This work provides biochemical and genetic evidence to demonstrate that a type II formin protein regulates rice morphogenesis through its influence over the formation of actin cytoskeleton and the organization of microtubules.
Abstract
Multicellular organisms contain a large number of formins; however, their physiological roles in plants remain poorly understood. Here, we reveal that formin homology 5 (FH5), a type II formin mutated in rice morphology determinant (rmd), plays a crucial role in determining rice (Oryza sativa) morphology. FH5/RMD encodes a formin-like protein consisting of an N-terminal phosphatase tensin (PTEN)-like domain, an FH1 domain, and an FH2 domain. The rmd mutants display a bending growth pattern in seedlings, are stunted as adult plants, and have aberrant inflorescence (panicle) and seed shape. Cytological analysis showed that rmd mutants have severe cell elongation defects and abnormal microtubule and microfilament arrays. FH5/RMD is ubiquitously expressed in rice tissues, and its protein localization to the chloroplast surface is mediated by the PTEN domain. Biochemical assays demonstrated that recombinant FH5 protein can nucleate actin polymerization from monomeric G-actin or actin/profilin complexes, cap the barbed end of actin filaments, and bundle actin filaments in vitro. Moreover, FH5 can directly bind to and bundle microtubules through its FH2 domain in vitro. Our findings suggest that the rice formin protein FH5 plays a critical role in determining plant morphology by regulating actin dynamics and proper spatial organization of microtubules and microfilaments.
INTRODUCTION
Variation in organ morphology contributes to the diversified architecture of flowering plants. The shape of each plant organ is determined by the type of cells it contains, and the specialized morphology for each cell is supported by the microtubule and microfilament cytoskeletal systems. The microfilament cytoskeleton, which is composed of the filamentous actin assembled from G-actin monomers, is a well-organized and dynamic system. It plays crucial roles in diverse cellular processes, including cell shape and polarity establishment and maintenance, cell division, cytoplasmic streaming, organelle movement, and responses to external signals (Wasteneys and Galway, 2003; Smith and Oppenheimer, 2005; Hussey et al., 2006; Staiger and Blanchoin, 2006; Paul and Pollard, 2009). Microtubule arrays consist of highly ordered and polymerized heterodimers of α-tubulin and β-tubulin and contribute to plant morphogenesis (Lloyd and Chan, 2004).
The organization and dynamics of the microfilament cytoskeleton are regulated by various actin binding proteins, which bind to monomeric or filamentous actin or both types of actin. Actin filaments have polar structures and elongate mainly at their barbed ends (Casella et al., 1981; Fox and Phillips, 1981; Tellam and Frieden, 1982; Yahara et al., 1982; Symons and Mitchison, 1991; Redmond et al., 1994). Thus, the formation of new barbed ends by the de novo nucleation of actin filaments is a crucial step in actin assembly (Blanchoin et al., 2000a; Wear et al., 2000; Pollard et al., 2001). During spontaneous filament assembly, the formation of actin nuclei is a rate-limiting step. Within the cell, actin assembly has been shown to be regulated by actin nucleation factors including the Arp2/3 complex (Pollard, 2007), Spire (Quinlan et al., 2005), cofilin (Andrianantoandro and Pollard, 2006), leiomodin (Chereau et al., 2008), and formins (Goode and Eck, 2007), proteins responsible for the formation of actin nuclei (Welch et al., 1997; Pruyne et al., 2002; Quinlan et al., 2005; Chereau et al., 2008). The Arp2/3 complex stimulates branched filament formation from preexisting actin filaments (Mullins et al., 1998; Pollard and Beltzner, 2002), whereas formins regulate the assembly of parallel actin filaments into actin bundles or cables (Kleinebrecht et al., 1982; Kovar and Pollard, 2004; Romero et al., 2004; Kovar, 2006).
Formins are known to be morphoregulatory proteins that direct the assembly of unbranched actin filaments in eukaryotes such as fungi, animals, and plants (Paul and Pollard, 2009). Most formins have two highly conserved domains, the Pro-rich domain (i.e., formin homology 1 [FH1]) and the formin homology domain (i.e., FH2). The FH1 domain in different formins contains varied number of polyproline stretches that are known to interact with the small actin monomer binding protein, profilin, or the actin/profilin complex, to promote actin polymerization from the barbed end (Chang et al., 1997; Pruyne et al., 2002; Kovar et al., 2006). This domain can concentrate and transfer multiple profilin-actin complexes onto the barbed end of the filament, allowing quick elongation (Paul and Pollard, 2009). On the other hand, the FH2 domain, which is the most conserved region in formins, is able to bind to the fast-growing filament end after nucleating a filament and inhibit actin filament elongation. The FH2 domain is involved in controlling actin organization and dynamics and may also coordinate organization of the microtubule and actin cytoskeleton (Basu and Chang, 2007; Paul and Pollard, 2009; Bartolini and Gundersen, 2010; Deeks et al., 2010; Li et al., 2010). Because eukaryotes have a large number of formin genes, which possibly have redundant functions, the in vivo functions of these formins remain largely unknown.
Plant formins are grouped into two subfamilies, type I and type II. Type I formins contain an N-terminal transmembrane domain, which is believed to be involved in the association of formin proteins with the plasma membrane, whereas this feature is absent from type II formins (Cvrcková et al., 2004). Some plant type II formins have an N-terminal phosphatase tensin (PTEN)-like domain, which was suggested to play a role in mediating localization of the protein to the apical side of the cells in the moss Physcomitrella patens (Vidali et al., 2009). Most of the reported type I formins of Arabidopsis thaliana, including AFH1, AFH3, At FH4, At FH5, At FH6, and At FH8 (Cheung and Wu, 2004; Favery et al., 2004; Deeks et al., 2005; Ingouff et al., 2005; Michelot et al., 2005; Yi et al., 2005; Ye et al., 2009; Cheung et al., 2010), were demonstrated to be microfilament regulators that affect the polarized growth of pollen tubes or root hairs or embryo development (Blanchoin and Staiger, 2010). Interestingly, At-FH4 was shown to be associated with the microtubule cytoskeleton via a plant-specific GOE region near its N terminus and able to nucleate F-actin, suggesting that At-FH4 may link membranes to both major cytoskeletal networks (Deeks et al., 2010).
Unlike the many well-studied type I formins, the functional role of type II formins has not been characterized in details in plants. In Physcomitrella, reducing the expression of all type II formins led to stunted plants that contain spherical cells with disrupted actin organization, whereas silencing of all the other formins did not result in apparent alteration to the cell morphology and actin organization (Vidali et al., 2009), suggesting the importance of type II formins in polarized cell growth and in the organization of the actin cytoskeleton. Moreover, domain swapping experiments in the same study showed that the FH1-FH2 domain of type II formins cannot be replaced by that of type I formins (Vidali et al., 2009), suggesting that the function of these domains may have diverged during the evolution of the two formin lineages. More recently, we revealed that AFH14, a type II formin from Arabidopsis, is able to regulate both microtubule and microfilament arrays. The afh14 mutant displays abnormal microtubule arrays in meiosis during microspore formation, and the AFH14 protein can bind to microtubules and microfilaments directly via the FH2 domain (Li et al., 2010).
Rice (Oryza sativa) is an important food crop worldwide and an excellent model monocot system for developmental biology. In this study, we show that FH5, a type II formin-like protein encoded by Rice Morphology Determinant (RMD), functions as an actin-nucleating protein that regulates rice plant morphology by modulating the microtubule- and microfilament-based cytoskeletal systems.
RESULTS
Identification and Morphological Characterization of the rmd Mutants
To understand the molecular mechanisms underlying rice plant morphogenesis, we isolated two mutants with altered growth architecture from a rice mutant population (O. sativa ssp Japonica) generated by 60Co γ-ray treatment (Chen et al., 2006). We named these two mutants, which share similar phenotypes, rmd-1 and rmd-2 (Figure 1; see Supplemental Figure 1 online). The two mutants were backcrossed to the wild-type plants three times. All F1 progenies from each cross displayed wild-type morphology, indicating that the rmd mutations are recessive. F2 progeny of the rmd-1 mutant showed a segregation of 277 normal and 83 mutant plants (χ2 = 0.725, 0.1 < P < 0.5), suggesting monofactorial recessive inheritance of the mutant phenotype. In addition, all F1 progeny from a cross between rmd-1 and rmd-2 displayed morphological defects similar to their parents, suggesting that rmd-1 and rmd-2 are allelic.
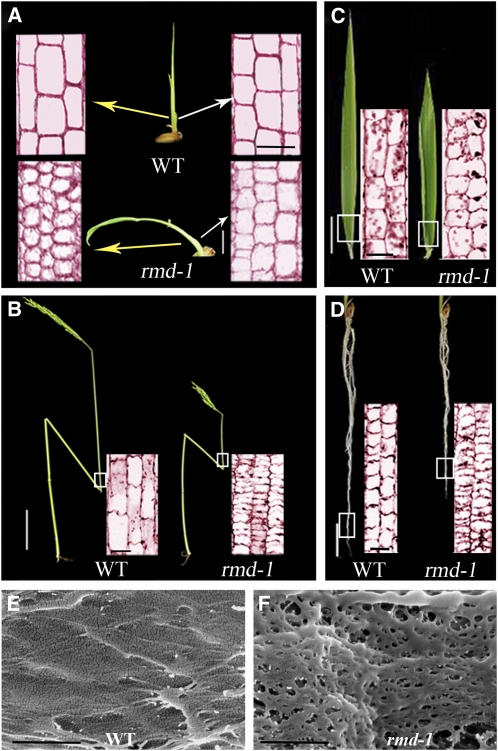
Figure 1.
rmd Mutants Display Morphological Defects from Vegetative to Reproductive Development.
(A) Seedlings; days after germination are indicated. WT, wild type. Bar = 8 mm.
(B) Rice plants at heading stage. Bar = 20 cm.
(C) Panicle and rachis of plants at heading stage. Bar =2 cm.
(D) Individual flowers from plants at the heading stage. Bar =2 mm.
(E) Seeds. Bar = 2 mm.
Compared with wild-type plants, rmd mutants exhibited changed architecture in both vegetative and reproductive phases. Unlike the straight growth of wild-type seedlings, the rmd-1 seedlings showed bending growth after germination (Figure 1A). This phenotype became gradually attenuated as the plants grew older; at ~30 d after germination, the rmd-1 seedlings grew straight (Figure 1A). Even though the rmd-1 mutants had no obvious changes in flowering time compared with the wild type, the height and morphology of the adult plants were significantly changed. At the seed setting stage, the culm length in rmd-1 was determined to be shorter (i.e., 48.6 ± 3.5 cm compared with 80.7 ± 3.0 cm in the wild type; n = 76, P < 0.001 by Student’s t test) (see Supplemental Figure 2A online). The decreased culm length of rmd-1 was attributable mainly to the reduced length of the four uppermost internodes (Figure 2B; see Supplemental Figure 2A online). The lengths of internodes I to IV in rmd-1 were 18, 5, 1, and 0.1 cm, respectively, shorter than those of the wild type (see Supplemental Figure 2A online). In addition, the rmd-1 mutant had shorter and curled flag leaves (Figure 2C; see Supplemental Figure 2B online) and panicles (Figure 1C; see Supplemental Figure 2A online). The flower filaments and roots of rmd-1 were also shorter than those of the wild type (Figures 1D and 2D; see Supplemental Figures 2C and 2D online), and the seeds showed an irregular and curled appearance (Figure 1E). The rmd-2 mutant plants exhibited phenotypic defects similar to rmd-1 (see Supplemental Figures 1 and 2 online). These data suggest that RMD plays a critical role in rice plant morphogenesis.
Figure 2.
rmd-1 Mutants Display Defects in Cell Elongation and Polarity as Revealed by Longitudinal Section Analysis.
(A) Leaf sheath from 12-d-old wild-type (WT) and rmd-1 seedlings. Seedling bar = 8 mm; sectioning bar = 50 μm.
(B) Culm from plants at the heading stage. Stem bar = 8 cm; sectioning bar = 100 μm.
(C) Leaf from plants at the heading stage. Leaf bar = 1.6 cm; sectioning bar = 100 μm.
(D) Roots from 14-d-old seedlings. Root bar = 5 mm; sectioning bar = 100 μm.
(E) and (F) Field emission scanning electron microscopy of the cell wall surface from epidermal cells in the root elongation zone from 14-d-old wild-type (E) and rmd-1 seedlings (F). Bars = 500 nm.
[See online article for color version of this figure.]
rmd Mutants Display Severe Defects in Cell Elongation
To examine the cellular alteration of the rmd mutants in more detail, we performed tissue section analysis. rmd-1 mutant seedlings displayed abnormal cell growth, for example, the average cell number per mm2 in the 12-d-old leaf sheath in rmd-1 was 1480 (n = 61), which was significantly higher than that of the wild-type plants (820, n = 60, P < 0.001). Moreover, longitudinal section analysis showed that cells on one flank of the rmd-1 leaf sheath (left side, Figure 2A) were much smaller than the wild-type cells, were irregularly shaped, and had obscure boundaries. In 12-d-old seedlings, the average length of a cell in the long axis in this region was 17 ± 2 μm in rmd-1 and 50 ± 2 μm in the wild type (n = 20, P < 0.005). Although cells on the opposite flank in the mutant were smaller (each was 24 ± 2 μm, n = 20, P < 0.005) than those of wild type (right of Figure 2A), they were still larger than cells on the other side of the same mutant, thus causing unbalanced and bending growth of the mutant seedling (Figure 2A).
At the mature stage, internode I was much shorter in rmd-1 (9.4 ± 2.5 cm, n = 76) than in the wild type (27.6 ± 3.1 cm, n = 76, P < 0.001), a phenotype resulting from a marked reduction in cell length (Figure 2B). There was no obvious difference in cell number from the base to the top of internode I between the wild type and rmd-1 (~5000 cells in each file, n = 49), suggesting that the phenotype observed in rmd-1 is caused mostly by defects in cell elongation rather than division. Moreover, cell elongation was also reduced in mature leaf and root cortical cells in rmd-1 (Figures 2C and 2D). Consistent with the observation that rmd-1 cells were irregularly shaped and had obscure cell boundaries (Figures 2A to 2D), field emission scanning electron microscope analysis of the rmd-1 root epidermal cells in the elongation zone revealed a disorganized cell wall surface (Figures 2E and 2F). The size of the rmd-1 pollen grains was also significantly reduced (35 ± 3 μm in diameter, n = 17, P < 0.005) compared with the wild type (45 ± 3 μm in diameter, n = 18) (Figure 3G), although pollen fertility in the rmd mutants appeared normal. Analysis of the rmd-2 mutant plants revealed cellular defects very similar to those of rmd-1 (see Supplemental Figures 3B and 3E online). These results together suggest that the rmd mutants are severely deficient in cell growth, particularly in cell elongation and shape formation.
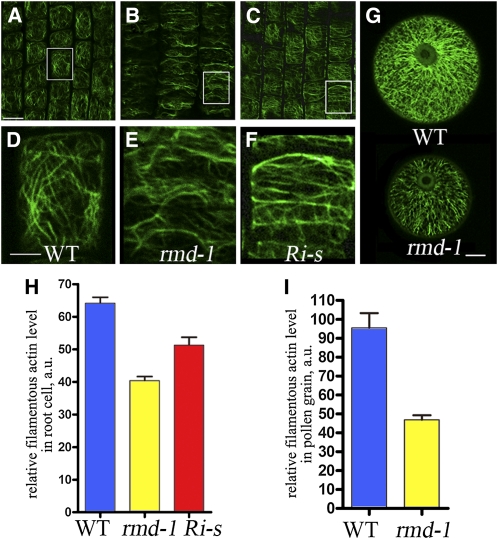
Figure 3.
rmd Mutants and FH5/RMD RNAi Lines Display Defective Actin Microfilament Networks.
(A) to (F) Actin filaments of cells from the root elongation zone of 7-d- old rice seedlings, stained by Alexa Fluor 488-phalloidin.
(A) Wild-type cells.
(B) rmd-1 cells.
(C) Cells of FH5/RMD RNAi lines with strong defects (Ri-s).
(D) A close-up of boxed region of (A). WT, wild type.
(E) A close-up of boxed region of (B).
(F) A close-up of boxed region of (C).
(G) F-actin pattern in pollen grains of the wild type and rmd-1 at stage 13 of anther development.
(H) Fluorescent pixel intensities (±se) of wild-type, rmd-1, and Ri-s root cells were 64.16 ± 3.72 (n = 32), 40.42 ± 2.46 (n = 30), and 51.3 ± 5.05 (n = 20).
(I) The average fluorescence pixel intensity (±se) in each pollen grain was 95.48 ± 7.76 (n = 18) in the wild type and 46.87 ± 2.36 (n = 17) in rmd-1 (P < 0.001).
Bars = 50 μm in (A) to (C), 25 μm in (D) to (F), and 10 μm in (G).
[See online article for color version of this figure.]
rmd-1 Mutants Are Impaired in Microfilament Organization
The severe defect in cell growth in the rmd mutants led us to speculate that RMD may be involved in organization of the cytoskeleton. To test this hypothesis, we observed the actin cytoskeleton in root tips and pollen grains after staining the filamentous (F-) actin with Alexa Fluor 488-phalloidin, a dye that specifically labels F-actin. In the wild type, the cortical cells in the root elongation zone displayed a fine F-actin network with abundant longitudinally oriented filaments (Figures 3A and 3D). By contrast, the rmd-1 mutant showed disordered F-actin organization and more transverse than longitudinal filaments in the cell (Figures 3B and 3E), suggesting that RMD plays a role in polarized actin organization.
Given that the abundance of F-actin is positively correlated with the fluorescence intensity, we quantified the relative amount of F-actin using fluorescent quantification analysis. Our data showed that the average fluorescence pixel intensity (±se) in each rmd-1 root cell was lower than that of the wild type (Figure 3H; see Supplemental Figure 4 online), suggesting a reduced amount of F-actin in rmd-1. In addition, a scan of 50 slices of the stained root cortical cells found a reduced number of actin bundles in rmd-1 (see Supplemental Figure 4 online). Consistent with these results from root cells, in pollen grains, we also observed significant differences in the organization of the F-actin network between wild-type and mutant plants (Figures 3G and 3I). Likewise, rmd-2 mutant roots and pollen grains displayed F-actin defects similar to those in rmd-1 (see Supplemental Figures 3H, 3K, and 4 online). These data suggest that the rice RMD protein is critical for the organization of actin filaments, especially in the formation of longitudinal actin cables.
rmd-1 Mutants Display Defective Microtubule Organization
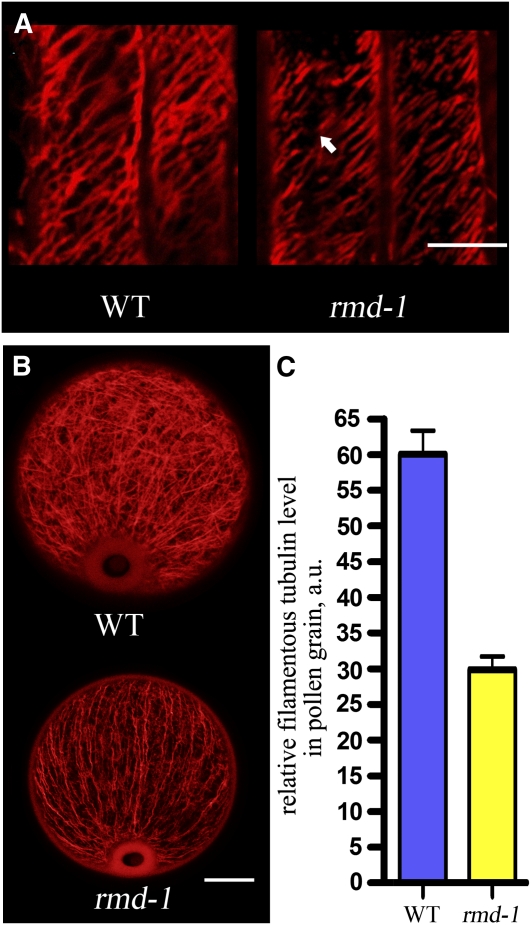
Microtubules are required for the maintenance of cell shape (Lloyd and Chan, 2004). To observe microtubule organizations in rmd mutants, we stained microtubules in root cells with anti-β-tubulin antibody (Sigma-Aldrich). Along the long axis of root cortical cells, wild-type plants had obliquely and transversely aligned microtubules, whereas rmd-1 formed discontinuous and fragmented microtubule arrays (Figure 4A). About 67% of microtubules in root cortical cells in rmd-1 were discontinuous (n = 30 cells). Moreover, rmd-1 pollen grains contained less abundant microtubule arrays compared with the well-developed and complex microtubule network in the wild type (Figure 4B). The average fluorescence pixel intensity in each rmd-1 pollen grain was lower than that of the wild type (Figure 4C). Similarly, defective microtubule arrays were observed in rmd-2 mutant roots (see Supplemental Figure 3N online). These results led us to the conclusion that RMD also affects the formation of microtubule networks in rice plants.
Figure 4.
The rmd Mutant Has Defective Microtubule Array.
(A) Microtubule arrays in the wild-type (WT) and rmd-1 root cortical cells. The arrow in rmd-1 indicates fragmented microtubule array. Bar = 20 μm.
(B) Microtubule arrays in the wild-type and rmd-1 pollen grains at stage 13 of anther development. Bar = 10 μm.
(C) The average fluorescence pixel intensity (±se) in each pollen grain was 60.07 ± 3.26 (n = 27) in the wild type and 29.87 ± 1.86 (n = 29) in rmd-1 (P < 0.001)
[See online article for color version of this figure.]
Map-Based Cloning and Characterization of RMD
Using markers from the 12 rice chromosomes and 50 mutants, we first mapped the RMD gene to chromosome 7, within a genetic distance between the two markers, WY705-1 (83.3 centimorgan) and WY706-4 (84.1 centimorgan) (see Supplemental Figure 5A online). To locate the gene further, 360 additional mutants from an F2 mapping population were analyzed using nine InDel markers in the region (see Supplemental Table 1 online). The mutation was mapped between markers WY706-5 and WY706-33 on the BAC clone AP004275, within a 15-kb region. By sequencing genomic DNA in this region in the mutant (see Supplemental Table 2 online), we found a T-to-C transition at nucleotide position 7694 and a four-nucleotide (AAGG) deletion from 7695 to 7698 in the predicted 11th exon of Os07g0596300. These changes led to a frameshift mutation in the 1442nd amino acid, resulting in the premature termination of the protein at the 1465th amino acid (see Supplemental Figure 5B online). The rmd-2 mutant had a four-nucleotide (ATGG) deletion from 1996 to 1999 in the predicted fourth exon of Os07g0596300, causing a frameshift at the 388th amino acid and premature termination at the 392nd amino acid (see Supplemental Figure 5B online). Os07g0596300 has been predicted to encode a type II formin protein called FH5, which was shown to be very close in sequence to AFH14 from Arabidopsis and FH12 from rice (Cvrcková et al., 2004).
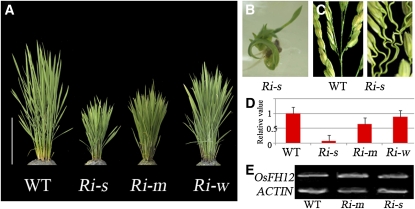
To verify the identity of RMD further, we generated an RNA interference (RNAi) construct using a 740-bp fragment from a gene-specific region of the FH5/RMD cDNA and transformed the construct into calli induced from young embryos of wild-type rice. Among the 70 RNAi plants (T0 transgenic lines) obtained, 25 displayed strong morphological defects (designated as Ri-s) similar to those of the rmd mutants (Figures 5A to 5D; see Supplemental Figures 3C, 3F, 3I, 3L, and 3O online), 34 showed a moderate phenotype (Ri-m), and 11 had weak defects (Ri-w) (Figures 5A and 5D). Using RT-PCR analysis, we confirmed that the RNAi construct specifically reduced the expression of FH5/RMD, whereas the expression of its closest homolog, FH12, was not affected in the RNAi lines (Figures 5D and 5E). Furthermore, the level of gene silencing correlated with the severity of the mutant phenotypes (Figures 5A and 5D). These data further confirmed that the FH5 gene we cloned is responsible for the phenotypes displayed in the rmd mutants.
Figure 5.
Phenotypes of FH5/RMD RNAi Lines.
(A) The FH5/RMD RNAi plants display varied levels of reduced height. Ri-s, strong phenotype, 35.9 ± 2.5 cm, n = 25; Ri-m, moderate phenotype, 44.9 ± 2.7 cm, n = 34; and Ri-w, weak phenotype, 80.3 ± 5.4 cm, n = 11. WT, wild type. Bar = 20 cm.
(B) and (C) The bending growth of seedling (B) and curled panicle branches (C) shown in Ri-s.
(D) qRT-PCR analysis of FH5/RMD in RNAi plants (error bars represent sd).
(E) RT-PCR analysis of FH12, a gene sharing high sequence similarity to FH5 in the RNAi lines, ACTIN as control.
[See online article for color version of this figure.]
We determined the expressed regions of the FH5/Os07g0596300 gene by comparing the genomic sequence with the released partial cDNA sequence (AK120222), the full-length protein sequence (Q84ZL0) from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/protein/172046705), and the sequence of our RT-PCR products. The predicted FH5 protein is 1627 amino acids long and contains three domains: a C2 domain (amino acids 200 to 337) that belongs to the PTEN (phosphatase and tensin-related) motif, a Pro-rich FH1 domain (amino acids 825 to 1170; see Supplemental Figure 6 online), and a conserved C-terminal FH2 domain (amino acids 1188 to 1588) (see Supplemental Figure 6 online). The PTEN and FH1 domains in FH5 are highly divergent from those in nonplant formins but share sequence similarities with the equivalent domains predicted to be present in type II rice formins (Cvrcková et al., 2004). The C2 domain of FH5 has 34% sequence similarity to human PTEN_C2, but it contains an Arg-to-Gly change at the 58th amino acid, which was assumed to abolish the lipid phosphatase activity (Cvrcková et al., 2004). Among the 345 amino acids in the FH1 domain in FH5, there are 190 Pro residues (~55%) that form 22 Pro-rich stretches (see Supplemental Figure 6 online), each of which contains 3 to 14 consecutive Pro residues. The FH2 domain of FH5 has 34 to 99% sequence identity to the equivalent region in 15 other rice formin-like proteins, with many highly conserved residues (Cvrcková et al., 2004). The fact that mutations in rmd-1 and rmd-2 both caused an abnormal FH2 domain and that these two mutants had similar phenotypic defects suggest that the FH2 domain is important for the function of FH5 (see Supplemental Figure 5B online).
To gain further information on the function of FH5, we examined the expression pattern of FH5 during rice development, using RT-PCR, quantitative RT-PCR (qRT-PCR), and promoter-β-glucuronidase (GUS) fusion analysis. Consistent with the important role of FH5 in proper growth and development, FH5 is ubiquitously expressed in roots, stems, leaves, and flowers at the heading stage (see Supplemental Figure 7A online). In seedlings, we observed stronger GUS staining in the coleoptile, root tip, and lateral root primordia (see Supplemental Figures 7B to 7H online). In mature plants, GUS expression was observed in floral organs (see Supplemental Figures 7I and 7J online). Moreover, recent expression profile data from laser microdissection–mediated microarray analysis showed that FH5 was expressed in microspores at various stages (Hobo et al., 2008).
Localization of FH5
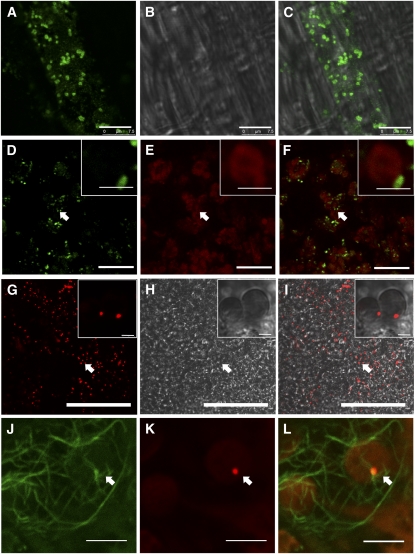
Unlike most Arabidopsis and rice type I formins, which contain putative secretory or membrane targeting signals, or transmembrane segments, FH5 and other type II formins do not have putative transmembrane domains. FH5 was predicted to be localized to the plastid (http://compbio.dfci.harvard.edu/cgi-bin/tgi/tc_report.pl?tc=TC471465andspecies=rice). To determine the subcellular localization of FH5, we generated a FH5 polyclonal antibody using a gene-specific region of FH5 (i.e., amino acid residues 534 to 709; see Supplemental Figure 5C online) with low similarity with other rice sequences. The specificity of the purified FH5 antibody was confirmed by the detection of a specific band with the expected size in protein extract from 14-d-old wild-type rice plants (see Supplemental Figure 8 online). Immunostaining analysis using rice leaf cells revealed that the FH5 protein is localized to small dots in the cytoplasm (Figures 6A to 6C; see Supplemental Figure 9 online). We speculated that these dots may be associated with the membrane of subcellular organelles. To test this prediction, we digested fresh rice leaves briefly with pectinase and cellulase to release cellular organelles and performed immunological analysis. Our data demonstrated that FH5 is localized to the chloroplast surface and that it seems to be anchored to a specific yet undetermined position on the organelle. We observed that all 70 FH5-detectable signal spots were associated with chloroplasts (Figures 6D to 6F and insets). Moreover, immunoblotting assays using proteins from isolated chloroplasts and nuclei from wild-type leaves detected FH5, together with the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit, mainly in chloroplasts (see Supplemental Figure 9F, lanes 1 and 3, online). These results confirmed the association of FH5 with chloroplasts and also suggested the possibility that specific spots on the chloroplast surface may serve as a FH5 depot for the cell.
Figure 6.
Subcellular Localization of FH5.
(A) to (C) Immunostaining analysis of FH5 localization in rice leaf cells.
(A) Immunological assay of FH5 showing the signal distribution in the cytoplasm.
(B) Bright field of (A).
(C) Merged signals of (A) and (B). Bars = 7.5 μm in (A) to (C).
(D) to (F) Immunostaining analysis of FH5 localization using rice leaf cells digested by cellulase and pectinase.
(D) Immunological assay showing FH5 signal at the outer membrane of chloroplasts (green signal).
(E) Chloroplast autofluorescence (red signal).
(F) Merged signals of (D) and (E). Insets in (D) to (F) are the close-ups of regions pointed out by the arrows. Bars = 10 μm in (D) to (F) and 1 μm in the insets.
(G) to (I) Analysis of FH5 PTEN-RFP subcellular localization in tobacco leaf cells. Insets are close-ups of the regions pointed out by the arrows.
(G) FH5 PTEN-RFP signals in the cytoplasm.
(H) Bright field of (G) showing chloroplasts.
(I) Merged signals of (G) and (H). Bars = 50 μm in (G) to (I) and 2.5 μm in insets. The control images are in Supplemental Figure 10 online.
(J) to (L) Analysis of the association between FH5 PTEN-RFP and microfilaments in tobacco leaf cells. Bars = 5 μm.
(J) Microfilaments (green signals) stained by Alexa Fluor 488-phalloidin.
(K) FH5 PTEN-RFP (red signals) indicated by the arrow.
(L) Merged signals of (J) and (K) showing the localization of FH5 PTEN-RFP on the outer envelope membrane of chloroplasts and its association with the microfilaments in the cytoplasm.
Like other plant type II formins, FH5 has a characteristic N-terminal PTEN-related domain that was predicted to be associated with membranes (Cvrcková et al., 2004). Recent investigations also indicated that the PTEN-like domain of moss (Physcomitrella) type II formins mediates the apical localization of these formins (Vidali et al., 2009). To test whether the PTEN-like domain of FH5 is sufficient for the localization of the protein to the chloroplast surface, we made a translational fusion between PTEN (amino acids 200 to 337) and red fluorescent protein (RFP) and transiently expressed the fusion protein in tobacco (Nicotiana tabacum) cells. Consistent with results of the immunostaining analysis, transiently expressed PTEN-RFP is also closely associated with a specific region on the chloroplast outer surface in tobacco cells (Figures 6G to 6I and insets; see Supplemental Figure 10 online). We observed that all 45 FH5 PTEN-RFP signal spots were associated with chloroplasts, suggesting that the PTEN-like domain of FH5 is sufficient to confer anchorage of the protein to the chloroplast outer surface. Moreover, PTEN-RFP signals were observed to be associated with the F-actin near the chloroplast (Figures 6J to 6L; see Supplemental Figure 11 online), suggesting that the PTEN domain on FH5 may provide a link between chloroplasts and the actin cytoskeleton.
FH5 Nucleates Actin Polymerization
To characterize further the functional properties of FH5 and determine whether FH5 has the ability to polymerize actin, we attempted to express a protein containing the entire FH1 and FH2 domains. However, after a number of attempts we failed to express the recombinant protein in bacterial, yeast, or baculovirus expression systems. Our inability to produce this recombinant protein is likely due to the fact that the FH1 domain of FH5 contains more polyproline stretches compared with other rice formins (see Supplemental Figure 6 online). As an alternative approach, we expressed in Escherichia coli a peptide (i.e., FH5 FH1FH2), which is a fusion of six polyproline stretches from the FH5 FH1 domain (amino acids 1098 to 1170), the FH2 domain (amino acids 1188 to 1588 on FH5), and a 6×His tag (see Supplemental Figure 12 online). In addition, we also fused a 6×His tag to the C terminus of the FH5 FH2 domain and expressed the recombinant protein (FH5 FH2) in E. coli. The recombinant FH5 FH1FH2 and FH2 domains were each affinity purified to ~98% in purity (see Supplemental Figure 12 online).
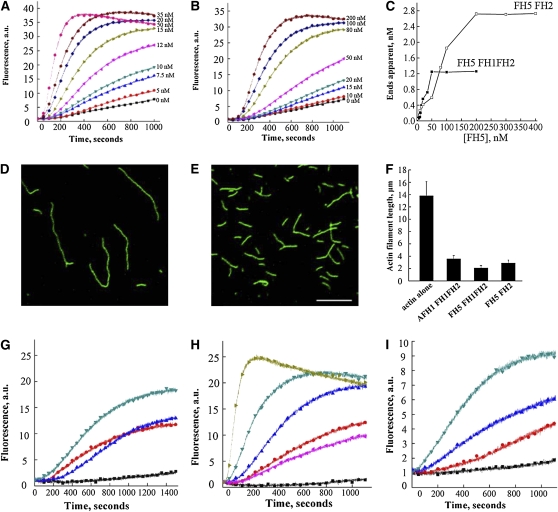
We then performed in vitro analysis using the recombinant FH5 FH1FH2 and FH5 FH2 domains to test whether FH5 functions as a formin to nucleate actin assembly. Actin monomers (10% pyrene labeled) were incubated with various concentrations of FH5 FH1FH2 and FH5 FH2, and actin assembly was monitored by pyrene fluorescence measurement. Both FH5 FH1FH2 and FH5 FH2 decreased the initial lag of actin polymerization curve in a dose-dependent manner, which is indicative of their active roles in actin nucleation (Figures 7A and 7B). The behavior of FH5 FH2 is similar to that of the FH2 domain of AFH1 (Michelot et al., 2005) but distinct from those of the FH2 domains from At-FH5 and AFH3, which do not promote actin nucleation from a pool of actin monomers (Ingouff et al., 2005; Ye et al., 2009).The concentration of newly barbed ends was strongly dependent on the amount of FH5 FH1FH2 and FH5 FH2. We also estimated the nucleation efficiency of FH5 FH1FH2 and FH5 FH2 using data obtained from three independent experiments. As shown in Figure 7C, the maximal nucleation efficiency of FH5 FH1FH2 was 0.029 filament barbed ends per formin molecule, which is close to that previously reported for AFH1 FH1FH2 (Michelot et al., 2005), but ~20 times higher than that of the equivalent fusion protein of AFH3 (Ye et al., 2009), further suggesting that FH5 can efficiently nucleate actin polymerization from actin monomers. Because filament length is inversely correlated with the number of nuclei generated during the polymerization, the actin nucleation activity can be further tested by measuring the length of actin filaments. We observed that the filament length was reduced in the presence of FH5 FH2 (Figure 7E) than that of actin monomer alone (Figure 7D). The average lengths of actin filaments in the presence of 100 nM AFH1 FH1FH2, FH5 FH1FH2, or FH5 FH2 were significantly shorter than that in the absence of formins (P < 0.01) (Figure 7F).
Figure 7.
Recombinant FH5 FH2 and FH5 FH1FH2 Nucleate Actin Polymerization from Monomeric Actin and Actin/Profilin Complexes.
(A) Time course of actin polymerization in the presence of FH5 FH1FH2 monitored by pyrene fluorescence. The signal is proportional to actin polymer concentration (Cooper et al., 1983). Different concentrations of FH5 FH1FH2 were added to 2 μM actin (10% pyrene labeled) before initiation of polymerization (from bottom to top: 0, 5, 7.5, 10, 12, 15, 20, 35, and 50 nM). a.u., arbitrary units.
(B) Time course of actin polymerization in the presence of FH5 FH2. Increasing amounts of FH5 FH2 were added to 2 μM actin (10% pyrene labeled) before initiation of polymerization (from bottom to top: 0, 10, 15, 20, 50, 80, 100, and 200 nM).
(C) Nucleation efficiency of FH5 FH2 and FH5 FH1FH2.
(D) and (E) Micrographs of actin filaments in the absence or presence of FH5 FH2. Actin alone (D); actin incubated with 100 nM FH5 FH2 (E). Bar = 5 μm.
(F) Mean length (±se) of actin filaments in the absence or presence of 100 nM formin.
(G) Time course of actin polymerization in the presence of AFH1 FH1FH2 and/or human profilin, monitored by pyrene fluorescence. From bottom to top: actin+profilin (black), actin alone (blue), actin+profilin+100 nM AFH1 FH1FH2 (red), and actin+100 nM AFH1 FH1FH2 (dark cyan).
(H) Time course of actin polymerization in the presence of FH5 FH1FH2 and/or human profilin, monitored by pyrene fluorescence. From bottom to top: actin+profilin (black), actin alone (pink), actin+profilin+10 nM FH5 FH1FH2 (red), actin+profilin+50 nM FH5 FH1FH2 (blue), actin+profilin+100 nM FH5 FH1FH2 (dark cyan), and actin+100 nM FH5 FH1FH2 (dark yellow).
(I) Time course of actin polymerization in the presence of FH5 FH1FH2 and lily profilin1 (Ll PRO1), monitored by pyrene fluorescence. From bottom to top: actin+LlPRO1 (black), actin+LlPRO1+10 nM FH5 FH1FH2 (red), actin+LlPRO1+20 nM FH5 FH1FH2 (blue), and actin+LlPRO1+50 nM FH5 FH1FH2 (dark cyan).
It has been reported that the majority of actin monomers is bound by a high concentration of profilin in some plant cells and that the actin/profilin complex represents the physiologically relevant source of actin monomers in plants (Wang et al., 2005; Chaudhry et al., 2007). We used recombinant FH5 FH1FH2 and FH5 FH2 to test whether FH5 has a physiological function in using the pool of actin bound to profilin for efficient actin filament nucleation. The well-characterized FH1FH2 domain of AFH1, which was previously shown to have the ability to nucleate actin alone and from the actin/profilin complex, was used as the positive control (Michelot et al., 2005). Compared with AFH1 FH1FH2 (Figure 7G), FH5 FH1FH2 had a more efficient nucleation activity using actin bound to human profilin as substrate (Figure 7H). When the human profilin was replaced with lily (Lilium longiflorum) profilin, FH5 FH1FH2 also showed active actin nucleating ability (Figure 7I). However, no obvious nucleating activity was observed for FH5 FH2 on the actin/profilin complex (see Supplemental Figure 13 online). Given that the number of polyproline stretches within the FH1 domain is only quantitatively, but not qualitatively, associated with its function on actin polymerization from profilin-bound actin (Kovar and Pollard, 2004), our data therefore indicate that FH5 can effectively nucleate actin polymerization from the actin/profilin complex and that the polyproline-rich FH1 domain is necessary for this property.
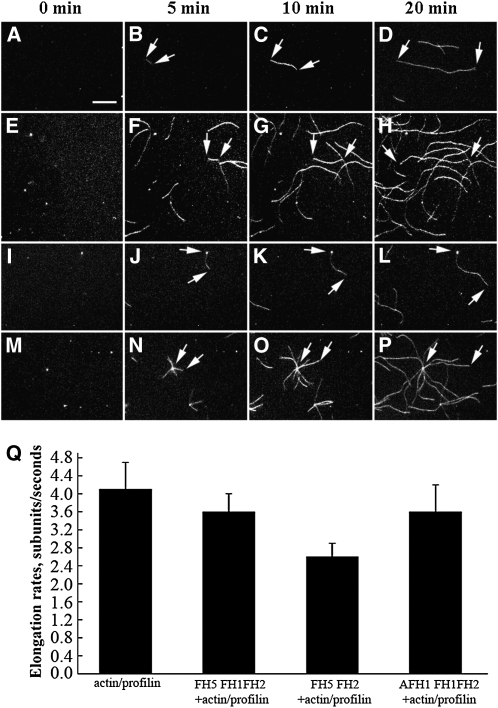
To examine further the effect of FH5 on actin assembly, we used time-lapse fluorescence microscopy to visualize directly actin polymerization from Oregon-green-actin/profilin complexes. Actin polymerization was observed near the surface of the cover glass. As the negative control, actin filaments of the actin/profilin complex alone could elongate several microns in 20 min with an average elongation rate of 4.1 ± 0.6 subunits per second (n = 18) in three independent experiments (Figures 8A to 8D and 8Q; see Supplemental Movie 1 online). In the presence of FH5 FH1FH2, more actin filaments were detectable, and the rate of actin elongation was 3.6 ± 0.4 subunits per second (n = 15) (Figures 8E to 8H and 8Q; see Supplemental Movie 2 online). However, the elongation rate of actin filaments was significantly reduced to 2.6 ± 0.3 subunits per second (P < 0.01; n = 15) upon addition of FH5 FH2 (Figures 8I to 8L and 8Q; see Supplemental Movie 3 online) compared with that of actin/profilin complex alone. This result suggested that FH5 FH2 can block actin filament elongation by capping barbed ends of the filaments, therefore preventing additional actin/profilin complexes from being added onto the capped barbed ends. The elongation rate conferred by the control protein AFH1 FH1FH2 was 3.6 ± 0.6 subunits per second (Figures 8M to 8Q; see Supplemental Movie 4 online), which is close to the published value (Michelot et al., 2005). Overall, the elongation rate of the actin filament in the presence of FH5 FH1FH2 (P = 0.07) or AFH1 FH1FH2 (P = 0.08) had no statistically significant difference from that of actin/profilin complex alone.
Figure 8.
Time-Lapse Fluorescence Microscopy Visualization of the Effect of FH5 Protein on Actin Filament Elongation from the Actin/Profilin Complex.
Oregon-green-actin/profilin complex (1 μM actin and 2 μM profilin) was polymerized in the absence or presence of 200 nM varoius formin proteins. Frames were taken at the indicated time during polymerization.
(A) to (P) Time-lapse micrographs of Oregon-green-actin/profilin polymerization with no formin ([A] to [D]; see Supplemental Movie 1 online), FH5 FH1FH2 ([E] to [H]; see Supplemental Movie 2 online), FH5 FH2 ([I] to [L]; see Supplemental Movie 3 online), and AFH1 FH1FH2 ([M] to [P]; see Supplemental Movie 4 online). Bars = 5 μm, and arrows indicate the position of actin filament ends over time.
(Q) Elongation rates (±se) of actin filaments in the absence or presence of FH5 FH1FH2, FH5 FH2, or AFH1 FH1FH2.
FH5 Binds to the Barbed Ends of F-Actin and Protects F-Actin from Dilution-Mediated Depolymerization
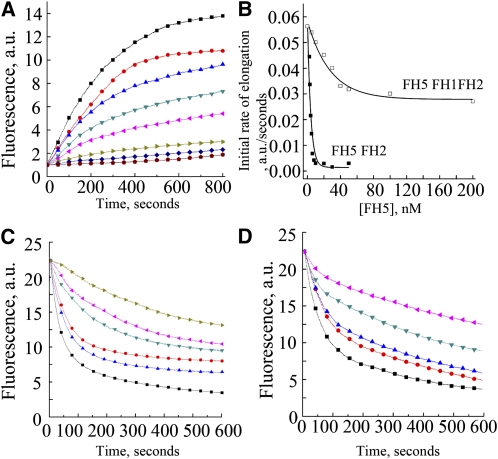
It has been demonstrated that several plant formins, such as AFH1, AFH3, At-FH5, and At-FH8, have barbed-end capping activity on actin filaments (Ingouff et al., 2005; Michelot et al., 2005; Yi et al., 2005; Ye et al., 2009). To examine barbed-end dynamics in the presence of FH5, seed elongation assays were performed using 0.8 μM actin filaments incubated with various concentrations of FH5 proteins before the addition of labeled actin monomers bound to profilin. We observed that the initial elongation rate decreased as the concentration of FH5 FH2 was increased (Figure 9A), indicating that FH5 FH2 caps the barbed ends of actin filaments. Subsequently, the apparent Kd values of binding to the barbed ends of actin filaments were determined to be 19.6 ± 2.2 nM for FH5 FH1FH2 and 3.4 ± 0.5 nM for FH5 FH2, based on three independent assays (Figure 9B). To verify the capping activity of FH5 further, we performed dilution-mediated actin filament depolymerization assays. The initial depolymerization rate decreased in a dose-dependent manner with the addition of FH5 FH1FH2 (Figure 9C) or FH5 FH2 (Figure 9D), further supporting their barbed-end capping activity. Taken together, these data suggest that both FH5 FH1FH2 and FH2 can bind to the barbed end of actin filaments and prevent actin polymerization and depolymerization from barbed ends.
Figure 9.
Recombinant FH5 FH2 and FH5 FH1FH2 Cap Actin Microfilaments.
(A) Kinetics of elongation of the barbed-end of actin filaments in the presence of FH5 FH2. Preformed actin filament seeds (0.8 μM) were incubated with various concentrations of FH5 FH2 (from top to bottom: 0, 2.5, 3, 4, 5, 6, 8, and 10 nM) before addition of 1.0 μM pyrene-actin monomers. a.u., arbitrary units.
(B) Variation in the initial rate of elongation as a function of FH5 FH2 or FH5 FH1FH2 concentration. The representative Kd is 19.1 nM for FH5 FH1FH2 and 2.9 nM for FH5 FH2.
(C) and (D) Kinetics of actin filament depolymerization in the presence of various concentrations of FH5 FH1FH2 (C) (from bottom to top: 0, 10, 20, 50, 100, and 200 nM) and FH5 FH2 (D) (from bottom to top: 0, 5, 10, 20, and 50 nM). FH5 FH1FH2 or FH5 FH2 were incubated with 5 μM actin filaments (80% pyrene-labeled) for 5 min before dilution of the solution 25-fold in buffer G.
FH5 Induces Bundling of Actin Filaments
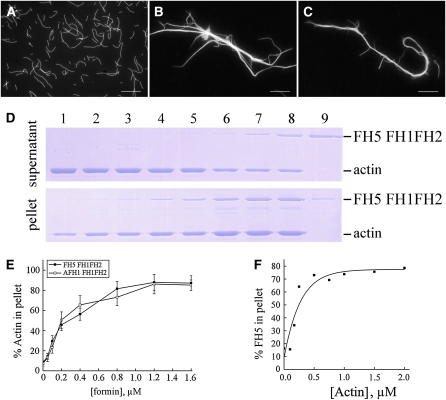
Several plant formins have been shown to be able to bundle actin filaments (Harris et al., 2004; Michelot et al., 2005; Moseley and Goode, 2005). To test the effect of FH5 FH1FH2 on bundling actin filaments, we visualized actin filaments directly using fluorescence microscopy. The negative control (i.e., actin filaments in the absence of formin proteins) appeared to be scattered individually (Figure 10A). After the addition of FH5 FH1FH2 (Figure 10B) or AFH1 FH1FH2 (Figure 10C), obvious actin bundles were observed. To examine further the bundling ability of FH5 FH1FH2 on actin filaments, we performed low-speed cosedimentation assays. At low speed, actin filaments and FH5 FH1FH2 were recovered mostly in the supernatant when used alone (Figure 10D, lanes 1 and 9). When these two components were mixed together, the percentage of actin in the pellet increased in a dose-dependent manner (Figures 10D, lanes 2 to 8, and 10E), indicating bundle formation. The actin bundling ability of FH5 FH1FH2 appeared to be comparable to that of AFH1 FH1FH2 (Figure 10E). Furthermore, to determine the apparent binding affinity between FH5 FH1FH2 and actin filaments, we also performed high-speed cosedimentation assays. After three independent experiments, the Kd value for FH5 FH1FH2 binding to the side of actin filaments was determined to be 0.18 ± 0.04 μM (means ± sd, n = 3; Figure 10F), which is close to the value for AFH1 FH1FH2 (~0.13 μM) (Michelot et al., 2005). Finally, we showed that FH5 FH2 is also capable of efficiently bundling actin filaments (see Supplemental Figure 14 online), an ability previously not found for the FH2 domain of AFH1 (Michelot et al., 2005).
Figure 10.
FH5 FH1FH2 Bundles Actin Filaments in Vitro.
(A) to (C) Direct visualization of actin filament bundles in the presence of FH5 FH1FH2 or AFH1 FH1FH2. Micrographs were taken from reactions with actin filaments alone (A) or actin plus 800 nM of FH5 FH1FH2 (B) or AFH1 FH1FH2 (C).
(D) Low-speed cosedimentation assays to determine the bundling activity of FH5 FH1FH2. Lane 1, actin alone; lanes 2 to 8, actin plus 0.05, 0.1, 0.2, 0.4, 0.8, 1.2, or 1.6 μM FH5 FH1FH2, respectively; lane 9, 1.2 μM FH5 FH1FH2 alone.
(E) Percentage of actin filaments recovered in the low-speed pellet as a function of the concentration of FH5 FH1FH2 (closed squares) or AFH1 FH1FH2 (open squares).
(F) FH5 FH1FH2 binds to actin filaments. High-speed cosedimentation assays were performed to determine the apparent binding affinity of FH5 FH1FH2 to actin filaments. Various amounts of phalloidin-stabilized actin filaments were mixed with 0.5 μM FH5 FH1FH2. Percentage of F-actin–bound FH5 FH1FH2 was plotted against the concentration of actin and fitted with a hyperbolic function. For this representative experiment, Kd = 0.15 μM.
[See online article for color version of this figure.]
FH5 Binds to and Bundles Microtubules
Recently, formins have been recognized as the prominent regulators of the microtubule cytoskeleton (Bartolini and Gundersen, 2010; Deeks et al., 2010; Li et al., 2010); At-FH4 and AFH14, in particular, were shown to interact with both F-actin and microtubules (Deeks et al., 2010; Li et al., 2010). To test whether FH5 binds to microtubules in vivo, we introduced the translationally fused construct of FH5 FH1FH2 (amino acids 825 to 1588)-RFP into onion epidermal cells, using green fluorescent protein (GFP)-MAP4 (Microtubule-associated protein 4) as the marker for the microtubule cytoskeleton (Marc et al., 1998). The FH5 FH1FH2-RFP signals colocalized with microtubule signals (Figures 11A to 11C), suggesting that FH5 FH1FH2 binds to microtubules in plant cells.
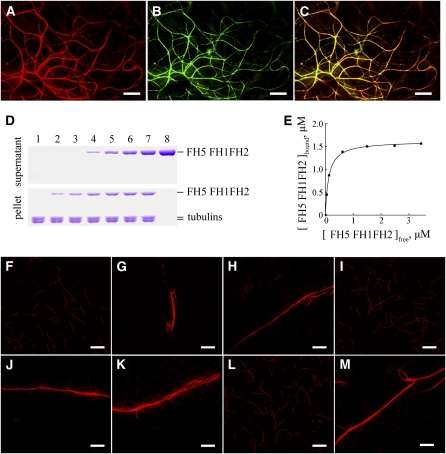
Figure 11.
Colocalization of FH5 FH1FH2-RFP with Microtubules in Vivo and Binding and Bundling of FH5 FH1FH2 to Microtubules in Vitro.
(A) to (C) Confocal images from onion epidermal cells expressing FH5 FH1FH2-RFP (A) and a microtubule marker, GFP-MAP4 (B). (C) shows merged signals from (A) and (B). Bars = 10 μm.
(D) Recombinant FH5 FH1FH2 protein is cosedimented with 1 μM taxol-stabilized microtubules. Recombinant FH5 FH1FH2 protein appeared mainly in the supernatant after centrifugation in the absence of microtubules and cosedimented with microtubules into the pellets. The concentration of FH5 FH1FH2 in lanes 1 to 8 was 0, 0.5, 1.0, 2.0, 3.0, 4.0, 5.0, and 5.0 μM, respectively.
(E) Increasing concentrations of FH5 FH1FH2 (0.5 to 5.0 μM) were incubated and cosedimented with 1.0 μM microtubules. The concentration of microtubule-bound FH5 FH1FH2 was plotted against the concentration of free FH5 FH1FH2 and fitted with a hyperbolic function. For this representative experiment, the Kd was 0.12 μM.
(F) to (M) Micrographs of rhodamine-conjugated microtubules in the absence or presence of formins. (F) Microtubules alone; (G) microtubules plus 3.0 μM FH5 FH2; (H) microtubules plus 5.0 μM FH5 FH2; (I) microtubules plus 5.0 μM denatured FH5 FH2; (J) microtubules plus 1.5 μM FH5 FH1FH2; (K) microtubules plus 2.5 μM FH5 FH1FH2; (L) microtubules plus 2.5 μM denatured FH5 FH1FH2; (M) microtubules plus 1.5 μM AFH14 FH1FH2. Bars = 20 μm.
To assess further the binding activity of FH5 FH1FH2 to microtubules, cosedimentation assays were performed by incubating taxol-stabilized microtubules with various concentrations of FH5 FH1FH2. In the absence of microtubules, very little FH5 FH1FH2 was detected in the pellet (Figure 11D, lane 8). By contrast, the amount of sedimented FH5 FH1FH2 increased as microtubules were added (Figure 11D, lanes 2 to 7), confirming that FH5 FH1FH2 directly binds to microtubules. To determine the binding affinity between FH5 FH1FH2 and microtubules, the amount of FH5 FH1FH2 in the pellet ([FH5 FH1FH2]bound) was plotted against the amount of FH5 FH1FH2 in the supernatant ([FH5 FH1FH2]free), and the resulting data were fitted with a hyperbolic function. In a representative experiment, the Kd value was determined to be 0.12 μM (Figure 11E). The mean Kd value for the binding between FH5 FH1FH2 and microtubules was 0.13 ± 0.02 μM (±sd, n = 3). Similarly, FH5 FH2 was also confirmed to cosediment with microtubules (see Supplemental Figure 15 online). These results suggest that, similar to the AFH14 protein (Li et al., 2010) (Figure 11M), FH5 binds to taxol-stabilized microtubules through its FH2 domain in vitro.
To examine further the effect of FH5 protein on microtubules, fluorescence microscopy was employed. In the absence of the recombinant FH5 FH2 protein, microtubules appeared to be scattered individually throughout the solution (Figure 11F). When adding the FH5 FH2, densely packed bundles appeared (Figure 11G). With an increasing amount of FH5 FH2 added, more high-order microtubule bundles formed (Figure 11H). By contrast, when we added denatured FH5 FH2, which had been boiled for 1 min, no microtubule bundling was observed (Figure 11I). Similar to FH5 FH2, FH5 FH1FH2 could also bundle microtubules; its bundling efficiency seemed to be higher than the former (Figures 11J to 11L). From these results we conclude that FH5 can bind to and bundle microtubules through its FH2 domain.
DISCUSSION
FH5 Modulates Rice Morphogenesis
Understanding the molecular basis of plant architecture is not only important for fundamental biology, but also useful for agricultural improvement of crops for higher yields. Plant morphogenesis is under tight control by a broad range of developmental and environmental signals (Wang and Li, 2008). In this study, we show the critical role of the type II formin FH5 in determining rice plant morphogenesis. Loss of function of FH5 in rmd mutants disrupts proper morphogenesis in both rice seedlings and mature plants. Thus, this report establishes that the mutation of a single angiosperm formin can have dramatic morphogenetic defects. Furthermore, in the companion article, Yang et al. (2011) characterized BENT UPPERMOST INTERNODE1, which also encodes FH5 and found similar results.
Formins are known to be actin-nucleating proteins that control the rate-limiting step of actin polymerization (Wasserman, 1998; Pollard et al., 2000; Sawin, 2002), but little is known about the biological role of formins in plant morphogenesis. Arabidopsis and rice genomes encode 21 and 16 putative formins, respectively (Cvrcková et al., 2004). The fewer number of formins in rice may be due to fewer gene duplications in its genome during evolution. Microarray data indicated that rice formins are expressed in most of the tissues throughout the life cycle (http://bioinformatics.med.yale.edu/riceatlas/). It is possible that they participate in the generation of cytoskeleton in a redundant manner or that each of these formins has a distinct role in the formation of cytoskeleton. Supporting the latter scenario, three formins from fission yeast have been shown to be responsible for the formation of distinct actin arrays (Petersen et al., 1998; Feierbach and Chang, 2001; Kovar et al., 2003).
It has been reported that Arabidopsis AFH1, AFH3, At FH4, At FH5, and At FH8 function in polarized growth of pollen tubes or root hairs or in embryo development (Cheung and Wu, 2004; Deeks et al., 2005; Ingouff et al., 2005; Michelot et al., 2005; Yi et al., 2005; Ye et al., 2009; Cheung et al., 2010). Among these proteins, only At-FH5 caused severe morphological defect when it was mutated (Ingouff et al., 2005). At-FH6 encodes a plasma membrane–associated formin protein, which can partially substitute for the function of its homologs in yeast and nematode cells (Favery et al., 2004).
In this study, we show a dramatic effect of the type II rice formin FH5 on the organization of microfilaments and microtubules and plant morphogenesis, suggesting the essential role of type II formins in plants. In agreement with this, knockdown of moss type II formins causes stunted plants, and knockout of AFH14 leads to abnormal meiosis (Vidali et al., 2009; Li et al., 2010).
FH5 Regulates Microfilament Organization in Rice
The cytoskeleton consists of microtubule- and microfilament-based systems, which together form a structurally rigid lattice that functions as a supporting scaffold in the organism. Instead of having longitudinal microfilament cables like those in wild-type cells, the rmd mutants contain more transverse microfilament cables, indicating that FH5/RMD is a key regulator in the formation of actin cytoskeleton in rice cells. Like other eukaryotic formins, FH5 has the conserved FH1 and FH2 domains. In this study, we show that FH5 can nucleate actin assembly from free or profilin-bound G-actin monomers, in a way similar to AFH1, AFH3, At-FH5, and At-FH8 proteins (Ingouff et al., 2005; Michelot et al., 2005; Yi et al., 2005; Ye et al., 2009). Moreover, our in vitro biochemical assays suggest that the FH5 FH2 domain itself can induce actin assembly, which is different from AFH3 and At-FH5, whose actin nucleating activity depends on the presence of the FH1 domain (Ingouff et al., 2005; Ye et al., 2009). Similar to the FH2 domain in AFH1 (Michelot et al., 2005; Yi et al., 2005; Ye et al., 2009), FH5 FH2 can also inhibit elongation of the actin filaments by capping the barbed ends of F-actin filaments.
In the 345–amino acid FH5 FH1 domain, there are 190 Pro residues, which form 22 polyproline stretches (i.e., the longest polyproline stretches found in rice formins) (see Supplemental Figure 6 online; Cvrcková et al., 2004). The conserved polyproline stretches within the formin FH1 domains have been shown to enhance filament elongation (Sagot et al., 2002; Kovar and Pollard, 2004; Romero et al., 2004, 2007; Paul and Pollard, 2008). Our time-lapse fluorescence microscopy analysis showed that the partial Pro residues in the FH5 FH1 domain are involved in the promotion of filament elongation in the presence of actin/profilin, suggesting that the combination of FH1 and FH2 in FH5 is required for formin-mediated microfilament organization and dynamics in plant cells. We also showed that FH5 FH1FH2’s activity in concentrating multiple actin/profilin complexes near the end of the filament is close to that of the well-characterized Arabidopsis AFH1 (Michelot et al., 2005), yet lower than those of the moss type II formins (Vidali et al., 2009). Consistent with the subcellular phenotype observed in rmd mutants, which have reduced numbers of longitudinal actin cables, the FH2 domain of FH5/RMD was shown in our study to be able to bundle actin filaments, an activity shared with AFH1 and Bnr1 (Michelot et al., 2005; Moseley and Goode, 2005).
FH5 Affects the Formation of Microtubule Arrays in Rice
Plant cells have a highly dynamic microtubule system that is organized into different arrays and essential for many important cellular processes. In response to environmental and developmental stimuli, the microtubule system can be reoriented, such as undergoing clockwise or counterclockwise rotation in plant cells (Lloyd and Chan, 2004; Chan et al., 2007). In plants, components of the microtubule system have been shown to be critical for plant architecture (Thitamadee et al., 2002; Twell et al., 2002; Nakajima et al., 2004; Sedbrook, 2004; Sunohara et al., 2009). For example, the rice TWISTED DWARF1 (TID1) gene encodes an α-tubulin that influences plant morphology; a dominant mutation of TID1 caused disrupted cellular microtubule arrays, severe dwarfism, and right helical growth (Sunohara et al., 2009).
In this study, we have shown that FH5/RMD affects microtubule organization. Microtubule arrays are transversely aligned along the long axis in wild-type cortical cells, whereas rmd mutant cells have discontinuous and fragmented microtubules. Transverse microtubules were thought to influence plant elongation growth and development by determining the length and orientation of cellulose microfibrils, essential contributors to the mechanical properties of the plant cell wall (Wasteneys, 2004). Consistent with this notion, rmd mutants display abnormal cell growth, in particular, reduced elongation, irregular cell shape, and defective cell walls. How FH5/RMD affects cell wall development remains to be uncovered.
We have also shown that FH5 FH1FH2 is colocalized with microtubules in vivo and that recombinant FH5 FH2 binds to and bundles microtubules in vitro. This microtubule bundling activity was also found for other formins, including AFH14, mDia1, mDia2, Formin1-Ib, INF1, and Capu (Rosales-Nieves et al., 2006; Zhou et al., 2006; Bartolini et al., 2008; Young et al., 2008; Li et al., 2010), suggesting that the FH2 domain may have a conserved function in some formins in coordinating the organization of microtubules and microfilaments in diverse species. Consistent with this view, several lines of evidence suggested that many cytoskeleton dependent biological processes require the coordination of actin filaments and microtubules (Fu et al., 2005; Crowell et al., 2009; Deeks et al., 2010; Li et al., 2010). How FH5 modulates cellular processes by changing the organization of microfilaments and microtubules remains to be elucidated. Based on its binding activity on both actin filaments and microtubules, it is most likely that FH5 directly affects the organization of both microfilaments and microtubules. At this point we cannot exclude the possibility that FH5 indirectly affects microtubule organization through its impact on the orientation of F-actin or that it indirectly affects microfilament organization through its direct role on microtubule orientation.
Most of the plant formins identified to date have been shown to be microfilament regulators (Ingouff et al., 2005; Michelot et al., 2005; Yi et al., 2005; Vidali et al., 2009; Ye et al., 2009; Blanchoin and Staiger, 2010; Deeks et al., 2010; Li et al., 2010). However, a new plant-specific domain (the GOE domain) in the type I formin At-FH4 was shown to bind directly to microtubules. Overexpression analysis revealed the accumulation of At-FH4 at the endoplasmic reticulum membrane and its coalignment with microtubules, providing the first evidence that a plant type I formin likely mediates the interaction between membranes and both major cytoskeletal networks (Deeks et al., 2010). FH5 shares strong sequence similarities with Arabidopsis AFH14 (Cvrcková et al., 2004), a type II formin capable of linking microtubules and microfilaments (Li et al., 2010). The recombinant FH1FH2 fragment of AFH14 can directly bind to purified microtubules and microfilaments and promotes the formation of microtubule or microfilament bundles, suggesting that this protein may function in recruiting microfilaments to microtubules (Li et al., 2010).
The PTEN Domain Mediates the Association of FH5 with Chloroplasts
As an organelle unique to plants, the chloroplast is involved in a variety of processes crucial for plant growth and development. It has been postulated that in plants the intracellular mobility and positioning of chloroplasts depend on microfilament- and microtubule-based cytoskeletons (Dong et al., 1998; Sato et al., 2001; Shimmen and Yokota, 2004; Wada and Suetsugu, 2004; Chuong et al., 2006; Paves and Truve, 2007; Reisen and Hanson, 2007; Takagi et al., 2009). Close links of chloroplasts with actin filaments were shown in vascular plants and algae (Takagi, 2003). Actin filaments are attached to the surface of chloroplasts and regulate chloroplast positioning and plastid stromule movement (Jouhet and Gray, 2009). Chloroplasts also display microtubule-mediated subcellular distribution; for example, the aggregation of chloroplasts in some C4 plants is dependent on microtubules (Chuong et al., 2006), and in the moss Physcomitrella, plastid movement in response to blue light is modulated by both microfilaments and microtubules (Sato et al., 2001). However, the molecular mechanism underlying the association of chloroplasts with the cytoskeleton remains unknown.
Using immunostaining analysis in this study, we observed the localization of FH5 at the chloroplast outer surface. This result suggests that FH5 may act as a key mediator between chloroplasts and the cytoskeletal system, although the physiological significance of this interaction remains unknown. We propose that this interaction may be involved, at least in part, in modulating the intracellular positioning and mobility of chloroplasts. Furthermore, we showed that the PTEN domain confers the distribution of FH5 to the chloroplast surface, supporting the view that the conserved PTEN-related domain of plant type II formins plays a structural rather than catalytic role (Cvrcková et al., 2004). Consistent with this, the N-terminal PTEN-like domain of moss type II formins was shown to be responsible for mediating apical localization of the formin protein and the formation of apical filamentous actin required for polarized growth (Vidali et al., 2009). Interestingly, our previous results from immunological assays of AFH14 and fluorescence microscopy analysis of AFH14-GFP revealed that, in addition to its localization to the cytoplasm, the preprophase band, the spindle, and the phragmoplast during interphase in dividing cells, AFH14, a type II formin, is also associated with dots of undetermined identity in the cytoplasm (Li et al., 2010). Moreover, the AtFH5-GFP fusion protein was also found to localize to unidentified dot-like structures in the cytoplasm, in addition to its localization to the cell plate (Ingouff et al., 2005). Whether AFH14 and At-FH5 are also physically associated with chloroplasts remains to be determined. Further investigation of the rmd mutants may provide more insights into the crosstalk between chloroplasts and the cytoskeleton in plants.
In conclusion, in this study, we provide biochemical and genetic evidence to demonstrate that a type II formin protein regulates rice morphogenesis through its influence over the formation of actin cytoskeleton and the organization of microtubules. FH5 is able to nucleate actin filaments de novo, cap the barbed end of F-actin, stabilize F-actin, and bundle microtubules. Mutations in FH5/RMD lead to aberrant microfilament and microtubule networks, which result in abnormal cell elongation and altered plant morphology. This work provides new insight into the physiological and developmental role of type II formins in higher plants.
METHODS
Mutant Materials and Growth Conditions
All plants were grown in the paddy field of Shanghai Jiao Tong University. The F2 mapping population was generated from a cross between the rmd-1 mutant in the 9522 background (japonica) and Guan Lu Ai (indica) (Chu et al., 2005; Liu et al., 2005; Chen et al., 2006; Wang et al., 2006).
Characterization of the Mutant Phenotype
Plants were photographed with an Olympus e-410 digital camera. For cross section observations, the materials were fixed as described by Li et al. (2006), and semithin (4 μm) sections were made using an Ultracut E ultramicrotome (Leica Microsystems) and stained with 1% safranin (Li et al., 2006).
Field Emission Scanning Electron Microscopy
The roots of 14-d-old rice seedlings were cut into thin strips. Cell wall preparation was performed using the method by Sugimoto et al. (2000). Then, the epidermal cell wall surface was observed with a field emission scanning electron microscope (FEI SIRION 200) in the Instrumental Analysis Center of Shanghai Jiao Tong University, using the method described by Wanner and Formanek (1995).
Construction of RNAi Vector and Rice Transformation
A gene-specific DNA fragment from FH5/RMD (amino acids 1486 to 2226) was amplified from the Nipponbare BAC clone, OSJNBap004275, which contains FH5/Os07g0596300, using the PCR primer pair RNAiF (KpnI and SpeI) and RNAiR (BamHI and SacI) (see Supplemental Table 3 online). The PCR product was then digested with SpeI and SacI and KpnI and BamHI, respectively, and the two fragments were individually cloned into the digested vector pTCK303, which contains a maize (Zea mays) ubiquitin promoter (kindly provided by Kong Chong) (Wang et al., 2004) to make the final construct, plasmid pTCK303:FHRNAi. This construct was then introduced into calli induced from the wild-type rice seeds by Agrobacterium tumifaciens transformation (Hiei et al., 1997).
Fluorescence Microscopy of Actin Filaments and F-Actin Quantification
F-actin was stained using the glycerol method (Olyslaegers and Verbelen, 1998). Roots from 5-d-old seedlings and fresh mature pollen grains were incubated in PEM buffer (100 mM PIPES, 10 mM EGTA, 5 mM MgSO4, and 0.3 M mannitol, pH 6.9) that contains 2% (w/v) glycerol (Sigma-Aldrich) and 6.6 μM Alexa Fluor 488-phalloidin staining (Invitrogen). After a 35-min incubation, root tips were observed in 50% glycerol (Sigma-Aldrich) with a Leica TCS SP5 confocal laser scanning microscope equipped with a ×63 1.46–numerical aperture HC PLANs objective. To quantify F-actin in root cells and pollen grains, images from wild-type, rmd, and RNAi transgenic plants were used to determine the average pixel intensity with LAS AF software described by Ye et al. (2009) and Thomas et al. (2006). Three independent lines for each genotype were observed and at least 10 images for each line.
Preparation of FH5-Specific Polyclonal Antibody
A FH5/RMD-specific fragment (amino acids 1600 to 2127) was amplified with primers FHatF (NdeI) and FHatR (XhoI) (see Supplemental Table 3 online), digested with NdeI and XhoI, and cloned into the pET-32a(+) vector digested with the same enzymes (Novagen). The recombinant protein was expressed in Escherichia coli strain BL21DE3 (Novagen) and used as an antigen to raise polyclonal FH5 antibodies in rabbit as described by Huang et al. (2003). The antibody was purified as described by Ritter (1991).
Chloroplast Purification and Immunoblotting
Chloroplasts were isolated from the leaves of 14-d-old rice seedlings using the method described by Joyard et al. (1990). Nuclear extracts were produced as the described by Zhang et al. (2010). Total cellular proteins were isolated according to the method used by Huang et al. (2003). Subsequently, immunoblotting was performed using the purified FH5 antibody or polyclonal antibody against the large subunit of Rubisco from tobacco (Nicotiana tabacum; kindly provided by Genyun Chen) (Chen et al., 2010) as the primary antibody (1:200 dilution), horseradish peroxidase–labeled goat anti-rabbit IgG(H+L) (diluted 1:500) as the secondary antibody, BeyoECL plus protein gel blotting detection reagents (Shanghai), and methods as previously described by Fan et al. (2004). Total proteins were prepared as described by Dai and Xue (2010).
Immunostaining of Microtubules and FH5
The leaves and roots of 5-d-old rice seedlings were cut into thin strips and pollen grains were treated with the method by Sugimoto et al. (2000). Specimens were observed in 50% glycerol with a Leica TCS SP5 confocal laser scanning microscope equipped with an ×63 1.46–numerical aperture HC PLANs objective. For microtubule staining, anti-β tubulin (Sigma-Aldrich) was used as the primary antibody (diluted 1:200) and fluorescein isothiocyanate anti-mouse IgG (Jackson) as the secondary antibody (diluted 1:200). For FH5 staining, the purified FH5 antibody was used as the primary antibody (diluted 1:200), and fluorescein isothiocyanate anti-rabbit IgG (Jackson) was used as the secondary antibody (diluted 1:200). The staging of anther development is referred to by Zhang and Wilson (2009). Three independent lines for each genotype were observed.
Agrobacterium Infiltration of Tobacco Leaves
The DNA fragment encoding PTEN (from amino acids 200 to 337) was amplified by the primers 13PTENf (NcoI) and 13PTENr (see Supplemental Table 3 online). The RFP DNA fragment was amplified by the primers 13RFPf and 13RFPr (PmacI) from the pBSK-RFP vector (Mette et al., 2002; Zhang et al., 2010) (see Supplemental Table 3 online). PTEN-RFP was constructed using the primers 13PTENf (NcoI) and 13RFPr (PmacI) and templates of cDNA fragments of FH5-PTEN and RFP (see Supplemental Table 3 online), digested with NcoI and PmacI, and cloned into pCAMBIA1301 (CAMBIA; hygromycin resistance, kindly provided by Richard Jefferson). The Agrobacterium strain EHA105 containing 1301-35s-PTEN-RFP was transformed into tobacco using the method described by He et al. (2008). Tobacco leaf cells were observed in 50% glycerol with a Leica TCS SP5 confocal laser scanning microscope.
RT-PCR, qRT-PCR, and Promoter Fusions
Total RNA was isolated from roots, stems, leaves, and mature flowers using Trizol reagent (Generay) as instructed by the supplier. After treatment with DNase (Promega), 0.3 μg RNA was employed to synthesize the oligo(dT) primed first-strand cDNA using the ReverTra Ace-α-First Strand cDNA synthesis kit (TOYOBO).
qRT-PCR analysis was performed using SYBR Premix EX Taq (TaKaRa) on a Rotor-Gene RG3000A detection system (Corbett Research) as described by Zhang et al. (2010) with the exception of the annealing temperature at 58°C when using the primers RMDRTF and RMDRTR (see Supplemental Table 3 online). RT-PCR analysis was performed as qRT-PCR without SYBR Green I. Three biological replicates were used, and each with three technical repeats.
A 3-kb fragment containing the upstream region of the FH5/RMD gene was amplified from the Nipponbare BAC clone (OSJNBap004275) using the primer pair RMDPF (HindIII) and RMDPR (NcoI) (see Supplemental Table 3 online). The amplified FH5 promoter fragment was digested by HindIII and NcoI and cloned into pCAMBIA1301 (CAMBIA; hygromycin resistance, kindly provided by Richard Jefferson), which expresses the GUS reporter gene. The pFH5-GUS construct was introduced into calli induced from the wild-type rice seeds by Agrobacterium transformation. Seedlings, roots, leaves, and flowers from heterozygous spikelets of transgenic lines were stained as described by Willemsen et al. (1998) and photographed using a phase-contrast microscope (Leica DM2500).
Protein Production
The DNA fragment encoding FH5 FH2 (from 3499 to 4881 with no Pro-rich region) was amplified from a cDNA clone (AK120222) with the primer pair VitroFH2F (NcoI) and VitroFH2R (HindIII; see Supplemental Table 3 online). A DNA fragment encoding FH5 FH1FH2 (from 3292 to 4881 with six Pro-rich regions) was amplified from a cDNA clone (AK120222) with the primer pair FH1aF (NcoI) and FH1aR (HindIII) (see Supplemental Table 3 online). These two fragments were cloned with the same sites in frame with 6x His in the pET-32a(+) vector (Novagen). The resulting expression vector was confirmed by DNA sequencing.
The error-free FH5-FH2-pET-32a(+) and FH5-FH1FH2-pET-32a(+) constructs were expressed in E. coli strain Origami B (DE3) (Novagen). Cells were grown at 37°C to an OD600 of 0.7 and treated with 0.5 mM isopropylthio-β-galactoside at 20°C overnight to induce protein expression. Cultures were collected by centrifugation and resuspended in binding buffer (400 mM NaCl and 40 mM PBS, pH 8.0). This was followed by purification using a Ni-NTA His bind resin following the protocol in the manufacturer’s manual (Novagen). The purified proteins were dialyzed overnight against buffer TK (5 mM Tris, 50 mM KCl, 0.5 mM DTT, and 0.5 mM EDTA) and frozen immediately in liquid nitrogen. Protein concentrations were determined using the Bradford reagent (Bio-Rad) and BSA as a standard.
Actin was isolated from rabbit skeletal muscle acetone powder using the method described by Pardee and Spudich (1982) and labeled on Cys-374 with pyrene iodoacetamide (Pollard, 1983) and Oregon-Green 488 iodoacetamide (Kuhn and Pollard, 2005). AFH1 FH1FH2 and human profilin 1 were expressed and purified as described (Fedorov et al., 1994; Michelot et al., 2005). The coding region of lily (Lilium longiflorum) profilin 1 (LlPRO1) was amplified by RT-PCR from pollen RNAs using primer pairs LlPRO1F (BamHI) and LlPRO1R (SacI) (see Supplemental Table 3 online) and subcloned in frame with 6x His into pET-30a(+) with the same sites. Lily profilin 1 was expressed and purified using a Ni-NTA His bind resin.
Actin Nucleation Assay
Monomeric actin or actin/profilin complex (10% pyrene-labeled) was incubated with different concentrations of recombinant formins at room temperature for 5 min. Pyrene fluorescence was monitored after the supplementation of one-tenth volume of 10× KMEI buffer (500 mM KCl, 10 mM MgCl2, 10 mM EGTA, and 100 mM imidazole-HCl, pH 7.0) with a fluorescence spectrophotometer (Fluoro Max-2; Instruments SA). The nucleation efficiency of FH5 FH2 and FH5 FH1FH2 was calculated according to Blanchoin et al. (2000b) using a rate constant of association 10 μM−1 s−1 for FH5 FH1FH2 and 1.3 μM−1 s−1 for FH5 FH2 (Michelot et al., 2005).
Time-Lapse Fluorescence Microscopy
The effect of FH5 on actin filament elongation was measured as described by Michelot et al. (2005). Glass flow cells, with a capacity of ~10 μL, were prepared each day according to Kuhn and Pollard (2005). The chambers were coated with a mixture of 10 nM NEM-myosin alone or with FH5 FH1FH2 (200 nM), FH5 FH2 (200 nM), or AFH1 FH1FH2 (200 nM) for 2 min and washed with 1% BSA in fluorescence buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 100 mM DTT, 0.2 mM ATP, 15 mM glucose, 20 μg/mL catalase, 100 μg/mL glucose oxidase, and 0.5% methylcellulose). A mixture of Oregon-Green–labeled actin (1 μM, 75% labeled) and profilin (2 μM) in fluorescence buffer was introduced into the flow cells. Actin filaments were observed under an Observer Z1 microscope (Carl Zeiss) equipped with an alphaPlanApo ×100/1.46–numerical aperture oil objective. Actin filament elongation was photographed at 10-s intervals for 20 min using an AxioCam Hsm camera and AxioVision software. Exposure time was 900 ms. To calculate the elongation rate, actin filament lengths were measured with ImageJ software (http://rsb.info.nih.gov/ij). Rates were converted from μm/s to subunits/s using 333 actin monomers per micrometer.
Actin Filament Elongation Assay to Determine the Affinity of FH5 FH1FH2 and FH5 FH2 to the Barbed End of Actin Filament
This assay was performed as described by Michelot et al. (2005). Freshly prepared actin filament seeds (0.8 μM) were incubated with different concentrations of FH5 proteins at room temperature for 5 min. Actin elongation at the barbed end of actin filaments was initiated by adding 1 μM G-actin (10% pyrene labeled) saturated with 4 μM human profilin 1 and one-tenth volume of 10× KMEI buffer. The change in pyrene fluorescence accompanying actin polymerization was monitored after actin elongation was initiated. The Kd value for FH5 binding to the barbed end of actin filaments was determined by fitting the initial rate of actin filament elongation against the concentration of FH5 according to the method described by Michelot et al. (2005).
Dynamics of Actin Filament Depolymerization
Dilution assays were performed to examine the effect of FH5 on F-actin depolymerization as described by Ingouff et al. (2005). Five micromolar preformed F-actin (80% pyrene-labeled) was incubated with different concentrations of recombinant FH5 FH1FH2 or FH5 FH2 at room temperature for 5 min, before the solution was diluted 25-fold into G buffer (2 mM Tris-HCl, pH 8.0, 0.1 mM CaCl2, 0.2 mM ATP, and 0.5 mM DTT). The decrease of pyrene fluorescence intensity accompanying actin depolymerization was monitored after dilution.
Low-Speed and High-Speed Cosedimentation Assays
Low-speed cosedimentation assays were performed to determine the bundling activity of FH5 as described by Michelot et al. (2005). All proteins were preclarified at 200,000g before the experiment. G-actin (3.0 μM) alone or G-actin with FH5 FH1FH2 or FH5 FH2 was incubated in 1× KMEI buffer for 30 min at room temperature. The samples were centrifuged at 13,500g at 4°C for 30 min, and the resulting supernatants and pellets were separated by 12% SDS-PAGE. The gels were stained with Coomassie Brilliant Blue R 250 (Sigma-Aldrich), and the amount of actin in the pellets or supernatants was quantified using Quantity One v4.6.5 software (Bio-Rad).
High-speed cosedimentation assays were employed to determine the F-actin binding activity of FH5 FH1FH2 as described by Khurana et al. (2010). Increasing amounts of phalloidin-stabilized F-actin (0, 0.1, 0.15, 0.25, 0.5, 0.75, 1.0, 1.5, and 2.0 μM) were incubated with 0.5 μM FH5 FH1FH2 for 30 min at room temperature. The samples were centrifuged at 200,000g for 1 h at 4°C. To determine the Kd value, the percentage of F-actin–bound FH5 FH1FH2 as a function of actin concentration was fitted to a hyperbolic function using Prism 5 software (GraphPad Software).
Fluorescence Microscopy Visualization of Actin Filaments and Bundles
To visualize the effect of FH5 and AFH1 proteins on the generation of actin filaments during nucleation, actin (3.0 μM) alone or together with 100 nM FH5 FH1FH2, FH5 FH2, or AFH1 FH1FH2 was incubated with 1× KMEI buffer at room temperature for 30 min and labeled with an equimolar amount of Alexa 488–phalloidin (Molecular Probes) during polymerization. In the experiment to test the bundling activity of FH5 FH1FH2 and FH5 FH2, 3.0 μM prepolymerized F-actin was incubated with different concentrations of FH5 proteins at room temperature for 30 min. All of the polymerized F-actin was diluted to 50 nM with 1× KMEI buffer, and 1 μL of the diluted sample was added to a 22 × 22-mm cover slip coated with poly-Lys (0.01%). F-actin was observed using a microscope (Carl Zeiss 200M) equipped with a ×100/1.5–numerical aperture Planapo objective. Digital images were collected with an Axio CamMR charge-coupled device camera using Axiovision software.
Particle Bombardment–Mediated Transient Protein Expression in Onion Epidermis
The RFP cDNA was amplified from the pBSK-RFP vector (Mette et al., 2002; Zhang et al., 2010) with the primer pair RFPF (KpnI) and RFPR (BamHI). The FH1FH2 cDNA (from 2224 to 4881) was amplified from the cDNA clone (AK120222) and the Nipponbare BAC clone (OSJNBap004275) with the primer pair VivoFH1FH2F (NcoI) and VivoFH1FH2R (KpnI) (see Supplemental Table 3 online). These two fragments were subcloned into the NcoI-BamHI–digested pRTL2 vector (Restrepo et al., 1990), which contains a dual cauliflower mosaic virus 35S promoter, to generate pRTL2-FH1FH2-RFP. Transient expression of the pRTL2-FH1FH2-RFP fusion in onion epidermis was performed as previously described (Collings et al., 2000) using a helium biolistic device (Bio-Rad PDS-1000). After 18 h incubation, the samples were observed with a confocal laser microscope (Leica TCS SP5).
Microtubule Cosedimentation Assays
Porcine brain tubulin and FH5 FH1FH2 or FH5 FH2 proteins were centrifuged at 60,000g at 4°C for 30 min before use. The binding reaction contains 1.0 μM preformed taxol-stabilized microtubules and various concentrations of FH5 FH1FH2 or FH5 FH2 proteins in 100 μL PEM buffer (100 mM PIPES, pH 6.9, 1mM EGTA, and 1 mM MgCl2). After 30 min incubation at room temperature, the samples were centrifuged at 25,000g for 30 min at 25°C. The pellet was resuspended with one volume of SDS loading buffer (v/v). The samples were applied onto 8% SDS-PAGE gels, which were later stained with Coomassie Brilliant Blue R 250. The amount of FH5 proteins bound to microtubules was determined by gel scanning with an Alpha_2200 scanner. The Kd value for the binding of FH5 FH1FH2 to microtubules was calculated by fitting the data of bound protein versus free protein to a hyperbolic function using Prism 5 software (GraphPad Software).
Microtubule Bundling Assays
Samples containing 1.0 μM preformed taxol-stabilized rhodamine-conjugated microtubules were incubated with FH5 proteins at room temperature for 30 min. Aliquots (1 μL) of the samples were placed onto a slide and observed using a confocal laser scanning microscope (Olympus FV-300).
Accession Numbers
Sequence data from this article for the cDNA and genomic DNA of FH5/RMD can be found in the GenBank/EMBL data libraries under accession numbers AK120222, Os07g0596300, and OSJNBap004275, respectively. The full-length FH5/RMD protein sequence can be found in the NCBI data library with the accession number Q84ZL0. Other sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: lily profilin 1 (AF200184.1), human profilin 1 (BC057828), AFH14 (At1g31810), and AFH1 (AT2G43800).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. rmd-2 Mutants Display Morphological Defects Similar to rmd-1.
Supplemental Figure 2. Analysis of the Height of Wild-Type, rmd-1, and rmd-2 Plants, Leaves, Roots, and Flower Filaments.
Supplemental Figure 3. Defects in Cell Development and Microfilament and Microtubule Organization in rmd-2 and Ri-s Lines.
Supplemental Figure 4. Actin Bundles from 50 Slices of the Stained Root Cortical Cells from 7-d-Old Plants.
Supplemental Figure 5. Molecular Identification of RMD.
Supplemental Figure 6. FH5/RMD Amino Acid Sequence.
Supplemental Figure 7. FH5 Is Expressed throughout the Plant.
Supplemental Figure 8. Specificity Analysis of the Purified FH5 Polyclonal Antibody.
Supplemental Figure 9. Subcellular Localization of FH5 in Rice Leaf Cells.
Supplemental Figure 10. Controls for FH5 PTEN-RFP Localization in Tobacco Cells.
Supplemental Figure 11. Analysis of FH5 PTEN-RFP Localization in Tobacco Cells.
Supplemental Figure 12. Generation of Recombinant FH5 FH2 and FH5 FH1FH2 Proteins.
Supplemental Figure 13. Recombinant FH2 Domain of FH5 Cannot Nucleate Actin Filament from Actin/Profilin Complexes in Vitro.
Supplemental Figure 14. Recombinant FH5 FH2 Bundles Actin Filaments in Vitro.
Supplemental Figure 15. Binding of FH5 FH2 to Microtubules in Vitro.
Supplemental Table 1. Primers Used for Mapping.
Supplemental Table 2. FH5/RMD Sequencing Primers.
Supplemental Table 3. Primers for Constructing Vectors.
Supplemental Movie 1. Time Lapse of Oregon-Green-Actin/Profilin Polymerization.
Supplemental Movie 2. Time Llapse of the Effect of 200 nM FH5 FH1FH2 on Oregon-Green-Actin/Profilin Polymerization.
Supplemental Movie 3. Time Lapse of the Effect of 200 nM FH5 FH2 on Oregon-Green-Actin/Profilin Polymerization.
Supplemental Movie 4. Time Lapse of the Effect of 200 nM AFH1 FH1FH2 on Oregon-Green-Actin/Profilin Polymerization.
Acknowledgments
We thank the anonymous reviewers for very helpful comments and Zhijing Luo and Mingjiao Chen for performing mutant screens and generation of F2 populations, Ming Yuan and Tonglin Mao for providing purified tubulin, Bo Liu for helpful comments and discussion, Bin Han and the Rice Genome Resource Center for providing BAC and cDNA clones, Kong Chong for the pTCK303 vector, Shanjin Huang for the AFH1 vector and the help with actin quantification, Genyun Chen for providing the Rubisco large subunit polyclonal antibody, and Richard Jefferson for providing the pCAMBIA1301 vector. This work was supported by Funds from the National Basic Research Program of China (2007CB108700 and 2009CB941500), the National “863” High-Tech Project (2006AA10A102 and 2007AA10Z112), the National Natural Science Foundation of China (30725022, 30830014, 90717109, and 30970174), the Chinese Transgenic Project (2009ZX08009-108B and 2009ZX08009-059B), and the Shanghai Leading Academic Discipline Project (B205).
References
- Andrianantoandro E., Pollard T.D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24: 13–23 [DOI] [PubMed] [Google Scholar]
- Bartolini F., Gundersen G.G. (2010). Formins and microtubules. Biochim. Biophys. Acta 1803: 164–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F., Moseley J.B., Schmoranzer J., Cassimeris L., Goode B.L., Gundersen G.G. (2008). The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J. Cell Biol. 181: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R., Chang F. (2007). Shaping the actin cytoskeleton using microtubule tips. Curr. Opin. Cell Biol. 19: 88–94 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Amann K.J., Higgs H.N., Marchand J.B., Kaiser D.A., Pollard T.D. (2000b). Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404: 1007–1011 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T.D., Mullins R.D. (2000a). Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 10: 1273–1282 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Staiger C.J. (2010). Plant formins: Diverse isoforms and unique molecular mechanism. Biochim. Biophys. Acta 1803: 201–206 [DOI] [PubMed] [Google Scholar]
- Casella J.F., Flanagan M.D., Lin S. (1981). Cytochalasin D inhibits actin polymerization and induces depolymerization of actin filaments formed during platelet shape change. Nature 293: 302–305 [DOI] [PubMed] [Google Scholar]
- Chan J., Calder G., Fox S., Lloyd C. (2007). Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat. Cell Biol. 9: 171–175 [DOI] [PubMed] [Google Scholar]
- Chang F., Drubin D., Nurse P. (1997). cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 137: 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry F., Guérin C., von Witsch M., Blanchoin L., Staiger C.J. (2007). Identification of Arabidopsis cyclase-associated protein 1 as the first nucleotide exchange factor for plant actin. Mol. Biol. Cell 18: 3002–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang P., Mi H.L., Chen G.Y., Xu D.Q. (2010). Reversible association of ribulose-1, 5-bisphosphate carboxylase/oxygenase activase with the thylakoid membrane depends upon the ATP level and pH in rice without heat stress. J. Exp. Bot. 61: 2939–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chu H.W., Yuan Z., Pan A.H., Liang W.Q., Huang H., Shen M.S., Zhang D.B., Chen L. (2006). Isolation and genetic analysis for rice mutants treated with 60Co γ-ray. J. Xiamen Univ. 45: 82–85 [Google Scholar]
- Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D.B., Rebowski G., Lappalainen P., Pollard T.D., Dominguez R. (2008). Leiomodin is an actin filament nucleator in muscle cells. Science 320: 239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Niroomand S., Zou Y., Wu H.M. (2010). A transmembrane formin nucleates subapical actin assembly and controls tip-focused growth in pollen tubes. Proc. Natl. Acad. Sci. USA 107: 16390–16395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2004). Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.W., et al. (2005). Genetic analysis and mapping of the rice leafy-hull mutant Oslh. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 31: 594–598 [PubMed] [Google Scholar]
- Chuong S.D., Franceschi V.R., Edwards G.E. (2006). The cytoskeleton maintains organelle partitioning required for single-cell C4 photosynthesis in Chenopodiaceae species. Plant Cell 18: 2207–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings D.A., Carter C.N., Rink J.C., Scott A.C., Wyatt S.E., Allen N.S. (2000). Plant nuclei can contain extensive grooves and invaginations. Plant Cell 12: 2425–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.A., Walker S.B., Pollard T.D. (1983). Pyrene actin: Documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 4: 253–262 [DOI] [PubMed] [Google Scholar]
- Crowell E.F., Bischoff V., Desprez T., Rolland A., Stierhof Y.D., Schumacher K., Gonneau M., Höfte H., Vernhettes S. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrcková F., Novotný M., Pícková D., Zárský V. (2004). Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genomics 5: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Xue H.W. (2010). Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 29: 1916–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks M.J., Cvrcková F., Machesky L.M., Mikitová V., Ketelaar T., Zársky V., Davies B., Hussey P.J. (2005). Arabidopsis group Ie formins localize to specific cell membrane domains, interact with actin-binding proteins and cause defects in cell expansion upon aberrant expression. New Phytol. 168: 529–540 [DOI] [PubMed] [Google Scholar]
- Deeks M.J., Fendrych M., Smertenko A., Bell K.S., Oparka K., Cvrcková F., Zársky V., Hussey P.J. (2010). The plant formin AtFH4 interacts with both actin and microtubules, and contains a newly identified microtubule-binding domain. J. Cell Sci. 123: 1209–1215 [DOI] [PubMed] [Google Scholar]
- Dong X.J., Nagai R., Takagi S. (1998). Microfilaments anchor chloroplasts along the outer periclinal wall in Vallisneria epidermal cells through cooperation of P-FR and photosynthesis. Plant Cell Physiol. 39: 1299–1306 [Google Scholar]
- Fan X., Hou J., Chen X., Chaudhry F., Staiger C.J., Ren H. (2004). Identification and characterization of a Ca2+-dependent actin filament-severing protein from lily pollen. Plant Physiol. 136: 3979–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B., Chelysheva L.A., Lebris M., Jammes F., Marmagne A., De Almeida-Engler J., Lecomte P., Vaury C., Arkowitz R.A., Abad P. (2004). Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell 16: 2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A.A., Pollard T.D., Almo S.C. (1994). Purification, characterization and crystallization of human platelet profilin expressed in Escherichia coli. J. Mol. Biol. 241: 480–482 [DOI] [PubMed] [Google Scholar]
- Feierbach B., Chang F. (2001). Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 11: 1656–1665 [DOI] [PubMed] [Google Scholar]
- Fox J.E., Phillips D.R. (1981). Inhibition of actin polymerization in blood platelets by cytochalasins. Nature 292: 650–652 [DOI] [PubMed] [Google Scholar]
- Fu Y., Gu Y., Zheng Z., Wasteneys G., Yang Z. (2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Goode B.L., Eck M.J. (2007). Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76: 593–627 [DOI] [PubMed] [Google Scholar]
- Harris E.S., Li F., Higgs H.N. (2004). The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem. 279: 20076–20087 [DOI] [PubMed] [Google Scholar]
- He X.F., Fang Y.Y., Feng L., Guo H.S. (2008). Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett. 582: 2445–2452 [DOI] [PubMed] [Google Scholar]
- Hiei Y., Komari T., Kubo T. (1997). Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 35: 205–218 [PubMed] [Google Scholar]
- Hobo T., et al. (2008). Various spatiotemporal expression profiles of anther-expressed genes in rice. Plant Cell Physiol. 49: 1417–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Liang W., Pan A., Zhou Z., Huang C., Chen J., Zhang D. (2003). Production of FaeG, the major subunit of K88 fimbriae, in transgenic tobacco plants and its immunogenicity in mice. Infect. Immun. 71: 5436–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey P.J., Ketelaar T., Deeks M.J. (2006). Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57: 109–125 [DOI] [PubMed] [Google Scholar]
- Ingouff M., Fitz Gerald J.N., Guérin C., Robert H., Sørensen M.B., Van Damme D., Geelen D., Blanchoin L., Berger F. (2005). Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 7: 374–380 [DOI] [PubMed] [Google Scholar]
- Jouhet J., Gray J.C. (2009). Interaction of actin and the chloroplast protein import apparatus. J. Biol. Chem. 284: 19132–19141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J., Block M.A., Pineau B., Albrieux C., Douce R. (1990). Envelope membranes from mature spinach chloroplasts contain a NADPH:protochlorophyllide reductase on the cytosolic side of the outer membrane. J. Biol. Chem. 265: 21820–21827 [PubMed] [Google Scholar]
- Khurana P., Henty J.L., Huang S., Staiger A.M., Blanchoin L., Staiger C.J. (2010). Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 22: 2727–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinebrecht J., Selow J., Winkler W. (1982). The mouse mutant limb-deformity (ld). Anat. Anz. 152: 313–324 [PubMed] [Google Scholar]
- Kovar D.R. (2006). Molecular details of formin-mediated actin assembly. Curr. Opin. Cell Biol. 18: 11–17 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Harris E.S., Mahaffy R., Higgs H.N., Pollard T.D. (2006). Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124: 423–435 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Kuhn J.R., Tichy A.L., Pollard T.D. (2003). The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 161: 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R., Pollard T.D. (2004). Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. USA 101: 14725–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.R., Pollard T.D. (2005). Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88: 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Shen Y., Cai C., Zhong C., Zhu L., Yuan M., Ren H. (2010). The type II Arabidopsis formin14 interacts with microtubules and microfilaments to regulate cell division. Plant Cell 22: 2710–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.S., et al. (2005). Genetic analysis and mapping of rice (Oryza sativa L.) male-sterile (OsMS-L) mutant. Chin. Sci. Bull. 50: 122–125 [Google Scholar]
- Lloyd C., Chan J. (2004). Microtubules and the shape of plants to come. Nat. Rev. Mol. Cell Biol. 5: 13–22 [DOI] [PubMed] [Google Scholar]
- Marc J., Granger C.L., Brincat J., Fisher D.D., Kao Th., McCubbin A.G., Cyr R.J. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette M.F., Kanno T., Aufsatz W., Jakowitsch J., van der Winden J., Matzke M.A., Matzke A.J.M. (2002). Endogenous viral sequences and their potential contribution to heritable virus resistance in plants. EMBO J. 21: 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A., Guérin C., Huang S.J., Ingouff M., Richard S., Rodiuc N., Staiger C.J., Blanchoin L. (2005). The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17: 2296–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.B., Goode B.L. (2005). Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J. Biol. Chem. 280: 28023–28033 [DOI] [PubMed] [Google Scholar]
- Mullins R.D., Heuser J.A., Pollard T.D. (1998). The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 95: 6181–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Furutani I., Tachimoto H., Matsubara H., Hashimoto T. (2004). SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell 16: 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olyslaegers G., Verbelen J.-P. (1998). Improved staining of F-actin and co-localization of mitochondria in plant cells. J. Microsc. 192: 73–77 [Google Scholar]
- Pardee J.D., Spudich J.A. (1982). Purification of muscle actin. Methods Cell Biol. 24: 271–289 [DOI] [PubMed] [Google Scholar]
- Paul A.S., Pollard T.D. (2008). The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 18: 9–19 Erratum. Curr. Biol. 18: 233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A.S., Pollard T.D. (2009). Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskeleton 66: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paves H., Truve E. (2007). Myosin inhibitors block accumulation movement of chloroplasts in Arabidopsis thaliana leaf cells. Protoplasma 230: 165–169 [DOI] [PubMed] [Google Scholar]
- Petersen J., Nielsen O., Egel R., Hagan I.M. (1998). FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 141: 1217–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. (1983). Measurement of rate constants for actin filament elongation in solution. Anal. Biochem. 134: 406–412 [DOI] [PubMed] [Google Scholar]
- Pollard T.D. (2007). Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36: 451–477 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Beltzner C.C. (2002). Structure and function of the Arp2/3 complex. Curr. Opin. Struct. Biol. 12: 768–774 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Blanchoin L., Mullins R.D. (2000). Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29: 545–576 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Blanchoin L., Mullins R.D. (2001). Actin dynamics. J. Cell Sci. 114: 3–4 [DOI] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science 297: 612–615 [DOI] [PubMed] [Google Scholar]
- Quinlan M.E., Heuser J.E., Kerkhoff E., Mullins R.D. (2005). Drosophila Spire is an actin nucleation factor. Nature 433: 382–388 [DOI] [PubMed] [Google Scholar]
- Redmond T., Tardif M., Zigmond S.H. (1994). Induction of actin polymerization in permeabilized neutrophils. Role of ATP. J. Biol. Chem. 269: 21657–21663 [PubMed] [Google Scholar]
- Reisen D., Hanson M.R. (2007). Association of six YFP-myosin XI-tail fusions with mobile plant cell organelles. BMC Plant Biol. 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo M.A., Freed D.D., Carrington J.C. (1990). Nuclear transport of plant potyviral proteins. Plant Cell 2: 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter K. (1991). Affinity purification of antibodies from sera using polyvinylidenedifluoride (PVDF) membranes as coupling matrices for antigens presented by autoantibodies to triosephosphate isomerase. J. Immunol. Methods 137: 209–215 [DOI] [PubMed] [Google Scholar]
- Romero S., Didry D., Larquet E., Boisset N., Pantaloni D., Carlier M.F. (2007). How ATP hydrolysis controls filament assembly from profilin-actin: Implication for formin processivity. J. Biol. Chem. 282: 8435–8445 [DOI] [PubMed] [Google Scholar]
- Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., Carlier M.F. (2004). Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 119: 419–429 [DOI] [PubMed] [Google Scholar]
- Rosales-Nieves A.E., Johndrow J.E., Keller L.C., Magie C.R., Pinto-Santini D.M., Parkhurst S.M. (2006). Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat. Cell Biol. 8: 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagot I., Klee S.K., Pellman D. (2002). Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4: 42–50 [DOI] [PubMed] [Google Scholar]
- Sato Y., Wada M., Kadota A. (2001). Choice of tracks, microtubules and/or actin filaments for chloroplast photo-movement is differentially controlled by phytochrome and a blue light receptor. J. Cell Sci. 114: 269–279 [DOI] [PubMed] [Google Scholar]
- Sawin K.E. (2002). Cell polarity: following formin function. Curr. Biol. 12: R6–R8 [DOI] [PubMed] [Google Scholar]
- Sedbrook J.C. (2004). MAPs in plant cells: Delineating microtubule growth dynamics and organization. Curr. Opin. Plant Biol. 7: 632–640 [DOI] [PubMed] [Google Scholar]
- Shimmen T., Yokota E. (2004). Cytoplasmic streaming in plants. Curr. Opin. Cell Biol. 16: 68–72 [DOI] [PubMed] [Google Scholar]
- Smith L.G., Oppenheimer D.G. (2005). Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 21: 271–295 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Blanchoin L. (2006). Actin dynamics: Old friends with new stories. Curr. Opin. Plant Biol. 9: 554–562 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Williamson R.E., Wasteneys G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124: 1493–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunohara H., Kawai T., Shimizu-Sato S., Sato Y., Sato K., Kitano H. (2009). A dominant mutation of TWISTED DWARF 1 encoding an alpha-tubulin protein causes severe dwarfism and right helical growth in rice. Genes Genet. Syst. 84: 209–218 [DOI] [PubMed] [Google Scholar]
- Symons M.H., Mitchison T.J. (1991). Control of actin polymerization in live and permeabilized fibroblasts. J. Cell Biol. 114: 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S. (2003). Actin-based photo-orientation movement of chloroplasts in plant cells. J. Exp. Biol. 206: 1963–1969 [DOI] [PubMed] [Google Scholar]
- Takagi S., Takamatsu H., Sakurai-Ozato N. (2009). Chloroplast anchoring: Its implications for the regulation of intracellular chloroplast distribution. J. Exp. Bot. 60: 3301–3310 [DOI] [PubMed] [Google Scholar]
- Tellam R., Frieden C. (1982). Cytochalasin D and platelet gelsolin accelerate actin polymer formation. A model for regulation of the extent of actin polymer formation in vivo. Biochemistry 21: 3207–3214 [DOI] [PubMed] [Google Scholar]
- Thitamadee S., Tuchihara K., Hashimoto T. (2002). Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193–196 [DOI] [PubMed] [Google Scholar]
- Thomas S.G., Huang S., Li S., Staiger C.J., Franklin-Tong V.E. (2006). Actin depolymerization is sufficient to induce programmed cell death in self-incompatible pollen. J. Cell Biol. 174: 221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D., Park S.K., Hawkins T.J., Schubert D., Schmidt R., Smertenko A., Hussey P.J. (2002). MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat. Cell Biol. 4: 711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., van Gisbergen P.A., Guérin C., Franco P., Li M., Burkart G.M., Augustine R.C., Blanchoin L., Bezanilla M. (2009). Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc. Natl. Acad. Sci. USA 106: 13341–13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M., Suetsugu N. (2004). Plant organelle positioning. Curr. Opin. Plant Biol. 7: 626–631 [DOI] [PubMed] [Google Scholar]
- Wang H.Y., Yu Y., Chen Z.L., Xia G.X. (2005). Functional characterization of Gossypium hirsutum profilin 1 gene (GhPFN1) in tobacco suspension cells. Characterization of in vivo functions of a cotton profilin gene. Planta 222: 594–603 [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59: 253–279 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang Y.F., Zhang D.B. (2006). Identification of the rice (Oryza sativa L.) mutant msp1-4 and expression analysis of its UDT1 and GAMYB genes. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 32: 527–534 [PubMed] [Google Scholar]
- Wang Z., Chen C., Xu Y., Jiang R., Han Y., Xu Z., Chong K. (2004). A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 22: 409–417 [Google Scholar]
- Wanner G., Formanek H. (1995). Imaging of DNA in human and plant chromosomes by high-resolution scanning electron microscopy. Chromosome Res. 3: 368–374 [DOI] [PubMed] [Google Scholar]
- Wasserman S. (1998). FH proteins as cytoskeletal organizers. Cell Biol. 8: 111–115 [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O. (2004). Progress in understanding the role of microtubules in plant cells. Curr. Opin. Plant Biol. 7: 651–660 [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O., Galway M.E. (2003). Remodeling the cytoskeleton for growth and form: An overview with some new views. Annu. Rev. Plant Biol. 54: 691–722 [DOI] [PubMed] [Google Scholar]
- Wear M.A., Schafer D.A., Cooper J.A. (2000). Actin dynamics: Assembly and disassembly of actin networks. Curr. Biol. 10: R891–R895 [DOI] [PubMed] [Google Scholar]
- Welch M.D., Iwamatsu A., Mitchison T.J. (1997). Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature 385: 265–269 [DOI] [PubMed] [Google Scholar]
- Willemsen V., Wolkenfelt H., de Vrieze G., Weisbeek P., Scheres B. (1998). The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125: 521–531 [DOI] [PubMed] [Google Scholar]
- Yahara I., Harada F., Sekita S., Yoshihira K., Natori S. (1982). Correlation between effects of 24 different cytochalasins on cellular structures and cellular events and those on actin in vitro. J. Cell Biol. 92: 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Ren S., Zhang X., Gao M., Ye S., Qi Y., Zheng Y., Wang J., Zeng L., Li Q., Huang S., He Z. (2011). BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23: 661–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Zheng Y., Yan A., Chen N., Wang Z., Huang S., Yang Z. (2009). Arabidopsis Formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 21: 3868–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K., Guo C., Chen D., Zhao B., Yang B., Ren H. (2005). Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis. Plant Physiol. 138: 1071–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K.G., Thurston S.F., Copeland S., Smallwood C., Copeland J.W. (2008). INF1 is a novel microtubule-associated formin. Mol. Biol. Cell 19: 5168–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.B., Wilson Z.A. (2009). Stamen specification and anther development in rice. Chin. Sci. Bull. 54: 1–12 [Google Scholar]
- Zhang H., Liang W., Yang X., Luo X., Jiang N., Ma H., Zhang D. (2010). Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22: 672–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Leder P., Martin S.S. (2006). Formin-1 protein associates with microtubules through a peptide domain encoded by exon-2. Exp. Cell Res. 312: 1119–1126 [DOI] [PubMed] [Google Scholar]