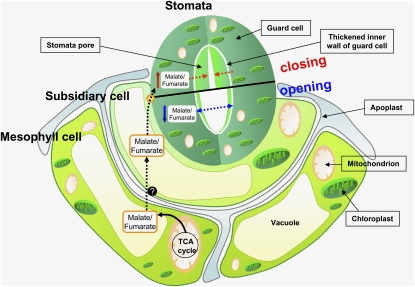
The antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase in tomato increases photosynthesis and biomass via an organic acid–mediated effect on stomatal aperture. This finding reinforces earlier suggestions that malate plays a crucial role in stomatal opening and supports the hypothesis that stomatal function can be regulated remotely via mesophyll-generated cues.
Abstract
Transgenic tomato (Solanum lycopersicum) plants expressing a fragment of the Sl SDH2-2 gene encoding the iron sulfur subunit of the succinate dehydrogenase protein complex in the antisense orientation under the control of the 35S promoter exhibit an enhanced rate of photosynthesis. The rate of the tricarboxylic acid (TCA) cycle was reduced in these transformants, and there were changes in the levels of metabolites associated with the TCA cycle. Furthermore, in comparison to wild-type plants, carbon dioxide assimilation was enhanced by up to 25% in the transgenic plants under ambient conditions, and mature plants were characterized by an increased biomass. Analysis of additional photosynthetic parameters revealed that the rate of transpiration and stomatal conductance were markedly elevated in the transgenic plants. The transformants displayed a strongly enhanced assimilation rate under both ambient and suboptimal environmental conditions, as well as an elevated maximal stomatal aperture. By contrast, when the Sl SDH2-2 gene was repressed by antisense RNA in a guard cell–specific manner, changes in neither stomatal aperture nor photosynthesis were observed. The data obtained are discussed in the context of the role of TCA cycle intermediates both generally with respect to photosynthetic metabolism and specifically with respect to their role in the regulation of stomatal aperture.
INTRODUCTION
Succinate dehydrogenase (EC 1.3.5.1), often referred to as complex II, has a dual function, being important in both the tricarboxylic acid (TCA) cycle and the aerobic respiratory chain, via the catalysis of the oxidation of succinate to fumarate and the reduction of ubiquinone to ubiquinol, respectively (Hägerhäll, 1997; Figueroa et al., 2001). Complex II is the simplest of all the complexes of the electron transport chain and displays a similar composition to the closely related fumarate reductases or succinate dehydrogenases of bacteria, yeast, and mammals (Scheffler, 1998). The conserved elements of this complex, which consists of a mere four polypeptides, comprise two peripheral membrane proteins, a flavoprotein (SDH1), and an iron sulfur binding protein (SDH2), as well as two integral membrane proteins (SDH3 and SDH4) (Vedel et al., 1999; Rasmusson et al., 2008). In plants, complex II has been demonstrated to contain four additional subunits (Eubel et al., 2003; Millar et al., 2004); however, no clear biological function of these additional subunits has been described. Moreover, in contrast with the situation for complexes I, III, and IV, for which multiple gene functional analyses have allowed direct evaluation of the physiological functions of their constituent subunits (Newton et al., 1990; Marienfeld and Newton, 1994; Pla et al., 1995; Howad and Kempken, 1997; Dutilleul et al., 2005; Vidal et al., 2007), as yet only two forward or reverse genetic strategies have been employed to study the function of complex II in plants (León et al., 2007; Roschzttardtz et al., 2009). These studies revealed that disruption of the expression of the SDH1-1 gene results in alterations in gametophyte development, pollen abortion, and reduced seed set (León et al., 2007) and that the absence of SDH2-3 in Arabidopsis thaliana seeds appears to slow their germination (Roschzttardtz et al., 2009). Furthermore, again by contrast with the other complexes of the inner mitochondrial membrane (see for example, Millar et al., 1993; Raghavendra et al., 1994; Sweetlove et al., 2002; Garmier et al., 2008), relatively few specific inhibitors of this complex have been found (Miyadera et al., 2003; Horsefield et al., 2006; Mogi et al., 2009). To our knowledge, none of these inhibitors, which have been used extensively to assess the function of complex II, has as yet been used in studies on plant mitochondria, rendering the study of Arabidopsis gametophyte and seed development the only functional studies of complex II in plants to date. In contrast with the situation observed in plants, complex II of nonplant systems has been the subject of somewhat greater scrutiny, with mutagenic studies being performed in Saccharomyces cerevisiae, Neurosporra crassa, Chinese hamster cells, and Caenorhabditis elegans (reviewed in Vedel et al., 1999). The study in C. elegans revealed that a mis-sense mutation in SDH cytochrome b (the 15-kD SDH3 anchoring protein) resulted in oxidative stress and premature aging in nematodes (Ishii et al., 1998). The targeted disruption of SDH1, SDH2, or SDH3 in S. cerevisiae, however, confers severe growth defects when cells are cultured on nonfermentable carbon sources (Lombardo et al., 1990; Daignan-Fornier et al., 1994). In humans, complex II deficiency has been reported both in isolation and in combination with other genetic defects (Morris et al., 1994; Bourgeron et al., 1995; Schon, 2000; Rustin and Rötig, 2002; Rutter et al., 2010), although the precise molecular bases of these deficiencies is often poorly characterized.
In contrast with the paucity of gene functional data concerning succinate dehydrogenase in plants, considerable information has been compiled concerning the majority of other steps of the TCA cycle (for review, see Sweetlove et al., 2007). With the exception of a handful of studies concerning root metabolism (Koyama et al., 2000; López-Bucio et al., 2000, 2003; van der Merwe et al., 2009), the majority of studies have focused on leaf tissue, despite the fact that the role of the TCA cycle in the illuminated leaf remains somewhat contentious (Tcherkez et al., 2005; Nunes-Nesi et al., 2007a). Intriguingly, quite diverse effects were observed upon downregulation of the various steps of the cycle, with deficiency of expression of aconitase and the mitochondrial malate dehydrogenase resulting in enhanced photosynthetic rates (Carrari et al., 2003; Nunes-Nesi et al., 2005), whereas inhibition of either citrate synthase, succinyl CoA ligase, or isocitrate dehydrogenase had little effect on the rates of photosynthesis itself and relatively minor consequences on photosynthetic metabolism in general (Studart-Guimarães et al., 2007; Sienkiewicz-Porzucek et al., 2008, 2010; Sulpice et al., 2010). By contrast, downregulation of the expression of fumarase restricted photosynthesis and plant growth. Detailed biochemical and physiological studies delimited this phenotype as being a consequence of a perturbation of stomatal function (Nunes-Nesi et al., 2007b) but were unable to define the exact mechanism underlying this phenomenon.
Here, we extend our characterization of the importance of the TCA cycle in tomato (Solanum lycopersicum) leaf function (for details, see the recent reviews of Sweetlove et al., 2010 and Nunes-Nesi et al., 2011) by describing the generation of transgenic tomato plants deficient in the expression of the iron-sulfur subunit of succinate dehydrogenase. These plants displayed increased rates of net photosynthesis and growth under normal greenhouse conditions as well as enhanced rates of net photosynthesis under suboptimal carbon dioxide concentrations. Physical measurement of stomatal aperture revealed that this was greater in the transgenics, although the number of stomata per leaf area remained constant. Analysis of stomatal apertures following incubation of wild-type leaf discs in physiological concentrations of malate or fumarate confirmed that the stomatal effect was organic acid mediated and independent of the abscisic acid (ABA) signal transduction pathway. Moreover, measurement of the levels of this phytohormone and the genes associated with its signal transduction revealed no changes in the transformants. While the results of these analyses were consistent with a predominant role of the mesophyll in determining the observed phenotypes, they did not categorically prove this. For this reason, we created a second set of transgenics in which the Sl SDH2-2 gene was repressed under the control of the guard cell–specific MYB60 promoter. The resultant transformants, by contrast with those expressing the construct under the control of the 35S promoter, revealed neither changes in stomatal aperture nor rates of photosynthesis. The results are discussed both generally in terms of the importance of succinate dehydrogenase in mitochondrial and photosynthetic metabolism and specifically with regard to the contribution it plays in mesophyll-mediated regulation of stomatal function.
RESULTS
Cloning of a cDNA Encoding the Iron-Sulfur Subunit of Succinate Dehydrogenase of Tomato
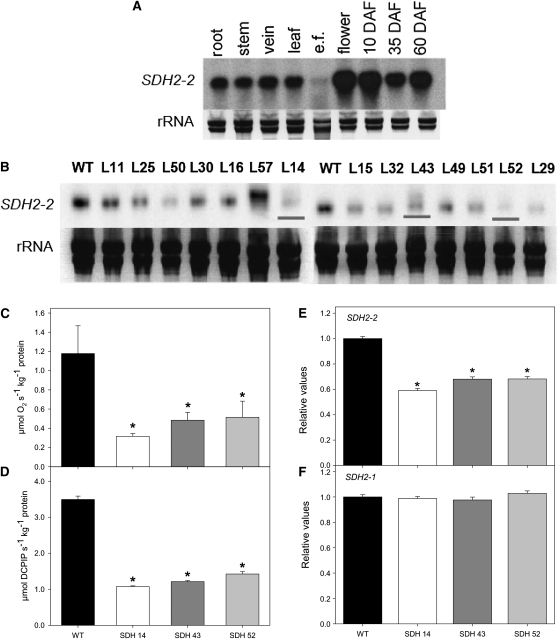
Searching tomato EST collections (Van der Hoeven et al., 2002) revealed the presence of 71 ESTs encoding the iron-sulfur subunit of succinate dehydrogenase. These ESTs belonged to two tentative consensus sequences (SlSDH2-1, SGN-U563725 and SlSDH2-2, SGN-U563726). This observation suggests that, in contrast with Arabidopsis, which contains three genes (Elorza et al., 2004), tomato, like maize (Zea mays) and rice (Oryza sativa) (Figueroa et al.,1999; Kubo et al., 1999), contains fewer genes encoding the iron subunit of succinate dehydrogenase Sl SDH2. Sequence analysis of the Sl SDH2-1 and Sl SDH2-2 genes revealed an open reading frame of 221 and 274 amino acids, respectively, and 81% of amino acid identity between them. Comparison with previously functionally characterized subunits of the iron-sulfur subunit of succinate dehydrogenase Sl SDH2-2 revealed relatively high identity with Arabidopsis SDH2-1 (77%) and SDH2-2 (77%), while it also showed amino acid identity (54%) to SDH2-3 of Arabidopsis and weak homology to human (58%), Drosophila (57%), yeast (56%), the diatom Phaeodactylum tricornutum (56%), and Chlamydomonas reinhardtii (55%) proteins (see Supplemental Figure 1A and Supplemental Data Set 1 online). Comparing Sl SDH2-2 with SDH2-2 homologs from more closely related species, higher identities were observed for the related species potato (Solanum tuberosum; 98%) and tobacco (Nicotiana tabacum; 91%), while lower identities were observed for the monocots maize (69%), rice (72%), and sugarcane (Saccharum officinarum; 64%) (see Supplemental Figure 1A online). Sl SDH2-2 bears characteristics of a mitochondrial transit peptide signal (von Heijne, 1986), while Sl SDH2-1 does not. Analysis of mRNA RNA gel blots indicates a near constitutive expression of tomato SDH2-2, with the transcript present at approximately equivalent levels in leaves, stems, roots, and fruits but only at low levels in leaf epidermal fragments (Figure 1A). In addition, the transcript is apparently ubiquitous during fruit development (Figure 1A).
Figure 1.
Characterization and Expression of Tomato Succinate Dehydrogenase (SDH2-2).
(A) RNA gel blot containing total RNA extracted from different organs of tomato plants. Total RNA was obtained from root, stem, vein, leaf, epidermal fragments (e.f.), flowers, and fruits 10, 35, and 60 d after flowering (DAF).
(B) RNA gel blot analysis of leaves of 4-week-old transgenic tomato plants with altered expression of SDH2-2 compared with the wild type (WT). The full-length 825-bp cDNA encoding the iron-sulfur subunit of succinate dehydrogenase was cloned in the antisense orientation into the transformation vector pK2WG7 between the CaMV promoter and the ocs terminator (see Supplemental Figure 1B online), and 15 transgenic tomato plants were obtained. Screening of the lines (L) by RNA gel blot yielded three lines that displayed a considerable reduction of Sl SDH2-2 (shown in red lines).
(C) and (D) Succinate-dependent oxygen consumption in freshly isolated mitochondria of green fruits 35 DAF (C) and succinate-dependent DCPIP reduction determined in enriched mitochondria from tomato leaves (D). Data are presented as mean values ± se and are averages from three to five different mitochondrial isolations.
(E) and (F) Relative transcript abundance of mitochondrial Complex II subunits (SDH2-2 and SDH2-1, respectively). The abundance of SDH mRNAs was measured by qRT-PCR, and values are presented as mean ± se of six individual plants per line. Asterisks indicate values that were determined by the Student’s t test to be significantly different from the wild type (P < 0.05).
Antisense SDH Transgenic Tomato Plants Exhibited Elevated Aerial Growth and Fruit Yield
The full-length 825-bp cDNA encoding the iron-sulfur subunit of succinate dehydrogenase was cloned in the antisense orientation into the transformation vector pK2WG7 between the cauliflower mosaic virus (CaMV) promoter and the ocs terminator (see Supplemental Figure 1B online). We then transferred 15 transgenic tomato plants obtained by Agrobacterium tumefaciens–mediated transformation to the greenhouse. Screening of the lines by RNA gel blot yielded three lines that displayed a considerable reduction of the Sl SDH2-2 (Figure 1B). These lines were clonally propagated in tissue culture and then transferred to the greenhouse. The succinate-dependent oxygen consumption was determined using a Clark-type electrode, after mitochondrial isolation from the green fruits of both wild-type and transformant plants using a Percoll gradient purification method (Millar et al., 2001). Statistical analysis revealed that three lines, SDH14, SDH43, and SDH52, exhibited reductions in enzyme activity that rendered them suitable for further analysis (Figure 1C). Using the same approach, we measured the mitochondrial rate of respiration, on provision of NADH, malate, citrate, or 2-oxoglutarate as substrate, in the wild type and the transformants (Table 1). Using those substrates, the rate of oxygen consumption was not altered in the transformants, confirming the findings of the above experiments and providing further evidence for the specificity of the inhibition. Additionally, we observed that the succinate-dependent dichlorophenolindophenol (DCPIP) reduction in the succinate dehydrogenase antisense lines (Figure 1D) was in good agreement with the succinate-dependent oxygen consumption. To verify the specificity of the constructs as well as to ensure that no compensatory effect occurred via the expression of the other isoforms, a secondary screen was performed at the mRNA level, using an established quantitative RT-PCR protocol (Czechowski et al., 2004). This revealed that only SDH2-2 expression was significantly reduced in the leaves of the transgenic lines (Figure 1E). Moreover, the expression of the nontargeted isoform SDH2-1 was unaltered in the transformants (Figure 1F). Interestingly, by contrast with the situation observed in Arabidopsis (Elorza et al., 2004), the expression of SDH2-1 was relatively low in lower epidermal fragments, with similarly low expression levels of the target isoform SDH2-2 (see Supplemental Figure 2 online). Furthermore, the expression of both isoforms was unaltered in lower epidermal fragments of the transformants. When taken together, the combined evidence presented demonstrates that these three lines (SDH14, SDH43, and SDH52) were suitable for assessing the effects of a mild reduction in the mitochondrial succinate dehydrogenase activity on mesophyll tissues.
Table 1.
Respiratory State 3 Activities of Mitochondria Isolated from Tomato Fruits from Wild-Type or Succinate Dehydrogenase Antisense Lines
| O2 Consumption (nmol O2 min−1 mg−1 Protein) |
||||
| Substrate | Wild Type | SDH14 | SDH43 | SDH52 |
| NADH | 142.41 ± 21.32 | 131.16 ± 16.57 | 127.61 ± 19.14 | 136.72 ± 24.88 |
| Malate | 108.23 ± 15.39 | 98.71 ± 14.27 | 102.36 ± 11.95 | 107.79 ± 16.52 |
| Citrate | 134.12 ± 12.45 | 127.29 ± 14.19 | 135.24 ± 16.01 | 133.14 ± 15.28 |
| 2-Oxoglutarate | 145.27 ± 8.71 | 138.97 ± 9.85 | 140.35 ± 10.15 | 134.52 ± 14.57 |
| RCR | 2.86 ± 0.28 | 2.45 ± 0.46 | 2.48 ± 0.25 | 2.94 ± 0.72 |
| ADP/O | 0.96 ± 0.19 | 0.85 ± 0.33 | 0.77 ± 0.27 | 0.92 ± 0.25 |
Mitochondrial protein (50 to 100 μg) was used for the respiratory measurements. Data are presented as mean values ± se and are averages from three to five different mitochondrial isolations from tomato fruits 35 d after flowering.
Since off-target effects of RNA interference constructs in plants have been suggested for fragments of 21 to 24 nucleotides or more (Thomas et al., 2001; Watson et al., 2005; Xu et al., 2006; Rossel et al., 2007) and it was computationally predicted that the chance for RNA interference off-target effects in plants is considerable, with around 50 to 70% of gene transcripts in plants having potential off-targets when used for posttranscriptional gene silencing that could obscure experimental results (Xu et al., 2006), we decided to confirm that nonspecific gene silencing had not taken place in our studies. The fragment used for the antisense construct was designed to have minimal complementarity with other genes; thus, a BLAST query against the Sol Genomics Network database (SGN unigenes) revealed few identical regions around 20 nucleotides. There were, however, no regions of homology to any other member of the succinate dehydrogenase complex family apart from the already tested SDH2-1 (Figure 1F) or, indeed, to any other transcript that could potentially be responsible for the phenotypes observed here. Thus, quantitative real-time RT-PCR was undertaken for the transcripts showing some short stretches of similarity: SGN-U579957 (glycolate oxidase); SGN-U580678 (ribulose-phosphate 3-epimerase); SGN-U566206 (similar to lipase in Arabidopsis); SGN-U584266 (UDP-sulfoquinovose:DAG sulfoquinovosyltransferase/sulfolipid synthase); SGN-U563031 (CTD phosphatase-like protein 3); SGN-U591223 (subtilase family protein); SGN-U595977 (hypothetical protein), and SGN-U573103 (unnamed) in lines that exhibited downregulation of either fumarase or succinate dehydrogenase. These assays revealed no significant alteration in the expression of any of those genes that could suggest off-target silencing due to the succinate dehydrogenase construct (see Supplemental Figure 3 online).
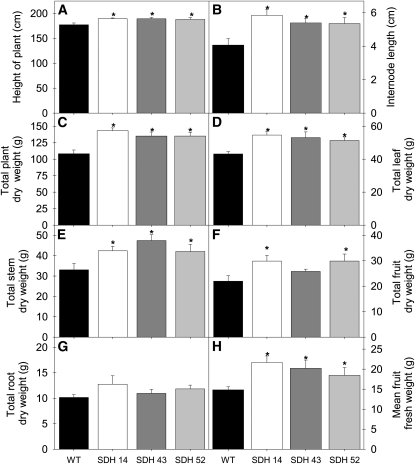
When we grew the transgenic plants in the greenhouse side by side with wild-type controls, a clear increase in the growth of the aerial parts of the transformants was observed during the later stages of growth (see Supplemental Figure 1C online). Close examination of the transgenic plants revealed that the most severely inhibited lines were significantly taller (Figure 2A) due to a larger internodal interval (Figure 2B). Increased total dry weight in the transformants (Figure 2C) was essentially associated with increases in leaf (Figure 2D), stem (Figure 2E), and fruit weight (significantly so in lines SDH14 and SDH52; Figure 2F), since no change in root weight (Figure 2G) was observed. When fruit weight was assessed on an individual fruit basis, it was apparent that the fruits of the transformants were significantly heavier (Figure 2H). In addition, there was no marked difference in leaf formation and leaf area, onset of senescence or flowering time, as well as on the fruit weight–to–whole plant weight ratio.
Figure 2.
Growth Phenotype of 10-Week-Old Antisense Succinate Dehydrogenase Tomato Plants.
Transgenic plants showed an enhanced aerial biomass with respect to the wild type in the later stages of growth. Height of plant (A); internode length (B); total plant dry weight (C); total leaf dry weight (D); total stem dry weight (E); total fruit dry weight (F); total root dry weight (G); and mean fruit weight (H). The lines used were as follows: the wild type (WT), black bars; SDH14, white bars; SDH43, dark-gray bars; SDH52, light-gray bars. Values are presented as means ± se of six individual plants per line; an asterisk indicates values that were determined by the Student’s t test to be significantly different (P < 0.05) from the wild type.
Considering that most of our results were obtained in 4- to 5-week-old tomato plants and that the most visible phenotype was observed in 10-week-old plants, we performed two complementary approaches to observe differences in gene expression and activity of succinate dehydrogenase. Briefly, we were able to show a decrease in both succinate-dependent DCPIP reduction determined in enriched mitochondria from tomato leaves and the relative transcript abundance of SDH2-2 over a 9-week period during leaf development (see Supplemental Figure 4 online). Additionally, we did not observe an age-dependent alteration in expression and activity of succinate dehydrogenase, providing further evidence of near constitutive expression of the gene, as shown in Figure 1A.
Analysis of the maximal catalytic activities of important enzymes of photosynthetic carbohydrate metabolism (ribulose-1,5-bisphosphate carboxylase/oxygenase [Rubisco], transketolase, transaldolase, fructose bisphosphatase, and glyceraldehyde 3-phosphate dehydrogenase), the TCA cycle and associated enzymes (NAD-dependent malate dehydrogenase, PEP carboxylase, and fumarase), or starch synthesis (AGPase; Table 2) revealed few consistent changes between the transgenic and wild-type lines. In addition, there were no changes in either the initial or total activities of NADP-dependent malate dehydrogenase of the chloroplast, a commonly used diagnostic marker for alterations in plastidial redox status (Scheibe et al., 2005). The exception to this statement is that the initial and total Rubisco activities of all three transgenic lines were significantly higher than those observed in the wild type.
Table 2.
Enzyme Activities in Leaves of Succinate Dehydrogenase Antisense Tomato Lines
| Wild Type | SDH14 | SDH43 | SDH52 | |
| Enzyme | nmol min−1 g−1 FW |
|||
| Rubisco initial | 2,507.9 ± 166.5 | 3,309.4 ± 95.2 | 3,374.2 ± 267.4 | 3,290.1 ± 252.1 |
| Rubisco total | 3,202.3 ± 23.8 | 3,911.6 ± 42.5 | 3,655.5 ± 85.5 | 3,516.2 ± 109.1 |
| Rubisco activation state (%) | 78.3 ± 5.1 | 84.7 ± 3.1 | 92.1 ± 5.9 | 87.2 ± 5.8 |
| PEP carboxylase | 607.1 ± 23.4 | 745.1 ± 68.7 | 708.2 ± 18.7 | 743.8 ± 38.8 |
| AGPase | 1,688.5 ± 367.3 | 1,961.7 ± 277.5 | 1,667.9 ± 101.7 | 1,355.7 ± 110.2 |
| NAD-MDH | 30,927.3 ± 4,223.2 | 36,601.4 ± 7738.5 | 30,999.1 ± 10,631.9 | 32,597.3 ± 8,151.7 |
| NADP-MDH initial | 165.2 ± 14.3 | 201.7 ± 8.2 | 171.4 ± 15.2 | 162.9 ± 15.5 |
| NADP-MDH total | 543.1 ± 52.7 | 564.8 ± 43.6 | 516.3 ± 16.6 | 528.8 ± 37.4 |
| NADP-MDH activation state (%) | 34.2 ± 8.7 | 36.3 ± 2.2 | 35.4 ± 4.7 | 30.5 ± 3.8 |
| NADP-GAPDH | 151.9 ± 10.0 | 182.4 ± 7.8 | 153.1 ± 25.4 | 157.0 ± 11.6 |
| FBPase | 178.6 ± 39.7 | 203.7 ± 27.9 | 191.4 ± 53.1 | 180.7 ± 52.1 |
| Fumarase | 2,362.68 ± 463.0 | 2,463.3 ± 649.2 | 2,487.7 ± 448.4 | 2,358.9 ± 531.9 |
| Transketolase | 582.7 ± 30.4 | 566.7 ± 37.5 | 507.5 ± 69.7 | 528.3 ± 33.4 |
| Transaldolase | 25.9 ± 2.7 | 31.4 ± 3.3 | 27.4 ± 4.2 | 28.5 ± 1.9 |
Activities were determined in 4-week-old fully expanded source leaves harvested 6 h into the photoperiod. Data presented are mean ± se (n = 6); values set in bold were determined by the Student's t test to be significantly different (P < 0.05) from the wild type. FW, fresh weight.
Inhibition of Succinate Dehydrogenase Results in a Reduced Flux through the TCA Cycle
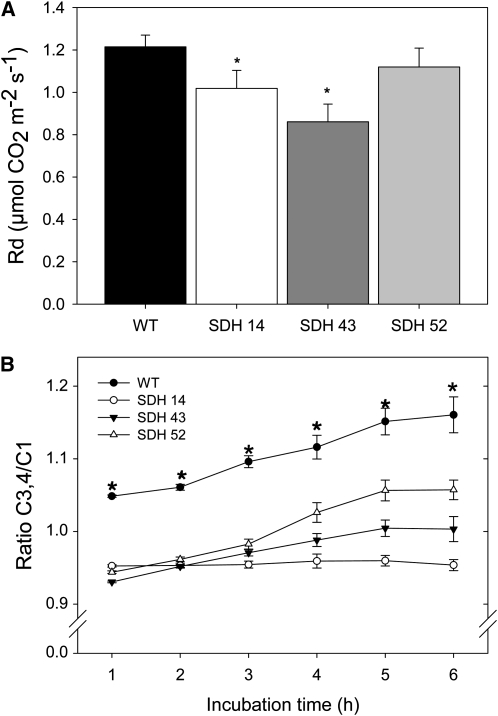
In a number of our previous studies (Carrari et al., 2003; Nunes-Nesi et al., 2005, 2007b), the analysis of the incorporation and subsequent metabolism of 14CO2 in genotypes deficient in the expression of TCA cycle enzymes suggested a reduction in flux through this cycle. However, analysis of the succinate dehydrogenase antisense plants revealed no such change in incorporation of radiolabel in TCA cycle intermediates or downstream metabolites thereof under CO2 saturating conditions (Table 3). To assess the rate of respiration more directly under normal growth conditions, we took two complementary approaches. First, we measured the rate of dark respiration via infra-red gas exchange analyses (Figure 3A). These measurements revealed a reduction in the rate of carbon dioxide evolution, with that of the transformants being >10% lower than that observed in the wild type. It is important to note that the degree of reduction in dark respiration is not entirely proportional to the reduction in succinate dehydrogenase activity (cf. Figures 1C, 1D, and 3A). However, perhaps this is unsurprising, given that recent studies have demonstrated that there are several functionally active, alternative donors to the plant mitochondrial electron transport chain (Ishizaki et al., 2005; Nunes-Nesi et al., 2005; Araújo et al., 2010). We next directly evaluated the rate of light respiration in the transformants. For this purpose, we recorded the evolution of 14CO2 following incubation of leaf discs in positional-labeled 14C-glucose molecules to assess the relative rate of flux through the TCA cycle. To do this, we incubated leaf discs taken from plants in the light and supplied these with [1-14C]-glucose, [2-14C]-glucose, [3,4-14C]-glucose, or [6-14C]-glucose over a period of 6 h. During this time, we collected the 14CO2 evolved at hourly intervals. Carbon dioxide can be released from the C1 position by the action of enzymes that are not associated with mitochondrial respiration, but carbon dioxide released from the C3,4 positions of glucose cannot (Nunes-Nesi et al., 2005). Thus, the ratio of carbon dioxide evolution from C1 to C3,4 positions of glucose provides a strong indication of the relative rate of the TCA cycle with respect to other processes of carbohydrate oxidation. When the relative 14CO2 release of the transgenic and wild-type lines is compared for the various fed substrates, an interesting pattern emerges, with a minor decrease in carbon dioxide evolution in the transgenics, irrespective of the label position in the substrate (see Supplemental Figure 5 online). In addition to these changes in absolute release, there was a shift in the evolution of 14CO2 from the variously labeled glucose molecules, with the relative release from the C3,4 positions being much lower in the transgenic lines than in the wild type (C3,4/C1 ratio shown in Figure 3B; after 6 h, the C3,4/C1 ratios were as follows: wild type = 1.16 ± 0.05; SDH14 = 0.95 ± 0.02; SDH43 = 1.01 ± 0.03; SDH52 = 1.02 ± 0.04). Thus, these data reveal that a lower proportion of carbohydrate oxidation is performed by the TCA cycle in the transgenic lines, and this result is in keeping with the observation of reduced dark respiration in these plants. The differences in release from C2 and C6 positions were far less marked, suggesting that there were no major alterations in metabolic fluxes involved in cycling through the pentose phosphate pathway or in pentan synthesis (Keeling et al., 1988).
Table 3.
Effect of Decreased Succinate Dehydrogenase Activity on Photosynthetic Carbon Partitioning at the Onset of Illumination of 4-Week-Old Fully Expanded Source Leaves
| Parameter | Wild Type | SDH14 | SDH43 | SDH52 |
| Label incorporated (Bq) | ||||
| Total uptake | 318.8 ± 30.3 | 320.1 ± 33.2 | 298.9 ± 8.2 | 321.1 ± 57.3 |
| Organic acids | 13.9 ± 1.1 | 13.9 ± 1.6 | 14.9 ± 2.1 | 12.9 ± 1.3 |
| Amino acids | 13.6 ± 1.9 | 12.2 ± 0.9 | 14.1 ± 1.1 | 13.7 ± 6.1 |
| Soluble sugars | 120.4 ± 20.3 | 116.2 ± 29.9 | 126.8 ± 4.5 | 124.3 ± 45.1 |
| Starch | 170.8 ± 15.3 | 177.7 ± 28.8 | 142.9 ± 6.3 | 170.3 ± 12.4 |
| Redistribution of radiolabel (as percentage of total assimilated) | ||||
| Organic acids | 4.6 ± 0.7 | 4.3 ± 0.1 | 4.9 ± 0.7 | 4.4 ± 0.8 |
| Amino acids | 4.4 ± 0.7 | 3.9 ± 0.4 | 4.7 ± 0.3 | 4.5 ± 0.5 |
| Soluble sugars | 36.8 ± 3.5 | 35.6 ± 6.7 | 42.5 ± 1.7 | 33.6 ± 7.8 |
| Starch | 54.2 ± 3.3 | 56.2 ± 6.6 | 47.7 ± 0.9 | 57.5 ± 6.9 |
Leaf discs were cut from six separate plants of each genotype at the end of the night and illuminated at 700 μmol photons m−2 s−1 of PAR in an oxygen electrode chamber containing air saturated with 14CO2. After 30 min, the leaf discs were extracted and fractionated. Values presented are the mean ± se of measurements from six individual plants per genotype.
Figure 3.
Respiratory Parameters in Leaves of Antisense Succinate Dehydrogenase Tomato Plants.
(A) Dark respiration measurements performed in 4- to 5-week-old plants. WT, wild type.
(B) Ratio of carbon dioxide evolution from C3,4 to C1 positions of glucose in leaves of antisense SDH tomato plants.
The leaf discs were taken from 4-week-old plants and were incubated in 10 mM MES-KOH solution, pH 6.5, 0.3 mM glucose supplemented with 2.32 kBq mL−1 of [1-14C]- or [3,4-14C]-glucose at an irradiance of 200 μmol m−2 s−1. The 14CO2 liberated was captured (at hourly intervals) in a KOH trap, and the amount of radiolabel released was subsequently quantified by liquid scintillation counting. Values are presented as means ± se of determinations on six individual plants per line. An asterisk indicates values that were determined by the Student’s t test to be significantly different (P < 0.05) from the wild type.
Photosynthetic Carbon Metabolism in Illuminated Leaves of the Succinate Dehydrogenase Transformants
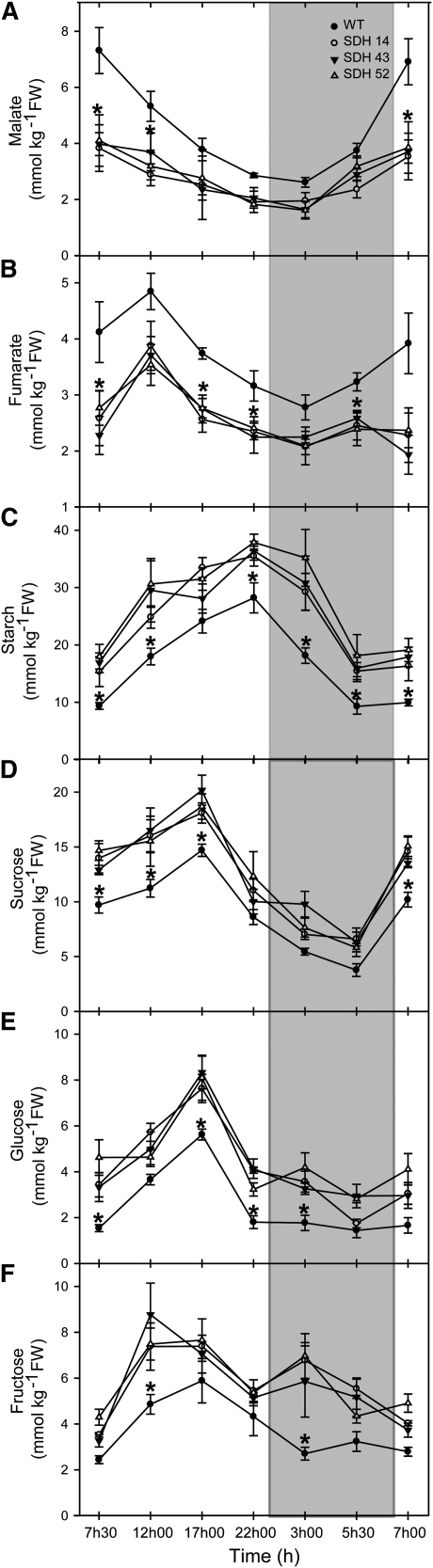
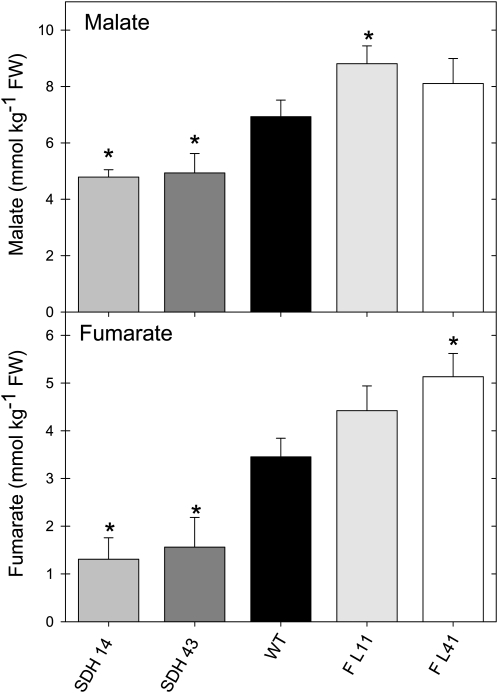
Analysis of the carbohydrate and organic acid content of leaves from 4-week-old plants during a diurnal cycle revealed that the transformants were characterized by a significant reduction in the levels of malate (Figure 4A) and fumarate (Figure 4B), but a significant increase in starch (Figure 4C), sucrose (Figure 4D), glucose (Figure 4E), and fructose (Figure 4C). While the rate of accumulation of carbohydrates during the day was essentially unaltered in the transformants, the absolute levels of all four carbohydrates were consistently higher across the diurnal cycle. Evaluation of the content of leaf pigments revealed that there were isolated changes in the levels of chlorophyll a, β-carotene, and antheraxanthin; however, they generally did not change in a manner consistent with the altered activity of succinate dehydrogenase (see Supplemental Table 1 online). The exception to this statement is violaxanthin, which was present at significantly lower levels in all transgenic lines than those observed for the wild type. In all cases, metabolite contents were in a similar range to those previously reported for tomato (Nunes-Nesi et al., 2005, 2007b).
Figure 4.
Diurnal Changes in Key Metabolite Content in Leaves of Antisense Succinate Dehydrogenase Tomato Plants.
Malate (A), fumarate (B), starch (C), sucrose (D), glucose (E), and fructose (F) were measured at each time point. Samples were taken from mature source leaves. The lines used were as follows: the wild type (WT), black circles; SDH14, open circles; SDH43, black triangles; SDH52, open triangles. The data presented are means ± se of measurements from six individual plants per line; an asterisk indicates values that were determined by the Student’s t test to be significantly different (P < 0.05) from the wild type. Gray bars indicate the dark period; white bars indicate the light period. FW, fresh weight.
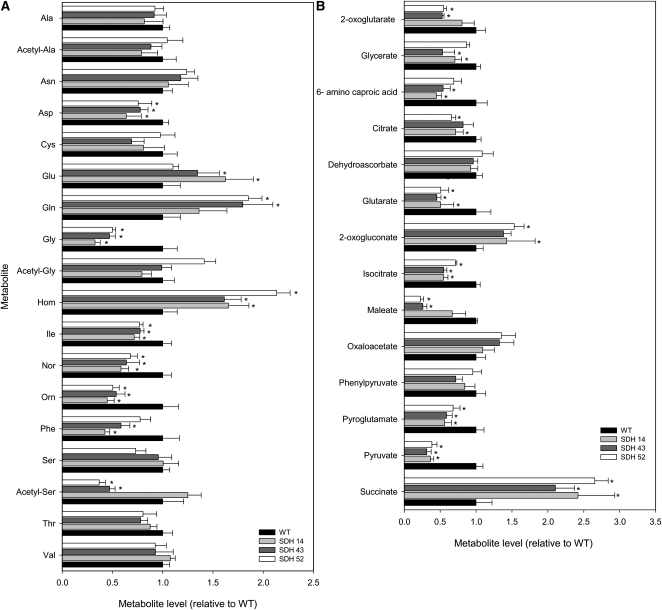
We next decided to extend this study to major primary pathways of plant photosynthetic metabolism by using an established gas chromatography–mass spectrometry (GC-MS) protocol for metabolic profiling (Fernie et al., 2004a). These studies revealed considerable changes in the levels of a wide range of amino acids (Figure 5A), organic acids (Figure 5B), and sugars (see Supplemental Table 2 online). Notably, Asp (all lines), Gly (all lines), Ile (all lines), norvaline (all lines), Orn (all lines), Phe (lines SDH14 and SDH43), acetyl-serine (lines SDH43 and SDH52), and Thr (line SDH43) were significantly decreased, while Glu (SDH14 and SDH43), Gln (lines SDH43 and SDH52), and homoserine (all lines) were significantly increased. As would perhaps be expected, and in line with the results for malate and fumarate described above, there was also considerable variation in the relative pool sizes of the organic acids measured. Pyruvate (all lines), maleate (lines SDH43 and SDH52), isocitrate (all lines), glutarate (all lines), citrate (lines SDH14 and SDH52), amino caproic acid (lines SDH14 and SDH43), glycerate (lines SDH14 and SDH43), and 2-oxoglutarate (lines SDH43 and SDH52) levels were significantly decreased, while succinate (all lines) and 2-oxogluconate (all lines) were significantly increased in the transformants. Other changes of note in the metabolite profiles were the significant increases in sucrose (all lines), maltose (lines SDH43 and SDH52), glucose (all lines), fructose (all lines), and putrescine (all lines), as well as the decreases in shikimate (all lines).
Figure 5.
Relative Metabolite Content of the Fully Expanded Leaves of Antisense Succinate Dehydrogenase Tomato Plants.
Amino acids (A) and organic acids (B) were determined as described in Methods. The full data sets from these metabolic profiling studies are available in Supplemental Table 2 online. Data are normalized with respect to the mean response calculated for the wild type (to allow statistical assessment, individual plants from this set were normalized in the same way). The lines used were as follows: the wild type (WT), black bars; SDH14, gray bars; SDH43, dark gray bars; SDH52, white bars. Values are presented as means ± se of six individual plants per line; an asterisk indicates values that were determined by the Student’s t test to be significantly different (P < 0.05) from the wild type.
Reduction in Succinate Dehydrogenase Activity Enhances Net Carbon Dioxide Assimilation via an Increased Stomatal Aperture
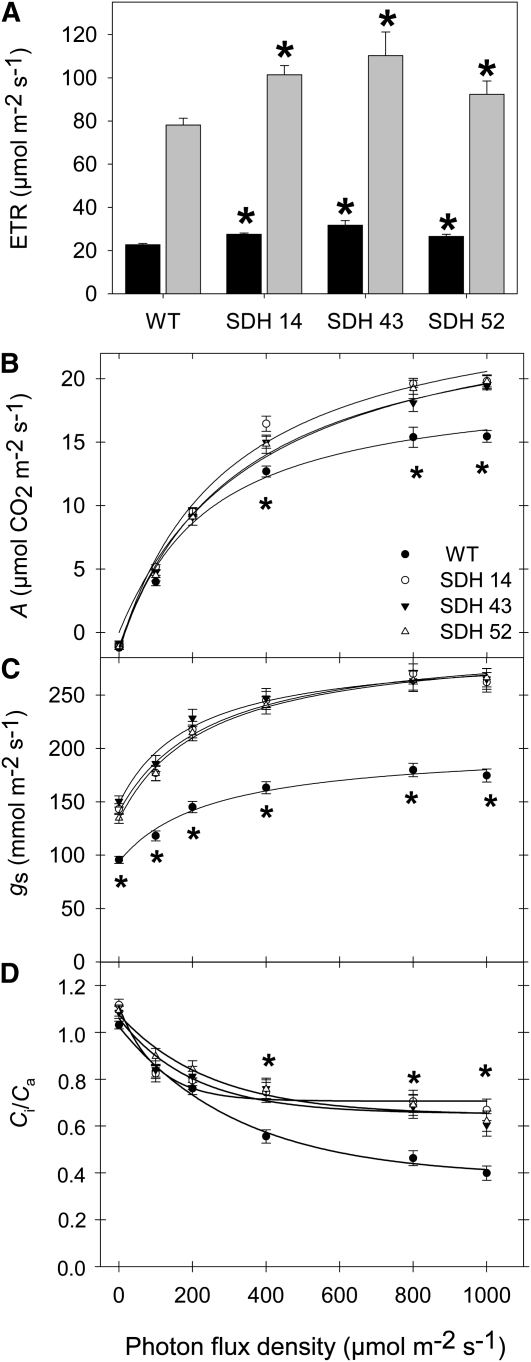
We next determined chlorophyll a fluorescence in vivo using a pulse amplitude modulation (PAM) fluorometer to calculate relative electron transport rates (ETRs) at both low (100 μmol m−2 s−1) and high (700 μmol m−2 s−1) photon flux density (PFD). These experiments revealed that the transformants displayed enhanced chloroplastic ETRs irrespective of the irradiance (Figure 6A). Gas exchange was additionally measured directly, in the same lines as used for the ETR measurements, under PFD that ranged from 0 to 1000 μmol m−2 s−1 (Figure 6B). The transformants exhibited assimilation rates that were as much as 25% higher than those of the wild type at PFDs of 400 μmol m−2 s−1 and above (Figure 6B). Analysis of other parameters of gas exchange revealed that the transformants were also characterized by enhanced stomatal conductance, regardless of the level of irradiance (Figure 6C), and an increased ratio of intercellular to ambient CO2 concentration, Ci/Ca, at PFDs of 400 μmol m−2 s−1 and above (Figure 6D).
Figure 6.
Effect of Decreased Mitochondrial Succinate Dehydrogenase Activity on Photosynthetic Parameters.
(A) In vivo chlorophyll a fluorescence was measured as an indicator of the ETR using a PAM fluorometer at PFD of 100 (black bars) and 700 (gray bars) μmol m−2 s−1.
(B) Assimilation rate (A) as a function of PFD intensities.
(C) Stomatal conductance (gs) as a function of the intensity of PFD.
(D) Internal-to-ambient CO2 concentration ratio (Ci/Ca) as a function of the intensity of PFD.
Values are presented as means ± se of six individual determinations per line. All measurements were performed in 4- to 5-week-old plants. The lines used were as follows: the wild type (WT), black circles; SDH14, white circles; SDH43, black triangles; SDH52, white triangles. An asterisk indicates values that were determined by the Student's t test to be significantly different (P < 0.05) from the wild type.
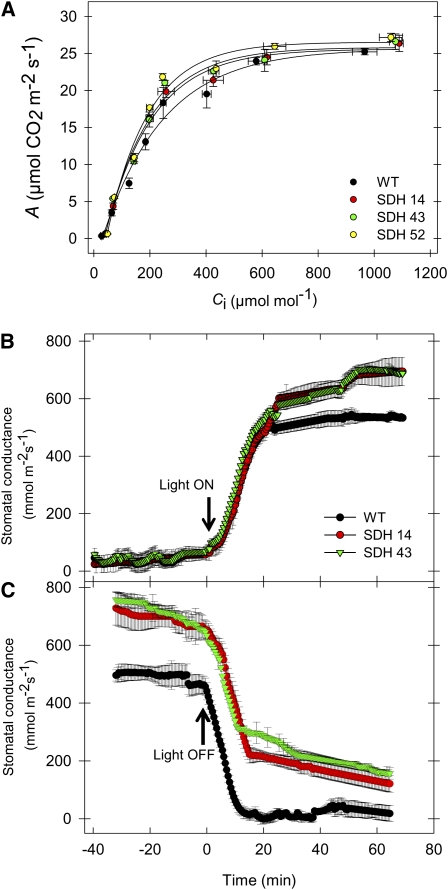
To further characterize photosynthesis in these lines, we next evaluated the response of the net carbon dioxide assimilation rate (A) to the internal carbon dioxide concentration (Ci) (ambient carbon dioxide ranging from 50 to 2000 μmol mol−1). Essentially, the A/Ci curves we obtained were quite similar regardless of the plant material analyzed (Figure 7A). As expected, therefore, analysis of these curves using the online fitting model developed by Sharkey et al. (2007) reveals that the maximum carboxylation velocity of Rubisco (Vcmax), maximum electron transport rate (Jmax), triose phosphate use (TPU), and Jmax/Vcmax ratio were generally unchanged with respect to alterations in the activity of succinate dehydrogenase (see Supplemental Table 3 online). These results support the findings that increased A, as described in Figure 6B, should be primarily associated with increased stomatal conductance rather than with increased mesophyll photosynthetic capacity to fix CO2 at a given Ci.
Figure 7.
Effect of Decreased Mitochondrial Succinate Dehydrogenase Activity on Photosynthetic Parameters.
(A) Rate of net CO2 assimilation as a function of internal leaf CO2 concentration (Ci). WT, wild type.
(B) and (C) Time taken for stomatal opening following a dark-to-light transition (B) and time taken for stomatal closure following a light-to-dark transition (C). Values are presented as means ± se of six individual determinations per line. All measurements were performed in 4- to 5-week-old plants.
In a final set of gas exchange experiments, we investigated the duration of stomatal opening and closing in the transgenic lines following light-to-dark and dark-to-light transitions and recorded the maximal aperture of the stomata. Interestingly, in contrast with the situation observed in transgenic lines deficient in the expression of fumarase (Nunes-Nesi et al., 2007b), in both instances the transformants exhibited unaltered rates of opening and closing (Figures 7B and 7C, respectively); however, the maximal aperture of the stomata of the transgenic lines was significantly enhanced. Finally, we calculated the stomatal density by examining abaxial epidermal impressions of the leaves of the wild type and transformants. However, this parameter was unaltered across the lines, even although the maximal aperture of the stomata was, by contrast with the situation previously observed in the fumarase antisense lines, significantly increased (Nunes-Nesi et al., 2007b; see Supplemental Figure 6 online). Additionally, the stomata index and length of stomatal pore had shown no statistical difference when compared with wild-type plants (see Supplemental Figure 6 online). Consistent with these data, water loss from leaves excised from succinate dehydrogenase antisense plants resulted in a 25% fresh weight loss after 180 min, whereas in leaves from wild-type and fumarase antisense plants, fresh weight loss was only 20 and 16% after 180 min, respectively (see Supplemental Figure 7 online). These differences in fresh weight loss are also in accordance with altered stomatal function, since the transgenic and wild-type leaves exhibit similar stomatal densities (Nunes-Nesi et al., 2007b; see Supplemental Figure 6 online.
Since the plants were grown in parallel in a greenhouse, with a minimum irradiance of 250 μmol photons m−2 s−1, it is supposed that they actually experienced a higher PFD during most of their life cycle. Thus, we expected that a higher photosynthetic rate would be coupled to a higher stomatal conductance throughout the plant growth period, as was indeed observed in SDH antisense plants when we applied PFDs of 400 μmol m-2s−1 and above (Figures 6B and 6C). Based on this, we decided to analyze the gas exchange parameters under natural growth conditions inside the greenhouse. Succinate dehydrogenase antisense plants exhibited assimilation rates that were significantly higher than those of the wild type, which was associated with higher stomatal conductance coupled to an increased Ci/Ca ratio (Table 4). In close agreement with these results, fumarase-deficient plants, which we previously demonstrated to have impaired stomatal function (Nunes-Nesi et al., 2007b), displayed an opposite phenotype for all parameters of gas exchange analyzed here (Table 4). In fact, when data for the wild-type and transgenic plants were pooled, a strong correlation (r = 0.96; P < 0.001) between assimilation rates and stomatal conductance was evident.
Table 4.
Gas Exchange Parameters of Wild-Type and Transgenic Tomato Plants
| A (μmol m−2 s−1) | gs (mmol m−2 s−1) | Ci/Ca | |
| FL11 | 10.6 ± 0.4 | 164.0 ± 4.2 | 0.61 ± 0.03 |
| FL41 | 9.9 ± 0.2 | 159.7 ± 5.9 | 0.63 ± 0.02 |
| Wild type | 13.1 ± 0.5 | 209.3 ± 7.8 | 0.68 ± 0.03 |
| SDH 14 | 17.9 ± 0.9 | 297.5 ± 11.3 | 0.77 ± 0.02 |
| SDH 43 | 16.9 ± 0.4 | 290.4 ± 2.2 | 0.75 ± 0.02 |
Instantaneous gas-exchange variables, namely, the net CO2 assimilation rate (A), stomatal conductance to water vapor (gs), and internal-to-ambient CO2 concentration ratio (Ci/Ca) were measured under natural irradiance. Values presented are means ± se of six individual plants per line. Values in bold were determined by the Student's t test to be significantly different (P < 0.05) from the wild type.
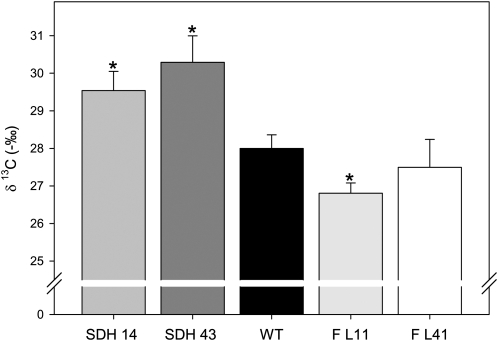
Compelling evidence for an increased stomata function is provided by the carbon isotope composition ratio (δ13C) pattern (Figure 8), since the succinate dehydrogenase antisense plants displayed lower (more negative) δ13C values than did the wild type. By contrast, fumarase antisense plants displayed higher δ13C. It should be emphasized that the isotope discrimination pattern provides a useful estimation of long-term gas exchange, since δ13C reflects the internal leaf carbon dioxide, which is dependent on stomatal conductance on the one hand and on the mesophyll capacity to fix CO2 on the other. Given that Rubisco preferentially fixes 12CO2 above 13CO2, when it has an abundant supply of carbon dioxide, considerably less 13C will be fixed, but when its supply is limited, Rubisco will increase 13C fixation (Stitt and Schulze, 1994). Taking into account that neither specific leaf area nor total leaf area were increased in the transformants (see Supplemental Figure 8 online), it follows that the increased whole plant biomass (Figure 2C) must have resulted from increased photosynthetic rates per unit leaf area as opposed to increased whole-plant photosynthesis due to a larger total leaf area. That said, the δ13C values will also be affected by respiratory fractionations. These fractionations are, however, likely to be very small or even negligible. In fact, it is important to note that small differences between genotypes could merely reflect changes in the respiratory rate. It is, however, highly unlikely that such changes could underlie the differences observed here. In addition, our δ13C results suggest that CO2 does not have to be recycled within the leaf, indicating that the biochemical discrimination by Rubisco is the predominant factor accounting for the observed values and that diffusion of CO2 within the leaf is relatively unimportant. We can, therefore, state that these results are in close accordance with the instantaneous gas exchange data, which revealed improved photosynthetic rates linked to increased stomatal conductance, which, in turn, led to improved carbon dioxide supply to the leaf tissues. In addition, these results are in good agreement with data presented in Tables 2 and 4 and Figure 7, which suggest that higher assimilation rates cannot be directly associated with a higher photosynthetic capacity, despite a higher Rubisco activity.
Figure 8.
Carbon Isotope Composition Ratio (δ13C) from Leaves of Wild-Type, Succinate Dehydrogenase, and Fumarase Antisense Lines.
Values are presented as means ± se of six individual determinations per line. All measurements were performed in 4-week-old plants. The lines used were as follows: the wild type (WT), black bars; SDH14, gray bars; SDH43, dark-gray bars; FL11, light-gray bars, FL41, white bars. An asterisk indicates values that were determined by the Student’s t test to be significantly different (P < 0.05) from the wild type.
Apoplastic Concentrations of Malate and Fumarate in Succinate Dehydrogenase and Fumarase Antisense Lines
While the reduced levels of whole leaf malate and fumarate in the succinate dehydrogenase antisense lines could theoretically provide a mechanism that could explain their altered stomatal function, we wanted to confirm that this phenomenon was also observed within the apoplastic fluid of the transformants. This is essential information, since it is the apoplastic concentration that is likely to be critical in driving stomatal opening and closure. Since we previously characterized antisense tomato plants deficient in fumarase activity as additionally displaying elevated cellular concentrations of malate and fumarate and altered stomatal function (Nunes-Nesi et al., 2007b), we also evaluated these lines here. There were significant decreases in the apoplastic levels of malate and fumarate in the succinate dehydrogenase antisense lines in comparison to the wild type. By contrast, the fumarase antisense lines showed increased apoplastic levels of malate (Figure 9A) and fumarate (Figure 9B). These results demonstrate a negative correlation between the concentrations of these metabolites and gas exchange through the stomata.
Figure 9.
Apoplastic Concentrations of Malate and Fumarate in Succinate Dehydrogenase and Fumarase Antisense Lines.
The apoplastic concentrations of malate (A) and fumarate (B) were determined as described in Methods. Values are presented as means ± se of six individual determinations per line. All measurements were performed in 4- to 5-week-old plants. The lines used were as follows: the wild type (WT), black bars; SDH14, gray bars; SDH43, dark-gray bars; FL11, light-gray bars; FL41, white bars. An asterisk indicates values that were determined by the Student’s t test to be significantly different (P < 0.05) from the wild type. FW, fresh weight.
In a first attempt to evaluate whether the phenotype observed here was due to the influence of the mesophyll on the guard cell or was a guard cell autonomous effect, we decided to isolate guard cell protoplasts (GCPs) and mesophyll cell protoplasts (MCPs) of succinate dehydrogenase and fumarase antisense lines as well as from wild-type tomato. Despite the technical complexity of preparing protoplasts from tomato, we were eventually able to generate and characterize appropriate protoplasts. Interestingly, as observed previously (Shimazaki et al., 1982, 1983; Gautier et al., 1991; Vani and Raghavendra, 1994), in all genotypes, the GCP revealed a much higher respiratory activity when compared with MCP (see Supplemental Figure 9 online). We also observed that both succinate dehydrogenase and fumarase lines had a significant reduced respiratory activity (see Supplemental Figure 9 online), in good agreement with previous results observed in leaf material (Figure 1C). When we analyzed the photosynthetic activity of GCP and MCP, we observed an increased O2 uptake in both cell protoplast types in succinate dehydrogenase antisense lines, while the opposite pattern was observed in fumarase antisense lines (see Supplemental Figure 9 online). We additionally measured the levels of malate and fumarate in the isolated protoplasts. There were significant decreases in the protoplastic levels of malate and fumarate in the succinate dehydrogenase antisense lines in comparison to the wild type (see Supplemental Figure 9 online). By contrast, the fumarase lines showed increased levels of malate and fumarate (see Supplemental Figure 9 online). While these data are somewhat difficult to interpret, we believe they are consistent with our previous suggestion that the stomatal effect is due to changes in mesophyll metabolism. Moreover, the changes in malate and fumarate in the guard cell protoplasts are directly proportional to that detected in the apoplastic washes. Furthermore, the fact that the protoplasts were isolated from transgenic lines displaying constitutive downregulation of the target gene means that the guard cells used for protoplast generation cannot be regarded as entirely independent entities, since their changes in photosynthetic and respiratory activities could be due to environmental reprogramming of guard cell gene expression.
Analysis of Stomatal Response of Wild-Type and Transgenic Lines
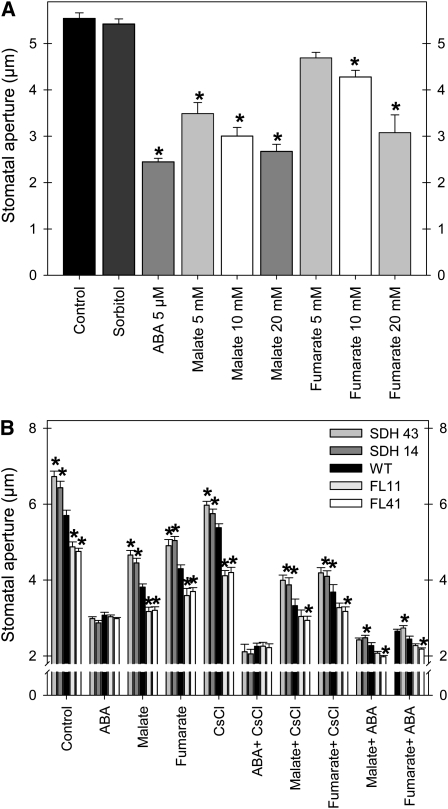
To further characterize these lines, we evaluated the response of intact leaves either from the wild type or succinate dehydrogenase antisense lines (SDH14 and SDH43) and fumarase antisense lines (FL11 and FL41) to the exogenous application of a range of physiologically relevant substances (ABA, malate, and fumarate), both in the presence and absence of the channel potassium transporter blocker CsCl (Ichida et al., 1997; Figure 10). Application of malate and fumarate restricted stomatal aperture in a concentration-dependent manner in all genotypes, and, as observed previously, this effect is more marked following malate feedings (Figure 10A; Lee et al., 2008). To control for possible osmotic effects, 20 mM sorbitol (the highest concentration used for malate and fumarate) was supplied to the medium. However, no apparent effect on guard cell movement was observed, thus ruling out osmotic effects being responsible for the altered stomatal function (Figure 10A). In the absence of experimental treatments, the succinate dehydrogenase lines exhibited an increased stomatal aperture, while the fumarase lines exhibited a decreased aperture with respect to the wild type. That said, application of ABA normalizes the aperture across the genotypes, providing further evidence that the effects observed in the transgenic lines are independent of ABA. This finding also contributes to our understanding of the establishment of the molecular hierarchy of stomatal movement (Fernie and Martinoia, 2009; Meyer et al., 2010a), suggesting that the ABA-mediated pathway exhibits a higher influence on stomatal function than does the organic acid–mediated pathway (Figure 10B). Intriguingly, the absolute stomatal aperture of the succinate dehydrogenase antisense lines following these feedings resembles that of the fumarase antisense lines in the absence of experimental treatment, suggesting that we were able to phenocopy their aperture by altering the apoplastic organic acid content. Application of the potassium transporter blocker CsCl (Ichida et al., 1997) resulted in a moderate reduction in stomatal aperture in all genotypes, albeit to a greater extent in the fumarase lines (Figure 10B). When the leaves were incubated in ABA and CsCl, malate and CsCl, or fumarate and CsCl, their stomatal apertures were further reduced in comparison to those of samples treated with ABA, malate, or fumarate alone. However, differences between the genotypes were essentially conserved. A similar situation was observed following incubation of leaves in ABA and malate, and ABA and fumarate. The combined data therefore suggest that the difference in stomatal behavior in the transgenic lines is independent both of potassium influx (for details, see Ichida et al., 1997) and ABA-mediated calcium influx (for details, see Schroeder and Hagiwara, 1990). We additionally analyzed the results in terms of relative values. For this purpose, we normalized the data with respect to the mean response calculated for the wild type in the control treatment (see Supplemental Figure 10 online). However, this data transformation essentially confirmed the results presented above and, as such, merely provides further support for our interpretations.
Figure 10.
Stomatal Movement in the Presence of Malate or Fumarate and in Combination with ABA and the Channel Blocker CsCl.
(A) The effects of malate and fumarate concentrations on stomatal closing of leaves of wild-type plants. For control treatments, we used the stomatal opening buffer and, additionally, 20 mM sorbitol.
(B) Effects of malate and fumarate alone and in combination with ABA and CsCl on transgenic plants.
Detached leaves were floated on stomatal opening buffer (10 mM MES-KOH, pH 6.15, 5 mM KCl, and 50 μM CaCl2) under illumination for 2 h to induce stomatal opening and then treated with different malate and fumarate concentrations for an additional hour (A) or with either 20 mM malate, 20 mM fumarate, 20 mM CsCl, or 5 μM ABA and the combinations shown in (B) for an additional hour. Thereafter, stomatal apertures were measured. Data are mean ± se of at least 120 stomata obtained from three independent experiments with comparable results. The lines used were as follows: the wild type (WT), black bars; SDH14, dark-gray bars; SDH43, gray bars; FL11, light-gray bars; FL41, white bars. An asterisk indicates values that were determined by the Student’s t test to be significantly different (P < 0.05) from the wild type or the corresponding control.
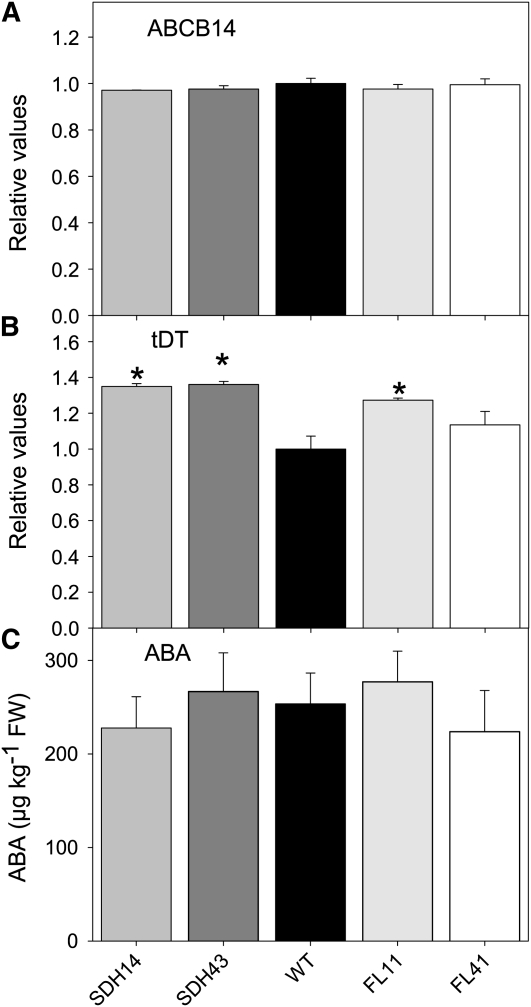
Given that malate has often been described as a component of mechanisms that sense high concentrations of CO2 (Vavasseur and Raghavendra, 2005; Geiger et al., 2009), we next analyzed whether the transgenic lines in addition exhibited differential expression of the currently known (and putative) malate transporters or if this response was mediated merely at the substrate level. Three malate transporters have been cloned that are responsible for cytosol to vacuole and cytosol to apoplast exchange (Emmerlich et al., 2003; Lee et al., 2008; Meyer et al., 2010b), and, by analogy to microbial systems, it had been thought likely that the SLOW ANION CHANNEL-ASSOCIATED1 (SLAC1) also transports malate (Camarasa et al., 2001; Negi et al., 2008). Recent evidence demonstrated by functional expression in Xenopus laevis oocytes that guard cell–expressed Arabidopsis SLAC1 encodes a weak voltage-dependent, anion-selective plasma membrane channel rather than a malate transporter (Geiger et al., 2009). To extend our characterization of the succinate dehydrogenase- and fumarase-deficient genotypes, we attempted to look at the level of gene expression of these three transporters. We were able to identify homologs of ABCB14 and the vacuolar malate transporter (tonoplast Dicarboxylate Transporter [tDT]) but not of SLAC1 when searching EST libraries and the currently available data from the tomato genome sequencing project (van der Hoeven et al., 2002; http://solgenomics.net/genomes/Solanum_lycopersicum/genome_data.pl). Quantitative real-time PCR (qRT-PCR) analysis of the transcript level of ABCB14 and tDT homologs revealed that the former was expressed at similar levels in the succinate dehydrogenase antisense lines and the wild type (Figure 11A), while the latter was upregulated in both the succinate dehydrogenase antisense and the fumarase antisense lines (Figure 11B), suggesting that the stomatal effects observed are also not mediated by an alteration in the efficiency of vacuolar malate export. This statement is in keeping with the fact that homozygous T-DNA insertional knockout mutants lacking a functional tDT did not show an obvious phenotype but contained less malate in leaves (Hurth et al., 2005) as observed in this work. In a further experiment, we evaluated the levels of ABA using a method recently established in our laboratory (van der Merwe et al., 2009); however, levels of the phytohormone were also invariant between genotypes (Figure 11C).
Figure 11.
Expression of Malate Transporters and Measurements of ABA Levels.
qRT-PCR analysis of the expression of ABCB14 (A) and the tonoplast dicarboxylate transporter (tDT) (B) and ABA analyses (C) in leaves of the wild-type and transgenic lines. All measurements were performed on 4-week-old plants. Values are presented as means ± se of six individual determinations per line. The lines used were as follows: the wild type (WT), black bars; SDH14, gray bars; SDH43, dark-gray bars; FL11, light-gray bars; FL41, white bars. An asterisk indicates values that were determined by the Student’s t test to be significantly different (P < 0.05) from the wild type. FW, fresh weight.
Analysis of Alterations in Gene Expression in Illuminated Leaves and in Epidermal Fragments
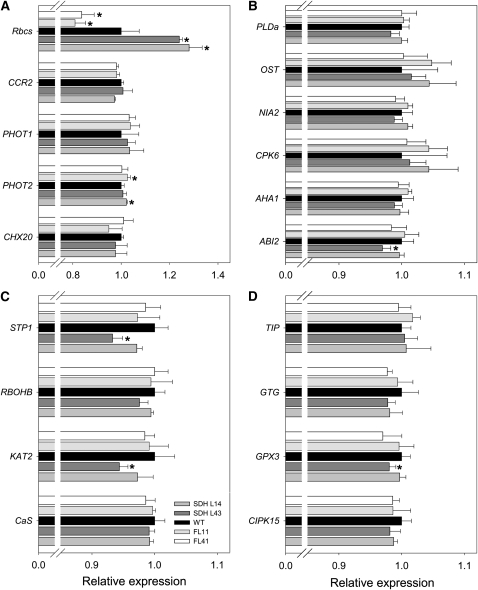
To broaden the characterization of the transgenic lines, we performed microarray analysis using TOM1 microarrays (Alba et al., 2004). For this purpose, we focused on the line SDH14 and the wild type and hybridized RNA both from whole leaf and epidermal fragments. Evaluation of epidermal fragments has proven highly informative in assessing the transcriptome of guard cells (Leonhardt et al., 2004), while the proteome of guard cell protoplasts has also recently been studied (Zhao et al., 2008). However, our studies revealed no significant changes in the expression of genes in the succinate dehydrogenase antisense line compared with the wild type after adjusting for multiple testing, in keeping with the few significant changes reported for the fumarase antisense lines (Nunes-Nesi et al., 2007b). For this reason, we decided to carry out a more focused analysis using a more sensitive qRT-PCR platform. Because different stimuli, such as CO2, humidity, light, and hormones, can regulate stomata opening (Schroeder et al., 2001; Pandey et al., 2007; Ward et al., 2009), we analyzed a range of genes involved in this process. We identified the tomato homologs of signature genes for stomatal signal cascade from the literature as previously shown, including the small subunit of Rubisco (Rbcs), light-responsive genes (Figure 12A), such as cation/H+ exchanger 20 (CHX20), phototropin 1 (PHOT1), PHOT2, and Cold Circadian Rhythm RNA Binding 2 (CCR2), as well as some ABA-responsive genes (Figure 12B), such as ABA insensitive 2 (ABI2), H+ ATPase (AHA2), calcium-dependent protein kinase 6 (CPK6), nitrate reductase 2 (NIA2), open stomata 1 (OST1), and phospholipase D α1 (PLDα1). Additionally, we also identified signaling and solute transporter related genes (Figures 12C and 12D) (e.g., calcium sensing receptor [CaS], inward-rectifying Shaker channel [KAT2], respiratory burst oxidase homolog B [RBOHB], sugar transporter 1 [STP1], calcineurin B-like interacting protein kinase [CIPK15], glutathione peroxidase 3 [GPX3], G-protein coupled recepetor type G protein [GTG1], and the tonoplast intrinsic protein [TIP]) and used these to probe changes in gene expression in either the succinate dehydrogenase or fumarase antisense lines at either the whole leaf or epidermal fragment levels (Figure 12). The levels of these genes were similar in the transgenic lines. As can be seen in the Figure 12A, the tranformants only showed clear opposite patterns in the expression of Rbcs, reflecting, to some extent, the higher initial and total Rubisco activities observed in succinate dehydrogenase antisense plants. Furthermore, the majority of the genes showed similar patterns of transcript accumulation, and while some quantitative differences were apparent and significant, none of those were consistent within the genotypes evaluated here.
Figure 12.
Expression of Genes Involved in the Stomatal Response of the Wild Type in Succinate Dehydrogenase and Fumarase Antisense Lines.
Light-responsive genes (A); ABA-responsive genes (B); signaling (C) and solute (D) transporter-related genes. The transcripts analyzed here were as follows: Rbcs, CCR2, PHOT1, PHOT2, CHX20, PLDα1, OST1, NIA2, CPK6, AHA2, ABI2, STP1, RBOHB, KAT2, CaS, TIP, GPCR1, GTG1, GPX3, and CIPK15. Analyses were done in epidermal fragments of fully expanded leaves of 4-week-old plants. Values are presented as means ± sd of six individual plants. The lines used were as follows: the wild type (WT), black bars; SDH14, gray bars; SDH43, dark-gray bars; FL11, light-gray bars; FL41, white bars. An asterisk indicates values determined by the Student’s t test to be significantly different (P < 0.05) from the wild type.
Analyzing the Regulation of Guard Cell Aperture by the Mesophyll
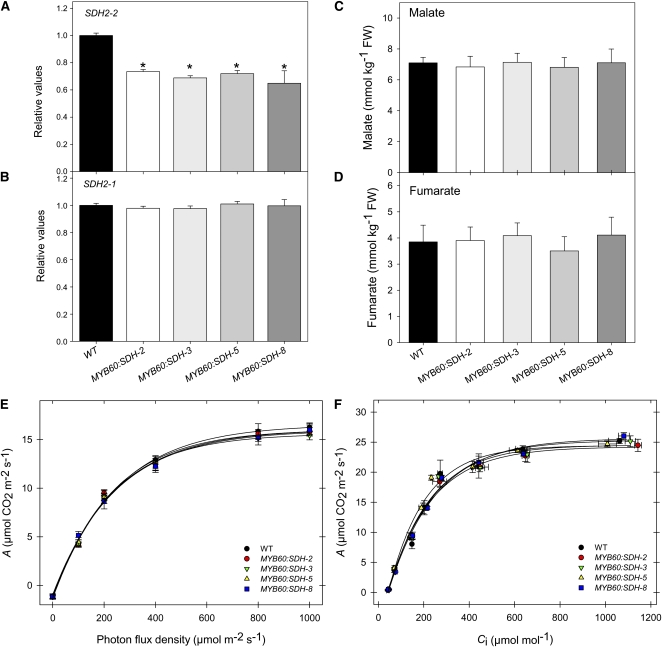
Since our results were obtained from transgenic lines displaying constitutive downregulation of SDH2-2, and considering that this gene has a relatively low expression in tomato guard cells, it is reasonable to hypothesize that the mesophyll regulates the stomatal aperture and that the stomatal effect observed in this study is due to changes in mesophyll metabolism. To address this question, we generated a series of lines of SDH2-2 in antisense orientation that had been independently transformed under the control of a guard cell–specific promoter, MYB60, which has been shown to be strongly expressed in guard cells but not in epidermal cells (Cominelli et al., 2005; Nagy et al., 2009). We then transferred nine transgenic lines obtained by Agrobacterium-mediated transformation to the greenhouse. Screening of the lines by qRT-PCR for SDH2-2 expression yielded four lines that displayed a considerable reduction in the level of SDH2-2 transcripts in epidermal fragments (Figure 13A). Moreover, the expression of the nontargeted isoform SDH2-1 in epidermal fragments was unaltered in the transformants (Figure 13B). We additionally verified that the expression of neither isoform was altered in total leaf extracts, confirming that these four lines (MYB60:SDH2, MYB60:SDH3; MYB60:SDH5, and MYB60:SDH8) were suitable for assessing the effects of a mild reduction in mitochondrial succinate dehydrogenase activity on guard cells. We additionally observed that the succinate-dependent DCPIP reduction (see Supplemental Figure 11 online) was not impaired in leaves of these transformants, further confirming the specificity of the guard cell inhibition. Detailed physiological analyses of the above-mentioned transgenic lines revealed that guard cell–targeted expression of SDH2-2 did not promote a similar stomatal phenotype as observed in lines in that SDH2-2 had been constitutively downregulated. First of all, changes in total leaf malate and fumarate contents (see Supplemental Figure 11 online) and in apoplastic concentration of both organic acids were not observed (Figures 13C and 13D). Second, we performed an extensive physiological characterization by gas exchange analysis, and we did not observe any alteration in assimilation rates or in stomatal conductance (Figures 13E and 13F). Furthermore, our studies demonstrated that the dynamics of stomatal opening and closing in response to light and dark, respectively, were not altered in the guard cell–specific transformants (see Supplemental Figure 11 online). Additionally, we did not observe any alteration in stomatal conductance, dark respiration, or Ci/Ca in the MYB60:SDH2-2 lines in both the light and CO2 response experiments. Consistent with the above described data, water loss from leaves excised from MYB60:SDH2 plants was invariant from the wild type with respect to fresh weight loss after 180 min. Furthermore, these plants revealed no clear differences in leaf formation, leaf area, onset of senescence or flowering time, growth phenotype, or their harvest index.
Figure 13.
Physiological Characterization of SDH2-2 Lines under the Control of the Guard Cell–Specific Promoter MYB60.
(A) and (B) Relative transcript abundance of mitochondrial Complex II subunits (SDH2-2 and SDH2-1, respectively). The abundance of SDH mRNAs was measured by quantitative RT-PCR. WT, wild type.
(C) and (D) Apoplastic concentrations of malate (C) and fumarate (D). Malate and fumarate were determined as described in Methods. FW, fresh weight.
(E) and (F) Assimilation rate as a function of PFD intensities (E) and net CO2 assimilation rate as a function of internal leaf CO2 concentration Ci (F). Values are presented as mean ± se of six individual plants per line. Asterisks indicate values that were determined by the Student’s t test to be significantly different from the wild type (P < 0.05).
DISCUSSION
Over many years, considerable research effort has been expended to better comprehend both the formation and physiology of stomata (Hedrich and Neher, 1987; Schroeder and Hagiwara, 1990; Berger and Altmann, 2000; Bergmann et al., 2004; Lee et al., 2008; Zhao et al., 2008; Casson and Hetherington, 2010). Given the relative ease of the isolation of guard cells, which gate stomata, our understanding of these cells exceeds that of many of the other 40 cell types described thus far in plants (Martin et al., 2001; Fernie, 2007). The adoption of forward genetic screens has seen massive advances in our understanding of the developmental processes that regulate the process of cell fate and subsequently underpin stomatal density (Berger et al., 1998; Nadeau and Sack, 2002; von Groll et al., 2002; Nadeau and Sack, 2003; Bergmann et al., 2004; Wang et al., 2007). By contrast, although the physiological roles of potassium, calcium, and the phytohormone ABA are well defined (Schroeder and Hagiwara, 1990; Schachtman et al., 1992; Pei et al., 2000), it is still a matter of debate as to whether guard cells function autonomously or are subject to regulation by their neighboring mesophyll cells (Lee et al., 2008; Mott, 2009). That said, both the data that we provide and the recent characterization of Arabidopsis plants deficient in the expression of ABCB14 (Lee et al., 2008) add further evidence to support the contention of Mott and others that the mesophyll harbors significant regulation over guard cell function (Lee et al., 2008; Fernie and Martinoia, 2009; Mott, 2009).
In this study, we were interested in establishing the importance of the TCA cycle enzyme succinate dehydrogenase, which catalyzes the conversion of succinate to fumarate, in the functioning of the illuminated leaf. After demonstrating that antisense succinate dehydrogenase lines had a decreased flux through the TCA cycle but elevated photosynthetic rates, as evidenced by feeding experiments, gas exchange measurements and GC-MS profiling, we concentrated our study on the stomatal function of the transformants. The link between mitochondrial metabolism and photosynthetic performance described here is by no means without precedence and has received much attention in the form of both reverse genetic and inhibitor studies (Padmasree et al., 2002; Fernie et al., 2004b; Plaxton and Podesta, 2006; Sweetlove et al., 2007). Evidence has accumulated that the operation of respiration can boost photosynthetic performance (Nunes-Nesi et al., 2005; Plaxton and Podesta, 2006; Tcherkez et al., 2009); however, the fact that the effects observed here were mediated by a modification in stomatal performance drove us to focus on this parameter. Intriguingly, tomato plants with a deficiency in fumarase expression were previously shown to have a reduced flux through the TCA cycle, a large reduction in carbon dioxide assimilation, and restricted growth, due to impaired stomatal aperture (Nunes-Nesi et al., 2007b). The succinate dehydrogenase lines characterized here display an essentially opposite phenotype, with enhanced carbon dioxide assimilation resulting in elevated aerial growth during the late stages of plant development. The delayed response at the whole-plant level is likely the cumulative effect of enhanced carbon assimilation over time.
There were few changes in other aspects of photosynthetic metabolism in the succinate dehydrogenase antisense lines, suggesting that the correct expression of SDH is of relatively small importance in terms of total cellular homeostasis. Interestingly, but not surprisingly, as was previously observed in other genotypes deficient in enzymes of the TCA cycle (Carrari et al., 2003; Nunes-Nesi et al., 2005, 2007a; Sienkiewicz-Porzucek et al., 2008), the activities of other key enzymes of carbon metabolism were largely unaltered, as were the levels of phosphorylated intermediates, suggesting that the increased CO2 assimilation rates were not mediated by a classical metabolic mechanism. Indeed, despite the fact that the previously characterized lines displayed large alterations in the expression of photosynthesis-related genes (Urbanczyk-Wochniak et al., 2006), the SDH lines described here were characterized by very few transcriptional changes. Furthermore, a comprehensive GC-MS–based metabolite profile revealed relatively few changes in metabolites, with those observed being relatively mild (Figure 5; see Supplemental Table 2 online).
Morphological analysis revealed that stomatal density was unaltered in the transformants. However, detailed gas exchange measurements revealed that the enhanced carbon dioxide assimilation was a function of enhanced stomatal conductance facilitated by the wider stomatal aperture of the transformants. Chloroplastic electron transport rate and both initial and total in vitro Rubisco activities were also enhanced in the transformants, and the activation state revealed a tendential increase across the lines. Given that the expression levels of the genes encoding the electron transport and Rubisco proteins were also elevated in the transgenics, this might suggest the existence of an adaptive mechanism that allows the available intracellular carbon dioxide to be used. This hypothesis is further supported by the results of metabolic control analysis studies in tobacco, which revealed that changes in the amount and activity of Rubisco did not universally correlate with alterations in the photosynthetic rates (Quick et al., 1991; Stitt et al., 1991). This suggests that the relatively minor (~25%) increase observed in the transgenics is unlikely to drive the changes in photosynthesis per se. Furthermore, similar changes in sugar content to those observed here were documented in the Aco1 mutant of the wild species of tomato, Solanum pennellii, which is deficient in the expression of aconitase but not in antisense lines of this species in which the expression of the mitochondrial malate dehydrogenase had been inhibited, despite the fact that both lines display elevated rates of photosynthesis and aerial growth (Carrari et al., 2003; Nunes-Nesi et al., 2005). However, the significance of this difference is currently unknown. Returning to the succinate dehydrogenase antisense plants, and given that Rubisco preferentially uses 12CO2 (Farquhar et al., 1982; Stitt and Schulze, 1994), an increased stomatal conductance would be expected to increase the amounts of carbon assimilated, especially since the activity of Rubisco is enhanced in parallel. The elevated growth rate of the transformants, in particular the increased fruit yield, provides further support for this statement. The increased fruit yield of the transformants additionally supplies more evidence in support of the theory that fruit yield is largely dependent on photoassimilate supply from the leaves (Fridman et al., 2004; Hackel et al., 2006; Schauer et al., 2006). When taken together, these observations suggest that manipulation of stomatal function is a promising approach for the improvement of crop yield. It is, however, important to highlight that this work was performed in a greenhouse under controlled conditions that precluded water stress. Given the twin roles of stomata as a conduit for CO2 and water, it is important to note that the adoption of the approach we describe here to field-grown crops (including tomato, itself, in certain climate zones) is unlikely to prove facile.
Having established that the elevated photosynthetic rates (and subsequent increased growth rates) were, at least, predominantly due to the altered stomatal function of the transformants, we concentrated on trying to understand the mechanisms underlying this change. To this end, we initially performed experiments to determine whether the changes in total cellular malate and fumarate content were reflected in the apoplastic concentrations of these metabolites. This was indeed the case for both the succinate dehydrogenase antisense lines, which displayed decreased apoplastic levels of both metabolites, and in the fumarase antisense lines, which displayed increased apoplastic levels of both metabolites. Additionally, the measurement of O2 uptake and evolution were consistent with the guard cell containing a high catabolic activity, but the mesophyll cell making a greater contribution to the anabolic process. In keeping with this, a remarkable correlation between malate (and fumarate) levels determined in the apoplast (Figure 9) and the protoplast preparations (see Supplemental Figure 9 online) is apparent. Given that the protoplasts are derived from transgenic plants, with constitutive downregulation of SDH2-2 by the transgene in the mesophyll cells, it follows that these changes in malate content in the guard cell result from the altered metabolism of its surrounding cellular environment. In a complementary approach, we evaluated the response of the wild type and both transgenic sets to the exogenous application of either metabolite at physiologically relevant concentrations. Application of both malate and fumarate to wild-type plants resulted in a concentration-dependent restriction of the stomatal aperture, albeit with malate being more potent at equivalent concentrations. This was also true in the transgenic lines, demonstrating that they are in no way impeded in their capacity to respond to this organic acid, thus providing further evidence that the effects we observed are independent of any alteration in the metabolic capacity of the guard cell. Given that malate is physiologically present in the apoplast at higher concentrations than fumarate (3 times as high, according to this study), it seems likely that malate exerts a greater in vivo influence on stomatal aperture than fumarate does. These studies, when considered alongside the recent identification of transporters that import malate (or fumarate) into the guard cell (Lee et al., 2008), or have been assumed to export it (Negi et al., 2008; Vahisalu et al., 2008), thus provide a mechanism by which these organic acids can influence stomatal function. When taken together, these studies provide a mechanism linking mesophyll and stomatal function and thus add further evidence to support the hypothesis of Mott (2009) that guard cells are not autonomously regulated.
Despite the fact that these data document the importance of organic acids in guard cell function, the experiments discussed thus far do not, in their own right, rule out an interaction with other well-characterized mechanisms of guard cell regulation, such as those that involve ABA, potassium, nitrous oxide, or calcium (Assmann and Shimazaki, 1999; Assmann and Wang, 2001; Hetherington, 2001; Coursol et al., 2003; Hetherington and Brownlee, 2004; Roelfsema and Hedrich, 2005; Israelsson et al., 2006; Mori et al., 2006; Sutter et al., 2007; Zhu et al., 2007; Kwak et al., 2008; Dreyer and Blatt, 2009; Li et al., 2009; Pandey et al., 2009; Ribeiro et al., 2009; Wilson et al., 2009). Although the accumulation of violaxanthin that was observed in the succinate dehydrogenase transgenic plants is reminiscent of the accumulation of this pigment in several mutants of ABA biosynthesis (Hurry et al., 1997; Galpaz et al., 2008), we were not able to detect any differences in the level of the phytohormone in the transformants. Moreover, analysis of the expression data did not uncover large changes in ABA-responsive genes (Hetherington and Quatrano, 1991; Hildmann et al., 1992; Abe et al., 1997) or of genes/proteins associated with ABA-mediated signal transduction relay (Leonhardt et al., 2004). The recent proteomic study of Zhao et al. (2008) highlighted a plethora of proteins that were differentially expressed in guard cells as opposed to mesophyll cells, many of which have previously been characterized as being ABA responsive. Similarly, the experiments in which leaves from the transgenic and wild-type lines were incubated in the presence of the potassium channel blocker CsCl (Ichida et al., 1997) yielded similar results. Moreover, the restriction in stomatal aperture was additive when leaves were incubated in CsCl and malate or in CsCl and fumarate. The efflux of both anions and K+ from guard cells via anion and K+out channels contributes to loss of guard cell turgor, which subsequently leads to stomatal closing (Schroeder and Hagiwara, 1989; Pei et al., 1997; Gaedeke et al., 2001; Wang et al., 2001; Vahisalu et al., 2008). The synthesis of these findings shaped the model that continuous membrane hyperpolarization by constitutively activated H+-ATPase counteracts ABA-induced stomatal closure (Merlot et al., 2007; Kwak et al., 2008). Since blocking of the potassium channels had the same effect irrespective of genotype, it is hard to believe that the metabolic shifts reported here mediated the stomatal phenotype through a direct effect on these transporters. Interestingly, the leaf disc experiments we described here suggest that the effect of ABA is potentially dominant to that of organic acids; however, the relevance of this finding in the context of the transgenics characterized here is minimal. Therefore, it is conceivable under certain cellular circumstances that stomatal function can be regulated by the rate of respiration of the neighboring mesophyll cells. Furthermore, the microarray data presented here display few of the trademark features of cell autonomous regulation. Finally, the lack of evidence for change in the levels of ascorbate or, in the evaluation of expression data, of reactive oxygen species, in this study leads us to conclude that the effects of stomatal aperture that we described are not mediated by hydrogen peroxide (Zhang et al., 2001; Chen and Gallie, 2004).
A process of elimination thus leaves us with malate (and fumarate) as the major mediator of the altered stomatal function exhibited in the transgenic plants. Our data, when taken alongside that recently obtained for Arabidopsis ABCB14 (Lee et al., 2008), provide strong evidence that modulation of the malate concentration in guard cells can greatly influence stomatal function. Considering the sensitivity of anion channels to extracellular anions, Hedrich and Marten (1993) suggested that guard cells might sense CO2 via changes in the apoplastic malate concentration. In keeping with this theory, the apoplastic malate concentration increases at high atmospheric CO2 concentrations (Hedrich et al., 1994). Furthermore, malate induces stomata closure in epidermal strips of fava bean (Vicia faba) with a half maximal concentration of 0.3 mM. In good agreement with these results, feeding malate to excised leaves reduces the transpiration rate in a dose-dependent manner (Hedrich et al., 2001). However, even at the highest concentration of malate used (40 mM), stomata still responded to CO2 in the atmosphere, indicating that guard cells must have at least one additional CO2 sensing system. Thus, further experimentation is required to tease out the molecular hierarchy shared between this mechanism and those mediated by ABA and K+ ions. It seems likely that the relative importance of each regulatory mechanism will vary with circumstances; however, a fuller understanding of this is surely required to engineer sustainable increases in crop yield. Finally, it is interesting to note that although neither malate nor fumarate exert their effects on stomata by affecting ABA, the phytohormone could, conditionally, act upstream of the organic acids, given that a recent study in Arabidopsis revealed the SDH2-3 gene to be upregulated by ABA (Roschzttardtz et al., 2009). It will be important to establish the functional significance of this observation in future studies.
In summary, the above questions notwithstanding, in this article, we demonstrated that antisense inhibition of succinate dehydrogenase in a guard cell–independent manner led to an alteration in the levels of organic acids in guard cells, while the antisense inhibition of fumarase led to a feedback inhibition of respiratory metabolism, which resulted in an increased concentration of malate (and, to a lesser extent, of fumarate) and, in turn, promoted stomatal closure. Thus, the results presented here show that alterations in mitochondrial metabolism of the mesophyll cells strongly impact stomatal function by regulating the levels of organic acids, as outlined in our model illustrated in Figure 14. This model describes not only the results of this work but also that of others (Lee et al., 2008; Sibbernsen and Mott, 2010), suggesting mesophyll regulation over guard cell aperture. Moreover, it is in strong accordance with the hypothesis of Hedrich and Marten (1993) that malate acts as a CO2 sensor and even implicates the mitochondrial TCA cycle and associated enzymes in this process. It follows that an increased concentration of CO2 would inhibit the decarboxylation reactions of the TCA cycle and, as such, restrict flux through pyruvate dehydrogenase and the NAD-malic enzyme. As a consequence, pyruvate and malate would be anticipated to accumulate, leading to a reduced stomatal aperture. By contrast, low CO2 would favor the decarboxylation reactions and promote an increase in the flux through pyruvate dehydrogenase and the NAD-malic enzyme, and, as such, a decrease in pyruvate and malate concentration would lead to an increased stomatal opening.
Figure 14.
Mitochondrial Function Triggers Stomatal Movement by Regulating Organic Acid Levels.
The malate (fumarate) produced by the TCA cycle is transported to the vacuole, where it is stored. By an unclear mechanism, the level of organic acid is altered in the subsidiary cells, leading to an increased (decreased) concentration in the guard cells that culminates with the closing (opening) of stomata. Stomatal movement is additionally regulated by other well-characterized mechanisms (K+, Cl−ABA, and Ca2+); therefore, future work is required to fully understand the molecular regulatory hierarchy of this highly specialized cell type.
The fact that the effects on stomatal aperture were not observed when a guard cell–specific promoter was used indicates that the mesophyll plays the predominant role in defining the phenotype observed in the transformants downregulated using the CaMV 35S promoter. Taken together with the fact that the downregulation of SDH2-1 under the control of the CaMV 35S promoter did not affect SDH expression in lower epidermal fragments allows us to exclude even a minor influence of this cell type on the results obtained. Future work is clearly required to further examine the crosstalk between malate and ABA, K+, and Cl− ions to fully understand the molecular regulatory hierarchy of this highly specialized cell type.
METHODS
Materials
Tomato (Solanum lycopersicum cv Moneymaker) was obtained from Meyer Beck. Plants were handled as described in the literature (Carrari et al., 2003; Nunes-Nesi et al., 2005). Briefly, plants were grown in parallel in the greenhouse, with a minimum irradiance of 250 μmol photons m−2 s−1, at 22°C, and under a 16-h-light/8-h-dark regime. Experiments were performed on mature fully expanded source leaves from 4- to 5-week-old plants. Unless specified otherwise, all chemicals and enzymes used in this study were obtained from Roche Diagnostics, with the exception of radiolabeled sodium bicarbonate and d-[1-14C]-, [2-14C]-, d-[3,4-14C]-, and d-[6-14C]- glucose, which were from Amersham International.
cDNA Cloning and Expression
First, an 825-bp full-length cDNA of Sl SDH2-2 was amplified using the primers 35S-SlSDH2-2 forward 5′-CACCATGGCGACTAGTTTAATCCGACG-3′ and 35S-SlSDH2-2 reverse 5′-AGGTGCCATCTCCAGCTTCTTG-3′ and cloned in antisense orientation into the vector pK2WG7 (Liu et al., 1990) between the CaMV 35S promoter and the ocs terminator. This construct was introduced into plants by an Agrobacterium tumefaciens–mediated transformation protocol, and plants were selected and maintained as described in the literature (Tauberger et al., 2000). Initial screening of 15 lines was performed using oxygen consumption analysis of the rate of respiration (Araújo et al., 2008) and RNA gel blot analysis. These screens allowed the selection of eight lines, which were taken to the next generation.
Second, guard cell–specific reduction of Sl SDH2-2 expression was obtained by the insertion of the 825-bp full-length Sl SDH2-2 cDNA in antisense orientation, under the control of the MYB60 promoter (Nagy et al., 2009) and nos terminator cloned into a Gateway plant-compatible transformation vector (Curtis and Grossniklaus, 2003). The following primers were used for this cloning: MYB60-SlSDH2-2 forward, 5′-TTGGCGCGCCATGGCGACTAGTTTAATC-3′, and MYB60-SlSDH2-2 reverse, 5′-CCTTAATTAAAGGTGCCATCTCCAGCTTC-3′. The construct obtained was introduced into plants by an Agrobacterium-mediated transformation protocol, and plants were selected and maintained as described by Tauberger et al. (2000). The screening of nine lines was performed by qRT-PCR analyses. These screens allowed for the selection of four lines, which were taken to the next generation.
Mitochondrial Respiration, Succinate-Dependent Oxygen Consumption, and DCPIP Reduction
Total succinate dehydrogenase activity was confirmed in the second harvest of these lines after which three lines were chosen for detailed physiological and biochemical analyses. The succinate dehydrogenase activity was determined using a Clark-type electrode, after mitochondrial isolation from fruits harvested at 35 d after flowering of both wild-type and transformant plants using a Percoll gradient purification method (Millar et al., 2001). The mitochondrial activity was subsequently determined by applying the same method to mitochondrial fractions that was described in the protocol for mitochondrial isolation described by Sweetlove et al. (2002). The purity of the mitochondrial preparations was confirmed as described previously (Carrari et al., 2003; Nunes-Nesi et al., 2007b). Protein was quantified using the Bio-Rad protein assay reagent (Bio-Rad Laboratories). Mitochondrial respiration was measured as oxygen consumption using a Clark-type electrode with the addition NADH (1 mM), malate (5 mM), citrate (5 mM), KCN (1 mM), ADP (0.5 mM), and salicylhydroxamic acid (SHAM) (10 mM) to determine mitochondrial respiration rates. Calibration of the electrode was performed by addition of sodium dithionite to remove all oxygen in the electrode chamber. All reactions were performed at 25°C using 1 mL of mitochondrial reaction medium (Sweetlove et al., 2002). To investigate the succinate-dependent O2 consumption, 10 mM succinate was added to the reaction solution. To confirm the purity of the mitochondrial preparations, the activity of cytochrome c oxidase (CCO) and UDP-glucose pyrophosphorylase (UGPase) (Carrari et al., 2003), which serve as marker enzymes for the mitochondria and cytoplasm, respectively, was determined. The contamination of the mitochondria was, in all instances, <10%, and recoveries of the marker enzymes were 91% for UGPase and 93% for CCO.
The mitochondria-enriched fraction from tomato leaves was obtained by homogenizing 250 mg of leaves in prechilled extraction medium containing 300 mM mannitol, 30 mM MOPS-KOH, pH 7.5, 1 mM EDTA, 0.1% (w/v) BSA-fraction V, 0.1% (w/v) polyvinylpyrrolidone, and 4 mM Cys. The homogenate was passed through three layers of miracloth (Calbiochem) and centrifuged at 1000g for 10 min. The supernatant was then transfered to new tubes and centrifuged at 12,500g for 20 min, and the pellet was resuspended using a soft paintbrush dipped in 300 mM mannitol, 30 mM MOPS-KOH, pH 7.5, and 1 mM EDTA. The succinate dehydrogenase activity was determined as described by Huang et al. (2010), with modifications. Briefly, the mitochondria-enriched fraction (50 to 100 μg protein) was assayed for activity spectrophotometrically at 600 nm and 25°C, in a reaction medium containing 50 mM potassium phosphate, pH 7.4, 10 mM sodium succinate, 0.1 mM EDTA, 0.1% (w/v) BSA, 10 mM potassium cyanide, 0.15 mM DCPIP, and 2 mM phenazine methosulfate.
Phylogenetic Analysis
Protein sequences were retrieved from GenBank through the BLASTp algorithm using the Sl SDH2-2 amino acid sequence as query. Sequences were aligned using the ClustalW software package (Higgins and Sharp, 1988) using default parameters. Neighbor-joining trees (Saitou and Nei, 1987) were constructed with MEGA4.1 Beta 2 software (Tamura et al., 2007) using midpoint rooting. Distances were calculated using pairwise deletion and Poisson correction for multiple hits, and bootstrap values were obtained with 1000 pseudoreplicates.
RNA Gel Blot Analysis
Total RNA was isolated using the commercially available Trizol kit (Gibco BRL) according to the manufacturer’s suggestions for extraction from plant material. Hybridization using standard conditions was performed using the ESTs for the iron-sulfur subunit of succinate dehydrogenase obtained from the Clemson University collection.
Analysis of Enzyme Activities
Enzyme extracts were prepared as described previously (Gibon et al., 2004), except that Triton X-100 was used at a concentration of 1% and glycerol at 20%. Fumarase, AGPase, PEP carboxylase, and sucrose phosphate synthase were determined as described by Gibon et al. (2004). Rubisco activity was determined as described by Sharkey et al. (1991). Malate dehydrogenase (NADP) was assayed as described by Scheibe and Stitt (1988) and malate dehydrogenase (NAD) as described by Jenner et al. (2001).
Determination of Metabolite Levels
Leaf samples were taken at the time points indicated, immediately frozen in liquid nitrogen, and stored at −80°C until further analysis. Extraction was performed by rapid grinding of tissue in liquid nitrogen and immediate addition of the appropriate extraction buffer. The levels of starch, sucrose, fructose, and glucose in the leaf tissue were determined exactly as described previously (Fernie et al., 2001). Malate and fumarate were determined exactly as detailed by Nunes-Nesi et al. (2007b). The levels of all other metabolites were quantified by GC-MS as described by Roessner et al. (2001), with the exception that the peak identification was optimized to tomato tissues (Roessner-Tunali et al., 2003), and the metabolites studied included recent additions to our mass spectral libraries (Kopka et al., 2005; Schauer et al., 2005). Photosynthetic pigments were determined exactly as described by Bender-Machado et al. (2004).
ABA Analysis
Extraction of ABA from leaves was performed exactly as described by van der Merwe et al. (2009).
Measurements of Photosynthetic Parameters
The 14C-labeling pattern of sucrose, starch, and other cellular constituents was performed by illuminating leaf discs (10-mm diameter) in a leaf-disc oxygen electrode (Hansatech) in saturating 14CO2 at a PFD of 700 μmol m−2 s−1 at 25°C for 30 min, and subsequent fractionation was performed exactly as detailed by Lytovchenko et al. (2002). Fluorescence emission was measured in vivo using a PAM fluorometer (Walz) on plants maintained at fixed irradiance (100 and 700 μmol photons m−2 s−1) for 30 min prior to measurement of chlorophyll a fluorescence yield and relative ETR, which were calculated using the WinControl software package (Walz). Gas-exchange measurements were performed with a LI-6400 open-flow gas exchange system (Li-Cor). Photosynthetic light response curves (A/PFD) were produced by increasing PFD from 0 to 1000 μmol (photons) m−2 s−1. The reference CO2 concentration (Ca) was set at 400 μmol CO2 mol−1 air. The responses of A to internal CO2 concentration (A/Ci curve) were determined at 700 μmol (photons) m−2 s−1, at 25°C. Measurements started at 350 μmol CO2 mol−1, and once the steady state was reached (within 3 to 5 min), CO2 concentration was gradually lowered to 50 μmol mol−1 and then increased stepwise up to 2000 μmol mol−1, exactly as described by Long and Bernacchi (2003). Estimation of the maximum carboxylation rate (Vcmax), electron transport rate (Jmax), and triose phosphate use (TPU) variables were computed from the A/Ci curves using the A/Ci curve fitting model developed by Sharkey et al. (2007). All measurements were performed at 25°C, and vapor pressure deficit was kept at 2.0 ± 0.2 kPa, while the amount of blue light was set to 10% PFD to optimize stomatal aperture.
Carbon Isotope Composition Ratio
Leaf tissue was collected between 11:00 and 13:00 h, and stable carbon isotope ratio (δ13C) was analyzed as described by DaMatta et al. (2002).
Measurement of Respiratory Parameters
Dark respiration was measured using the same gas exchange system as defined above. Estimations of the TCA cycle flux on the basis of 14CO2 evolution were performed following incubation of isolated leaf discs in 10 mM MES-KOH, pH 6.5, containing 2.32 KBq mL−1 of [1-14C]-, [2-14C]-,[3,4-14C]-, or [6-14C]Glc. Evolved 14CO2 was trapped in KOH and quantified by liquid scintillation counting. The results were interpreted following Rees and Beevers (1960).
Stomatal Analysis
After 2 h of illumination in the dark-light cycle as described above, dental resin imprints (Kagan et al., 1992; Berger and Altmann, 2000) were taken from the abaxial surface of two leaflets, the third and fourth fully developed leaves. Nail polish copies were prepared as described by von Groll et al. (2002), and the images were taken with a digital camera (Color View; Soft Imaging System) attached to a microscope (Olympus BX41). The measurements were performed on the images using the CellP software (Soft Imaging System). Stomatal density was determined in five to eight different fields of 0.55 mm2 per leaflet, and aperture measurements were determined in 90 to 120 guard cell pairs distributed in at least six separate fields of 0.14 mm2. For Figure 10, detached leaves were cut and floated in stomatal opening solution containing 10 mM MES-KOH, pH 6.15, 5 mM KCl, and 50 μM CaCl2 at 25°C. The described solutions were added to the opening solution after 2 h of illumination, and stomatal apertures were measured 2 h later.
Water Loss Measurements
For water loss measurements, the weight of detached leaves, incubated abaxial side up under greenhouse conditions, was measured at the indicated time points. Water loss was calculated as a percentage of the initial fresh weight.
Isolation of Apoplastic Fluid
Apoplastic fluid was isolated essentially by following the protocol of Sweetlove et al. (1998). Briefly, leaves were collected and washed in ice-cold milli-Q water (Milipore) and were then vacuum infiltrated in 100 mM KCl twice for 2 min each. The leaves were then blotted dry, placed between two funnels to hold them flat, and centrifuged for 10 min at 1000g at 4°C. The volume of the collected liquid was measured and stored at −80°C until required.
Preparation of Epidermal Fragments
Epidermal fragments from fully expanded leaves highly enriched for guard cells (>90%) were prepared using the blender method described by Scheibe et al. (1990).
Isolation of Guard Cell Protoplasts
Guard cell protoplasts from tomato plants were isolated and purified mainly as described in the protocol developed for Arabidopsis thaliana (Pandey et al., 2002; Yoo et al., 2007) with modifications. Fully expanded leaves (~10 g) with the main veins removed were surfaced sterilized in 0.5% (v/v) NaOCl and 0.12% (v/v) Tween 20 solution for 5 min, washed in 96% (v/v) ethanol for 2 s, followed by three washes in sterile distilled water. The leaves were then blended twice for 1 min in a waring blender in 100 mL of cold distilled water. The first enzyme digestion of epidermal peels was performed for 1 h at a shaking speed of 150 rpm. The second enzyme digestion was performed for 1 h at a speed of 50 rpm. The pore size of the nylon mesh used after the first and the second digestions was 60 and 30 μm, respectively. After Histopaque purification, the cells were resuspended in 1 mL of basic solution containing 5 mM MES-Tris, pH 5.5, 0.5 m M CaCl2, 0.5 mM MgCl 2, 10 μM KH 2PO4, 0.5 mM ascorbic acid, and 0.55 M sorbitol. Ten microliters of the suspension was then taken, and the number of protoplasts was estimated with a hemocytometer. The pellet was washed three times with 0.4 M mannitol containing 1 mM CaCl2. Isolated guard cell protoplasts were stored in 0.4 M mannitol containing 1 mM CaCl2 at 2 to 4°C in the dark until use. Protein concentrations were determined as described above and chlorophyll concentration was determined as described by Porra et al. (1989). The yield of guard cell protoplasts was on average 5 × 105 mL−1, which corresponds to ~30 μg of protein. The purity of the final guard cell preparation was consistently higher than 99.0% on a cell basis, with little contamination originating from mesophyll cells and epidermal cells.
Preparation of Mesophyll Cell Protoplasts
Mesophyll cell protoplasts were prepared as described (Shimazaki et al., 1982) with modifications. Fully expanded leaves (~3.0 g) were sterilized in 0.5% (v/v) NaOCl, 0.12% (v/v) Tween 20 solution for 5 min, washed in 96% (v/v) ethanol for 2 s, followed by three washes in sterile distilled water. The leaves were placed in 0.3 M sorbitol and 50 mM CaCl2 and sliced into ~1- to 2-mm strips. After 30 min of plasmolysis at room temperature, the strips were vacuum infiltrated three times for 1 min and treated with 25 mL of an enzyme solution containing 2% (w/v) Cellulase Onozuka R-10 (Yakult Honsha) and 0.5% (w/v) Macerozyme R-10 (Yakult Honsha) in a buffer containing 0.65 M mannitol, 2 mM CaCl2, 5 mM MES-KOH, pH 5.5, and 0.2% (w/v) BSA. Enzymatic digestion was performed for ~30 min at room temperature after vacuum infiltration. The second digestion was performed for 2.0 h at 25°C. The released mesophyll cell protoplasts were collected by low-speed centrifugation (100g for 7 min) and were washed twice with 0.6 M mannitol containing 1 mM CaCl2. Finally, the protoplasts were resuspended in standard uptake buffer (1 mM CaCl2, 1 mM MgCl2, 0.65 M mannitol, and 10 mM MES-Tris, pH 5.5). Isolated mesophyll cell protoplasts were stored on ice in the dark until use. Protein and chlorophyll concentrations were determined as stated above. The rate of O2 evolution and uptake was determined at 25°C as described elsewhere (Shimazaki et al., 1982) for both guard cell and mesophyll cell protoplasts.
Microarray Analysis
TOM1 glass slides containing arrayed tomato ESTs were obtained directly from the Center for Gene Expression Profiling at the Boyce Thompson Institute, Cornell University, the Geneva Agricultural Experiment Station, and the USDA Federal Plant and Nutrition Laboratory. The tomato array contains 13,440 spots randomly selected from cDNA libraries isolated from a range of tissues, including leaf, root, fruit, and flowers, and representing a broad range of metabolic and developmental processes. Further annotation of this file was performed to provide gene identities and putative functions for the ESTs described on the Solanaceae Genomics Network website (http://soldb.cit.cornell.edu). Fluorescent probe preparation and microarray hybridization were performed exactly as described previously (Urbanczyk-Wochniak et al., 2006). Five microarrays were hybridized with extracts from whole leaves from wild-type and SDH14 plants using a dye-swap strategy, such that wild-type plants were labeled with Cy3 three times. In the case of epidermal fragments, four slides were hybridized, where each genotype was labeled with Cy3 two times. Microarray experiment slides were normalized with print-tip loess and moving minimum background subtraction using the Bioconductor limma package framework (Gentleman et al., 2004). Microarray slides were subsequently scale normalized, with the log ratios being adjusted to have the same median absolute deviation across arrays (Yang et al., 2002; Smyth and Speed, 2003). Moderated t statistics were used to detect any genes likely to be differentially expressed between wild-type and SDH14 plants either in the whole leaf or in epidermal fragments (Smyth, 2004). Finally, the resulting P values were corrected for multiple testing using the Benjamini Hochberg procedure (Benjamini and Hochberg, 1995).
qRT-PCR
qRT- PCR was performed exactly as described by Zanor et al. (2009) using the fluorescent intercalating dye SYBR Green in an iCycler detection system (Bio-Rad). The primers used here are described in Supplemental Table 4 online. To normalize gene expression, the constitutively expressed ubiquitin3 (UBI3) was amplified using the following primers: forward, 5′-AGGTTGATGACACTGGAAAGGTT-3′, reverse, 5′-ATCGCCTCCAGCCTTGTTGTA-3′ (Wang et al., 2008).
Statistical Analysis
Data were statistically examined using analysis of variance and tested for significant (P < 0.05) differences using Student’s t tests. The term significant is used in the text only when the change in question has been confirmed to be significant (P < 0.05) with the Student’s t test. All the statistical analyses were performed using the algorithm embedded into Microsoft Excel.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: SDH2-2 (SGN-U563726); SDH 2-1 (SGN-U563725); Rbcs (SGN-U577358); CHX20 (SGN-U585825) ; PHOT1 (SGN-U563240); PHOT2 (SGN-U565446); CCR2 (SGN-U578513); ABI2 (SGN-U569923); AHA2 (SGN-U593808); CPK6 (SGN-U317299); NIA2 (SGN-U579543); OST1 (SGN-U568947); PLDα1 (SGN-U580437); CaS (SGN-U567109); KAT2 (SGN-U603185); RBOHB (SGN-U564615); STP1 (SGN-U579712); CIPK15 (SGN-U583600); GPX3 (SGN-U576688); GTG1 (SGN-U581783) ; TIP (SGN-U580398); ABCB 14 (SGN-U571503); tDT (SGN-U565207); UBI3 (X58253); SGN-U579957 (glycolate oxidase); SGN-U580678 (ribulose-phosphate 3-epimerase); SGN-U566206 (similar to lipase in Arabidopsis); SGN-U584266 (UDP-sulfoquinovose:DAG sulfoquinovosyltransferase/sulfolipid synthase); SGN-U563031 (CTD phosphatase-like protein 3); SGN-U591223 (subtilase family protein); SGN-U595977 (hypothetical protein); and SGN-U573103 (unnamed).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization and Expression of Tomato SDH2-2.
Supplemental Figure 2. Expression of Tomato Succinate Dehydrogenase in Epidermal Fragments.
Supplemental Figure 3. Relative Expression of Off Target Genes Supposed to Effect Succinate Dehydrogenase Sequence in Tomato.
Supplemental Figure 4. Analysis of Succinate Dehydrogenase Expression and Activity in Tomato Plants during Normal Plant Development.
Supplemental Figure 5. Carbon Dioxide Evolution from Positionally Labeled Glucose in Leaves of Antisense SDH Tomato Plants.
Supplemental Figure 6. Stomatal Properties of 4-Week-Old Transgenic and Wild-Type Tomato Plants.
Supplemental Figure 7. Fresh Weight Loss from Detached Leaves of Wild-Type and Transgenic Tomato Plants.
Supplemental Figure 8. Growth Variables of Leaves of 4-Week-Old Transgenic and Wild-Type Tomato Plants.
Supplemental Figure 9. Respiratory and Photosynthetic Activities of GCP and MCP.
Supplemental Figure 10. Relative Stomatal Movement under Malate or Fumarate and Its Combination with ABA and Channel Blocker.
Supplemental Figure 11. Physiological Characterization of SDH2-2 Lines under the Control of a Guard Cell–Specific Promoter, MYB60.
Supplemental Table 1. Pigment Contents in Antisense Succinate Dehydrogenase Tomato Plants.
Supplemental Table 2. Relative Metabolite Content of the Fully Expanded Leaves from 4-Week-Old Plants of the Antisense Succinate Dehydrogenase Plants.
Supplemental Table 3. Gas Exchange Parameters Derived by Fitting A/Ci Data.
Supplemental Table 4. Primers Used in the RT-PCR Analyses Performed in This Study.
Supplemental Data Set 1. Text File Corresponding to Alignment Shown in Supplemental Figure 1.
Acknowledgments
We thank Helga Kulka (Max-Planck-Institut für Molekulare Pflanzenphysiologie) for the excellent care of the plants as well as Lothar Willmitzer, Mark Stitt, Ralph Bock, and Joost T. van Dongen (same institute) for useful discussions of the results presented here. We thank Eugenia Maximova and Josef Bergstein (Max-Planck-Institut für Molekulare Pflanzenphysiologie) for technical support of the microscopy analyses and for excellent photographic work, respectively. We also thank Alexander Ivakov (Max-Planck-Institut für Molekulare Pflanzenphysiologie) and Anna M. Zbierzak (same institute) for help in performing gas-exchange and pigment measurements, respectively. This work was supported by grants from the Max Planck Gesellschaft (to W.L.A. and A.N.-N.) and the Konrad Adenauer Stiftung (to C.S.-W.) as well as ERAnet Plant Genomics projects TRIPTOP and STRESSNET funded by the German Research Council (DFG), an Alexander von Humboldt fellowship (to T.T.), the Millennium Nucleus for Plant Functional Genomics (P06-009-F), Millennium Scientific Initiative Program, and Fondecyt 1100601, Chile (to D.F. and X.J.).
References
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba R., et al. (2004). ESTs, cDNA microarrays, and gene expression profiling: Tools for dissecting plant physiology and development. Plant J. 39: 697–714 [DOI] [PubMed] [Google Scholar]
- Araújo W.L., Ishizaki K., Nunes-Nesi A., Larson T.R., Tohge T., Krahnert I., Witt S., Obata T., Schauer N., Graham I.A., Leaver C.J., Fernie A.R. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., Nunes-Nesi A., Trenkamp S., Bunik V.I., Fernie A.R. (2008). Inhibition of 2-oxoglutarate dehydrogenase in potato tuber suggests the enzyme is limiting for respiration and confirms its importance in nitrogen assimilation. Plant Physiol. 148: 1782–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann S.M., Shimazaki Ki. (1999). The multisensory guard cell. Stomatal responses to blue light and abscisic acid. Plant Physiol. 119: 809–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann S.M., Wang X.Q. (2001). From milliseconds to millions of years: Guard cells and environmental responses. Curr. Opin. Plant Biol. 4: 421–428 [DOI] [PubMed] [Google Scholar]
- Bender-Machado L., Bäuerlein M., Carrari F., Schauer N., Lytovchenko A., Gibon Y., Kelly A.A., Loureiro M., Müller-Röber B., Willmitzer L., Fernie A.R. (2004). Expression of a yeast acetyl CoA hydrolase in the mitochondrion of tobacco plants inhibits growth and restricts photosynthesis. Plant Mol. Biol. 55: 645–662 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300 [Google Scholar]
- Berger D., Altmann T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14: 1119–1131 [PMC free article] [PubMed] [Google Scholar]
- Berger F., Linstead P., Dolan L., Haseloff J. (1998). Stomata patterning on the hypocotyl of Arabidopsis thaliana is controlled by genes involved in the control of root epidermis patterning. Dev. Biol. 194: 226–234 [DOI] [PubMed] [Google Scholar]
- Bergmann D.C., Lukowitz W., Somerville C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 [DOI] [PubMed] [Google Scholar]
- Bourgeron T., Rustin P., Chretien D., Birch-Machin M., Bourgeois M., Viegas-Péquignot E., Munnich A., Rötig A. (1995). Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet. 11: 144–149 [DOI] [PubMed] [Google Scholar]
- Camarasa C., Bidard F., Bony M., Barre P., Dequin S. (2001). Characterization of Schizosaccharomyces pombe malate permease by expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 67: 4144–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F., Nunes-Nesi A., Gibon Y., Lytovchenko A., Loureiro M.E., Fernie A.R. (2003). Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol. 133: 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S.A., Hetherington A.M. (2010). Environmental regulation of stomatal development. Curr. Opin. Plant Biol. 13: 90–95 [DOI] [PubMed] [Google Scholar]
- Chen Z., Gallie D.R. (2004). The ascorbic acid redox state controls guard cell signaling and stomatal movement. Plant Cell 16: 1143–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E., Galbiati M., Vavasseur A., Conti L., Sala T., Vuylsteke M., Leonhardt N., Dellaporta S.L., Tonelli C. (2005). A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 15: 1196–1200 [DOI] [PubMed] [Google Scholar]
- Coursol S., Fan L.-M., Le Stunff H., Spiegel S., Gilroy S., Assmann S.M. (2003). Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423: 651–654 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Bari R.P., Stitt M., Scheible W.R., Udvardi M.K. (2004). Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Daignan-Fornier B., Valens M., Lemire B.D., Bolotin-Fukuhara M. (1994). Structure and regulation of SDH3, the yeast gene encoding the cytochrome b560 subunit of respiratory complex II. J. Biol. Chem. 269: 15469–15472 [PubMed] [Google Scholar]
- DaMatta F.M., Loos R.A., Silva E.A., Loureiro M.E., Ducatti C. (2002). Effects of soil water deficit and nitrogen nutrition on water relations and photosynthesis of pot-grown Coffea canephora Pierre. Trees (Berl.) 16: 555–558 [Google Scholar]
- Dreyer I., Blatt M.R. (2009). What makes a gate? The ins and outs of Kv-like K+ channels in plants. Trends Plant Sci. 14: 383–390 [DOI] [PubMed] [Google Scholar]
- Dutilleul C., Lelarge C., Prioul J.L., De Paepe R., Foyer C.H., Noctor G. (2005). Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol. 139: 64–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elorza A., León G., Gómez I., Mouras A., Holuigue L., Araya A., Jordana X. (2004). Nuclear SDH2-1 and SDH2-2 genes, encoding the iron-sulfur subunit of mitochondrial complex II in Arabidopsis, have distinct cell-specific expression patterns and promoter activities. Plant Physiol. 136: 4072–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerlich V., Linka N., Reinhold T., Hurth M.A., Traub M., Martinoia E., Neuhaus H.E. (2003). The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier. Proc. Natl. Acad. Sci. USA 100: 11122–11126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H., Jänsch L., Braun H.-P. (2003). New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiol. 133: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar G.D., O'Leary M.H.O., Berry J.A. (1982). On the relationship between carbon isotope discrimination and the intercellular carbon dioxide concentration in leaves. Aust. J. Plant Physiol. 9: 121–137 [Google Scholar]
- Fernie A.R. (2007). The future of metabolic phytochemistry: Larger numbers of metabolites, higher resolution, greater understanding. Phytochemistry 68: 2861–2880 [DOI] [PubMed] [Google Scholar]
- Fernie A.R., Carrari F., Sweetlove L.J. (2004b). Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 7: 254–261 [DOI] [PubMed] [Google Scholar]
- Fernie A.R., Martinoia E. (2009). Malate. Jack of all trades or master of a few? Phytochemistry 70: 828–832 [DOI] [PubMed] [Google Scholar]
- Fernie A.R., Roscher A., Ratcliffe R.G., Kruger N.J. (2001). Fructose 2,6-bisphosphate activates pyrophosphate: Fructose-6-phosphate 1-phosphotransferase and increases triose phosphate to hexose phosphate cycling in heterotrophic cells. Planta 212: 250–263 [DOI] [PubMed] [Google Scholar]
- Fernie A.R., Trethewey R.N., Krotzky A., Willmitzer L. (2004a). Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5: 763–769 [DOI] [PubMed] [Google Scholar]
- Figueroa P., Gómez I., Holuigue L., Araya A., Jordana X. (1999). Transfer of rps14 from the mitochondrion to the nucleus in maize implied integration within a gene encoding the iron-sulphur subunit of succinate dehydrogenase and expression by alternative splicing. Plant J. 18: 601–609 [DOI] [PubMed] [Google Scholar]
- Figueroa P., León G., Elorza A., Holuigue L., Jordana X. (2001). Three different genes encode the iron-sulfur subunit of succinate dehydrogenase in Arabidopsis thaliana. Plant Mol. Biol. 46: 241–250 [DOI] [PubMed] [Google Scholar]
- Fridman E., Carrari F., Liu Y.S., Fernie A.R., Zamir D. (2004). Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Gaedeke N., Klein M., Kolukisaoglu U., Forestier C., Müller A., Ansorge M., Becker D., Mamnun Y., Kuchler K., Schulz B., Mueller-Roeber B., Martinoia E. (2001). The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J. 20: 1875–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N., Wang Q., Menda N., Zamir D., Hirschberg J. (2008). Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J. 53: 717–730 [DOI] [PubMed] [Google Scholar]
- Garmier M., Carroll A.J., Delannoy E., Vallet C., Day D.A., Small I.D., Millar A.H. (2008). Complex I dysfunction redirects cellular and mitochondrial metabolism in Arabidopsis. Plant Physiol. 148: 1324–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier H., Vavasseur A., Gans P., Lascève G. (1991). Relationship between respiration and photosynthesis in guard gell and mesophyll cell protoplasts of Commelina communis L. Plant Physiol. 95: 636–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K.A., Romeis T., Hedrich R. (2009). Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y., Blaesing O.E., Hannemann J., Carillo P., Höhne M., Hendriks J.H.M., Palacios N., Cross J., Selbig J., Stitt M. (2004). A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: Comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel A., Schauer N., Carrari F., Fernie A.R., Grimm B., Kühn C. (2006). Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. Plant J. 45: 180–192 [DOI] [PubMed] [Google Scholar]
- Hägerhäll C. (1997). Succinate: Quinone oxidoreductases. Variations on a conserved theme. Biochim. Biophys. Acta 1320: 107–141 [DOI] [PubMed] [Google Scholar]
- Hedrich R., Marten I. (1993). Malate-induced feedback regulation of plasma membrane anion channels could provide a CO2 sensor to guard cells. EMBO J. 12: 897–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrich R., Neher E. (1987). Cytoplasmic calcium regulates voltage-dependent ion channels in plant. Nature 329: 833–836 [Google Scholar]
- Hedrich R., Marten I., Lohse G., Dietrich P., Winter H., Lohous G., Heldt H.-W. (1994). Malate-sensitive anion channels enable guard cells to sense changes in the ambient CO2 concentration. Plant J. 6: 741–748 [Google Scholar]
- Hedrich R., Neimanis S., Savchenko G., Felle H.H., Kaiser W.M., Heber U. (2001). Changes in apoplastic pH and membrane potential in leaves in relation to stomatal responses to CO2, malate, abscisic acid or interruption of water supply. Planta 213: 594–601 [DOI] [PubMed] [Google Scholar]
- Hetherington A.M. (2001). Guard cell signaling. Cell 107: 711–714 [DOI] [PubMed] [Google Scholar]
- Hetherington A.M., Brownlee C. (2004). The generation of Ca(2+) signals in plants. Annu. Rev. Plant Biol. 55: 401–427 [DOI] [PubMed] [Google Scholar]
- Hetherington A.M., Quatrano R.S. (1991). Mechanism of action of abscisic acid at the cellular level. New Phytol. 119: 9–32 [DOI] [PubMed] [Google Scholar]
- Higgins D.G., Sharp P.M. (1988). CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene 73: 237–244 [DOI] [PubMed] [Google Scholar]
- Hildmann T., Ebneth M., Peña-Cortés H., Sánchez-Serrano J.J., Willmitzer L., Prat S. (1992). General roles of abscisic and jasmonic acids in gene activation as a result of mechanical wounding. Plant Cell 4: 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsefield R., Yankovskaya V., Sexton G., Whittingham W., Shiomi K., Omura S., Byrne B., Cecchini G., Iwata S. (2006). Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): A mechanism of electron transfer and proton conduction during ubiquinone reduction. J. Biol. Chem. 281: 7309–7316 [DOI] [PubMed] [Google Scholar]
- Howad W., Kempken F. (1997). Cell type-specific loss of atp6 RNA editing in cytoplasmic male sterile Sorghum bicolor. Proc. Natl. Acad. Sci. USA 94: 11090–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Taylor N.L., Narsai R., Eubel H., Whelan J., Millar A.H. (2010). Functional and composition differences between mitochondrial complex II in Arabidopsis and rice are correlated with the complex genetic history of the enzyme. Plant Mol. Biol. 72: 331–342 [DOI] [PubMed] [Google Scholar]
- Hurry V., Anderson J.M., Chow W.S., Osmond C.B. (1997). Accumulation of zeaxanthin in abscisic acid–deficient mutants of Arabidopsis does not affect chlorophyll fluorescence quenching or sensitivity to photoinhibition in vivo. Plant Physiol. 113: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurth M.A., Suh S.J., Kretzschmar T., Geis T., Bregante M., Gambale F., Martinoia E., Neuhaus H.E. (2005). Impaired pH homeostasis in Arabidopsis lacking the vacuolar dicarboxylate transporter and analysis of carboxylic acid transport across the tonoplast. Plant Physiol. 137: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida A.M., Pei Z.M., Baizabal-Aguirre V.M., Turner K.J., Schroeder J.I. (1997). Expression of a Cs(+)-resistant guard cell K+ channel confers Cs(+)-resistant, light-induced stomatal opening in transgenic arabidopsis. Plant Cell 9: 1843–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N., Fujii M., Hartman P.S., Tsuda M., Yasuda K., Senoo-Matsuda N., Yanase S., Ayusawa D., Suzuki K. (1998). A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature 394: 694–697 [DOI] [PubMed] [Google Scholar]
- Ishizaki K., Larson T.R., Schauer N., Fernie A.R., Graham I.A., Leaver C.J. (2005). The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsson M., Siegel R.S., Young J., Hashimoto M., Iba K., Schroeder J.I. (2006). Guard cell ABA and CO2 signaling network updates and Ca2+ sensor priming hypothesis. Curr. Opin. Plant Biol. 9: 654–663 [DOI] [PubMed] [Google Scholar]
- Jenner H.L., Winning B.M., Millar A.H., Tomlinson K.L., Leaver C.J., Hill S.A. (2001). NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol. 126: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan M., Novoplansky N., Sachs T. (1992). Variable cell lineages form the functional pea epidermis. Ann. Bot. (Lond.) 69: 303–312 [Google Scholar]
- Keeling P.L., Wood J.R., Tyson R.H., Bridges I.G. (1988). Starch biosynthesis in developing wheat grain - Evidence against the direct involvement of triose phosphates in the metabolic pathway. Plant Physiol. 87: 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J., et al. (2005). GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Koyama H., Kawamura A., Kihara T., Hara T., Takita E., Shibata D. (2000). Overexpression of mitochondrial citrate synthase in Arabidopsis thaliana improved growth on a phosphorus-limited soil. Plant Cell Physiol. 41: 1030–1037 [DOI] [PubMed] [Google Scholar]
- Kubo N., Harada K., Hirai A., Kadowaki K. (1999). A single nuclear transcript encoding mitochondrial RPS14 and SDHB of rice is processed by alternative splicing: common use of the same mitochondrial targeting signal for different proteins. Proc. Natl. Acad. Sci. USA 96: 9207–9211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J.M., Mäser P., Schroeder J.I. (2008). The clickable guard cell, Version II: Interactive model of guard cell signal transduction mechanisms and pathways. The Arabidopsis Book 6:e0999 doi/10.1199/tab.0999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Choi Y., Burla B., Kim Y.Y., Jeon B., Maeshima M., Yoo J.Y., Martinoia E., Lee Y. (2008). The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat. Cell Biol. 10: 1217–1223 [DOI] [PubMed] [Google Scholar]
- León G., Holuigue L., Jordana X. (2007). Mitochondrial complex II is essential for gametophyte development in Arabidopsis. Plant Physiol. 143: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N., Kwak J.M., Robert N., Waner D., Leonhardt G., Schroeder J.I. (2004). Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.H., Liu Y.Q., Lü P., Lin H.F., Bai Y., Wang X.C., Chen Y.L. (2009). A signaling pathway linking nitric oxide production to heterotrimeric G protein and hydrogen peroxide regulates extracellular calmodulin induction of stomatal closure in Arabidopsis. Plant Physiol. 150: 114–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.J., Prat S., Willmitzer L., Frommer W.B. (1990). Cis regulatory elements directing tuber-specific and sucrose-inducible expression of a chimeric class I patatin promoter/GUS-gene fusion. Mol. Gen. Genet. 223: 401–406 [DOI] [PubMed] [Google Scholar]
- Lombardo A., Carine K., Scheffler I.E. (1990). Cloning and characterization of the iron-sulfur subunit gene of succinate dehydrogenase from Saccharomyces cerevisiae. J. Biol. Chem. 265: 10419–10423 [PubMed] [Google Scholar]
- Long S.P., Bernacchi C.J. (2003). Gas exchange measurements, what can they tell us about the underlying limitations to photosynthesis? Procedures and sources of error. J. Exp. Bot. 54: 2393–2401 [DOI] [PubMed] [Google Scholar]
- López-Bucio J., Cruz-Ramírez A., Herrera-Estrella L. (2003). The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6: 280–287 [DOI] [PubMed] [Google Scholar]
- López-Bucio J., Nieto-Jacobo M.F., Ramírez-Rodríguez V., Herrera-Estrella L. (2000). Organic acid metabolism in plants: from adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci. 160: 1–13 [DOI] [PubMed] [Google Scholar]
- Lytovchenko A., Sweetlove L.J., Pauly M., Fernie A.R. (2002). The influence of cytosolic phosphoglucomutase on photosynthetic carbohydrate metabolism. Planta 215: 1013–1021 [DOI] [PubMed] [Google Scholar]
- Marienfeld J.R., Newton K.J. (1994). The maize NCS2 abnormal growth mutant has a chimeric nad4-nad7 mitochondrial gene and is associated with reduced complex I function. Genetics 138: 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Bhatt K., Baumann K. (2001). Shaping in plant cells. Curr. Opin. Plant Biol. 4: 540–549 [DOI] [PubMed] [Google Scholar]
- Merlot S., Leonhardt N., Fenzi F., Valon C., Costa M., Piette L., Vavasseur A., Genty B., Boivin K., Mueller A., Giraudat J., Leung J. (2007). Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 26: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S., De Angeli A., Fernie A.R., Martinoia E. (2010a). Intra- and extra-cellular excretion of carboxylates. Trends Plant Sci. 15: 40–47 [DOI] [PubMed] [Google Scholar]
- Meyer S., Mumm P., Imes D., Endler A., Weder B., Al-Rasheid K.A., Geiger D., Marten I., Martinoia E., Hedrich R. (2010b). AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 63: 1054–1062 [DOI] [PubMed] [Google Scholar]
- Millar A.H., Eubel H., Jänsch L., Kruft V., Heazlewood J.L., Braun H.-P. (2004). Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol. Biol. 56: 77–90 [DOI] [PubMed] [Google Scholar]
- Millar A.H., Sweetlove L.J., Giegé P., Leaver C.J. (2001). Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Millar A.H., Wiskich J.T., Whelan J., Day D.A. (1993). Organic acid activation of the alternative oxidase of plant mitochondria. FEBS Lett. 329: 259–262 [DOI] [PubMed] [Google Scholar]
- Miyadera H., Shiomi K., Ui H., Yamaguchi Y., Masuma R., Tomoda H., Miyoshi H., Osanai A., Kita K., Omura S. (2003). Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase). Proc. Natl. Acad. Sci. USA 100: 473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi T., Kawakami T., Arai H., Igarashi Y., Matsushita K., Mori M., Shiomi K., Omura S., Harada S., Kita K. (2009). Siccanin rediscovered as a species-selective succinate dehydrogenase inhibitor. J. Biochem. 146: 383–387 [DOI] [PubMed] [Google Scholar]
- Mori I.C., Murata Y., Yang Y., Munemasa S., Wang Y.F., Andreoli S., Tiriac H., Alonso J.M., Harper J.F., Ecker J.R., Kwak J.M., Schroeder J.I. (2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol. 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A.A.M., Farnsworth L., Ackrell B.A.C., Turnbull D.M., Birch-Machin M.A. (1994). The cDNA sequence of the flavoprotein subunit of human heart succinate dehydrogenase. Biochim. Biophys. Acta 1185: 125–128 [DOI] [PubMed] [Google Scholar]
- Mott K.A. (2009). Opinion: Stomatal responses to light and CO(2) depend on the mesophyll. Plant Cell Environ. 32: 1479–1486 [DOI] [PubMed] [Google Scholar]
- Nadeau J.A., Sack F.D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Nadeau J.A., Sack F.D. (2003). Stomatal development: Cross talk puts mouths in place. Trends Plant Sci. 8: 294–299 [DOI] [PubMed] [Google Scholar]
- Nagy R., Grob H., Weder B., Green P., Klein M., Frelet-Barrand A., Schjoerring J.K., Brearley C., Martinoia E. (2009). The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J. Biol. Chem. 284: 33614–33622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi J., Matsuda O., Nagasawa T., Oba Y., Takahashi H., Kawai-Yamada M., Uchimiya H., Hashimoto M., Iba K. (2008). CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452: 483–486 [DOI] [PubMed] [Google Scholar]
- Newton K.J., Knudsen C., Gabay-Laughnan S., Laughnan J.R. (1990). An abnormal growth mutant in maize has a defective mitochondrial cytochrome oxidase gene. Plant Cell 2: 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A., Araújo W.L., Fernie A.R. (2011). Targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiol. 155: 101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A., Carrari F., Gibon Y., Sulpice R., Lytovchenko A., Fisahn J., Graham J., Ratcliffe R.G., Sweetlove L.J., Fernie A.R. (2007b). Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J. 50: 1093–1106 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A., Carrari F., Lytovchenko A., Smith A.M., Loureiro M.E., Ratcliffe R.G., Sweetlove L.J., Fernie A.R. (2005). Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 137: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A., Sweetlove L.J., Fernie A.R. (2007a). Operation and function of the tricarboxlic acid cycle in the illuminated leaf. Physiol. Plant. 129: 45–56 [Google Scholar]
- Padmasree K., Padmavathi L., Raghavendra A.S. (2002). Essentiality of mitochondrial oxidative metabolism for photosynthesis: optimization of carbon assimilation and protection against photoinhibition. Crit. Rev. Biochem. Mol. Biol. 37: 71–119 [DOI] [PubMed] [Google Scholar]
- Pandey S., Nelson D.C., Assmann S.M. (2009). Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Pandey S., Wang X.-Q., Coursol S.A., Assmann S.M. (2002). Preparation and applications of Arabidopsis thaliana guard cell protoplasts. New Phytol. 153: 517–526 [DOI] [PubMed] [Google Scholar]
- Pandey S., Zhang W., Assmann S.M. (2007). Roles of ion channels and transporters in guard cell signal transduction. FEBS Lett. 581: 2325–2336 [DOI] [PubMed] [Google Scholar]
- Pei Z-M., Kuchitsu K., Ward J.M., Schwarz M., Schroeder J.I. (1997). Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9: 409–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z.-M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G.J., Grill E., Schroeder J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Pla M., Mathieu C., De Paepe R., Chétrit P., Vedel F. (1995). Deletion of the last two exons of the mitochondrial nad7 gene results in lack of the NAD7 polypeptide in a Nicotiana sylvestris CMS mutant. Mol. Gen. Genet. 248: 79–88 [DOI] [PubMed] [Google Scholar]
- Plaxton W.C., Podesta F.E. (2006). The functional organization and control of plant respiration. Crit. Rev. Plant Sci. 25: 159–198 [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Quick W.P., Schurr U., Scheibe R., Schulze E.D., Rodermel S.R., Bogorad L., Stitt M. (1991). Decreased ribulose- 1,5-bisphosphate carboxylase-oxygenase in transgenic tobacco transformed with 'antisense' rbcS. I. Impact on photosynthesis in ambient growth conditions. Planta 183: 542–554 [DOI] [PubMed] [Google Scholar]
- Raghavendra A.S., Padmasree K., Saradadevi K. (1994). The functional interdependence of photosynthesis and respiration in plant cells: Interactions between chloroplast and mitochondria. Plant Sci. 97: 1–14 [Google Scholar]
- Rasmusson A.G., Geisler D.A., Møller I.M. (2008). The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 8: 47–60 [DOI] [PubMed] [Google Scholar]
- Rees T.A., Beevers H. (1960). Pathways of glucose dissimilation in carrot slices. Plant Physiol. 35: 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro D.M., Desikan R., Bright J., Confraria A., Harrison J., Hancock J.T., Barros R.S., Neill S.J., Wilson I.D. (2009). Differential requirement for NO during ABA-induced stomatal closure in turgid and wilted leaves. Plant Cell Environ. 32: 46–57 [DOI] [PubMed] [Google Scholar]
- Roelfsema M.R.G., Hedrich R. (2005). In the light of stomatal opening: New insights into ‘the Watergate’. New Phytol. 167: 665–691 [DOI] [PubMed] [Google Scholar]
- Roessner U., Luedemann A., Brust D., Fiehn O., Linke T., Willmitzer L., Fernie A.R. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U., Hegemann B., Lytovchenko A., Carrari F., Bruedigam C., Granot D., Fernie A.R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133: 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H., Fuentes I., Vásquez M., Corvalán C., León G., Gómez I., Araya A., Holuigue L., Vicente-Carbajosa J., Jordana X. (2009). A nuclear gene encoding the iron-sulfur subunit of mitochondrial complex II is regulated by B3 domain transcription factors during seed development in Arabidopsis. Plant Physiol. 150: 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel J.B., Wilson P.B., Hussain D., Woo N.S., Gordon M.J., Mewett O.P., Howell K.A., Whelan J., Kazan K., Pogson B.J. (2007). Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell 19: 4091–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustin P., Rötig A. (2002). Inborn errors of complex II—Unusual human mitochondrial diseases. Biochim. Biophys. Acta 1553: 117–122 [DOI] [PubMed] [Google Scholar]
- Rutter J., Winge D.R., Schiffman J.D. (2010). Succinate dehydrogenase - Assembly, regulation and role in human disease. Mitochondrion 10: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Schachtman D.P., Schroeder J.I., Lucas W.J., Anderson J.A., Gaber R.F. (1992). Expression of an inward-rectifying potassium channel by the Arabidopsis KAT1 cDNA. Science 258: 1654–1658 [DOI] [PubMed] [Google Scholar]
- Schauer N., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Schauer N., Steinhauser D., Strelkov S., Schomburg D., Allison G., Moritz T., Lundgren K., Roessner-Tunali U., Forbes M.G., Willmitzer L., Fernie A.R., Kopka J. (2005). GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 579: 1332–1337 [DOI] [PubMed] [Google Scholar]
- Scheffler I.E. (1998). Molecular genetics of succinate:quinone oxidoreductase in eukaryotes. Prog. Nucleic Acid Res. Mol. Biol. 60: 267–315 [DOI] [PubMed] [Google Scholar]
- Scheibe R., Backhausen J.E., Emmerlich V., Holtgrefe S. (2005). Strategies to maintain redox homeostasis during photosynthesis under changing conditions. J. Exp. Bot. 56: 1481–1489 [DOI] [PubMed] [Google Scholar]
- Scheibe R., Reckmann U., Hedrich R., Raschke K. (1990). Malate dehydrogenases in guard cells of Pisum sativum. Plant Physiol. 93: 1358–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R., Stitt M. (1988). Comparison of NADP-malate dehydrogenase activation QA reduction and O2 evolution in spinach leaves. Plant Physiol. Biochem. 26: 473–481 [Google Scholar]
- Schon E.A. (2000). Mitochondrial genetics and disease. Trends Biochem. Sci. 25: 555–560 [DOI] [PubMed] [Google Scholar]
- Schroeder J.I., Allen G.J., Hugouvieux V., Kwak J.M., Waner D. (2001). Guard cell signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Schroeder J.I., Hagiwara S. (1989). Cytosolic calcium regulates ion channels in the plasma membrane of Vicia faba guard cells. Nature 338: 427–430 [Google Scholar]
- Schroeder J.I., Hagiwara S. (1990). Repetitive increases in cytosolic Ca2+ of guard cells by abscisic acid activation of nonselective Ca2+ permeable channels. Proc. Natl. Acad. Sci. USA 87: 9305–9309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey T.D., Bernacchi C.J., Farquhar G.D., Singsaas E.L. (2007). Fitting photosynthetic carbon dioxide response curves for C(3) leaves. Plant Cell Environ. 30: 1035–1040 [DOI] [PubMed] [Google Scholar]
- Sharkey T.D., Savitch L.V., Butz N.D. (1991). Photometric method for routine determination of kcat and carbamylation of rubisco. Photosynth. Res. 28: 41–48 [DOI] [PubMed] [Google Scholar]
- Shimazaki K., Gotow K., Kondo N. (1982). Photosynthetic properties of guard cell protoplasts from Vicia faba L. Plant Cell Physiol. 23: 871–879 [Google Scholar]
- Shimazaki K., Gotow K., Sakaki T., Kondo N. (1983). High respiratory activity of guard cell protoplasts from Vicia faba L. Plant Cell Physiol. 24: 1049–1056 [Google Scholar]
- Sibbernsen E.D., Mott K.A. (2010). Stomatal responses to flooding of the intercellular air spaces suggest a vapor-phase signal between the mesophyll and the guard cells. Plant Physiol. 153: 1435–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A., Nunes-Nesi A., Sulpice R., Lisec J., Centeno D.C., Carillo P., Leisse A., Urbanczyk-Wochniak E., Fernie A.R. (2008). Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. Plant Physiol. 147: 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A., Sulpice R., Osorio S., Krahnert I., Leisse A., Urbanczyk-Wochniak E., Hodges M., Fernie A.R., Nunes-Nesi A. (2010). Mild reductions in mitochondrial NAD-dependent isocitrate dehydrogenase activity result in altered nitrate assimilation and pigmentation but do not impact growth. Mol. Plant 3: 156–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3. [DOI] [PubMed] [Google Scholar]
- Smyth G.K., Speed T. (2003). Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Stitt M., Quick W.P., Schurr U., Schulze E.D., Rodermel S.R., Bogorad L. (1991). Decreased ribulose-1,5- bisphosphate carboxylase-oxygenase in transgenic tobacco transformed with 'antisense' rbcS. II. Flux control coefficients for photosynthesis in varying light, CO2 and air humidity. Planta 183: 555–566 [DOI] [PubMed] [Google Scholar]
- Stitt M., Schulze E.D. (1994). Does Rubisco control the rate of photosynthesis and plant growth? An exercise in molecular ecophysiology. Plant Cell Environ. 17: 465–487 [Google Scholar]
- Studart-Guimarães C., Fait A., Nunes-Nesi A., Carrari F., Usadel B., Fernie A.R. (2007). Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the gamma-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol. 145: 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R., Sienkiewicz-Porzucek A., Osorio S., Krahnert I., Stitt M., Fernie A.R., Nunes-Nesi A. (2010). Mild reductions in cytosolic NADP-dependent isocitrate dehydrogenase activity result in lower amino acid contents and pigmentation without impacting growth. Amino Acids 39: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter J.U., Sieben C., Hartel A., Eisenach C., Thiel G., Blatt M.R. (2007). Abscisic acid triggers the endocytosis of the Arabidopsis KAT1 K+ channel and its recycling to the plasma membrane. Curr. Biol. 17: 1396–1402 [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Beard K.F., Nunes-Nesi A., Fernie A.R., Ratcliffe R.G. (2010). Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 15: 462–470 [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Fait A., Nunes-Nesi A., Williams T., Fernie A.R. (2007). The mitochondrion: An integration point in cellular metabolism and signalling. Crit. Rev. Plant Sci. 26: 17–43 [Google Scholar]
- Sweetlove L.J., Heazlewood J.L., Herald V., Holtzapffel R., Day D.A., Leaver C.J., Millar A.H. (2002). The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 32: 891–904 [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Kossmann J., Riesmeier J.W., Trethewey R.N., Hill S.A. (1998). The control of source to sink carbon flux during tuber development in potato. Plant J. 15: 697–706 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tauberger E., Fernie A.R., Emmermann M., Renz A., Kossmann J., Willmitzer L., Trethewey R.N. (2000). Antisense inhibition of plastidial phosphoglucomutase provides compelling evidence that potato tuber amyloplasts import carbon from the cytosol in the form of glucose-6-phosphate. Plant J. 23: 43–53 [DOI] [PubMed] [Google Scholar]
- Tcherkez G., Cornic G., Bligny R., Gout E., Ghashghaie J. (2005). In vivo respiratory metabolism of illuminated leaves. Plant Physiol. 138: 1596–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez G., Mahé A., Gauthier P., Mauve C., Gout E., Bligny R., Cornic G., Hodges M. (2009). In folio respiratory fluxomics revealed by 13C isotopic labeling and H/D isotope effects highlight the noncyclic nature of the tricarboxylic acid “cycle” in illuminated leaves. Plant Physiol. 151: 620–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C.L., Jones L., Baulcombe D.C., Maule A.J. (2001). Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J. 25: 417–425 [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E., et al. (2006). Conversion of MapMan to allow the analysis of transcript data from Solanaceous species: Effects of genetic and environmental alterations in energy metabolism in the leaf. Plant Mol. Biol. 60: 773–792 [DOI] [PubMed] [Google Scholar]
- Vahisalu T., Kollist H., Wang Y.F., Nishimura N., Chan W.Y., Valerio G., Lamminmäki A., Brosché M., Moldau H., Desikan R., Schroeder J.I., Kangasjärvi J. (2008). SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven R., Ronning C., Giovannoni J., Martin G., Tanksley S. (2002). Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14: 1441–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe M.J., Osorio S., Moritz T., Nunes-Nesi A., Fernie A.R. (2009). Decreased mitochondrial activities of malate dehydrogenase and fumarase in tomato lead to altered root growth and architecture via diverse mechanisms. Plant Physiol. 149: 653–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vani T., Raghavendra A.S. (1994). High mitochondrial activity but incomplete engagement of the cyanide-resistant alternative pathway in guard cell protoplasts of pea. Plant Physiol. 105: 1263–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavasseur A., Raghavendra A.S. (2005). Guard cell metabolism and CO2 sensing. New Phytol. 165: 665–682 [DOI] [PubMed] [Google Scholar]
- Vedel F., Lalanne E., Sabar M., Chetrit P., De Paepe R. (1999). The mitochondrial respiratory transport chain and ATP synthase complexes: Composition, structure and mutational studies. Plant Physiol. Biochem. 37: 629–643 [Google Scholar]
- Vidal G., Ribas-Carbo M., Garmier M., Dubertret G., Rasmusson A.G., Mathieu C., Foyer C.H., De Paepe R. (2007). Lack of respiratory chain complex I impairs alternative oxidase engagement and modulates redox signaling during elicitor-induced cell death in tobacco. Plant Cell 19: 640–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Groll U., Berger D., Altmann T. (2002). The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14: 1527–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. (1986). A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14: 4683–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.C., Ngwenyama N., Liu Y.D., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Liu J., Feng Y., Niu X., Giovannoni J., Liu Y. (2008). Altered plastid levels and potential for improved fruit nutrient content by downregulation of the tomato DDB1-interacting protein CUL4. Plant J. 55: 89–103 [DOI] [PubMed] [Google Scholar]
- Wang X.Q., Ullah H., Jones A.M., Assmann S.M. (2001). G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Ward J.M., Mäser P., Schroeder J.I. (2009). Plant ion channels: Gene families, physiology, and functional genomics analyses. Annu. Rev. Physiol. 71: 59–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J.M., Fusaro A.F., Wang M.B., Waterhouse P.M. (2005). RNA silencing platforms in plants. FEBS Lett. 579: 5982–5987 [DOI] [PubMed] [Google Scholar]
- Wilson I.D., Ribeiro D.M., Bright J., Confraria A., Harrison J., Barros R.S., Desikan R., Neill S.J., Hancock J.T. (2009). Role of nitric oxide in regulating stomatal apertures. Plant Signal. Behav. 4: 467–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Zhang Y.J., Kang L., Roossinck M.J., Mysore K.S. (2006). Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol. 142: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P. (2002). Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zanor M.I., et al. (2009). RNA interference of LIN5 in tomato confirms its role in controlling Brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiol. 150: 1204–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang L., Dong F., Gao J., Galbraith D.W., Song C.P. (2001). Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 126: 1438–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.X., Zhang W., Stanley B.A., Assmann S.M. (2008). Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20: 3210–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S.Y., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]