70s accumulate during hs, their constitutively expressed cognates (Hsc70) are essential for general cellular functions due to their involvement in the control of protein homeostasis (Young et al., 2004). They assist the folding of nascent polypeptides released from the ribosome (Hartl and Hayer-Hartl, 2002), sorting of proteins to cell organelles by interaction with mitochondrial and chloroplast protein import complexes (Zhang and Glaser, 2002; Mirus and Schleiff, 2009), and form a link to the ubiquitin-mediated proteasomal degradation pathway (Ballinger et al., 1999; Lüders et al., 2000; Lee et al., 2009). To govern this multitude of different tasks, Hsp70 function relies on interaction with different cochaperones and cofactors for substrate interaction and regulation of ATP hydrolysis, which in turn affects substrate turnover rates (Kampinga and Craig, 2010; Mayer, 2010).
A second important chaperone family involved in cellular homeostasis is the Hsp90 family. In vertebrates, this chaperone functions in the maturation and regulation of kinases, steroid hormone receptors, and transcription factors (Riggs et al., 2004; Whitesell and Lindquist, 2005; Pratt et al., 2008; Wandinger et al., 2008; Taipale et al., 2010). The recruitment and regulation of Hsp90 client proteins is connected to the function of Hsp70 and a multitude of other cochaperones. This is best exemplified for the mammalian glucocorticoid receptor (Hutchison et al., 1993). Both chaperones cooperate in client protein folding to an active conformation, protein stabilization, and protein turnover by the formation of multichaperone complexes (Pratt et al., 2010). The versatile function of Hsp90 in many signaling pathways defines it as a capacitor against phenotypical manifestation of mutational defects; thus, this chaperone is involved in the maintenance of phenotypic plasticity and developmental stability (Queitsch et al., 2002; Sangster and Queitsch, 2005; Sangster et al., 2007).
Besides their importance for protein folding, a function of Hsp70 and Hsp90 as negative regulators for heat stress transcription factors (Hsfs) has been proposed for mammalian Hsf1 (Abravaya et al., 1992; Baler et al., 1992). Under hs, partially denatured proteins compete with Hsf for Hsp70 binding, which leads to Hsf activation and induction of the HSR. This results in enhanced expression and accumulation of Hsp70, which in turn resumes its repressor function during the attenuation of the HSR (Mosser et al., 1993; Rabindran et al., 1994). More recent results support the widely accepted model that under normal growth conditions, the inactive state of Hsfs is maintained by Hsp70/Hsp90 multichaperone complexes (Nair et al., 1996; Shi et al., 1998; Zou et al., 1998; Guo et al., 2001). This exemplifies the dual function of Hsp70 and Hsp90 proteins as chaperones involved in the maintenance of cellular protein homeostasis, on the one hand, and as negative feedback regulators of the HSR, on the other hand.
Plants possess a complex HSR network including multiple Hsp and Hsf isoforms. For example, Arabidopsis thaliana contains 21, rice (Oryza sativa) 23, and tomato (Solanum lycopersicum) at least 25 Hsf-encoding genes (Baniwal et al., 2004). Hsfs have a modular structure comprising a DNA binding domain (DBD; Damberger et al., 1994), an oligomerization domain (Peteranderl et al., 1999), and a C-terminal activation domain (Treuter et al., 1993). Based on variations in these three domains, especially the oligomerization domain, plant Hsfs can be divided into three classes (A, B, and C). The most obvious differences are the insertion of additional amino acid residues in the oligomerization domain of class A and class C Hsfs and the absence of transcriptional activator motifs in the C-terminal domain of class B and C Hsfs (Nover et al., 2001; Bharti et al., 2004).
Tomato HsfA1a (termed HsfA1 here), HsfA2, and HsfB1 are some of the best-characterized plant Hsfs involved in the regulation of HSR (Baniwal et al., 2004). HsfA1 is constitutively expressed and has a unique function as “master regulator” of the HSR (Mishra et al., 2002). Activation by hs triggers nuclear retention of HsfA1 and leads to the expression of HsfA2 and HsfB1 and the formation of heterooligomeric complexes (termed superactivator complexes) between HsfA1 and HsfA2 (Scharf et al., 1998; Heerklotz et al., 2001). The latter drives HSR by enhanced activation of hs gene expression (Chan-Schaminet et al., 2009). During attenuation of the HSR, HsfA1 is redistributed to the cytoplasm (Scharf et al., 1998), while HsfA2 is kept in an inactive state by interaction with members of the sHsp17-CII subfamily (for small Hsps; Port et al., 2004). This imparts an induced thermotolerance state, since HsfA2 becomes the most prominent Hsf and can be rapidly rerecruited by HsfA1 upon further hs exposures. In contrast, HsfB1 accumulation is only transient, and its protein level declines rapidly during the early phase of HSR (Scharf et al., 1998). Class B Hsf members cannot activate themselves and are assumed to function as repressors of hs gene expression (Czarnecka-Verner et al., 2004; Ikeda and Ohme-Takagi, 2009; Kumar et al., 2009). However, tomato HsfB1 has a coregulator function, enhancing the activity of class A Hsfs and other housekeeping transcription factors in the context of the histone acetyl transferase–like protein1 (HAC1)–containing ternary coactivator complex (Bharti et al., 2004).
In spite of the increasing knowledge of functional Hsf interactions (Chan-Schaminet et al., 2009) and Hsf–Hsp crosstalk (Yamada et al., 2007, Meiri and Breiman, 2009), the mechanism of regulation of the Hsf network by chaperones in plants has remained elusive. Here, we demonstrate physical interactions for the three tomato Hsfs, A1, A2, and B1, with Hsp70 and Hsp90 chaperones. We further provide evidence that each of the Hsfs is regulated by either Hsp70 or Hsp90, or both, although at different levels. Thereby, we summarize the emerging picture of the complex interplay between Hsf and Hsp networks involved in the regulation of cellular HSR and thermotolerance.
RESULTS
Physical Interactions of Hsp90 and Hsp70 with Hsfs
The interaction of Hsfs with chaperones of the Hsp70 and Hsp90 families could be an integral part of the regulatory network controlling Hsf activity in plants. However, our understanding of the regulatory network is limited because of the complexity of the plant Hsf network. Moreover, interactions with either Hsp70 or Hsp90 have only been reported for a few Hsfs of Arabidopsis (Kim and Schöffl, 2002; Yamada et al., 2007; Meiri and Breiman, 2009). Thus, we have investigated putative interactions of tomato HsfA1, HsfA2, and HsfB1 with Hsp70 and Hsp90 at first by yeast two-hybrid analyses. Both HsfA1 and HsfA2, but not HsfB1, were found to interact with the C-terminal portion of Hsp90. This interaction is not dependent on the C-terminal MEEVD motif (Figure 1) known to be responsible for interaction with proteins containing tetratricopeptide repeat domains (Mirus and Schleiff, 2009; Taipale et al., 2010). Similarly, HsfA1 and HsfA2 interacted with Hsp70 (Figure 1). Here, we observed that HsfA1 interacts with the N-terminal region, whereas HsfA2 interacts with the C-terminal portion of Hsp70.
Figure 1.
Yeast Two-Hybrid Analysis of Hsp–Hsf Interactions.
Interactions between Hsp90 or Hsp70 and HsfA1, HsfA2, or HsfB1 were studied by yeast two-hybrid analyses, and the results are represented for Gal4DBD alone as a control and in fusion with C-terminal fragments of HsfA1 or HsfA2 or the full-length proteins as bait constructs. On the left, the names of Hsp prey constructs are given, including the amino acid residues covered. Block diagrams indicate the domain structures of the Gal4AD-Hsp fusion proteins. Full-length Hsp90 (top) and Hsp70 (bottom) are shown with the positions of amino acid residues indicated by numbers. DD, Dimerization domain; E/K, charged linker regions; L, hydrophobic linker; M, middle domain; M/IEEVD, conserved amino acid residues at the C-terminal ends of both chaperones; NBD, nucleotide binding domain; n.d., not determined; SBD, substrate binding domain; +, interaction observed; −, no interaction observed.
Next, we used tomato mesophyll protoplasts as a transient expression system (Mishra et al., 2002) to confirm the observed interactions in plant cells by coimmunoprecipitation (CoIP). Hemagglutinin (HA)-tagged Hsp70 and profilin-tagged Hsp90 were expressed either individually or together after cotransformation with expression constructs for HsfA1, HsfA2, or HsfB1. Subsequently, the formation of Hsf–Hsp complexes was analyzed by CoIP using Hsf-specific antibodies (Figures 2A–2C). Both Hsp70 and Hsp90 were detected in the elution fractions, irrespective of whether the Hsps were expressed individually or in combination (Figures 2A–2C, lanes 4). In contrast, neither of the two Hsps was detected in the elution fraction when the CoIP was performed using nonspecific preimmune serum (Figure 2D, lane 4). These results indicate that in plant cells, all three Hsfs form complexes with both Hsps based on Hsf–Hsp-specific interactions.
Figure 2.
Hsp70 and Hsp90 Physically Interact with Hsfs in Plant Cells.
Tomato protoplasts were cotransformed with expression constructs encoding HsfA1 (pA1; [A], [D], [F], and [G]), HsfA2 (pA2; [B], [D], [F], and [G]), or HsfB1 (pB1; [C], [D], [F], and [G]) and expression constructs for HA-tagged Hsp70 (+p70) and/or profilin-tagged Hsp90 (+p90).
(A) to (D) Matrices coated with antibodies against the Hsfs ([A]–[C]) or preimmune serum (D) were incubated with cell lysate. Five percent of the input (I; lane 1) and aliquots of the flow through (FT; lane 2), fourth wash step (W4; lane 3), and elution fraction (E; lane 4) were processed for immunodetection with the antibodies indicated on the right.
(E) and (F) CoIP using matrices coated with Hsp70 antibodies and lysates from protoplasts cotransformed with both Hsp70 and Hsp90 expression plasmids in combination with a plasmid coding either for GFP (E), or HsfA1 ([F], top), HsfA2 ([F], middle), or HsfB1 ([F], bottom).
(G) Protein extracts derived from the same number of protoplasts were subjected to SDS-PAGE and immunodetection with antibodies against the HA tag (for Hsp70) or the profilin tag (for Hsp90) to confirm similar expression levels in all samples.
To further support this observation, we transformed each Hsf together with both Hsps and used Hsp70 antibodies for CoIP. To verify the specificity of the Hsf–chaperone interactions, we also cotransformed a green fluorescent protein (GFP) expression construct as a control. The Hsp70 antibody efficiently precipitated HA-tagged Hsp70 but not GFP coexpressed in the same protoplasts (Figure 2E, lane 4). In contrast, all three Hsfs were detected in the corresponding elution fractions (Figure 2F, lane 4). With this approach, we confirmed the results obtained before when using Hsf-specific antibodies for CoIP.
Thus, when challenging the yeast two-hybrid result for HsfB1 by immunoprecipitation from the background of plant cells, we observed that the antibodies against HsfB1 are able to coimmunoprecipitate both Hsp90 and Hsp70 (Figure 2C, lane 4) and that the interaction of HsfB1 with Hsp70 is specific, because GFP cannot be precipitated with this approach (Figures 2E and 2F, lanes 4). This suggests that the interaction between the chaperones and HsfB1 in the yeast two-hybrid assays was compromised for unknown reasons.
Regulation of Hsf Activity by Hsp70 and Hsp90
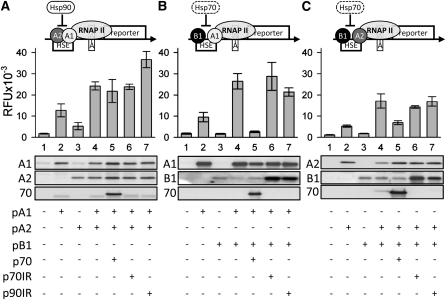
The identification of Hsf–Hsp complexes prompted us to investigate the functional significance of these interactions. We analyzed the influence of Hsps on the activity of Hsfs by monitoring β-glucuronidase (GUS) activity in protoplasts transformed with an Hsf-dependent GUS reporter construct (Treuter et al., 1993). We cotransformed the Hsfs individually, in combination, and together with a non-isoform-specific inverted repeat (IR) construct targeting either Hsp70 or Hsp90 and leading to the RNA interference (RNAi)–mediated knockdown of newly synthesized Hsp70 or Hsp90 (Tripp et al., 2009). To complement the analysis, we also determined the GUS activity in protoplasts cotransformed with an expression construct of Hsp70. As reported earlier (Scharf et al., 1998), the activation of GUS expression by HsfA1 (Figure 3A, sample 2) is strongly enhanced upon coexpression with HsfA2 (sample 4). The expression of Hsp70 or its silencing did not influence the activity of this Hsf combination (samples 5 and 6). In contrast, cotransformation of the IR construct targeting hsp90 transcripts further enhanced the GUS activity without any obvious alteration of the Hsf protein levels (sample 7 and immunoblot of this sample). We conclude that the observed effect is specific and Hsp90 negatively influences the activity of at least one of the two Hsfs, as none of the coexpressed Hsp constructs showed any influence on the GUS activity in the absence of the Hsfs (see Supplemental Figure 1A online).
Figure 3.
Regulation of Hsf Activity by Hsp70 and Hsp90.
Tomato protoplasts were cotransformed with the Hsf-dependent promoter GUS reporter plasmid pGmhsp17.3B-CI*:GUS together with expression constructs encoding Hsfs and Hsp70 under the control of the cauliflower mosaic virus 35S promoter or in combination with the non-isoform-specific IR constructs of Hsp70 (p70IR) or Hsp90 (p90IR). The pictograms illustrate the possible inhibitory influence of the chaperones on Hsf (A1, A2, and B1) activities as concluded from the data. RNAP II, RNA polymerase II.
(A) Activity of the GUS reporter (sample 1) was monitored in protoplasts cotransformed with HsfA1 (lanes 2 and 4–7), HsfA2 (lanes 3–7), a construct expressing HA-tagged Hsp70 (lane 5), or non-isoform-specific IR constructs of Hsp70 (lane 6) or Hsp90 (lane 7). The graph shows the GUS activities (in relative fluorescence units [RFU]). Protein levels were monitored in aliquots taken from the same protoplast samples by immunoblotting using the indicated antibodies. Error bars represent the sd of at least three independent experiments.
(B) and (C) Experiments for analyzing the combinations of HsfA1 and HsfB1 (B) or HsfA2 and HsfB1 (C) were performed as in (A).
Although HsfB1 does not activate transcription on its own, a stimulation of transcription activity of HsfA1 was reported by coexpression of HsfB1 (Bharti et al., 2004). Consistently, the combination of HsfA1 and HsfB1 resulted in an enhanced stimulation of the GUS activity (Figure 3B, samples 2 and 3 versus sample 4). Cotransformation of the IR construct of both Hsp70 and Hsp90 resulted in a strong accumulation of HsfB1 (immunoblot in Figure 3B, lanes 6 and 7), but GUS expression was not further stimulated (samples 6 and 7). By contrast, coexpression of Hsp70 led to the reduction of GUS activity to almost background levels (sample 1 versus sample 5) without a significant alteration of the HsfB1 protein level. As GUS activity in this case was reduced to background levels, the function of HsfA1 must be strongly affected. Hence, Hsp70 seems to compromise the function of HsfA1 if HsfB1 but not if HsfA2 is present (Figure 3A, sample 5, versus Figure 3B, sample 5).
Combining HsfA2 and HsfB1 results in a stimulation of GUS activity as well (Figure 3C, samples 2 and 3 versus sample 4), and cotransformation of the IR constructs again had no influence on the GUS activity (samples 6 and 7). However, in this Hsf combination, coexpression of Hsp70 resulted in reduction of the GUS activity to the level found for HsfA2 alone (sample 2 versus sample 5). From this, we conclude that in this case, the inhibitory effect of Hsp70 can be attributed to the interaction with HsfB1. The reasons for the differences between the two combinations with HsfB1 are not clear, but it is intriguing to recall that HsfA1 interacts with the nucleotide binding domain of Hsp70, whereas HsfA2 interacts with the C-terminal portion of this chaperone (Figure 1). This may also influence details of the binding of Hsp70 to the Hsfs in context with the transcriptional machinery.
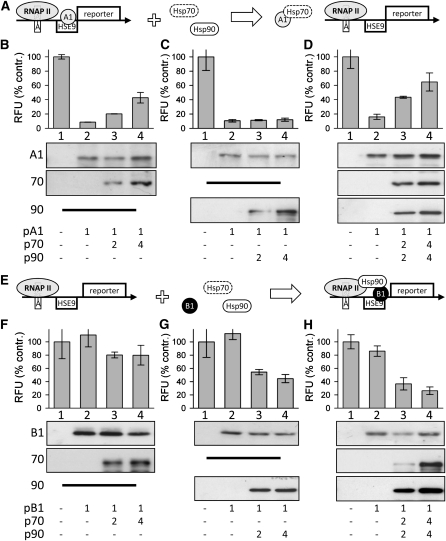
Influence of Hsp70 and Hsp90 on DNA Binding of HsfA1 and HsfB1
The function of the HsfA1/A2 superactivator complex seems to be affected by Hsp90 (Figure 3A, top), while the activity of the HsfA2/HsfB1 and HsfA1/HsfB1 combinations is repressed by Hsp70 functions (Figures 3B and 3C). Thus, we analyzed the influence of Hsp70 or Hsp90 on the DNA binding activity of HsfA1 and HsfB1 to distinguish between effects on the transcription activator function and DNA binding activities. Here, we did not analyze HsfA2, because coexpression of Hsp70 with HsfA2/HsfB1 resulted in a transcriptional activity comparable to that observed for HsfA2 alone (Figure 3C). The DNA binding activity was determined by the previously established GUS repressor assay, where GUS expression can only be monitored when no Hsf is bound to the heat shock element (HSE; Pelham and Bienz, 1982) motifs inserted downstream of the TATA box in the 5′ untranslated region of the GUS reporter gene (Figure 4A; Treuter et al., 1993). When HsfA1 was cotransformed with this reporter construct, only basal levels of GUS activity were detectable (Figures 4B–4D, samples 2), indicating strong binding of HsfA1 to the HSE motifs. When the expression constructs of Hsp70 and HsfA1 were cotransformed, we observed an increase of GUS activity in an Hsp70 concentration–dependent manner (Figure 4B, sample 2 versus samples 3 and 4). Evidently, Hsp70 inhibits the binding of HsfA1 to HSE. As shown before, the inactivation is not caused by protein degradation, as the expression of Hsp70 did not result in reduced levels of HsfA1 (immunoblots in Figures 4B and 4D) and expression of Hsp70 alone had no effect on the GUS activity in the repressor assay (see Supplemental Figure 1B online). In contrast to Hsp70, coexpression of Hsp90 had no effect on the DNA binding of HsfA1 (Figure 4C, sample 2 versus samples 3 and 4), and the combination of both chaperones gave effects similar to Hsp70 alone (Figures 4B and 4D, samples 3 and 4).
Figure 4.
Effects of Hsp70 and Hsp90 on the DNA Binding of HsfA1 and HsfB1.
(A) and (E) Putative models of functional interactions based on the results are illustrated for HsfA1 (A) and HsfB1 (E). See text for further discussion.
(B) to (D) and (F) to (H) The GUS repressor construct p35S:HSE9-GUS (samples 1) was cotransformed into tomato protoplasts with expression constructs (samples 2–4) encoding HsfA1 ([B]–[D]) or HsfB1 ([F]–[H]) in combination with Hsp70 ([B] and [F]; samples 3 and 4), Hsp90 ([C] and [G]; samples 3 and 4), or Hsp70 and Hsp90 ([D] and [H]; samples 3 and 4). GUS activities are given in percentage of control (contr.; top), and error bars represent the sd of multiple experiments. Immunoblots of the proteins indicated at the left are shown below for each sample. At the bottom, the combinations and amounts (μg) of expression constructs used for cotransformation of 105 protoplasts are given.
HSE9, synthetic heat shock element inserted downstream of the TATA box (TA); RFU, relative fluorescence units; RNAP II, RNA polymerase II.
Besides the effects of Hsp70 on HsfA1, we had also noticed an influence of this chaperone on HsfB1 activity (Figure 3C). In contrast to HsfA1, we did not observe any repression of the GUS activity by testing HsfB1 in the repressor assay (Figures 4E–4H, samples 1 and 2), and coexpression of Hsp70 had no significant influence on this result (Figure 4F, samples 3 and 4). The low DNA binding activity observed here is consistent with previous findings that HsfB1 DNA binding is dependent on the formation of the ternary coactivator complex with HsfA1 and HAC1 (Bharti et al., 2004). Furthermore, the data indicate that the influence of Hsp70 on the coactivator function (Figures 3B and 3C) is not directly connected to the DNA binding activity of HsfB1. In contrast, repression of GUS activity was observed by coexpression of HsfB1 and Hsp90 (Figure 4G, samples 3 and 4), and as seen for HsfA1, coexpression of HsfB1 with Hsp70 and Hsp90 did not lead to further reduced or enhanced GUS activities (Figure 4H, samples 3 and 4). Taken together, we conclude that the interaction between Hsp70 and HsfA1 inhibits DNA binding (Figure 4A), whereas the interaction of Hsp90 with HsfB1 supports the targeting of this Hsf to the HSE binding sites on the DNA (Figure 4E).
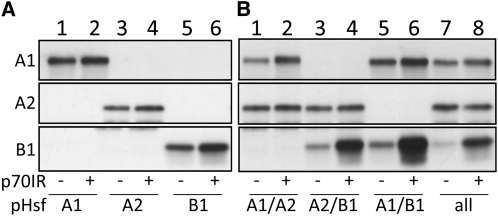
Hsp70 Together with HsfA1 and HsfA2 Modulates the Abundance of HsfB1
While analyzing the influence of the Hsps on Hsf activity, we observed a strong increase of HsfB1 levels in the presence of the IR constructs targeting either Hsp90 or Hsp70 (Figure 3). In contrast, the protein levels of coexpressed HsfA1 or HsfA2 were hardly affected (Figure 3). To further analyze this effect, we studied the influence of Hsp70 repression on the protein levels when the Hsfs were expressed individually, in different combinations, or all together. As shown before, in the presence of the Hsp70-specific IR construct, we did not observe changes in the HsfA1 or HsfA2 levels comparable to HsfB1 (Figure 5). In contrast, the influence of Hsp70 RNAi on the enhancement of the HsfB1 level was much stronger in the presence of the other Hsfs than in their absence (Figure 5A versus Figure 5B). Hence, we conclude that in the presence of HsfA1 and HsfA2, the HsfB1 protein level is tightly regulated in a negative manner by Hsp70. It is worth mentioning that the combination of all three Hsfs (Figure 5B, lane 7) is reminiscent of the situation in thermotolerant cells recovering from hs treatment.
Figure 5.
Hsp70 Is Involved in the Regulation of Hsf Protein Levels.
(A) Tomato protoplasts were transformed with expression constructs for HsfA1 (lanes 1 and 2), HsfA2 (lanes 3 and 4), or HsfB1 (lanes 5 and 6) in combination with the non-isoform-specific Hsp70 IR construct p70IR (lanes 2, 4, and 6).
(B) Combinations of Hsfs as indicated were expressed either in the absence (lanes 1, 3, 5, and 7) or in the presence (lanes 2, 4, 6, and 8) of p70IR.
Hsf protein levels were determined in total protein extracts by immunoblotting using the indicated Hsf-specific antibodies.
Hsp90 Contributes to the Control of Hsf Levels
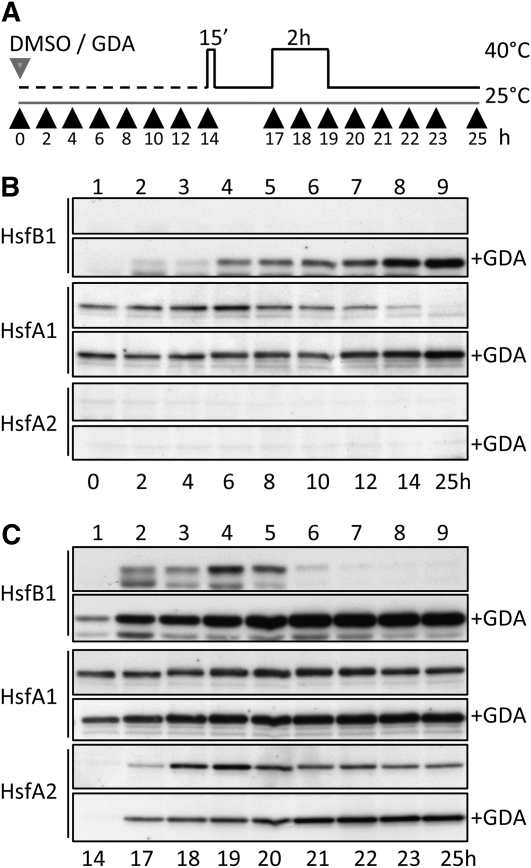
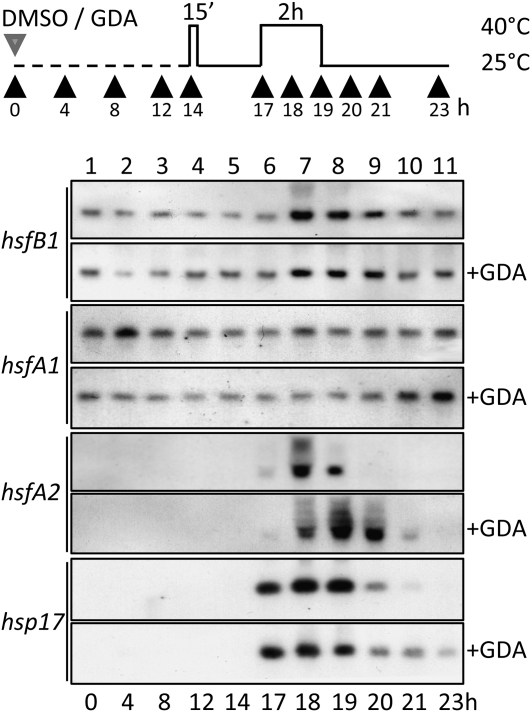
Because Hsp90 functions can be specifically inhibited by binding geldanamycin (GDA) to the ATP binding pocket of the chaperone and preventing the stabilization of client protein interaction (Whitesell and Lindquist, 2005; Pearl et al., 2008), we used this drug to analyze the endogenous Hsfs in tomato suspension culture cells (Figure 6). Under control conditions, HsfB1 is hardly detectable in exponentially growing cells (Figure 6B, lane 1). However, the low endogenous level was significantly enhanced in the presence of 1 μM GDA within the first 2 h of treatment (lane 2) and continuously increased further during longer incubation times (lanes 3–9). Similar to Hsp70 or tubulin monitored as controls, treatment with GDA did not exhibit any effect on the Hsp90 protein level (see Supplemental Figure 2A online). Interestingly, for the constitutively expressed HsfA1, a decline of the protein level was observed after longer incubation in the presence of DMSO alone used as a solvent control for GDA (Figure 6B, lanes 8 and 9), which was not observed in nontreated cells (data not shown). This effect was completely abrogated in the presence of GDA. In contrast to HsfB1, an increase of the hs-inducible HsfA2 was not detectable under these conditions. The observed results indicate that inhibition of Hsp90 seems not to be sufficient to trigger the HSR in these cells, which would have caused the expression of HsfA2, and that especially the HsfB1 level is controlled by the function of Hsp90, at least under normal growth conditions.
Figure 6.
Influence of Hsp90 on Hsf Protein Levels in Tomato Cell Cultures.
(A) The pictogram illustrates the addition of 0.1% DMSO or 0.1% DMSO and 1 μM GDA to tomato cell cultures (gray arrowhead) and the hs regime applied (black line; the gray line shows control temperature conditions). Black arrowheads indicate the time points of sample harvesting.
(B) and (C) Immunoblots of total protein extracts from control and GDA-treated cells (+GDA) harvested at the indicated time points after incubation at 25°C (B) or during the hs regime applied (C). For immunodetection of the indicated Hsfs (left), specific antibodies were used. For immunodetection of additionally monitored proteins on the same blots, see Supplemental Figure 2 online.
We next analyzed Hsf and Hsp protein levels during repeated cycles of hs exposure and recovery in suspension culture cells in the absence or in the presence of GDA to determine possible functions of Hsp90 in heat-stressed cells. By this method, we examined different stages of HSR (see pictogram in Figure 6A). After preinduction of HSR, accumulation of the hs-inducible HsfA2 and HsfB1 was observed, in the absence or in the presence of GDA (Figure 6C, lane 2), and the HsfA2 and HsfB1 levels were further enhanced during application of a second hs treatment (lanes 3 and 4). However, for both Hsfs, the increase of protein levels after preinduction was much stronger in the presence of the inhibitor. The influence of GDA treatment on Hsf protein abundance became even more prominent during recovery from the second hs treatment. After increasing to maximum levels during hs, HsfA2 and HsfB1 declined in the subsequent recovery period in the absence of GDA, whereas in the presence of the inhibitor, the protein level was further increased for HsfB1 and was maintained at high levels for HsfA2 (Figure 6C). The level of HsfA1, which was not affected by GDA, was slightly increased in combination with the hs treatment (Figure 6C). By comparison with the changes observed for the Hsfs under nonstress conditions (Figure 6B) and the proteins monitored for control (see Supplemental Figure 2 online), it is evident that inhibition of Hsp90 mainly influences the abundance of all three Hsfs during HSR, with the most prominent effect on HsfB1 levels.
Influence of Hsp90 on the Abundance of Hsf Transcripts
Next, we wanted to know whether the changes observed for the three Hsfs in the presence of GDA indicate influences of Hsp90 on the transcript level or on the stability of the proteins. We thus compared the transcript levels during GDA and hs treatments. As shown in Figure 7, the quantity of HsfA1 transcripts remained largely constant during all treatments (Figure 7, lanes 1–11). The accumulation of HsfA2 transcripts as well as of Hsp17-CI mRNA became only detectable after hs induction (lanes 6 and 7). This result provides further evidence that inhibition of Hsp90 by GDA is not sufficient to trigger the activation of these genes under nonstress conditions. However, inhibition of Hsp90 by GDA yielded an enhancement of the HsfA2 transcript level during hs and delayed the decline in the subsequent recovery period when compared with the situation in the absence of GDA (Figure 7). Evidently, HsfA2 transcript degradation requires functional Hsp90. The abundance of HsfB1 transcripts was much less affected. The basal transcript level in nonstressed cells (lanes 1–6) was increased moderately during hs (lanes 7 and 8) and rapidly declined to the normal level during recovery (lanes 9–11). However, GDA treatment had no effect on the HsfB1 transcript levels.
Figure 7.
Influence of GDA on Hsf Transcript Levels in Tomato Cell Cultures.
RNA gel blot analysis of hsfB1, hsfA1, hsfA2, and hsp17-CI (hsp17) transcript levels in cell suspension cultures treated according to the regimen illustrated at the top. The addition of DMSO/GDA and the time points of cell harvesting are indicated by gray and black arrowheads, respectively. For further details of inhibitor and hs treatment conditions, see legend to Figure 6A.
In summary, the expression analysis and inhibitor experiments performed in suspension culture cells revealed that Hsp90 significantly contributes to the control of HSR by influencing the accumulation of HsfA2 and HsfB1. For HsfB1, especially the stability of the protein is affected by inhibition of Hsp90 under hs as well as under normal growth conditions. By contrast, the influence of Hsp90 on the abundance of HsfA2 can be attributed mainly to the control of HsfA2 transcript levels during hs. Although inhibition by GDA was obviously not sufficient to induce the transcriptional activation of hs gene expression, contributions of Hsp90 can be attributed to the control of both the initial induction and the attenuation phase of HSR.
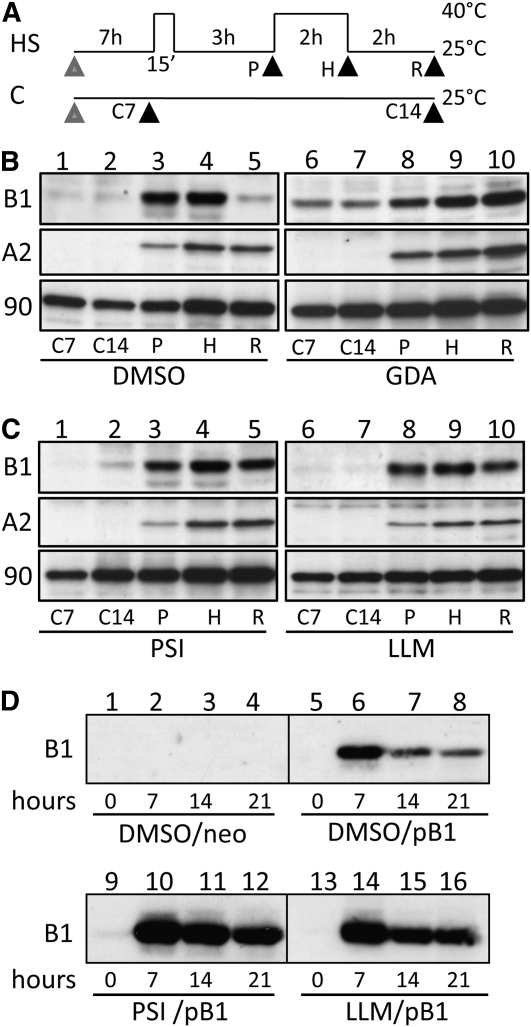
Hsp90 Acts in Targeting HsfB1 Protein for Proteasomal Degradation
All results observed so far suggest that Hsp90 regulates HsfB1 at two levels. On the one hand, in the absence of HsfA1 or HsfA2, Hsp90 targets HsfB1 to the DNA (Figure 4), and on the other hand, Hsp90 inhibition results in the stabilization of HsfB1 (Figure 6). Both effects could be explained by the physical interaction observed between HsfB1 and the chaperone (Figure 2). Especially the observed stabilization of HsfB1 by Hsp90 inhibition leads to the assumption that Hsp90 might be involved in targeting this Hsf for degradation. Therefore, we analyzed the degradation of HsfB1 in suspension culture cells in response to hs treatment, as shown in Figure 8A. Thereby, we compared the influence of GDA and two proteasome inhibitors, proteasome inhibitor I (PSI) and N-acetyl-l-leucinyl-l-leucinylmethional (LLM), which have been previously described as potent inhibitors of proteasome activity in vertebrate cells (Lee and Goldberg, 1998), on the accumulation of HsfB1. We further controlled the abundance of HsfA2, which is not regulated by Hsp90 at the protein level (Figure 6), and of Hsp90 itself. The usual decline of the HsfB1 level during hs recovery (Figure 8B, lane 5) was inhibited in the presence of GDA (lane 10). When cell cultures were treated with PSI or LLM, the decline of the HsfB1 level was inhibited to a similar extent (Figure 8B, lane 5, versus Figure 8C, lanes 5 and 10). Consistently, polyubiquitin-conjugated proteins accumulated upon addition of GDA, PSI, or LLM (see Supplemental Figure 3 online), which parallels previous observations while treating plant cells with GDA or the proteasome inhibitor MG132 (Kurepa et al., 2008; Nishizawa-Yokoi et al., 2010). Although the accumulation of HsfB1 was comparable in the presence of all three inhibitors at the concentrations applied (Figure 8), the accumulation of polyubiquitin-conjugated proteins was more pronounced for PSI and LLM (see Supplemental Figure 3 online). Thus, the accumulation of HsfB1 during GDA treatment can be attributed to malfunction of Hsp90, as Hsp90 clients are primarily affected by GDA treatment, while other proteins are only indirectly governed by this treatment as a consequence of the increased concentration of polyubiquitin-conjugated proteins in general. In contrast to HsfB1, the levels of HsfA2, Hsp90 (Figure 8), and HsfA1 (see Supplemental Figure 3 online) were not affected by any of the inhibitor treatments applied.
Figure 8.
Hsp90 and Proteasome Activity Are Required for Degradation of HsfB1.
(A) The hs treatment (HS) or control temperature regime (C) of cell cultures is shown. Black arrowheads indicate time points of sample preparations (P, preinduced; H, heat-stressed; R, recovery; C7, control after 7 h; C14, control after 14 h), and the gray arrowhead indicates the addition of DMSO (solute control for all inhibitors), GDA, or the proteasome inhibitors PSI or LLM.
(B) Abundance of HsfB1 (B1), HsfA2 (A2), and Hsp90 (90) proteins analyzed by immunodetection in total protein extracts of cells treated as indicated (at the bottom) in the presence of 0.1% DMSO (lanes 1–5) or 1 μM GDA (lanes 6–10).
(C) Same as (B) for treatments in the presence of 25 μM PSI (lanes 1–5) or 25 μM LLM (lanes 6–10).
(D) Tomato protoplasts were transformed with empty vector (lanes 1–4) or pB1 (lanes 5–16) followed by treatment with 0.1% DMSO (lanes 1–8), 25 μM PSI (lanes 9–12), or 25 μM LLM (lanes 13–16) and immunodetection of HsfB1 in protein extracts corresponding to 20,000 protoplasts harvested immediately (0 h; lanes 1, 5, 9, and 13), 7 h (lanes 2, 6, 10, and 14), 14 h (lanes 3, 7, 11, and 15), or 21 h (lanes 4, 8, 12, and 16) after transformation.
In protoplasts, HsfB1 also accumulated to higher levels when coexpressed with HsfA1 or HsfA2 and the IR construct of Hsp90 (Figure 3). To confirm that this effect is controlled by targeting the protein to degradation by proteasomes as well, we transformed protoplasts with the HsfB1 expression construct and incubated them in the presence of the two proteasome inhibitors described above. When the HsfB1 protein level was analyzed, we observed a strong enrichment of the protein after 7 h (Figure 8D, compare lanes 6, 10, and 14), and the protein was stabilized in the presence of the inhibitors at least for 14 h (compare lanes 7, 11, and 15). Hence, the stabilization of HsfB1 by treatment of cell cultures or protoplasts with GDA or proteasome inhibitors suggests an Hsp90-mediated targeting of HsfB1 for degradation by the proteasome.
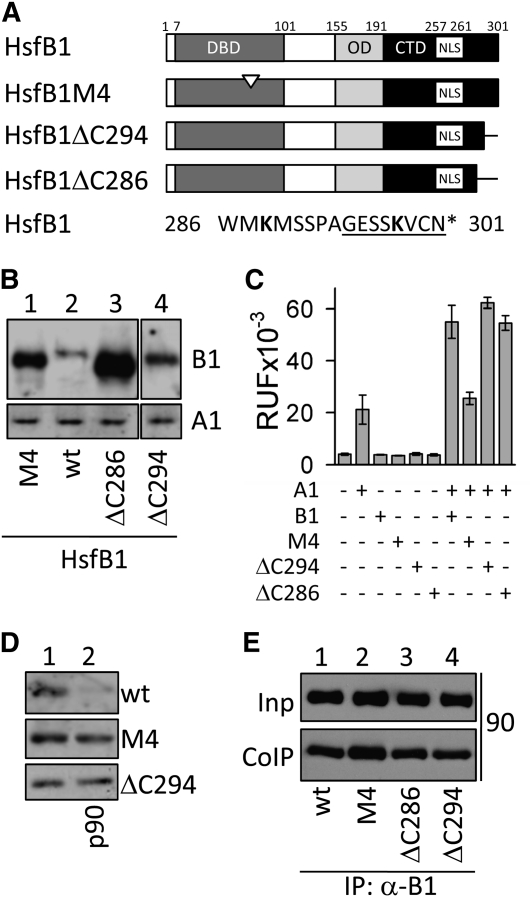
Degradation of HsfB1 Depends on DNA Binding and a C-Terminal Element of HsfB1
HsfB1 is targeted to the DNA in an Hsp90-dependent manner (Figure 4), but at the same time, Hsp90 is involved in the regulation of HsfB1 degradation (Figures 7 and 8). Thus, we made use of a mutant of HsfB1 that was shown to be impaired in DNA binding (M4; Boscheinen et al., 1997) to investigate the relation between these two processes. In addition, we analyzed two mutants with deletions at the C terminus of HsfB1 (Bharti et al., 2004) with respect to their stability (Figure 9A). To this aim, we first compared the protein abundance of wild-type HsfB1 and the three mutants after transformation and expression in the tomato protoplasts. For all three mutants, the protein level was higher than for wild-type HsfB1 (Figure 9B). In the GUS reporter assay, none of the HsfB1 forms showed activity on its own (Figure 9C). Due to the mutation in the DBD causing the loss of DNA binding activity, HsfB1M4 did not stimulate the activity of HsfA1. In contrast, deletion of 7 (ΔC294) or 15 (ΔC286) amino acid residues at the C-terminal end of HsfB1 had no influence on the coactivator function (Figure 9C). These results indicate that the HsfB1 protein stability is not directly connected to the activity of HsfB1 as coactivator and that impaired DNA binding (mutant M4) leads to hyperaccumulation of the protein.
Figure 9.
Hsp90-Controlled HsfB1 Degradation Involves DNA Binding and a C-Terminal Element.
(A) Domain structure of wild-type and mutant forms of HsfB1 are illustrated. The white arrowhead indicates the amino acid exchange KH55,56EL in the DBD of M4 and the C-terminal amino acid residues deleted in mutants ΔC294 (underlined) and ΔC286. CTD, C-terminal domain; NLS, nuclear localization signal; OD, oligomerization domain.
(B) Tomato protoplasts were transformed with expression constructs of wild-type (wt; lane 2) and mutant forms of HsfB1 (lanes 1, 3, and 4) and analyzed by immunodetection using HsfB1-specific antibodies (B1). Note that all samples were processed on the same blot, as well as the immunodetection of endogenous HsfA1 (A1), for which a 50 times longer exposure time than for HsfB1 was used.
(C) GUS reporter assay in protoplasts transformed with expression constructs coding for wild-type or mutant forms of HsfB1 as indicated below, either alone or in combination with HsfA1. Error bars represent sd of multiple experiments. RFU, Relative fluorescence units.
(D) Immunodetection of HsfB1 in protein extracts from protoplasts transformed with expression constructs coding for the indicated forms of HsfB1 (samples 1 and 2) and of Hsp90 (p90; lane 2) using HsfB1 antibodies.
(E) Profilin-tagged Hsp90 and HsfB1 forms were coexpressed in protoplasts (input [Inp]), immunoprecipitated with HsfB1-specific antibodies (CoIP), and the precipitated proteins were immunodetected with profilin tag–specific antibodies.
To analyze the influence of Hsp90 on the stability of the mutated HsfB1 proteins, wild-type HsfB1 and the mutant forms were coexpressed with Hsp90 (Figure 9D). The level of the wild-type protein was largely reduced when coexpressed with Hsp90. In contrast, the levels of the two mutant forms M4 and ΔC294 were not affected by coexpression of Hsp90 (lane 1 versus lane 2), although the mutant forms still interact with Hsp90, as confirmed by CoIP using HsfB1-specific antibodies (Figure 9E). Thus, the accumulation of the mutant form M4 demonstrates that Hsp90-dependent HsfB1 degradation requires DNA binding. In addition, we have identified a third element positioned at the C terminus of HsfB1, which is required for degradation but which does not influence DNA binding or interaction of the Hsf with Hsp90.
DISCUSSION
The widely accepted model of Hsf regulation assumes that molecular chaperones of the 70- and 90-kD families maintain Hsfs inactive but competent for activation under nonstress conditions and act in Hsf inactivation during attenuation of stress response. This model is mainly based on studies with Hsf1 in vertebrates and supports especially the role of Hsp90 and its cochaperones for signaling systems (Morimoto, 1998). For plants, which usually have more than 20 Hsf-encoding genes, our current knowledge about the control of Hsf activity by chaperones is still very limited. In tomato, the functional interplay of three Hsfs is central for the control of hs gene expression. The constitutively expressed HsfA1 is the master regulator of thermotolerance (Mishra et al., 2002), and exposure to hs conditions leads to its nuclear retention and induction of hs gene expression, including HsfA2 and HsfB1 (Scharf et al., 1998). The latter two Hsfs contribute to the enhancement of hs gene expression by functional interactions with HsfA1 (Bharti et al., 2004; Chan-Schaminet et al., 2009). In addition, HsfB1 is important for the recovery of housekeeping gene expression. Thus, the three Hsfs are required for the regulation of gene expression in response to hs and also under normal conditions.
We present several lines of evidence that in tomato cells, cytoplasmic Hsp70 and Hsp90 chaperones coregulate hs gene expression within a network of mutual, but factor-specific, interactions with Hsfs (summarized in Figure 10), and this regulatory network might also be valid in its main aspects for other plants. The functional interplay is manifested by physical interactions between Hsp90 and Hsp70 with Hsfs (Figures 1 and 2; Kim and Schöffl, 2002; Yamada et al., 2007; Meiri and Breiman, 2009; Meiri et al., 2010) and the Hsp90-dependent control of transcript levels as seen for HsfA2 (Figure 7). For the latter, inhibition of Hsp90 function leads to extended accumulation and stabilization of hsfA2 transcripts, which in turn results in the accumulation of HsfA2 protein (Figure 6). The physical association of the Hsfs with both chaperones indicates either the coexistence of distinct Hsf–chaperone complexes or close cooperation of Hsp70 and Hsp90 in the functional interplay with Hsfs. The latter is reminiscent of the situation described for several Hsp90 client proteins like the glucocorticoid receptor in vertebrate cells. Folding of the protein into premature receptor complexes and maintenance of competence for ligand binding followed by its final activation are tightly controlled by both chaperone functions (Pratt et al., 2008).
Figure 10.
Model of Multiple Regulatory Interactions between Hsfs and Chaperones during HSR in Tomato.
Under normal conditions, the inactive state of the HsfA1 (A1) as master regulator of the hs response is maintained by interaction with Hsp70 and Hsp90, and both chaperones contribute to the quantity control of HsfB1 (B1) protein, which is kept at a low level due to rapid degradation. During acute hs, the activation of HsfA1 and the rapid accumulation of HsfB1 are presumably induced by the demand of Hsp70 and Hsp90 due to increasing amounts of denatured proteins. HsfA2 expression is induced, but the protein interacts with sHsps (Hsp17) and becomes recruited into large cytoplasmic multichaperone complexes, known as hs granules (HSG). During attenuation/recovery, restoration of the free pool of Hsp70 and Hsp90 results in (1) inactivation of HsfA1 by Hsp70-induced release from DNA, (2) Hsp90-induced DNA binding of HsfB1 to function as an intermediate repressor for hs-inducible genes, and subsequently (3) Hsp90-dependent degradation of HsfB1. In parallel, HsfB1 is recruited to Hsf binding sites in the promoter regions of housekeeping genes (HK-gene) as coactivator of other transcription factors (X). After return to normal temperatures, HsfA2 is released from hs granules, but its activity is further controlled by interaction with sHsps. At repeated hs, the rapid formation of HsfA1/A2 superactivator complexes by HsfA2 release from sHsps enhances the expression of hs genes. Although most of the experimental details were elaborated for tomato, it is very likely that basic aspects of this model are also valid for other plants.
However, the mode of Hsf–chaperone interaction might be different in the individual complexes. By yeast two-hybrid analysis using both class A Hsfs without their own DBD, we realized that HsfA2 interacts with the C-terminal portions of both chaperones, which are commonly assumed to provide the binding sites for substrate proteins (Figure 1). HsfA1 also interacts with the C-terminal portion of Hsp90 but with the N-terminal portion of Hsp70. Unfortunately, the yeast-two-hybrid analysis failed to work with HsfB1; hence, the interaction sites within the chaperones could not yet be assigned. However, an interaction of HsfB1 with the N-terminal portion of Hsp90, which was lacking in the constructs, cannot be excluded, since full-length Hsp90 was not tested. The possible difference in chaperone interactions is also manifested by the distinct functional interplay between chaperones and Hsfs. It can be suggested that the interaction between HsfA1 and Hsp70 serves as a central regulatory module during HSR, because Hsp70, but not Hsp90, impairs HsfA1 binding to the HSE target sites on the DNA (Figure 4). Hence, this interaction possibly controls the inactive state in nonstressed cells (Figure 10, normal conditions) but also induces the repression of HsfA1 activity during attenuation of the stress response (Figure 10, attenuation/recovery). The latter is supported by the strong repression of activity of HsfA1/B1 coactivator complexes in the presence of Hsp70 (Figure 3). By contrast, the activity of the HsfA1/A2 complex is not influenced by coexpression of Hsp70 (Figure 3). This could be explained by the inaccessibility of HsfA1 for Hsp70 in the heterooligomeric HsfA1/A2 complex (Chan-Schaminet et al., 2009) and by the insensitivity of HsfA2 to the presence of Hsp70 (Figure 3). However, HsfA2 activity is controlled by interaction with Hsp17.4-CII (Port et al., 2004), which indicates the existence of additional, sHsp-dependent control mechanisms of HsfA2 activity during stress responses (Figure 10). Hsp90, in contrast to Hsp70, appears to influence the activity of the HsfA1/A2 complex (Figure 3A), which is consistent with recent findings on Arabidopsis HsfA2. Breiman and coworkers (Meiri and Breiman, 2009; Meiri et al., 2010) demonstrated that HsfA2 is nucleus-localized and active in the complex with Hsp90 and the cochaperone FKBP62 (ROF1) but becomes inactivated when the second cochaperone, FKBP65 (ROF2), becomes part of the complex in late stages of the HSR. Although it remains to be demonstrated, it is tempting to assume that a similar mechanism is involved in the control of tomato HsfA2.
In contrast to the HsfA1/HsfA2 scenario, the stimulation of HsfA2 activity by HsfB1 is repressed by coexpression of Hsp70 (Figure 3), which can be attributed to the function of HsfB1, because the activity of HsfA2 remains. However, the reduction cannot be explained by a direct influence of Hsp70 on the DNA binding activity of HsfB1 (Figure 4), which leads to the question of whether the proposed function of HsfB1 as a transcriptional coactivator that enforces the recruitment of class A Hsfs into ternary complexes with HAC1 (Bharti et al., 2004) could be targeted by Hsp70. Together with the repression of HsfA1 by Hsp70, such a mechanism would contribute to the repression of hs gene expression as long as sufficient amounts of Hsp70 are present. This is usually the case in nonstressed cells and during HSR attenuation. In addition, the DNA binding of HsfB1 strongly depends on the interaction with Hsp90 (Figure 4). Based on these observations, we conclude that the ratio between the two chaperones directly influences the composition and activity of transcriptional activator complexes formed by the three tomato Hsfs. Consequently, we propose the existence of a straight regulatory feedback loop of changing Hsp70/Hsp90 levels during HSR, where HsfB1 has a central function as modulator for the fine-tuning of hs gene expression, irrespective of whether a coactivator or a repressor function is assumed.
However, Hsp90 not only mediates the association of HsfB1 with DNA (Figure 10, attenuation/recovery) but also controls the abundance of the transcription factor (Figures 3, 5, 6, 8, and 9). Here, Hsp90 is needed for the degradation of HsfB1 by the proteasome (Figures 8 and 10, recovery). In this respect, Hsp90 function seems to be related to Hsp70 function, because HsfB1 is stabilized by coexpression of IR constructs targeting both chaperones (Figure 3). This may indicate their close cooperation, as reported for other Hsp90 client proteins. This scenario parallels the observation that in Escherichia coli, the hs-specific transcription factor σ32 is targeted in a DnaK-dependent manner to the FtsH protease complex during down-regulation of HSR (Rodriguez et al., 2008). This suggests that Hsf–Hsp interactions, as one regulatory principle involved in the control of the cellular HSR, might be conserved throughout all systems. Furthermore, targeting of eukaryotic DNA-bound transcription factors to proteasomal degradation is discussed as a mode for promoter clearance (Muratani and Tansey, 2003). Taking up this idea, the Hsp90-mediated DNA binding of HsfB1 would in turn promote its degradation for subsequent down-regulation of hs-induced transcription during recovery (Figure 10). Consistently, DNA binding of HsfB1 is required for degradation as well (Figure 9). However, DNA and Hsp90 binding of HsfB1 are not the only factors involved in the control of HsfB1 degradation. We identified a C-terminal element that is not involved in these two interactions but is required for degradation (Figure 9). This element contains two Lys residues and several Ser residues; thus, it might be speculated that this region is posttranslationally modified (e.g., by phosphorylation or ubiquitination) or recruits additional not yet identified factors.
The functional diversification of Hsfs is tightly connected with factor-specific interactions with members of different chaperone families (Hsp90, Hsp70, sHsps). The formation of distinct regulatory circuits provides the basis for an efficient fine-tuning of the cellular HSR during repeated cycles of hs and recovery (Figure 10). Besides the direct control of HsfA1 activity by Hsp70 and of HsfA2 by Hsp17.4-CII, we describe a functional interaction of HsfB1 with Hsp90. This interaction is required for DNA binding of HsfB1, which may stimulate its coactivator function in complexes with HAC1 in tomato (i.e., together with HsfA1 during induction of HSR and with housekeeping transcription factors during recovery; Bharti et al., 2004) or the repression of pathogen response–related genes in Arabidopsis (Kumar et al., 2009). The latter would be consistent with the observed HsfB1–Hsp90 interaction in Arabidopsis and the phenotype described in hsfB1/hsfB2b double knockout plants (Yamada et al., 2007; Kumar et al., 2009). Remarkably, HsfB1 of Arabidopsis does not interact with HAC1, an interaction required for the coactivator function (Bharti et al., 2004), which explains the lacking coactivator function as demonstrated in the hsfB1/hsfB2b mutant plants. Nevertheless, the Hsp90 dependence of HsfB1–DNA interaction is most likely conserved. However, tomato HsfB1 might also exhibit a repressor function comparable to the one of Arabidopsis HsfB1, namely after Hsp70-induced inactivation of HsfA1 during attenuation of HSR (Figure 10). Interestingly, a conserved repressor domain with the highly conserved tetrapeptide motif (LFGV) preceding the nuclear localization signal of HsfB1 was described, which was also identified in other plant transcription factors (Ikeda and Ohme-Takagi, 2009). One could speculate that this domain is involved in the interaction with the chaperones (or other corepressors), indicating the broad importance of chaperone regulation of plant transcription factor activities. However, this suggestion remains to be experimentally confirmed.
Remarkably, Hsp90 controls the level and the activity of HsfB1 in both nonstressed and thermotolerant cells. Until now, Hsp90 client proteins were considered to be stabilized by interaction with the chaperone, and a function of Hsp90 in protein degradation was discussed only for proteins that remain unfolded after several rounds of chaperone binding (reviewed in Taipale et al., 2010). Thus, to the best of our knowledge, HsfB1 represents the first example of a transcription factor that is targeted by Hsp90 to promote its function and to mediate its degradation at the same time. Considering a close functional cooperation of Hsp70 and Hsp90, it is intriguing to postulate that the HsfB1–Hsp90 interaction unit is directly involved in the control of the balance of chaperone homeostasis and its adaptation under changing environmental conditions. Unraveling the molecular mechanisms underlying this remarkable specification in more detail will be the challenge of future investigations.
METHODS
Plasmid Constructs
The Hsf-dependent reporter plasmid pGmhsp17.3B-CI*:GUS, the Hsf-dependent repressor plasmid p35S:HSE9-GUS, and the plant expression vectors for wild tomato (Solanum peruvianum [Sp]) HsfA1, HsfA2, and HsfB1 have been described (Treuter et al., 1993), and an overview of expression constructs, including sequences and accession numbers, is given in Supplemental Table 1 online. Sp HsfA1 is a homolog of Sl HsfA1a, which was described as master regulator of HSR in the cultivated tomato (Solanum lycopersicum var Moneymaker; Mishra et al., 2002). Triple HA-tagged Hsp70 (TC214904) and profilin-tagged Hsp90 (TC212970) expression constructs were created by amplification from cDNA using oligonucleotides listed in Supplemental Table 2 online. The sequence information from the Institute for Genomic Research tomato EST database (http://compbio.dfci.harvard.edu/tgi/plant.html) was used for oligonucleotide design. After restriction, the Hsp70 cDNA fragment was inserted into the pRT vector, providing a triple HA epitope tag (Siddique et al., 2003). For cloning of pProf-Hsp90, the p35dS:GUSxHsfA2 (Chan-Schaminet et al., 2009) plasmid was linearized with NcoI and BamHI, excising GUSxHsfA2. The profilin tag (residues 40–49 of birch [Betula spp] profilin; Fedorov et al., 1997) was produced by annealing oligonucleotides profT and profB (see Supplemental Table 2 online) followed by insertion into the linearized plasmid. Subsequently, the p35dS:prof was linearized with BamHI and the Hsp90 cDNA fragment was inserted.
Cloning of the IR construct for Hsp70 RNAi was described (Tripp et al., 2009). Two overlapping cDNA fragments were amplified to generate the Hsp90-specific IR construct for expression of double-stranded hairpin transcripts corresponding to the mostly conserved nucleotide sequence regions in the open reading frames of all cytoplasmic members of the tomato Hsp90 gene family.
The bait vectors for yeast two-hybrid studies encoding the Hsf fusion constructs with the Gal4p DBD were described (Scharf et al., 1998; Döring et al., 2000). The two prey constructs pADGal4xHsp90(aa376-699) and pADGal4xHsp70(aa4-651) were isolated by screening for Hsf interaction partners using the cDNA library prepared from heat-stressed S. peruvianum cell suspension cultures (Scharf et al., 1998). These parental plasmids were used as template DNA to generate further derivatives of prey constructs encoding shorter fragments of the two chaperones to perform the interaction studies shown in Figure 1.
Yeast Two-Hybrid Protein Interaction Assay
For two-hybrid interaction studies, the pBDGal4 bait and the pADGal4 prey vector system (Stratagene) were used as described (Scharf et al., 1998; Port et al., 2004). Protein interaction was monitored by testing colony growth of yeast cells cotransformed with both types of vector constructs for His prototrophy. For all combinations with the HsfA2 bait construct, the stringency was increased by colony growth in presence of 5 mM 3-aminotriazole (Döring et al., 2000).
Transient Expression Studies in Protoplasts
For transient gene expression studies, tomato (var Moneymaker) leaf mesophyll protoplasts were used. Polyethylene glycol–mediated cotransformation of reporter and Hsf expression plasmids was performed as described (Mishra et al., 2002). Reporter–GUS activities were determined as described (Treuter et al., 1993). For inhibitor studies, GDA (Sigma-Aldrich), PSI (Sigma-Aldrich), and LLM (Calbiochem) were dissolved in DMSO and added directly after transformation to a final concentration of 1 μM (GDA) and 25 μM (PSI and LLM) with DMSO adjusted to 0.1%. If not indicated otherwise, protoplasts were incubated afterward for 14 h before harvesting.
Immunoblots and Antibodies
Preparation of protein extracts was performed as described (Port et al., 2004). Aliquots corresponding to 20,000 protoplasts were subjected to SDS-PAGE and transferred to nitrocellulose membranes (Schleicher and Schuell) prior to probing with antisera and chemiluminescence detection following the manufacturer’s protocol (Perkin-Elmer Life Sciences). Immunodetection of tomato HsfA1, HsfA2, or HsfB1 was performed with polyclonal antisera (Lyck et al., 1997). For detection of HA-tagged Hsp70, monoclonal HA antibodies (clone 16B12) were obtained from Hiss Diagnostics, and the monoclonal anti-ubiquitin antibody P4D1 (Santa Cruz Biotechnology) was used for detection of polyubiquitin-conjugated proteins. Monoclonal antibodies against the profilin tag were kindly provided by Anna Starzinski-Powitz (Goethe University Frankfurt). Secondary horseradish peroxidase–conjugated antibodies were obtained from Sigma-Aldrich.
CoIP
Protein extracts from four samples of 1 × 105 protoplasts transformed with 5 μg of Hsf and Hsp expression plasmids were prepared in 200 μL of NEB250 lysis buffer containing 25 mM Hepes, pH 7.5, 250 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 10 mM NaF, 1 mM β-mercaptoethanol, 0.2% (w/v) Nonidet P-40, 10% (w/v) glycerol, 50 μg/mL Na-p-tosyl-l-lysyl chloromethyl ketone, and Complete protease inhibitor mixture (Roche) according to the manufacturer’s protocol. After removal of cellular debris by centrifugation for 10 min at 12,000 rpm at 4°C, the supernatant was diluted to 400 μL with NEB buffer without NaCl to adjust the final NaCl concentration to 125 mM. Aliquots of 20 μL were used as input controls. Hsf-specific or Hsp70-specific antisera cross-linked to NHS-activated Sepharose beads (GE Healthcare) according to the manufacturer’s protocol were added as 1:10 slurry to the residual samples (40 μL). After incubation at 4°C for 1 h with gentle agitation, the beads were separated and washed four times with 500 μL of 10 mM Tris buffer (pH 7.5) supplemented with 140 mM NaCl. Bound proteins were eluted with 60 μL of 2× SDS sample buffer, and aliquots of 20 μL were separated by SDS-PAGE and processed for immunoblotting.
Cell Culture Handling and Analysis
S. peruvianum cell suspension cultures were cultivated in modified Murashige and Skoog medium (Nover et al., 1982). For hs treatments and inhibitor studies, exponentially growing cell cultures were used 3 d after inoculation into fresh medium at cell densities corresponding to ~100 mg fresh weight/mL. The hs treatments were performed in a rotary shaking water bath prewarmed to the indicated temperatures. For preinduction, cultures were heat-stressed for 15 min at 40°C followed by a 3-h recovery at 25°C. If not indicated otherwise, GDA and proteasome inhibitors were added to final concentrations as indicated above for protoplasts. Preparation of whole cell protein extracts and analysis of Hsf and Hsp levels by immunodetection were performed as described before (Siddique et al., 2003).
Total RNA was prepared for RNA gel blot analysis as described (Mishra et al., 2002). For analysis of transcript levels by RT-PCR, RNA preparation and treatment with DNase I were performed by using the RNeasy plant mini kit (Qiagen) according to the manufacturer’s protocol. Primer combinations used for the generation of Hsf- and Hsp-specific digoxigenin-labeled RNA gel blot probes and for the amplification of transcript-specific cDNA fragments are summarized in Supplemental Table 2 online.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: tomato HsfA1, CAA47869; tomato HsfA2, CAA47870; tomato HsfB1, CAA39034; tomato Hsp90, AAB01376; tomato Hsp70, CAA37970; tomato Hsp17-CI, AJ225046.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Influence of Hsp70 and Hsp90 on the Expression of GUS Reporter Constructs.
Supplemental Figure 2. Influence of GDA and hs Treatment on Protein Expression Levels in Tomato Suspension Culture Cells.
Supplemental Figure 3. Influence of 26S Proteasome and Hsp90 Inhibition on the Levels of Hsfs and Polyubiquitin-Conjugated Proteins.
Supplemental Table 1. List of Constructs Used in This Work.
Supplemental Table 2. List of Oligonucleotides Used for Cloning of Expression Constructs in This Work and for Transcript Analyses.
Acknowledgments
We thank Lutz Nover for critical comments during preparation of the manuscript, Giesela Englich and Sascha Röth for technical assistance, and Shravan Kumar Mishra for material support. Financial support from the Deutsche Forschungsgemeinschaft (grant SCHA577/6-1 to K.-D.S.) and from the Volkswagenstiftung (to E.S.) is acknowledged. A.H. was supported by a fellowship from the Centre of Membrane Proteomics, Goethe University Frankfurt.
References
- Abravaya K., Myers M.P., Murphy S.P., Morimoto R.I. (1992). The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 6: 1153–1164 [DOI] [PubMed] [Google Scholar]
- Baler R., Welch W.J., Voellmy R. (1992). Heat shock gene regulation by nascent polypeptides and denatured proteins: hsp70 as a potential autoregulatory factor. J. Cell Biol. 117: 1151–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger C.A., Connell P., Wu Y., Hu Z., Thompson L.J., Yin L.Y., Patterson C. (1999). Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19: 4535–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniwal S.K., et al. (2004). Heat stress response in plants: A complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 29: 471–487 [DOI] [PubMed] [Google Scholar]
- Bharti K., Von Koskull-Döring P., Bharti S., Kumar P., Tintschl-Körbitzer A., Treuter E., Nover L. (2004). Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16: 1521–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscheinen O., Lyck R., Queitsch C., Treuter E., Zimarino V., Scharf K.-D. (1997). Heat stress transcription factors from tomato can functionally replace HSF1 in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 255: 322–331 [DOI] [PubMed] [Google Scholar]
- Bösl B., Grimminger V., Walter S. (2006). The molecular chaperone Hsp104—A molecular machine for protein disaggregation. J. Struct. Biol. 156: 139–148 [DOI] [PubMed] [Google Scholar]
- Boston R.S., Viitanen P.V., Vierling E. (1996). Molecular chaperones and protein folding in plants. Plant Mol. Biol. 32: 191–222 [DOI] [PubMed] [Google Scholar]
- Bukau B., Weissman J., Horwich A. (2006). Molecular chaperones and protein quality control. Cell 125: 443–451 [DOI] [PubMed] [Google Scholar]
- Chan-Schaminet K.Y., Baniwal S.K., Bublak D., Nover L., Scharf K.D. (2009). Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J. Biol. Chem. 284: 20848–20857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecka-Verner E., Pan S., Salem T., Gurley W.B. (2004). Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol. Biol. 56: 57–75 [DOI] [PubMed] [Google Scholar]
- Damberger F.F., Pelton J.G., Harrison C.J., Nelson H.C., Wemmer D.E. (1994). Solution structure of the DNA-binding domain of the heat shock transcription factor determined by multidimensional heteronuclear magnetic resonance spectroscopy. Protein Sci. 3: 1806–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring P., Treuter E., Kistner C., Lyck R., Chen A., Nover L. (2000). The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 12: 265–278 [PMC free article] [PubMed] [Google Scholar]
- Fedorov A.A., Ball T., Mahoney N.M., Valenta R., Almo S.C. (1997). The molecular basis for allergen cross-reactivity: crystal structure and IgE-epitope mapping of birch pollen profilin. Structure 5: 33–45 [DOI] [PubMed] [Google Scholar]
- Guo Y., Guettouche T., Fenna M., Boellmann F., Pratt W.B., Toft D.O., Smith D.F., Voellmy R. (2001). Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J. Biol. Chem. 276: 45791–45799 [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Hayer-Hartl M. (2002). Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Hayer-Hartl M. (2009). Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16: 574–581 [DOI] [PubMed] [Google Scholar]
- Heerklotz D., Döring P., Bonzelius F., Winkelhaus S., Nover L. (2001). The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol. Cell. Biol. 21: 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison K.A., Scherrer L.C., Czar M.J., Stancato L.F., Chow Y.H., Jove R., Pratt W.B. (1993). Regulation of glucocorticoid receptor function through assembly of a receptor-heat shock protein complex. Ann. N. Y. Acad. Sci. 684: 35–48 [DOI] [PubMed] [Google Scholar]
- Ikeda M., Ohme-Takagi M. (2009). A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50: 970–975 [DOI] [PubMed] [Google Scholar]
- Kampinga H.H., Craig E.A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11: 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.H., Schöffl F. (2002). Interaction between Arabidopsis heat shock transcription factor 1 and 70 kDa heat shock proteins. J. Exp. Bot. 53: 371–375 [DOI] [PubMed] [Google Scholar]
- Kumar M., Busch W., Birke H., Kemmerling B., Nürnberger T., Schöffl F. (2009). Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol Plant 2: 152–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J., Toh-E A., Smalle J.A. (2008). 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 53: 102–114 [DOI] [PubMed] [Google Scholar]
- Lee D.H., Goldberg A.L. (1998). Proteasome inhibitors: Valuable new tools for cell biologists. Trends Cell Biol. 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Lee S., Lee D.W., Lee Y., Mayer U., Stierhof Y.D., Lee S., Jürgens G., Hwang I. (2009). Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 21: 3984–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Demand J., Höhfeld J. (2000). The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J. Biol. Chem. 275: 4613–4617 [DOI] [PubMed] [Google Scholar]
- Lyck R., Harmening U., Höhfeld I., Treuter E., Scharf K.D., Nover L. (1997). Intracellular distribution and identification of the nuclear localization signals of two plant heat-stress transcription factors. Planta 202: 117–125 [DOI] [PubMed] [Google Scholar]
- Mayer M.P. (2010). Gymnastics of molecular chaperones. Mol. Cell 39: 321–331 [DOI] [PubMed] [Google Scholar]
- Mayer M.P., Bukau B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri D., Breiman A. (2009). Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 59: 387–399 [DOI] [PubMed] [Google Scholar]
- Meiri D., Tazat K., Cohen-Peer R., Farchi-Pisanty O., Aviezer-Hagai K., Avni A., Breiman A. (2010). Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol. Biol. 72: 191–203 [DOI] [PubMed] [Google Scholar]
- Mirus O., Schleiff E. (2009). The evolution of tetratricopeptide repeat domain containing receptors involved in protein translocation. Endocytobiosis Cell Res. 19: 31–50 [Google Scholar]
- Mishra S.K., Tripp J., Winkelhaus S., Tschiersch B., Theres K., Nover L., Scharf K.D. (2002). In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev. 16: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto R.I. (1998). Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12: 3788–3796 [DOI] [PubMed] [Google Scholar]
- Morimoto R.I. (2008). Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser D.D., Duchaine J., Massie B. (1993). The DNA-binding activity of the human heat shock transcription factor is regulated in vivo by hsp70. Mol. Cell. Biol. 13: 5427–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M., Tansey W.P. (2003). How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4: 192–201 [DOI] [PubMed] [Google Scholar]
- Nair S.C., Toran E.J., Rimerman R.A., Hjermstad S., Smithgall T.E., Smith D.F. (1996). A pathway of multi-chaperone interactions common to diverse regulatory proteins: Estrogen receptor, Fes tyrosine kinase, heat shock transcription factor Hsf1, and the aryl hydrocarbon receptor. Cell Stress Chaperones 1: 237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto H., Vígh L. (2007). The small heat shock proteins and their clients. Cell. Mol. Life Sci. 64: 294–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A., Tainaka H., Yoshida E., Tamoi M., Yabuta Y., Shigeoka S. (2010). The 26S proteasome function and Hsp90 activity involved in the regulation of HsfA2 expression in response to oxidative stress. Plant Cell Physiol. 51: 486–496 [DOI] [PubMed] [Google Scholar]
- Nover L., Bharti K., Döring P., Mishra S.K., Ganguli A., Scharf K.D. (2001). Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L., Kranz E., Scharf K.D. (1982). Growth cycle of suspension cultures of Lycopersicon esculentum and L. peruvianum. Biochem. Physiol. Pflanz. 177: 483–499 [Google Scholar]
- Pearl L.H., Prodromou C., Workman P. (2008). The Hsp90 molecular chaperone: An open and shut case for treatment. Biochem. J. 410: 439–453 [DOI] [PubMed] [Google Scholar]
- Pelham H.R., Bienz M. (1982). A synthetic heat-shock promoter element confers heat-inducibility on the herpes simplex virus thymidine kinase gene. EMBO J. 1: 1473–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peteranderl R., Rabenstein M., Shin Y.K., Liu C.W., Wemmer D.E., King D.S., Nelson H.C. (1999). Biochemical and biophysical characterization of the trimerization domain from the heat shock transcription factor. Biochemistry 38: 3559–3569 [DOI] [PubMed] [Google Scholar]
- Port M., Tripp J., Zielinski D., Weber C., Heerklotz D., Winkelhaus S., Bublak D., Scharf K.D. (2004). Role of Hsp17.4-CII as coregulator and cytoplasmic retention factor of tomato heat stress transcription factor HsfA2. Plant Physiol. 135: 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt W.B., Morishima Y., Osawa Y. (2008). The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J. Biol. Chem. 283: 22885–22889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt W.B., Morishima Y., Peng H.M., Osawa Y. (2010). Proposal for a role of the Hsp90/Hsp70-based chaperone machinery in making triage decisions when proteins undergo oxidative and toxic damage. Exp. Biol. Med. (Maywood) 235: 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C., Sangster T.A., Lindquist S. (2002). Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624 [DOI] [PubMed] [Google Scholar]
- Rabindran S.K., Wisniewski J., Li L., Li G.C., Wu C. (1994). Interaction between heat shock factor and hsp70 is insufficient to suppress induction of DNA-binding activity in vivo. Mol. Cell. Biol. 14: 6552–6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K., Haslbeck M., Buchner J. (2010). The heat shock response: Life on the verge of death. Mol. Cell 40: 253–266 [DOI] [PubMed] [Google Scholar]
- Riggs D.L., Cox M.B., Cheung-Flynn J., Prapapanich V., Carrigan P.E., Smith D.F. (2004). Functional specificity of co-chaperone interactions with Hsp90 client proteins. Crit. Rev. Biochem. Mol. Biol. 39: 279–295 [DOI] [PubMed] [Google Scholar]
- Rodriguez F., Arsène-Ploetze F., Rist W., Rüdiger S., Schneider-Mergener J., Mayer M.P., Bukau B. (2008). Molecular basis for regulation of the heat shock transcription factor σ32 by the DnaK and DnaJ chaperones. Mol. Cell 32: 347–358 [DOI] [PubMed] [Google Scholar]
- Sangster T.A., Bahrami A., Wilczek A., Watanabe E., Schellenberg K., McLellan C., Kelley A., Kong S.W., Queitsch C., Lindquist S. (2007). Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PLoS ONE 2: e648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster T.A., Queitsch C. (2005). The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr. Opin. Plant Biol. 8: 86–92 [DOI] [PubMed] [Google Scholar]
- Scharf K.D., Heider H., Höhfeld I., Lyck R., Schmidt E., Nover L. (1998). The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell. Biol. 18: 2240–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Mosser D.D., Morimoto R.I. (1998). Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 12: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M., Port M., Tripp J., Weber C., Zielinski D., Calligaris R., Winkelhaus S., Scharf K.D. (2003). Tomato heat stress protein Hsp16.1-CIII represents a member of a new class of nucleocytoplasmic small heat stress proteins in plants. Cell Stress Chaperones 8: 381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M., Jarosz D.F., Lindquist S. (2010). HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11: 515–528 [DOI] [PubMed] [Google Scholar]
- Tripp J., Mishra S.K., Scharf K.D. (2009). Functional dissection of the cytosolic chaperone network in tomato mesophyll protoplasts. Plant Cell Environ. 32: 123–133 [DOI] [PubMed] [Google Scholar]
- Treuter E., Nover L., Ohme K., Scharf K.D. (1993). Promoter specificity and deletion analysis of three heat stress transcription factors of tomato. Mol. Gen. Genet. 240: 113–125 [DOI] [PubMed] [Google Scholar]
- Wandinger S.K., Richter K., Buchner J. (2008). The Hsp90 chaperone machinery. J. Biol. Chem. 283: 18473–18477 [DOI] [PubMed] [Google Scholar]
- Whitesell L., Lindquist S.L. (2005). HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5: 761–772 [DOI] [PubMed] [Google Scholar]
- Yamada K., Fukao Y., Hayashi M., Fukazawa M., Suzuki I., Nishimura M. (2007). Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J. Biol. Chem. 282: 37794–37804 [DOI] [PubMed] [Google Scholar]
- Young J.C., Agashe V.R., Siegers K., Hartl F.U. (2004). Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5: 781–791 [DOI] [PubMed] [Google Scholar]
- Zhang X.P., Glaser E. (2002). Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci. 7: 14–21 [DOI] [PubMed] [Google Scholar]
- Zou J., Guo Y., Guettouche T., Smith D.F., Voellmy R. (1998). Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94: 471–480 [DOI] [PubMed] [Google Scholar]