This study establishes that plant TSPO, an enigmatic stress-induced membrane protein, binds porphyrins, including heme in vitro and in vivo, and that heme binding regulates TSPO degradation through autophagy. This work indicates that TSPO expression may, at least in part, transiently protect plant cell from porphyrin-induced damages.
Abstract
TSPO, a stress-induced, posttranslationally regulated, early secretory pathway-localized plant cell membrane protein, belongs to the TspO/MBR family of regulatory proteins, which can bind porphyrins. This work finds that boosting tetrapyrrole biosynthesis enhanced TSPO degradation in Arabidopsis thaliana and that TSPO could bind heme in vitro and in vivo. This binding required the His residue at position 91 (H91), but not that at position 115 (H115). The H91A and double H91A/H115A substitutions stabilized TSPO and rendered the protein insensitive to heme-regulated degradation, suggesting that heme binding regulates At-TSPO degradation. TSPO degradation was inhibited in the autophagy-defective atg5 mutant and was sensitive to inhibitors of type III phosphoinositide 3-kinases, which regulate autophagy in eukaryotic cells. Mutation of the two Tyr residues in a putative ubiquitin-like ATG8 interacting motif of At-TSPO did not affect heme binding in vitro but stabilized the protein in vivo, suggesting that downregulation of At-TSPO requires an active autophagy pathway, in addition to heme. Abscisic acid–dependent TSPO induction was accompanied by an increase in unbound heme levels, and downregulation of TSPO coincided with the return to steady state levels of unbound heme, suggesting that a physiological consequence of active TSPO downregulation may be heme scavenging. In addition, overexpression of TSPO attenuated aminolevulinic acid–induced porphyria in plant cells. Taken together, these data support a role for TSPO in porphyrin binding and scavenging during stress in plants.
INTRODUCTION
Abiotic stresses, including salinity, drought, high light, high temperature, and freezing can be perceived by plants, in part, as a transient or permanent water deficit. Sensing and signaling events that detect abiotic stress–induced changes in plant water status and initiate downstream responses, such as abscisic acid (ABA) accumulation and osmoregulation, remain uncharacterized in plants (Verslues and Zhu, 2007). The stress phytohormone ABA regulates fundamental growth and developmental processes in the plant, including seed dormancy and germination, seedling establishment and growth, and plant water status through regulation of stomatal closure (Finkelstein et al., 2002; Nambara and Marion-Poll, 2005). The increase in active ABA levels in plant cells during water-related stress regulates the expression of ABA-responsive genes by interacting with cognate cytosolic and/or organelle-bound receptors and downstream effectors modulating the activity of defined transcriptional regulators (Shen et al., 2006; Fujii and Zhu, 2009; Ma et al., 2009; Park et al., 2009; Wu et al., 2009; Shang et al., 2010).
A subset of plant ABA-responsive genes is strictly ABA dependent in that their expression is almost undetectable in the absence of elevated levels of cellular ABA. Their biological role may be required only transiently, and the plant cell under stress therefore needs an efficient regulatory mechanism to transcriptionally and/or posttranslationally regulate their expression. Although many stress-specific genes have been characterized in plants, how plants readjust levels of a stress-induced protein when normal physiological conditions resume has not been addressed. In particular, the questions of how, when, and where the induced proteins are targeted for degradation when their activities become irrelevant await answers.
Arabidopsis thaliana TSPO, a tryptophan-rich sensory protein (TSPO)–related membrane protein (Guillaumot et al., 2009a), is a potential multiple abiotic stress regulator (Kant et al., 2008) encoded by a single, intronless locus, At2g47770. At-TSPO belongs to the Trp-rich sensory protein/peripheral-type benzodiazepine receptor (TspO/MBR) protein family, which are membrane-anchored proteins, found, with few exceptions, in organisms ranging from Archaea to metazoans (reviewed in Gavish et al., 1999; Lacapère and Papadopoulos, 2003; Papadopoulos et al., 2006). Since their identification in the late 70s (Braestrup et al., 1977), TSPOs have been the subject of intensive research, almost exclusively in animal cells, to pinpoint their function. In mammals, TSPO1 (Fan et al., 2009), also known as the 18-kD Translocator protein, is an essential, widely expressed, evolutionarily conserved mitochondrial outer membrane protein that is involved in a wide range of physiological functions and pathologies, including neurodegeneration and cancer (reviewed in Papadopoulos et al., 2006). The pharmacology of TSPOs has been extensively studied. For instance, TSPO1 is known to bind a plethora of structurally unrelated compounds, including promising candidates for fast-acting anxiolytic drugs with less severe side effects than benzodiazepines (Rupprecht et al., 2009). However, little is known about the mode of action of TSPOs (i.e., whether they operate as pumps, transporters, or channels) (Korkhov et al., 2010).
TSPO-related proteins were recently described in plants (Corsi et al., 2004; Lindemann et al., 2004; Frank et al., 2007; Guillaumot et al., 2009a). Plant TSPOs appear to be nonessential, and their biological functions are not yet defined, although their induction by abiotic stress and ABA seems to be established (Frank et al., 2007; Kant et al., 2008; Guillaumot et al., 2009a). At-TSPO is transcriptionally regulated by the master bZIP-type transcription factors AREB1, AREB2, and ABF3, which are involved in ABA-responsive element-dependent ABA signaling (Yoshida et al., 2010). At-TSPO transcripts are detected mainly in desiccation-resistant plant structures, such as seeds and pollen grains, and, to some extent, in senescing leaves but can be induced in vegetative tissues by abiotic stresses, including osmotic and salt stress, magnesium deficiency, high light, and ABA treatment (Kreps et al., 2002; Seki et al., 2002; Zimmermann et al., 2004; Brady et al., 2007; Catala et al., 2007; Kleine et al., 2007; Winter et al., 2007; Dinneny et al., 2008; Hermans et al., 2010). We previously showed that overexpression of At-TSPO can be detrimental to plant cells (Guillaumot et al., 2009a). ABA-induced expression of TSPO in vegetative tissue is transient, and the degradation of induced or constitutively expressed TSPO is enhanced by boosting tetrapyrrole biosynthesis (Guillaumot et al., 2009a, 2009b), suggesting that posttranslational regulation of TSPO may depend on tetrapyrrole metabolism.
In mammalian cells, the endogenous ligands of TSPO1 include cholesterol and porphyrins (Papadopoulos et al., 2006). Rat TSPO1 can bind protoporphyrin IX (PPIX) in vivo (Ozaki et al., 2010). Detergent-purified TSPO1 can bind porphyrins in vitro, including PPIX, hemin (Fe3+-PPIX, oxidized heme), and coproporphyrin III (Taketani et al., 1995; Wendler et al., 2003; Korkhov et al., 2010). However, the mechanism of this binding and its biological significance in vivo are not yet clear. It is thought that TSPOs might be involved in the translocation of porphyrins, including heme (Fe2+-PPIX), across the mitochondrial outer membrane in mammalian cells (Taketani et al., 1995; Wendler et al., 2003), although the movement of anionic and hydrophobic porphyrins, such as heme, into a negatively charged environment requires energy (Krishnamurthy et al., 2006). TSPO from the facultative photosynthetic bacterium Rhodobacter sphaeroides acts as an oxygen-dependent signal generator that negatively regulates photosynthetic gene expression when the environment favors aerobic (oxidative) growth (Yeliseev and Kaplan, 1995; Yeliseev et al., 1997; Yeliseev and Kaplan, 2000). This function may be related to the efflux of specific porphyrins that are intermediates in photopigment biosynthesis (Yeliseev and Kaplan, 1999). Plant tetrapyrroles (chlorophylls, bilins, siroheme, and hemes) are synthesized in plastids but are also required for vital cellular activities in numerous organelles (Takahashi et al., 2008; Severance and Hamza, 2009; Mochizuki et al., 2010). For instance, hemes play a critical role in diverse biological processes in eukaryotic cells, including respiration, protein targeting, transcription and translation regulation, ion channel regulation and signaling, microRNA processing, and protein degradation (Severance and Hamza, 2009; Mochizuki et al., 2010).
Here, we present evidence that (1) At-TSPO is a heme binding membrane protein, (2) its downregulation by plant cells requires heme binding, and (3) a functional macroautophagy pathway is also required for the cell to degrade this stress-induced membrane protein.
RESULTS
Heme Metabolism Affects TSPO Stability in the Plant Cell
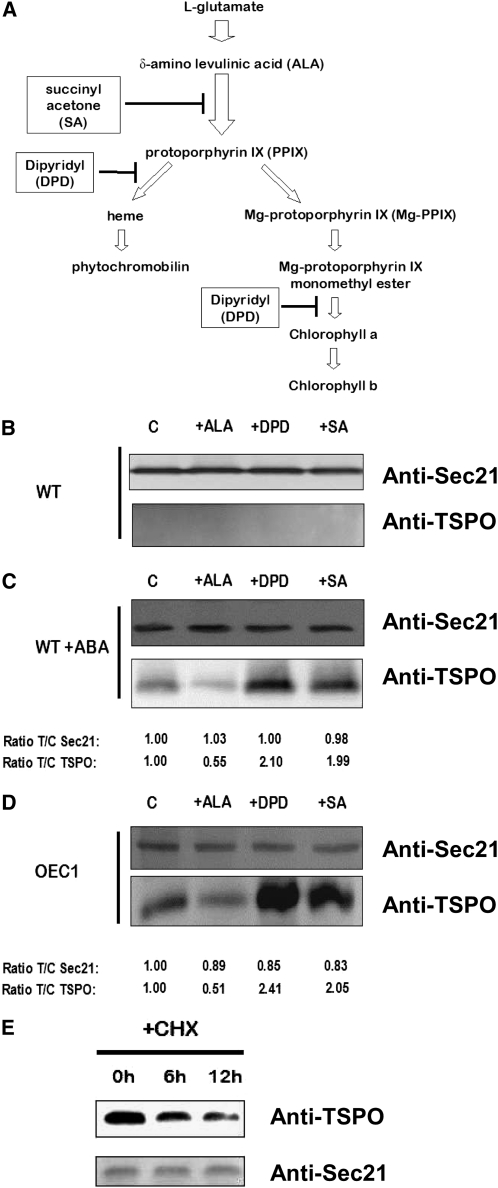
Circumstantial evidence suggests that Arabidopsis cells strictly regulate their levels of TSPO (Guillaumot et al., 2009a). At the transcriptional level, TSPO expression appears to be cell type specific as well as environmentally and developmentally regulated (see Supplemental Figure 1 online). Even when constitutively expressed, TSPO fails to accumulate evenly in all cell types and, in particular, does not accumulate in mesophyll cells (Guillaumot et al., 2009a). Overexpression of the protein can be detrimental under certain physiological conditions, and boosting tetrapyrrole biosynthesis through δ-aminolevulinic acid (ALA) feeding enhances plant cell degradation of At-TSPO, suggesting that posttranslational regulation of TSPO may involve tetrapyrroles (Guillaumot et al., 2009a, 2009b). To test this possibility and to identify the key player(s) involved, we used defined pharmacological agents to tamper with tetrapyrrole biosynthesis. A simplified scheme of tetrapyrrole biosynthesis in plant plastids is shown in Figure 1A. Succinyl acetone (SA), a potent inhibitor of 5-aminolevulinate dehydratase, is commonly used as a heme biosynthesis inhibitor in eukaryotic cells (Krishnamurthy et al., 2006), although it also inhibits the biosynthesis of all the cyclic porphyrins downstream of ALA. The herbicide and ferrous iron chelator 2,2’-dipyridyl (DPD) inhibits heme biosynthesis from PPIX and the chlorophyll branch of the pathway downstream of magnesium PPIX monomethyl ester (Figure 1A) (Nott et al., 2006). Before analyzing TSPO stability, we checked whether these pharmacological treatments had any impact on TSPO expression. Figure 1B shows that TSPO is not induced in wild-type Arabidopsis suspension cell cultures treated with ALA, DPD, or SA and incubated for 24 h in the dark. We previously showed that TSPO can be induced in Arabidopsis wild-type suspension cell cultures by ABA treatment (Guillaumot et al., 2009a). Wild-type cells were preincubated with ABA for 24 h, then with ALA, DPD, or SA in the continuous presence of ABA for 24 h and the relative amount of SEC21 (a Golgi-localized membrane protein used as loading control) protein and TSPO quantified as the treatment/control signal ratio (Figure 1C, T/C ratio). Figure 1C shows that these pharmacological treatments had different effects on the relative amount of TSPO detected by immunoblotting. Compared with control cells (lane C), ALA feeding (+ALA) caused about a 50% reduction in detectable TSPO, suggesting that boosting tetrapyrrole biosynthesis accelerated TSPO downregulation by the cells. By contrast, DPD or SA treatment almost doubled the amount of TSPO compared with control cells. However, intracellular levels of a protein are dependent on both its rate of synthesis and its rate of degradation (Callis, 1995). It is possible that TSPO accumulation is affected at the transcriptional level by these pharmacological treatments, but this is unlikely, since similar results were obtained with a transgenic suspension cell culture (OEC1) overexpressing At-TSPO (Figure 1D), and ALA, DPD, and SA are not known to affect translation in eukaryotic cells. Inhibition of protein synthesis by cycloheximide (CHX) revealed that TSPO is actively degraded in OEC1 plant cells (Figure 1E). These results therefore suggest increased stability of the protein as a consequence of DPD and SA treatments and accelerated degradation after CHX or ALA treatment.
Figure 1.
At-TSPO Is Regulated by Tetrapyrrole Metabolism.
(A) Simplified scheme of the tetrapyrrole biosynthetic pathway in the plant chloroplast, the pharmacological agents used, and the specific steps inhibited.
(B) Arabidopsis suspension-cultured cells were incubated for 24 h in the dark in the presence of 5 mM ALA, DPD, or SA. Control cells (C) were incubated in the presence of DMSO (mock, solvent used to dissolve the chemicals). Total proteins were extracted and analyzed by immunoblotting (IB) for TSPO using the Arabidopsis Golgi protein At-SEC21 (γ COPI) as loading control.
(C) Arabidopsis suspension cells were preincubated in 50 μM ABA for 24 h before being subjected to the same treatments and analyses as in (B), and the signal for both SEC21 and TSPO in each treated sample was compared with that in the control as a treated/control ratio (T/C) (bottom lines). The TSPO/SEC21 ratio (ratio of the ratios) was 0.5 with ALA, 2.1 with DPD, and 2 with SA.
(D) A transgenic line (OEC1) overexpressing At-TSPO was treated as in (C). The TSPO/SEC21 ratio was 0.6 with ALA, 2.8 with DPD, and 2.5 with SA.
(E) A transgenic cell line as in (D) was incubated in presence of the protein synthesis inhibitor CHX (100 μM) and immunoblotted for TSPO and SEC21 analyzed at different time points.
The results in (B) to (D) are representative of those obtained in at least three separate experiments. The results in (E) were reproduced twice. WT, Wild type.
The DPD results suggest that possible accumulation of PPIX, magnesium PPIX, and magnesium PPX monomethyl ester was not responsible for the ALA-dependent enhanced degradation of TSPO and that heme, the synthesis of which is inhibited by DPD, or its downstream catabolic products could be responsible for TSPO downregulation. Since cyclic porphyrins bind to TSPO proteins and are potential endogenous ligands, we wondered if At-TSPO could bind heme.
At-TSPO Binds Heme in Vitro
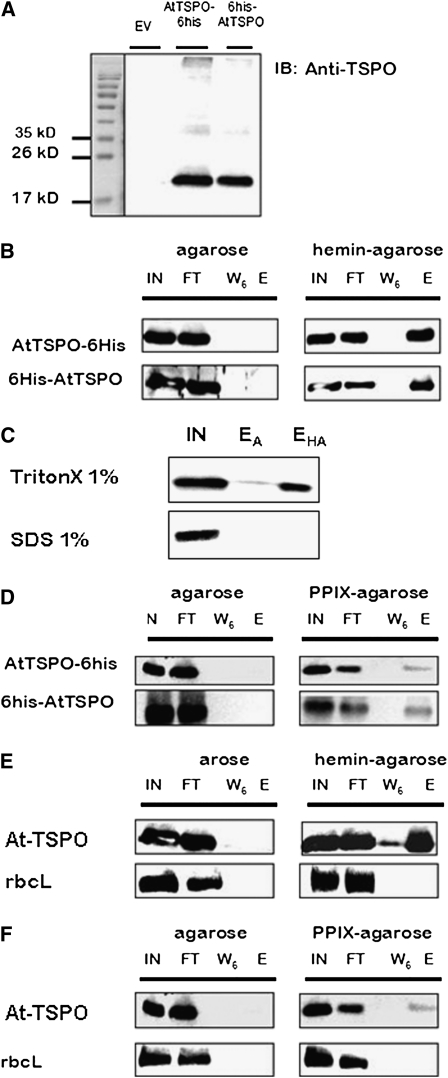
To determine whether At-TSPO could bind porphyrins and, in particular heme, in vitro, we heterologously expressed the protein in Saccharomyces cerevisiae, a yeast lacking TSPO-related proteins. To facilitate purification, the expressed At-TSPO was provided with a 6-His tag at the N terminus (6His-AtTSPO) or C terminus (AtTSPO-6His). Figure 2A shows At-TSPO purified by nickel nitrilotriacetic acid (Ni-NTA) chromatography from yeast microsomes, using the identically treated empty vector (EV)–transformed extract as a control to assess the specificity of the anti-AtTSPO antibodies, which have been previously checked using plant extracts but not yeast extracts (Guillaumot et al., 2009a). Affinity chromatography using hemin-agarose was performed to test the binding of At-TSPO to covalently immobilized hemin, using the same agarose matrix without hemin as a negative control, and examining the input, the flow-through, the last wash (W6, wash #6), and eluted material by immunoblotting. As shown in Figure 2B, purified and dialyzed (to remove the imidazole used during protein purification) AtTSPO-6His or 6His-AtTSPO was not retained strikingly by the non-hemin-coupled matrix (left panel, lane E), whereas both were bound to hemin-agarose and detected in the eluted fraction (right panel, lane E), showing that detergent-solubilized At-TSPO could bind to immobilized hemin in vitro. Similar results were obtained with dialyzed or nondialyzed purified At-TSPO. It was possible that the observed hemin binding could be due to the 6-His tag added to the recombinant protein. To exclude this possibility, we first compared the heme binding of the untagged protein solubilized from microsomes using SDS or the mild detergent Triton X-100. Figure 2C shows that At-TSPO was retained by the hemin-agarose column when solubilized with 1% (v/v) Triton X-100 but not with 1% (v/v) SDS (lane EHA), while neither were retained by the plain agarose column (lane EA). These results show that hemin binding requires structured At-TSPO. Second, we checked whether the binding was specific to metalloporphyrins by substituting PPIX for hemin. Figure 2D shows that, although AtTSPO-6His and 6His-AtTSPO also bind to a PPIX-agarose column, the amount of protein retained was less than that retained by the hemin-agarose column, suggesting that the affinity for hemin is higher than that for PPIX. Third, we overexpressed untagged At-TSPO in Arabidopsis cultured cells, solubilized it from the microsomes, and tested its ability to bind to the hemin-agarose affinity column. Figure 2E shows that the untagged protein bound to hemin, whereas the abundant large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcL) did not. Again, as shown in Figure 2F, the binding to the PPIX-agarose column was lower.
Figure 2.
At-TSPO Binds Heme in Vitro: Evidence from Affinity Chromatography.
(A) At-TSPO with a His tag (AtTSPO-6His or 6His-AtTSPO) heterologously expressed in yeast was detergent solubilized from microsomes and subjected to Ni-NTA purification, with the corresponding sample from EV-transformed (EV with no At-TSPO cDNA) yeast being used as the control, and the eluted fractions were analyzed by immunoblotting for At-TSPO; the anti-AtTSPO antibodies did not cross-react with yeast proteins (the faint signal occasionally detected around 40 kD represents a dimer of At-TSPO).
(B) Binding of At-TSPO to hemin-agarose or agarose alone, as negative control. The input (IN), flow-through (FT), last wash (W6, wash #6), and eluted (E) fractions were analyzed by immunoblotting for At-TSPO.
(C) The same experiment as in (B), but the untagged protein expressed in yeast was solubilized with either 1% Triton X-100 or 1% SDS and the solubilized material tested for binding to control agarose (EA) or agarose coupled to hemin (EHA).
(D) The same experiment as in (B) but using PPIX-agarose.
(E) and (F) Untagged At-TSPO was constitutively expressed in Arabidopsis cultured cells and the solubilized protein incubated with hemin-agarose or PPIX-agarose, with agarose alone as negative control, and the same samples as in (B) were analyzed using rbcL as a control for nonspecific binding.
(A) is a representative purification result, (B) and (D) were independently reproduced at least five times, (C) was repeated twice with similar results, and (E) and (F) were reproduced three times.
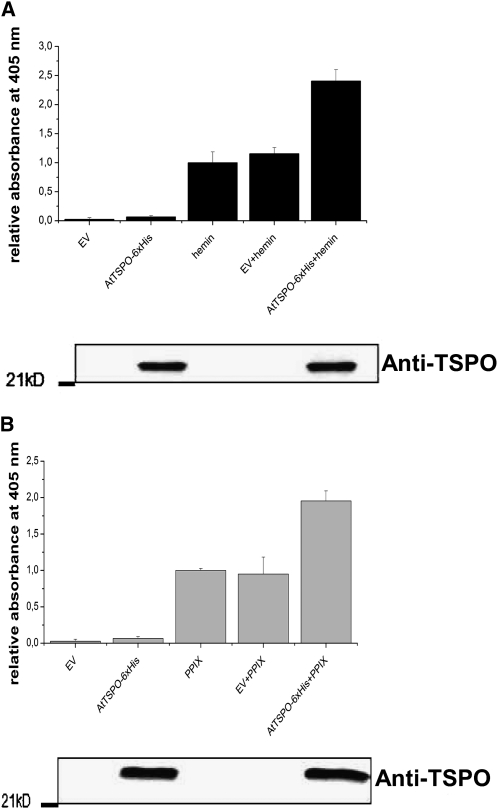
We also designed a pull-down assay to examine porphyrin binding in vitro. AtTSPO-6His purified from yeast was dialyzed, mixed with hemin or PPIX, and reimmobilized on a Ni-NTA column; then, after a second round of protein purification, the relative amount of copurified porphyrin was assessed spectrophotometrically at 405 nm. The data presented in Figure 3 (bottom panels) show that preincubating AtTSPO-6His with hemin (Figure 3A) or PPIX (Figure 3B) did not interfere with its binding to the Ni-NTA matrix, as revealed by immunoblot analysis of the purified fractions. The eluate of the solubilized EV control material from Ni-NTA column or AtTSPO-6His in the absence of porphyrin (AtTSPO-6His) showed little absorbance at 405 nm (Figures 3A and 3B). However, in the presence of At-TSPO (AtTSPO-6His + hemin/PPIX), a twofold increase in relative absorbance of porphyrin was seen compared with porphyrin alone (hemin/PPIX) or porphyrin mixed with the EV sample (EV + hemin/PPIX). These consistent affinity chromatography and pull-down assay results demonstrate that, at least in vitro, At-TSPO binds hemin.
Figure 3.
At-TSPO Binds to Heme in Vitro: Evidence from the Pull-Down Assay.
Purified AtTSPO-6His from yeast microsomes was dialyzed to remove the imidazole used in the Ni-NTA purification and then mixed with hemin (A) or PPIX (B) and the sample repurified on the Ni-NTA column. The affinity chromatography eluates of solubilized proteins from EV-transformed yeast or hemin or PPIX alone were used as controls. The absorbance at 405 nm of the eluted Ni-NTA fraction was recorded and normalized to the values for hemin or PPIX alone passed though the Ni-NTA column. Immunoblot analyses (bottom panel in [A] and [B]) were used to assess the presence of At-TSPO in the various fractions. The experiments were repeated at least three times, each in duplicate, and the error bars represent se.
YFP-AtTSPO Colocalizes in Vivo with Palladium Mesoprotoporphyrin IX
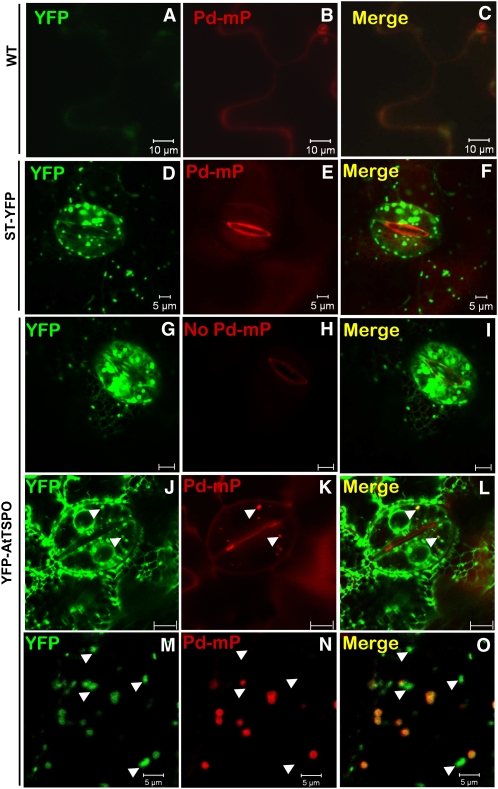
Palladium mesoprotoporphyrin IX (Pd-mP) is a fluorescent analog of heme and can be used to follow subcellular heme dynamics (Lara et al., 2005). We reasoned that, if At-TSPO binds heme in vivo, this should result in colocalization of Pd-mP with yellow fluorescent protein–tagged At-TSPO (YFP-AtTSPO) expressed in plant cells. We used transgenic tobacco (Nicotiana tabacum) plants constitutively expressing YFP-AtTSPO to test this hypothesis; tobacco was preferred to Arabidopsis because we already had a transgenic plant expressing a control Golgi membrane marker construct sialyl transferase (ST)-YFP, a membrane fusion protein already shown to colocalize with YFP-AtTSPO (Guillaumot et al., 2009a). In addition, the expression pattern of YFP-AtTSPO is the same in Arabidopsis and tobacco, with YFP-AtTSPO fluorescence being stronger in the guard cells and epidermal cells. A 10 μM solution of Pd-mP was infiltrated into a leaf from wild-type tobacco plants and ST-YFP and YFP-AtTSPO transgenic plants, and then leaf epidermal cells were imaged by confocal microscopy. Representative images are shown in Figure 4. A diffuse Pd-mP signal (red channel) was detected in the wild-type control sample (Figure 4B) and the ST-YFP sample (Figure 4E), whereas the Golgi stacks highlighted by ST-YFP (Figure 4D) contained no Pd-mP signal (Figure 4F). Interestingly, some YFP-AtTSPO–highlighted Golgi stacks showed Pd-mP fluorescence (arrowheads in Figures 4J to 4L). Surprisingly, in the presence of YFP-AtTSPO, which could be seen in the endoplasmic reticulum (ER), no Pd-mP signal was seen in this organelle (Figures 4J to 4L), and <50% of the Golgi stacks were labeled with Pd-mP (arrowheads in Figures 4J to 4L). To determine whether this labeling pattern was a peculiarity of the guard cells, we also imaged pavement cells and again found that <50% of the Golgi stacks were stained with Pd-mP (Figures 4M to 4O, arrowheads indicating unstained Golgi stacks). This pattern was not modified by a longer incubation period before imaging and/or increasing the Pd-mP concentration up to 100 μM. It may be that the stained Golgi stacks contain YFP-AtTSPO that is not bound to endogenous porphyrin. However, the partial colocalization of Pd-mP and YFP-AtTSPO strongly suggests that At-TSPO can also bind heme in vivo.
Figure 4.
YFP-AtTSPO Colocalizes with Pd-mP in Vivo.
Heme binding in vivo by At-TSPO was indirectly investigated by confocal imaging of YFP-AtTSPO–expressing plant cells treated with the heme fluorescent analog Pd-mP. Leaves from a wild-type (WT) tobacco plant ([A] to [C]) or a transgenic line expressing a Golgi membrane marker (ST-YFP; [D] to [F]) were used as controls. The controls and a transgenic line expressing YFP-AtTSPO (YFP-AtTSPO; [G] to [O]) were infiltrated with 10 μM Pd-mP, and the YFP fluorescence (green channel) and Pd-mP fluorescence (red channel) were imaged simultaneously. Representative images are shown. The arrowheads in (D) to (F) point to Golgi stacks showing both YFP and Pd-mP fluorescence, and those in (G) to (I) point to Golgi stacks with YFP fluorescence but without Pd-mP fluorescence. Bars 5 μm in (A) to (F), 10 μm in (G) to (L), and 5 μm in (M) to (O).
Heterologously Expressed At-TSPO Is Downregulated and Sensitive to Heme Metabolism
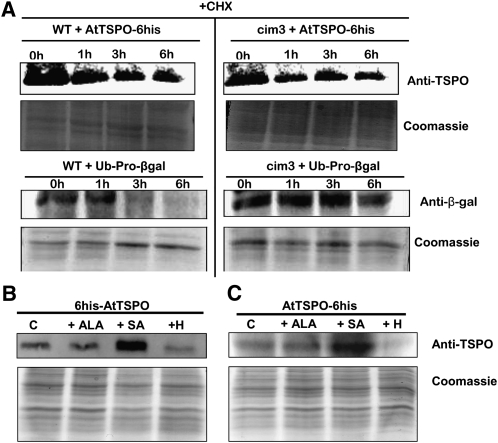
As shown with the plant cells, CHX treatment of yeast cells expressing At-TSPO showed that the protein is also downregulated in this heterologous host (Figure 5A). The degradation rate of At-TSPO in yeast was not affected by the 26S protease cim3-1 mutation (Ghislain et al., 1993) (cf. wild-type cells, top left panel, to cim3-1 cells, top right panel). To verify that the degradation of short-lived proteins was indeed less efficient in the cim3-1 mutant cells, we expressed the synthetic reporter construct ubiquitin-Pro-β-galactosidase fusion protein as an in vivo control substrate (Ghislain et al., 1993, 1996). The reporter fusion protein was rapidly degraded in wild-type cells but stabilized in the cim3-1 mutant cells (cf. wild-type cells, bottom left panel, to cim3-1 cells, bottom right panel). These results show that heterologously expressed At-TSPO in yeast is downregulated, and this degradation process may not require an active proteasome pathway.
Figure 5.
At-TSPO Heterologously Expressed in Yeast Is Sensitive to Heme Metabolism.
(A) Yeast cells (wild type [WT] or 26S protease mutant cim3-1) expressing AtTSPO-6His were incubated in presence of 366 μM CHX, and At-TSPO stability was analyzed by immmunoblot at different time points (top panels). As a control for the 26S protease–dependent degradation, a short-lived Ub-Pro-βgal reporter was used (bottom panels). The Coomassie blue–stained gels below each blot show protein loading levels.
(B) and (C) Yeast cells expressing 6His-AtTSPO (B) or AtTSPO-6His (C) were fed ALA (2.5 mM), SA (1 mM), or hemin (0.5 mM) for 48 h, and then the amount of At-TSPO was assessed by immunoblotting with the untreated cells as control (C) (top panel) and replica gels were stained with Coomassie blue for loading comparisons (bottom panel).
The results presented are representative of those in four independent experiments ([B] and [C]) or two independent experiments (A).
Having generated yeast strains expressing At-TSPO, we wondered if the protein stability in yeast might also be sensitive to tetrapyrrole metabolism as in plants and in particular to heme. If heme sensitivity happened to be conserved in yeast, this could be a simple tool for the rapid characterization of various mutant forms of At-TSPO. In contrast with plant cells, yeast cells are not sensitive to ALA feeding, probably because of ALA presence and metabolism in the cytosol (Hoffman et al., 2003) and a possible unidirectional transport of this metabolite across the mitochondrial membranes. In yeast, heme biosynthesis is compartmentalized, and the part of heme biosynthetic pathway performed in the cytosol starts with the export of ALA from the mitochondria (Severance and Hamza, 2009). Translocation of ALA-derived intermediates from the cytosol into the mitochondria is a key limiting step in heme biosynthesis in yeast. In plant cells, ALA metabolism and heme biosynthesis take place in the plastids (Mochizuki et al., 2010). In addition, yeast cells can take up exogenous heme under anaerobic growth conditions, which is not the case in plant cells. To evaluate the suitability of yeast as a tool to investigate heme-dependent At-TSPO degradation, we fed His-tagged At-TSPO–expressing cells with ALA, SA, or hemin and analyzed At-TSPO stability by immunoblotting. Figures 5B and 5C show that the levels of AtTSPO-6His and 6His-AtTSPO expressed in yeast were almost unaffected by ALA feeding (lanes +ALA). As expected, SA stabilized At-TSPO (lanes +SA), and feeding hemin accelerated the degradation of At-TSPO (lanes +H). Although the yeast S. cerevisiae is devoid of a TSPO-related protein, it seems that the pathway required for the active degradation of At-TSPO is not specific to this protein or to plant cells. We therefore conclude that the heme-dependent downregulation of At-TSPO observed in plant cells is conserved in yeast.
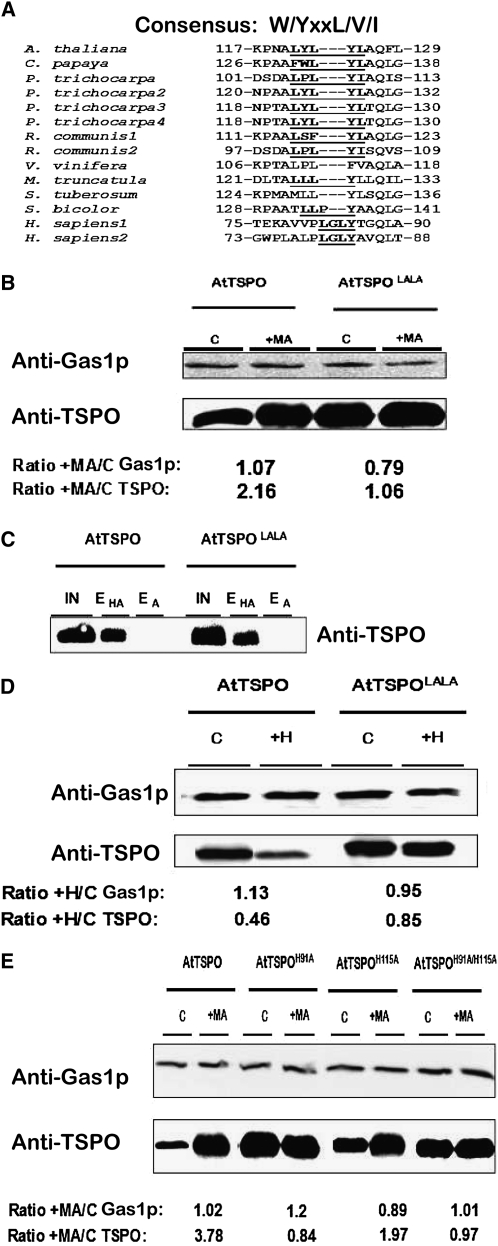
Heme Binding by At-TSPO Requires the His Residue at Position 91 but Not That at Position 115
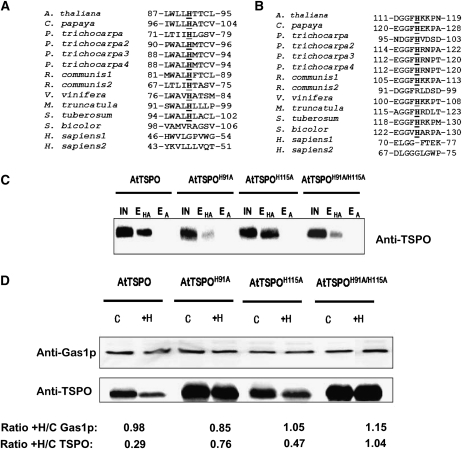
To determine whether At-TSPO degradation was directly linked to heme binding in vivo, we searched for residues in the primary sequence of At-TSPO potentially involved in heme binding. Many heme binding proteins share common motifs, such as CxxCH, CxxC, or the conserved heme regulatory CP motif (Schulz et al., 1998; Stevens et al., 2003; Yang et al., 2008), but no such motif is present in At-TSPO. However, it takes only a single residue in the right environment to bind heme, and this residue is almost always a His, as evidenced by the cytochrome c-type heme chaperone (CcmE) (Schulz et al., 1998). At-TSPO contains two His residues at positions 91 (H91) and 115 (H115). As shown in the local alignments in Figures 6A and 6B, both appear to be relatively well conserved in angiosperm TSPOs but not in human TSPO1 and TSPO2. We generated At-TSPO mutants with either a H91A substitution (AtTSPOH91A) or a H115A substitution (AtTSPOH115A) and the double mutant H91A/H115A (AtTSPOH91A/H115A), expressed them in yeast, and compared their binding activity to that of the wild-type protein using hemin-agarose affinity chromatography. Compared with that of the wild type, the hemin binding ability of mutant H91A was drastically reduced (Figure 6C, compare lanes EHA for AtTSPO and AtTSPOH91A), whereas the H115A substitution had no striking effect (Figure 6C, compare lanes EHA for AtTSPO and AtTSPOH115A), and the double mutant AtTSPOH91A/H115A showed the same reduced hemin binding ability as the single AtTSPOH91A mutant (Figure 6C, compare lanes EHA for AtTSPO and AtTSPOH91A/H115A), suggesting that H91, but not H115, may be the axial coordinator of heme in At-TSPO. If heme binding by At-TSPO does require H91 and this binding is linked to At-TSPO downregulation, then the H91A mutant forms would be expected to be less sensitive to heme-induced degradation of the protein in vivo. We fed yeast strains expressing the mutant forms with hemin and compared the relative stability of the mutant to that of the wild-type protein, using the yeast glycophospholipid-anchored surface protein (Gas1p) as a normalizing control for immunoblot signals. As shown in Figure 6D, the AtTSPOH91A and AtTSPOH91A/H115A mutant forms failed to respond to hemin-induced degradation (AtTSPO/Gas1p ratio ≈ 1), while AtTSPOH115A was still sensitive to hemin-induced rapid degradation just as the wild-type protein (AtTSPO/Gas1p ratio of 0.45 and 0.3, respectively). These data support the conclusions that heme binding by At-TSPO requires His at position 91 and that porphyrin binding enhances At-TSPO degradation in vivo.
Figure 6.
Heme Binding by At-TSPO Requires His at Position 91.
(A) and (B) ClustalW local alignment of His at positions 91 (A) and 115 (B) (At-TSPO numbering) in some angiosperm TSPOs. The only two His residues in At-TSPO are relatively well conserved in angiosperm TSPOs but not in human TSPO1 and TSPO2.
(C) The His residues were mutated to Ala to generate the single mutant forms AtTSPOH91A and AtTSPOH91A and the double mutant AtTSPOH91A/H115A, which were expressed in yeast (without a His tag) and their hemin binding capacity assessed as in Figure 2 in comparison to the wild-type At-TSPO. IN, input; EHA, eluate from hemin-agarose column; EA, eluate from agarose column.
(D) The sensitivity of the mutants in (C) to exogenous hemin was investigated as in Figure 5. The yeast membrane protein Gas1p was used as loading control. The signals for Gas1p and At-TSPO in each treated sample were compared with those in the control (treated/control ratio; +H/C) (underneath the figure). The AtTSPO/Gas1p ratio (ratio of ratios) was 0.3 for wild-type At-TSPO, 0.9 for AtTSPOH91A, 0.45 for AtTSPOH115A, and 0.9 for AtTSPOH91A/H115A. (C) and (D) were reproduced at least three times.
ABA Transiently Increases Unbound Heme Levels in the Plant Cell
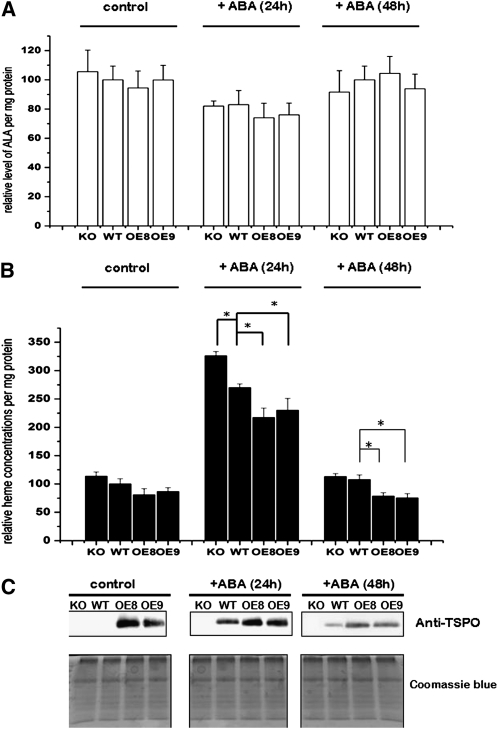
The observed heme binding by At-TSPO suggests that, when expressed, this protein could regulate heme metabolism in the plant cell, either directly or indirectly. To examine this possibility, we measured both the ALA content (precursor of all tetrapyrroles and a key limiting step in the biosynthetic pathway) and the unbound heme content of plant seedlings incubated with ABA in the wild type, a homozygote TDNA insertional knockout line (KO), and two independent homozygote AtTSPO-overexpressing lines (OE8 and OE9). As shown in Figure 7, ABA treatment for up to 48 h had no statistical effect on ALA levels in any of the genotypes tested (Figure 7A) but transiently upregulated unbound heme in all analyzed samples (Figure 7B). Unbound heme levels increased at least twofold after 24 h of incubation before returning to steady state levels after 48 h of incubation. After 24 h of ABA treatment, levels of unbound heme were highest in the KO plants, followed by the wild-type plants, then the two transgenic overexpressing lines. As shown in Figure 7C, parallel immunoblot analyses showed that the reduction in unbound heme levels coincided with the time-dependent downregulation of induced or constitutively expressed At-TSPO, suggesting a possible link between the two observations. Similar results were obtained with Arabidopsis cultured cells (see Supplemental Figure 2 online).
Figure 7.
ABA Transiently Upregulates Unbound Heme Levels in Plant Cells.
(A) The tetrapyrrole precursor ALA was quantified in a wild-type (WT) plant, an At-TSPO homozygote KO plant, and two homozygote At-TSPO–overexpressing lines (OE8 and OE9) either left untreated or incubated with 50 μM ABA for 24 and 48 h. Then, ALA levels were measured and normalized to the total protein levels. No statistical (Student’s t test, α = 0.05) difference is seen.
(B) Unbound heme was quantified in the same samples as in (A). A statistically significant difference (Student’s t test, α = 0.05) compared with the wild type is indicated by an asterisk.
(C) At-TSPO expression in the same samples as in (A) analyzed by immunoblotting and replica gels stained with Coomassie blue for loading comparisons.
The errors bars in (A) and (B) are standard deviations for the mean of six independent samples; (C) is a representative result from one of these six samples.
At-TSPO Is Degraded in Plant and Yeast Cells through the Autophagy Pathway
We then addressed the question of which biochemical pathway is responsible for At-TSPO degradation in eukaryotic cells. There are two major proteolytic systems for degradation in eukaryotes: the proteasome pathway for short-lived proteins and the lysosomal or vacuolar system of degradation by autophagy for long-lived proteins, such as At-TSPO (Xiong et al., 2007a, 2007b; Bassham, 2007; Yang and Klionsky, 2010).
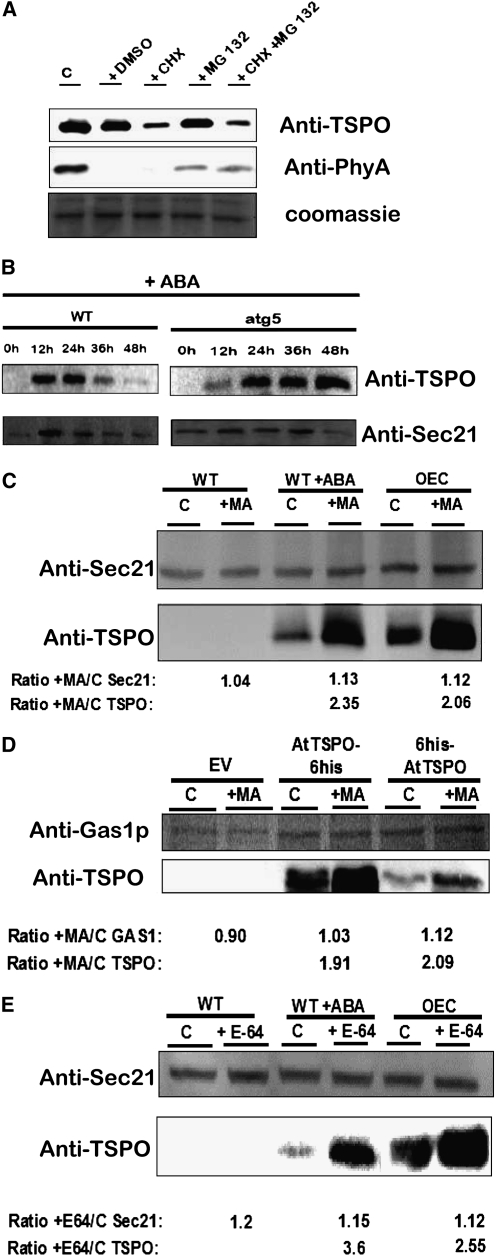
We first checked the possible involvement of the proteasome pathway in At-TSPO downregulation in the plant cell. OE8 seedlings grown in the dark for 10 d were incubated under continuous light for 6 h in presence of CHX and/or the proteasome inhibitor MG132. The light- and proteasome-sensitive Phytochrome A (PhyA) (Shanklin et al., 1987) was used as a control in immunoblotting (Figure 8A). Inhibition of protein synthesis by CHX treatment resulted in At-TSPO downregulation (cf. lanes C [control] and mock [DMSO] to lane CHX). Interestingly, the proteasome inhibitor alone or combined with CHX had no effect on the degradation rate of At-TSPO in contrast with the proteasome-sensitive PhyA, which is partially stabilized in presence of MG132 (middle panel). We showed previously that the degradation rates of At-TSPO in wild-type yeast cells and in the 26S protease mutant cim3-1 cells were similar (Figure 5A). These results show that the downregulation of At-TSPO in plant cells and in yeast is not dependent upon the proteasome degradation pathway.
Figure 8.
At-TSPO Is Degraded through the Autophagy Pathway.
(A) An Arabidopsis transgenic line overexpressing At-TSPO (OE8) was germinated and grown in the dark for 10 d on MS/2 plates and then transferred to liquid medium supplemented with 100 μM CHX and/or 50 μM MG 132 and incubated for 6 h under continuous light. Total proteins were analyzed by immunoblotting for the stability of At-TSPO before the light incubation (C, control) in presence of DSMO (mock) (top panel); PhyA was used as a light-sensitive, proteasome-sensitive control (middle panel). A replica gel was stained to check for equivalent protein loading (bottom panel).
(B) ABA-induced At-TSPO is relatively more stable in an autophagy-deficient mutant. At-TSPO was induced in wild-type and atg5 Arabidopsis seedlings by incubating them in MS/2 liquid medium containing 50 μM ABA. The seedlings were sampled at different time points and the stability of TSPO (top panels) analyzed by immunoblotting, with SEC21 as loading control (bottom panels).
(C) Arabidopsis suspension cell lines wild type, wild type +ABA (wild type treated with ABA to induce TSPO expression), and OEC (an At-TSPO–overexpressing line) were left untreated or were treated with an inhibitor of type III phosphoinositide 3-kinases, MA (5 mM), and their TSPO content assessed by immunoblotting. Arabidopsis SEC21 was used as loading control, and the signals for SEC21 and TSPO in each treated sample were compared with those in the control (C) as the MA-treated/control ratio (+MA/C). The AtTSPO/SEC21 ratio was 2.1 for wild type + ABA and 1.84 for OEC.
(D) Same experiment as in (C) using yeast expressing AtTSPO-6His, 6His-AtTSPO, and the EV control; the MA concentration was 10 mM, and Gas1p was used as loading control. The AtTSPO/Gas1p ratio was 1.85 for AtTSPO-6His and 1.86 for 6His-AtTSPO.
(E) The same experiment as in (C) but using the vacuolar Cys protease inhibitor E64d (E64, 100 μM) instead of MA. The AtTSPO/SEC21 ratio was 3.1 for wild type + ABA and 2.3 for OEC.
The results in (A) and (B) were reproduced twice and in (C) to (E) are representative of those in at least three separate experiments. WT, Wild type.
Oxidative stress in plant cells is enhanced by abiotic stress or ABA treatment, and reactive oxygen species (ROS) activate autophagy to degrade damaged organelles and oxidized proteins rapidly and effectively in plant cells (Xiong et al., 2007a, 2007b). Autophagy is a general term for the processes (macroautophagy, microautophagy, and chaperone-mediated autophagy) by which cytoplasmic materials, including organelles, reach the lysosome/vacuole for degradation (Rubinsztein, 2006; Bassham, 2007; Levine and Kroemer, 2008; Mizushima et al., 2008; Yang and Klionsky, 2010); however, macroautophagy is the best studied of these processes and will be referred to hereafter simply as autophagy.
To test the possibility that At-TSPO might be degraded in vivo through autophagy, we reasoned that if this is the case, At-TSPO should be more stable in an autophagy-deficient mutant compared with wild-type plants. To test this hypothesis, we induced At-TSPO in wild-type Arabidopsis and in the At-ATG5 knockout (atg5) mutant seedlings (Yoshimoto et al., 2009; Kwon et al., 2010) and followed the stability of the ABA-induced At-TSPO with time. The ABA-dependent induction of At-TSPO in wild-type seedlings was transient, peaking around 24 h after induction and declining thereafter (Figure 8B, wild-type panels), consistent with our previous findings (Guillaumot et al., 2009a). However, when atg5 mutant seedlings were subjected to the same treatment, the induced At-TSPO appeared to be stabilized by 24 h after induction (Figure 8B, atg5 panels). These results indicate that At-TSPO degradation by plant tissues requires a functional autophagic pathway.
Autophagy is a catabolic pathway that is evolutionarily conserved from yeast to mammals, and central to this process is the formation of autophagosomes, which are double-membrane vesicles responsible for delivering long-lived proteins and excess or damaged organelles to the lysosome/vacuole for degradation and for the reuse of the resulting macromolecules (Yang and Klionsky, 2010). As autophagosome formation requires class III phosphoinositide 3-kinase (PI3K) activity, PI3K inhibitors, such as 3-methyladenine (MA) or wortmannin, are commonly used to inhibit autophagy (Blommaart et al., 1997; Takatsuka et al., 2004; Itakura et al., 2008; Klionsky et al., 2008; Matsunaga et al., 2009). We reasoned that inhibiting autophagy should result in increased stability of the protein. First, we treated wild-type Arabidopsis cultured cells (with or without preincubation with ABA to induce TSPO) or a transgenic cell line overexpressing At-TSPO (OEC) with MA. Figure 8C shows that MA treatment on its own did not induce TSPO expression in wild-type cells. However, ABA-induced TSPO was more stable in the presence of MA than in the non-MA-treated sample (bottom panel, compare lanes C [control] and lane +MA in the wild type +ABA group). In addition, enhanced stability was also observed for constitutively expressed TSPO after MA treatment (bottom panel, compare lanes C [control] and lane +MA in the OEC group). Wortmannin treatment had similar effects (see Supplemental Figure 3 online). Second, to check whether this phenomenon was also seen in non-plant cells, we tested MA sensitivity of At-TSPO heterologously expressed in yeast, and, again, the degradation of both AtTSPO-6His and 6His-AtTSPO was slowed after treating the yeast cells with MA (Figure 8D). Although MA was thought to be an autophagy-specific inhibitor (Seglen and Gordon, 1982), in addition to regulating autophagy, type III PI3K enzymes regulate diverse cell signaling and membrane trafficking processes; therefore, these PI3K inhibitors are not strictly autophagy specific (Klionsky et al., 2008; Mizushima et al., 2010). To verify independently that At-TSPO was transported to the vacuole for degradation, we reasoned that inhibiting vacuolar proteases should enhance At-TSPO stability. As shown in Figure 8E, when Arabidopsis cultured cells were treated with the commonly used Cys protease inhibitor E64d, the downregulation of ABA-induced or constitutively expressed TSPO was reduced. Taken together, these data strongly support the idea that At-TSPO is mainly, if not exclusively, degraded via autophagy in plant cells and in yeast.
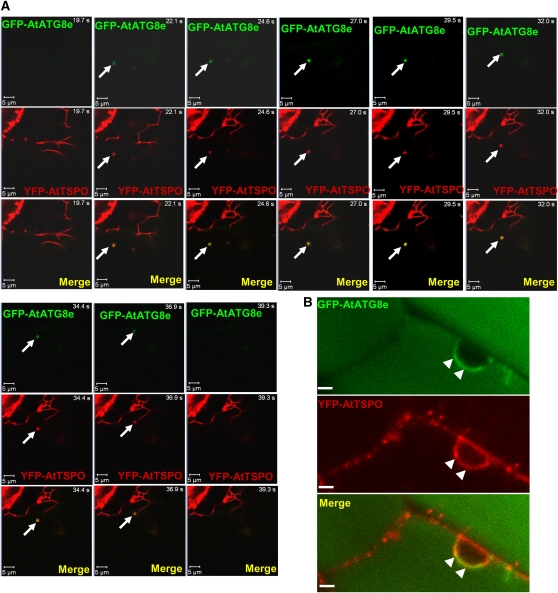
YFP-AtTSPO Partially Colocalizes with GFP-AtATG8e in Plant Cells
The process of autophagy involves a set of evolutionarily conserved gene products, known as the ATG proteins, which are required for the formation of the isolation membrane and the autophagosome (Mizushima et al., 2010). The regulatory conserved ATG8 family proteins are ubiquitin-like proteins that are conjugated to phosphatidylethanolamine and attached to both faces of the double-membrane autophagosome from its formation to its degradation in the vacuole and are therefore commonly used in eukaryotic cells as autophagosome markers, for example, fused to a fluorescent protein (Contento et al., 2005; Mizushima et al., 2010).
Since YFP-AtTSPO was also sensitive to heme metabolism and autophagy inhibitors (see Supplemental Figure 4 online) and to senescence (see Supplemental Figure 5 online), we reasoned that coexpression of YFP-AtTSPO and green fluorescent protein (GFP)-AtATG8 should occasionally result in colocalization of the two fusion proteins even in the absence of stress or starvation. To test this idea, we retransformed a transgenic tobacco line overexpressing YFP-AtTSPO with GFP-AtATG8e (Contento et al., 2005). Figure 9 shows representative confocal images of root cells coexpressing YFP-AtTSPO and GFP-AtATG8e. As seen in the time-lapse series in Figure 9A, a moving autophagosome was detected in the field of view at time point 22.1 s (arrow, green channel), and this punctate structure also contained YFP-AtTSPO (arrows, red channel and merged image). This moving autophagosome was visible up to time point 36.9 s, with perfect colocalization of the YFP and GFP fluorescence. This interesting observation suggests that a YFP-AtTSPO–containing organelle or the fusion protein itself can be engulfed by a GFP-AtATG8e–positive autophagosome in the plant cell, strengthening the idea that At-TSPO can be degraded through autophagy. In the experiments shown in Figure 9B, we treated the root cells before imaging with concanamycin A, a vacuolar H+-ATPase inhibitor used to increase the pH and indirectly reduce vacuolar protease activity (Matsuoka et al., 1997; Xiong et al., 2007b; Kleine-Vehn et al., 2008). As expected, diffuse GFP fluorescence from GFP-AtATG8e accumulating in the vacuole could be detected (green channel, top panel). We did not detect the YFP fluorescence in the vacuolar compartment probably because of YFP signal dilution. Interestingly, and probably as a result of reduced autophagic flux (Mizushima et al., 2010), brighter GFP-AtATG8e–highlighted structures could also be visualized, for example around the nucleus, locally colocalizing with YFP-AtTSPO fluorescence (arrowheads).
Figure 9.
YFP-AtTSPO Partially Colocalizes with GFP-AtATG8e in Plant Cells.
(A) Confocal images of a time-lapse study on root cells of a tobacco plant stably coexpressing YFP-AtTSPO and the autophagy marker GFP-AtATG8e. The arrow indicates a moving autophagosome (GFP-AtATG8e fluorescence, green channel, and merged) containing YFP-AtTSPO fluorescence (red channel and merged) seen from time point 22.1 to 36.9 s. Bar = 5 μm.
(B) Similar sample to that in (A) but imaged after concanamycin A treatment of the root cells. Diffuse GFP-AtATG8e fluorescence is visible in the vacuole (green channel), and brighter areas are seen around the nucleus (arrowheads) colocalizing with YFP-AtTSPO (red channel and merged image). Bar = 5 μm.
At-TSPO Degradation Requires a Putative ATG8 Interacting Motif in Addition to Heme
Although autophagy is, in principle, a nonselective degradation process, recent studies have shed light on other autophagy modes that selectively degrade proteins, organelles, and even invasive pathogens (reviewed in Noda et al., 2010). Structural studies have uncovered conserved specific interactions between ATG8 family proteins and either autophagic cargo receptors or defined autophagic cargoes through a consensus W/YxxL/V/I sequence termed the ATG8 family interacting motif (AIM) on the binding partner of the ATG8 protein (Noda et al., 2008, 2010). The results presented in Figure 9 suggested that At-TSPO or organelles containing At-TSPO might be engulfed by autophagosomes through an ATG8-mediated interaction. This possibility was increased by the finding that At-TSPO and other angiosperm TSPOs contain a relatively conserved AIM-related motif outside the predicted transmembrane domains (Figure 10A). To investigate the possibility that this motif could function as an AIM and may be required for At-TSPO degradation, we mutated the two Tyr residues in 121LYLYL125 into Ala residues, generating AtTSPOLALA. This was expressed in yeast and its sensitivity to the autophagy inhibitor MA compared with that of the wild-type protein. As shown in Figure 10B, in the absence of MA, AtTSPOLALA was more stable than the wild-type protein (cf. the control [C] lanes in AtTSPO and AtTSPOLALA). In addition, AtTSPOLALA was far less sensitive to MA treatment, suggesting that autophagy of AtTSPOLALA was compromised compared with that of the wild-type protein. This could have been due to the Tyr substitutions indirectly affecting heme binding; however, as shown in Figure 10C, this was not the case, since heme binding was not affected by the mutations (cf. lanes EHA for AtTSPO and AtTSPOLALA). AtTSPOLALA was less sensitive to heme added to the culture medium, as shown in Figure 10D. These data show that At-TSPO degradation by autophagy requires the AIM-like motif, in addition to heme. To determine whether heme binding directly regulated autophagy, we evaluated the sensitivity of the previously generated His mutants to MA treatment, and the results in Figure 10E confirm that deficiency in heme binding, as exemplified by the two mutants AtTSPOH91A and AtTSPOH91A/H115A, correlated with reduced sensitivity to MA. Thus, we conclude that heme binding and a functional AIM in At-TSPO are required for autophagy-dependent degradation of the protein in vivo.
Figure 10.
At-TSPO Degradation Requires an ATG8 Family Interacting Motif.
(A) ClustalW local alignment of a putative ATG8-family interacting motif (AIM; in bold and underlined) in some angiosperm TSPOs but absent in human TSPO1 and TSPO2.
(B) To assess the functionality of the At-TSPO putative AIM (121LYLYL125), the two Tyr residues were mutated to Ala and the resulting mutant (AtTSPOLALA) expressed in yeast. Its sensitivity to MA was compared with that of the wild-type protein as in Figure 8B. The AtTSPO/Gas1p ratio was 2.02 for At-TSPO and 1.34 for AtTSPOLALA.
(C) The hemin binding ability of AtTSPOLALA was assessed as in Figure 6C.
(D) The hemin sensitivity of AtTSPOLALA was assessed as in Figure 6D; the AtTSPO/Gas1p ratio was 0.41 for wild-type At-TSPO and 0.9 for AtTSPOLALA.
(E) The sensitivity of the His mutants was investigated as in (B). The AtTSPO/Gas1p ratio was 3.7 for wild-type At-TSPO, 0.7 for AtTSPOH91A, 2.21 for AtTSPOH115A, and 0.96 for AtTSPOH91A/H115A.
The results in (B) to (D) are representative of those in two independent experiments; those in (E) are representative of those in three experiments.
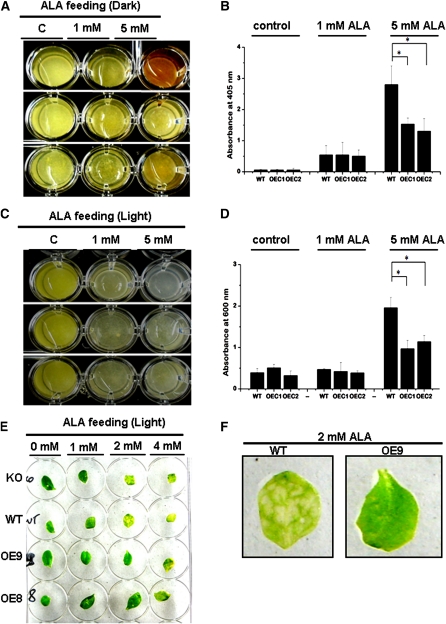
At-TSPO Can Alleviate ALA-Induced Porphyria in Plant Cells
Since At-TSPO can bind porphyrins and is efficiently degraded through autophagy by the cell, it might act as a potential porphyrin (including heme) scavenger in vivo. To test this potential At-TSPO function during stress, we increased tetrapyrrole biosynthesis in Arabidopsis plant cells by ALA feeding. When Arabidopsis cultured cells were incubated with ALA for 4 d in the dark, wild-type cells accumulated porphyrins, as shown by their brownish-red coloration (Figure 11A, wild type treated with 5 mM ALA). Interestingly, the independent At-TSPO–overexpressing cell lines OEC1 and, to a lesser extent, OEC2 were less colored at the same ALA concentration, suggesting that their content of harmful porphyrins was reduced. The absorbance at 405 nm of crude cells extracts from 5 mM ALA-treated cells indicates that the wild-type cells accumulated more porphyrin-like compounds as compared with the transgenic cells overexpressing At-TSPO (Figure 11B). Free porphyrins, including heme, are extremely harmful compounds as they can become cytotoxic by producing powerful radicals in the presence of light (Wagner et al., 2004; Severance and Hamza, 2009; Mochizuki et al., 2010). When ALA feeding was performed under continuous illumination, wild-type cells became completely photobleached after 4 d of incubation (Figure 11C, wild type treated with 5 mM ALA), while OEC1 and OEC2 were markedly less affected by the same treatment, suggesting that overexpression of At-TSPO might protect the cell from porphyrin-induced cytotoxicity. This conclusion was verified by measuring cell viability after ALA treatment (Figure 11D). In presence of 5 mM ALA, the wild-type cells were less viable compared with the transgenic cells overexpressing At-TSPO. A similar conclusion can be extended to the plant: whereas Arabidopsis KO and wild-type mature leaves were severely chlorotic when incubated with ALA at concentrations of 2 and 4 mM, leaves from overexpressing plants OE8 and OE9 appeared macroscopically unaffected or less affected at the same ALA concentrations (Figures 11E and 11F).
Figure 11.
Overexpression of At-TSPO Can Alleviate Porphyria in Plant Cells.
(A) and (B) Tetrapyrrole biosynthesis was boosted in Arabidopsis cultured cells by incubating them with ALA for 4 d in the dark. The resulting coloration of the wild-type (WT) cells and two independent At-TSPO–overexpressing lines (OEC1 and OEC2) indicate a differential accumulation of uncommitted porphyrins (A) confirmed by the comparative absorbance at 405 nm of the respective extracts (B). The error bars in (B) represent standard errors, and statistically significant differences (Student’s t test at α = 0.05) are highlighted by an asterisk.
(C) and (D) The same experiment as in (A) but with the cells incubated 24 h under continuous light. The imaged cells show differential bleaching between the wild type on the one hand and OEC1 and OEC2 on the other and were confirmed by an Evans blue residual cell viability test (D).
(E) and (F) Similar experiment to that in (C) but using cauline leaves from a wild-type plant, an At-TSPO KO plant, and two independent and homozygote At-TSPO–overexpressing lines (OE8 and OE9). At 2 mM ALA, the imaged leaves show variations in porphyrin-induced damages between the wild-type and KO sample on the one hand and OE8 and OE9 on the other, with visible chlorotic areas (wild type) in the magnified images in (F).
DISCUSSION
In this work, we present evidence that At-TSPO is a heme binding protein, that heme binding is required for At-TSPO downregulation through the autophagic pathway, and that At-TSPO degradation through autophagy is conserved even after heterologous expression of At-TSPO in yeast and requires a functional AIM-related motif. These findings raise a number of questions.
Where and How Does At-TSPO Acquire Heme in Plant Cells?
At-TSPO is an ER-Golgi–localized membrane protein in the plant cell (Guillaumot et al., 2009a). Despite the fact that heme biosynthesis and its regulation have been extensively studied, very little is known about the intracellular trafficking of heme (or any other porphyrin species), but it stands to reason that specific molecules and pathways transport heme from the site of synthesis to other cellular compartments and assist its incorporation into hemoproteins (Severance and Hamza, 2009; Mochizuki et al., 2010). The steady state rate of heme efflux from isolated chloroplasts has been estimated to be 0.12 to 0.45 pmol/min, based on a free heme estimate in the plastid of ~6 μM (Thomas and Weinstein, 1990). It is not clear when and how hemoproteins acquire their heme in eukaryotic cells (i.e., cotranslationally or posttranslationally). Heme is a hydrophobic and potential powerful cytotoxic molecule, with a tendency to aggregate at neutral pH, so understandably, levels of intracellular uncommitted heme are strictly maintained at below 109 M (Taketani, 2005). This level may be even lower in the cytosol. We showed that a transient increase in unbound heme levels occurred concurrently with ABA-induced expression of At-TSPO (Figure 7). Although the method used for this quantification supposedly measures free heme, it is likely that the measured heme pool includes uncommitted heme and a portion of the noncovalently bound heme released from proteins during the extraction procedure. Nevertheless, these data clearly show that, during ABA perception and signaling, the plant cell transiently increases its heme content. It may be that an intracellular increase in ABA (during water-related stress or from exogenous ABA) reduces the flux of PPIX toward the chlorophyll branch of the tetrapyrrole biosynthetic pathway, since the expression of chlorophyll binding proteins is inhibited (Koussevitzky et al., 2007), thus channeling the available PPIX toward the heme branch, and this may explain why we found little modification of the levels of the precursor ALA (Figure 7). The increased heme levels may fulfill two physiological purposes related to the inevitable oxidative stress accompanying abiotic stress. First, part of the uncommitted heme may be degraded in the plastid by heme oxygenase (HO) to generate biliverdin-IXα, a known antioxidant molecule (Otterbein et al., 2003; Balestrasse et al., 2005). The highly expressed HO1 isoform in plants is upregulated by various abiotic stresses and ABA (reviewed in Shekhawat and Verma, 2010). Second, some of the uncommitted heme may leave the plastid by an as yet uncharacterized mechanism and be allocated to heme binding proteins, including ROS scavenging enzymes, in various compartments of the cell. The in vivo heme analog binding assay suggested that a small fraction of the Golgi-localized At-TSPO may be devoid of porphyrins (Figure 4). It may be that the ER-bound and some of the Golgi-localized At-TSPO molecules are complexed with heme (or other porphyrins).
We showed that heme binding by At-TSPO requires H91 but not H115 (Figure 6). Although the two His residues are relatively well conserved in plant TSPOs, only H91 is surrounded by the hydrophobic residues required to form a potential heme binding groove (Lucana et al., 2004). H91 may function as a heme iron axial coordinator, suggesting that the interaction between At-TSPO and heme is noncovalent and most likely reversible. It is not yet clear how heme is transferred to At-TSPO (in the ER or the Golgi) (i.e., directly from the plastid or from the cytosol). While it is possible that soluble heme binding proteins, such as members of the p22HBP/Soul family (Takahashi et al., 2008), may transport heme within, or to, defined organelles, their function in porphyrin trafficking, if any, remains to be determined. Stromules are tubular structures, stroma-filled protrusions, of plant plastids and are thought, besides functioning to aiding the exchange of material between plastids, to play a role in facilitating the exchange of material between plastids and other organelles (Kwok and Hanson, 2004; Ishida et al., 2008). A trafficking pathway for proteins exists between the Golgi and plastids (Villarejo et al., 2005; Kitajima et al., 2009), and another for lipids exists between plastids and the ER (Awai et al., 2006), probably through direct organelle contact. A possible direct transfer of heme by the tethering of physical interorganellar membranes, as described between mitochondria and the ER in yeast (Kornmann et al., 2009), could explain how heme from the plastid could be incorporated into proteins that are processed in the secretory pathway (Severance and Hamza, 2009), such as At-TSPO.
Is At-TSPO a Heme/Porphyrin Transporter/Scavenger?
Although the biological role of TSPOs have been extensively investigated in mammalian cells for more than three decades, we know nothing about the mode of action of any member of this fairly conserved family of membrane proteins (i.e., whether they operate as pumps, transporters, or channels). Since it is a hydrophobic anionic porphyrin, transport of heme across a lipid bilayer should consume energy. Hence, most of the heme transporters characterized so far belong to the ATP binding cassette protein superfamily (Severance and Hamza, 2009; Mochizuki et al., 2010). However, transmembrane proteins can function as heme chaperones, as exemplified by CcmE in bacteria and Arabidopsis, which delivers heme to apocytochrome C (Spielewoy et al., 2001; Ahuja and Thöny-Meyer, 2006). The reduction in cellular unbound heme levels concomitant with At-TSPO degradation (Figure 7) might be due to the transfer of uncommitted heme to yet unidentified hemoproteins in the ER and/or the Golgi, via At-TSPO. However, this possible hemochaperone activity does not explain why, in a heme-dependent manner, the protein, and probably the whole Golgi stack containing At-TSPO, is then targeted for degradation. We favor the alternative explanation that At-TSPO regulates unbound heme levels in the cell during stress (Figure 12). During stress, the ABA-dependent heme increase in the plastid is required to protect cellular structures from ROS damage. Part of the unbound heme must be incorporated into a multitude of ROS scavengers distributed in other compartments of the cell and also into other hemoproteins, including ABA regulators, such as the ER-localized catabolic enzyme ABA 8’-hydroxylase. It is vital for the plant cell to regulate levels of active ABA strictly. To avoid excess unbound heme accumulating in the membranes or in the cytosol, At-TSPO is also induced by ABA as a porphyrin scavenger. Frank et al. (2007) found that a knockout mutant for one of the Physcomitrella patens TSPOs showed PPIX accumulation and increased sensitivity to oxidative stress. It is also possible that binding of heme to At-TSPO can enhance ABA-dependent oxidative stress signaling, as At-TSPO enhanced the intrinsic peroxidase activity of heme (see Supplemental Figure 6 online). This may result from the bound heme interacting with H2O2 and generating powerful radicals (Balla et al., 2000). Consistent with this hypothesis, At-TSPO overexpression substantially increased ROS levels in plant cells in the presence or absence of ABA (see Supplemental Figures 7 and 8 online). These results have to be taken with caution since the absence of functional chloroplasts in cultured plant cells grown in the dark results in the overexpressed At-TSPO being more stable. The potential role of At-TSPO in ROS homeostasis in the plant cell is still far from clear. However, a role of At-TSPO as a heme/porphyrins scavenger is in accordance with the finding that overexpression of this protein alleviated ALA-induced porphyria in plant cells (Figure 11). TSPO2, a divergent TSPO isoform exclusively expressed in avian and mammalian primitive erythrocytes, is localized to the ER and not the mitochondria, and its expression is tightly correlated with that of hemoglobin genes (Fan et al., 2009; Nakazawa et al., 2009), suggesting a role in heme and hemoprotein accumulation during erythrocyte maturation. In addition, TSPO knockout in zebra fish resulted in embryos with specific erythropoetic cell depletion, and this lack of blood cells was independent of the TSPO function in cholesterol transport and steroidogenesis (Rampon et al., 2009).
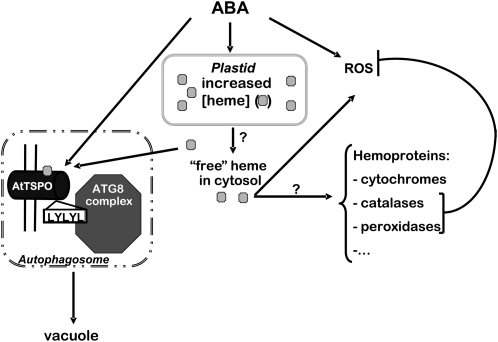
Figure 12.
Hypothetic Model of At-TSPO Function and Regulation in Plant Cells.
Increased ABA levels in the plant cell transiently upregulate uncommitted heme levels. The uncommitted hemes are used to increase the activity of ROS scavengers inside and outside the chloroplast (required for the cell to cope with the accompanying oxidative stress). ABA also induces TSPO as a heme scavenger to cope with possible excess and deleterious heme in the cell. TSPO binds heme and interacts with ATG8 family regulators via a conserved AIM motif (121LYLYL125) and is targeted for degradation through autophagy. How heme is transported out of the plastid is not known, and how and when hemoproteins acquire their heme is not yet clear (question marks).
Why Do Plant Cells Need an Efficient At-TSPO Downregulation Mechanism?
At-TSPO might be required for a short time in enhancing the stress-induced oxidative stress and preventing the accumulation of uncommitted heme/porphyrins in the cell. However, in the long term, these activities are likely to be detrimental by affecting ROS homeostasis and competing for heme allocation to ROS scavengers or other hemoproteins, including ABA catabolic enzymes (Figure 12). In contrast with TSPO1 in animal cells, At-TSPO appears not to be essential for plant growth and development, since the knockout mutant is viable. We previously found that overexpression of At-TSPO renders cultured Arabidopsis cells more sensitive to salt stress (Guillaumot et al., 2009a). Enzymatic degradation of heme is restricted to the plastids in the plant because of the specific localization of HO in this organelle. It is not known whether heme or porphyrins in general can be retranslocated in the plastids. One way of getting rid of the potential deleterious effects of heme bound to At-TSPO would be to degrade the complex via the vacuole, a degradation site for various hemoproteins and organelles containing hemoproteins. The autophagosomes are formed in mammalian cells from ER, Golgi, or even mitochondrial membranes (Hayashi-Nishino et al., 2009; Ylä-Anttila et al., 2009; Hailey et al., 2010; Yen et al., 2010). This potential mechanism is reminiscent of heme binding to the prion protein PrPC, which generates ROS and induces PrPC internalization and turnover (Lee et al., 2007). Heme is known to regulate the turnover of various regulatory proteins (Severance and Hamza, 2009), but none have yet been characterized in plants.
We found that At-TSPO was degraded through autophagy and that this degradation required the AIM-like motif 121LYLYL125. Structural studies suggest that AIM motifs adopt an extended β-conformation and form an intermolecular parallel β-sheet with the β2 domain of ATG8 family proteins (Noda et al., 2008). It is possible that the consensus W/YxxL/V/I could be recognized in both orientations in the primary sequence. We showed that mutating the hydrophobic residues possibly required for ATG8 binding resulted in inhibition of At-TSPO degradation. We cannot exclude the possibility that a fraction of At-TSPO might be degraded by other cellular process than autophagy. However, genetic and pharmacological inhibition of the proteasome pathway did not affect the stability of At-TSPO, and the autophagy-dependent degradation pathway was conserved even after heterologous expression of At-TSPO in yeast. These findings suggest that the cellular mechanism for At-TSPO recognition and degradation by eukaryotic cells is a housekeeping mechanism. It would be interesting to determine whether any of the degradation-deficient mutant forms of At-TSPO has a phenotypic consequence in the plant.
METHODS
Plant Materials
Arabidopsis thaliana Columbia-0 ecotype was used. The wild type suspension cultured cells and the generation of the transgenic cell lines overexpressing At-TSPO were described by Guillaumot et al. (2009a). OEC1, OEC2, and OEC are three independent transgenic cell lines expressing At-TSPO under the control of the p35S promoter. An Arabidopsis homozygous TDNA insertional line (KO) in TSPO and the TSPO-overexpressing transgenic plant lines were described by Guillaumot et al. (2009a). Transgenic tobacco plant expressing ST-YFP (Geelen et al., 2002) was also previously described (Guillaumot et al., 2009a).
Genetic Constructs and Transformations
Standard molecular biology protocols were followed, the generated plasmids were amplified in Escherichia coli strain DH5α, and all coding sequences were validated by sequencing. To generate yeast expression plasmids encoding At-TSPO with or without the 6-His tag, specific primers were used to PCR amplify the At-TSPO coding sequence as described by Guillaumot et al. (2009a). The primer pairs AtTSPO5′ (5′-AAAATCTAGAATGGATTCTCAGGACATC-3′) and AtTSPO3′ (5′-AAAACTGAGTTATCACGCGACTGCAAGCTTTA-3′), AtTSPO5′ and AtTSPO-6xHis3′ (5′-AAAACTCGAGTTAGTGATGGTGATGGTGATG-3′), and 6xHis-AtTSPO5′ (5′-AAAATCTAGAATGCATCACCATCACCATCAC-3′) and AtTSPO3′ were used to generate AtTSPO, AtTSPO-6His, and 6His-AtTSPO, respectively. These amplified PCR products were digested with XbaI/XhoI and cloned into the vector p426GAL1 (Mumberg et al., 1994), previously opened with SpeI/XhoI and the generated plasmids named p426GAL1-AtTSPO, p426GAL1-AtTSPO-6His, and p426GAL1-6His-AtTSPO, respectively. The p426GAL1-AtTSPO plasmid was used as the template for the generation of AtTSPOLALA, AtTSPOH91A, and AtTSPOH115A. These constructs were prepared by overlapping PCR. To generate AtTSPOLALA, the first PCR product was amplified using the primers AtTSPO5′ and AtTSPOLALA3′ (5′-AAACTGAGCTAAAGCAAGAGCCAGAGCATTGGG-3′) and the second using the primers AtTSPOLALA5′ (5′-CCCAATGCTCTGGCTCTTGCTTTAGCTCAGTTT-3′) and AtTSPO3′. To generate AtTSPOH91A and AtTSPOH115A, the first PCR products were generated using the primer pairs AtTSPO5′ and either AtTSPOH91A3′ (5′-GCGAGACACGTTGTTGCTAGGAGCCACA-3′) or AtTSPOH115A3′ (5′-AGCATTGGGCTTCTTTGCGAAGCCACC-3′), respectively, and the second PCR products were obtained using the pairs AtTSPO3′ and either AtTSPOH91A5′ (5′-CCTCTGTGGCTCCTAGCAACAACGTGTC-3′) or AtTSPOH115A5′ (5′-GACGGTGGCTTCGCAAAGAAGCCCAATG-3′). The final amplified PCR products were digested with XbaI/XhoI and cloned into p426GAL1, previously opened with SpeI/XhoI, and the generated plasmids named p426GAL1-AtTSPOLALA, p426GAL1-AtTSPOH91A, and p426GAL1-AtTSPOH115A, respectively. AtTSPOH91A/H115A was generated by PCR combining the appropriate fragments from AtTSPOH91A and AtTSPOH115A and also cloned into p426GAL1 to give p426GAL1-AtTSPOH91A/H115A. Yeast strains CMY394 and Y762 (Ghislain et al., 1993) were transformed as described by Gietz and Woods (2002). The generation of the YFP-AtTSPO–expressing tobacco (Nicotiana tabacum) and Arabidopsis plants and the transgenic suspension Arabidopsis cultured cells has been described by Guillaumot et al. (2009a). The binary vector containing GFP-AtATG8e (Contento et al., 2005) was a kind gift from D. Bassham (Iowa State University). Arabidopsis and tobacco plant transformations were performed using standard methods using Agrobacterium tumefaciens–mediated transformation as described by Guillaumot et al. (2009a).
Plant and Yeast Treatment
Seed disinfection, plant growth conditions, and ABA treatment were performed as described by Guillaumot et al. (2009a). Plant cultured cells were grown as described previously (Guillaumot et al., 2009a). Plant seedlings grown for 14 d in vitro were transferred to liquid MS/2 (half-strength Murashige and Skoog culture medium) containing ALA, DPD, or MA at the indicated concentration and incubated for 1 to 4 d in the dark with or without a prior 24 h ABA induction. Plant cultured cells were harvested 3 d after subculture and then transferred to fresh culture media containing ALA, DPD, MA, E-64d, wortmannin, or CHX at the indicated concentration. A 50 mM ABA stock solution was prepared in ethanol, DPD, MA, SA, CHX, and wortmannin stock solutions were prepared in DMSO, and a 10 mM E-64d stock solution was prepared in methanol. The controls contained the appropriate solvent at the highest final concentration used. Porphyria was induced in plant cells by incubating the cultured plant cells with ALA at the indicated concentration either in the dark or under continuous light for up to 4 d, and then the cells were thoroughly washed or resuspended in the culture medium without ALA for imaging. Total porphyrin extraction and quantification were conducted as described by Dixon et al. (2008). Cell viability was assessed as described by Frank et al. (2007). The first cauline leaves of soil-grown plants at the bolting stage were also incubated in MS/2 liquid medium containing ALA at room temperature under continuous light and imaged after 24 h of incubation. At-TSPO induction was achieved in yeast cells by growing them for 2 d at 30°C in liquid synthetic medium without the auxotrophic marker uracil and with galactose as carbon source. When required, the medium was supplemented with ALA, DPD, SA, or hemin (freshly prepared in 20 mM NaOH), CHX, and/or MG132 (prepared in DSMO) at the indicated concentration.
Protein Extraction and Analyses
Plant protein extraction was performed as described by Guillaumot et al. (2009a). Total yeast proteins were prepared in buffer A (20 mM Na-phosphate buffer, pH 7.8, 150 mM NaCl, and 2% [w/v] dodecyl-β-d-maltoside [DDM] containing 1 mM PMSF and a protease inhibitor cocktail [final concentration 2 μg/mL each of leupeptin, aprotinin, antipain, pepstatin, and chymostatin]). When required, DDM was replaced by SDS (1% [v/v]) or Triton X-100 (1% [v/v]) in the solubilization buffer A. Disruption of yeast cells was performed at room temperature in a Precellys24 apparatus (Bertin Technologies) using 500- to 750-μm glass beads. After homogenization, cell debris was pelleted by centrifugation for 5 min at 5000g in a cooled microcentrifuge and the supernatant used immediately for protein quantification using the BCA method (Sigma-Aldrich). Microsomes were obtained and solubilized according to Kanczewska et al. (2005), except that 0.1% polyoxyethylene 8-mistryl ether (C14E8) was used for stripping and 2% DDM for membrane protein solubilization. The solubilized proteins were incubated for a minimum of 2 h at room temperature with prewashed Ni-Sepharose high performance beads (GE Healthcare) suspended in buffer A containing 20 mM imidazole and the beads poured into a column that was then washed with increasing concentrations of imidazole (20, 40, and 60 mM) in washing buffer A (with 0.1% [w/v] DDM instead of 2% [w/v] DDM). Then, bound proteins were eluted with 500 mM imidazole in washing buffer. If required, the eluted proteins were dialyzed extensively at 4°C against 20 mM Na-phosphate, 150 mM NaCl, and 0.1% (w/v) DDM and concentrated using an Amicon ultracentrifugal filter device (Millipore). Protein electrophoresis, immunoblotting, and signal quantifications were performed as described by Guillaumot et al. (2009a). Antibodies against At-SEC21, PhyA, and At-rbcL were purchased from AgriSera, the anti-Gas1p antibodies (Morsomme et al., 2003) were a kind gift from Pierre Morsomme (Institute of Life Sciences, Université Catholique de Louvain, Belgium), anti-β-galactosidase was purchased from Promega, the horseradish peroxidase–coupled anti-rabbit and anti-mouse secondary antibodies were from Sigma-Aldrich, and the anti-AtTSPO and anti-YFP antibodies have been described previously (Guillaumot et al., 2009a). For immunoblotting, the anti-YFP antibodies were used at a dilution of 1:500, the anti-AtSEC21, anti-AtrbcL, anti-Gas1p antibodies at 1:1000, anti-β-galactosidase at1/1000, anti-AtTSPO antibodies at 1:5000, and anti-rabbit IgG antibodies at 1:10,000.
Porphyrin Affinity Chromatography and Pull-Down Assays
For the hemin affinity binding assay, we used commercially available hemin-agarose (Sigma-Aldrich). For the PPIX affinity binding assay, we cross-linked PPIX in the dark to Sepharose CL4B beads using a Carboxylink kit (Pierce) according to the manufacturer’s instructions. Cross-linking to the beads was verified by the coloration of the column by PPIX (dark brownish-red). Untreated Sepharose CL4B beads were used as a negative control for nonspecific interactions between the agarose and membrane proteins. Approximately 500 μg of total membrane proteins was solubilized on a rotating wheel at 4°C for 1 h in 1 mL of solubilization buffer (20 mM Na-phosphate, pH 7.8, 500 mM NaCl, and 2% DDM), and then after centrifugation (15,000g, 20 min, 4°C), the supernatant was recovered and N-laurylsarcosine was added to a final concentration of 1% (v/v). Routinely, 200 μL of this mixture was then added to 100 μL of the porphyrin-agarose slurry in a 1 mL column prewashed with solubilization buffer and the suspension incubated at room temperature for 1 h on a rotating wheel (20 rotations per minute), and then the nonbound material was collected and the beads washed three times with solubilization buffer and three times with PBS. Then, the bound protein was eluted by addition of 4× SDS protein loading buffer and incubation at room temperature for 10 min. The input, flow-through, last wash, and elution were subjected to SDS-PAGE and immunoblot analyses.
For the pull-down assay, purified heterologously expressed AtTSPO-6His and 6His-AtTSPO were intensely dialyzed against buffer B (20 mM Na-phosphate, pH 7.8, 500 mM NaCl, and 0.1% DDM) to remove the imidazole used to elute the protein from the Ni-NTA matrix. The dialyzed fraction (150 μL) was made to a final concentration of 1 mM hemin (from a 10 mM stock in 20 mM NaOH) or 1 mM PPIX (from a 10 mM stock in DMSO), and the mixtures and the respective mock samples containing 20 mM NaOH or DMSO were incubated in the dark for 30 min on a rotating wheel at room temperature. The samples were then applied to a suspension of prewashed Ni-NTA beads and incubated 1 h at room temperature, and then the beads were poured into a column, the flow-through was collected, and the column washed twice with 5 column volumes of buffer B and bound proteins eluted with 2 column volumes of buffer B containing 500 mM imidazole. The eluted material was transferred to a multiplate and spectrophotometrically analyzed for the presence of porphyrins (absorption at 405 nm). Buffer solution and the eluate from affinity chromatography of solubilized proteins from yeast transformed with the EV were used as negative controls. All experiments were performed at least three times.
ALA and Heme Extraction and Quantification
Plant cell suspensions were filtered on a Miracloth membrane and washed with 20 mL of deionized sterile water. For the determination of ALA and heme, 500 mg fresh weight of cells was used as the starting material. All extractions were performed in the dark. ALA extraction and measurement were performed as described by Mauzerall and Granick (1956). Heme concentrations were determined using a hemin assay kit from Biovision. Heme extractions were performed based on Weinstein and Beale (1983). Briefly, the washed cells were extracted at least five times by resuspension in cold 90% (v/v) aqueous acetone containing 10 mM NH4OH and the extracts discarded. Noncovalently bound heme was then extracted twice in cold 90% aqueous acetone containing 5% (v/v) 12.1 M HCl and the two HCl extracts combined and diluted 100-fold in Biovision kit buffer and heme levels determined according to the manufacturer’s instructions.
For the determination of ALA and heme levels in plant tissues, 6-week-old plants grown on soil under short-day conditions were harvested and ground in liquid nitrogen, and then 100 mg of the powder was used for determination of ALA and heme levels. All experiments were performed with at least three times with six independent samplings per experiment.
Analysis of At-TSPO Degradation in a Yeast Strain Impaired in Proteasome Activity
Yeast RPN4 wild-type (CMY394) and rpn4 temperature-sensitive (cim3-1ts) mutant (Y762) strains were transformed with p426GAL1-AtTSPO and the pUB23 plasmid expressing a ubiquitin-Pro-β-galactosidase fusion from the GAL1 promoter (Ghislain et al., 1993). Exponential growing cells (optical density at 600 nm of 0.5 to 1.0) from 10 mL cultures in SGAL medium were incubated in presence of 366 μM CHX. Samples were withdrawn at different time points, and the stability of At-TSPO and Ub-Pro-βgal was analyzed by immunoblotting.
Cell Imaging
GFP and YFP confocal imaging was performed as described by Guillaumot et al. (2009a). Concanamycin A treatment prior to imaging was performed as described by Sláviková et al. (2005). Pd-mP (Frontier Scientific Europe) was prepared in 20 mM PBS containing 0.1% (v/v) N-laurylsarcosine and infiltrated through the stomata of a tobacco leaf (Batoko et al., 2000), followed by confocal imaging. The fluorescent porphyrin was excited with a 405-nm diode laser (4% attenuation) and the amplified emitted light recorded between 425 and 510 nm. YFP fluorescence was simultaneously excited with a 514-nm argon multiline laser and the amplified emitted light recorded between 530 and 610 nm. The confocal images were processed using Imaris software (Bitplane).
Peroxidase Assay
The peroxidase-like activity of heme was determined as described by Lee et al. (2007) and repeated at least five times. Briefly, 20 μM hemin was added to the purified recombinant TSPO fraction (6His-ATSPO or AtTSPO-6His). The peroxidase activity of the mixture was measured by the oxidation of 3,3′,5,5′-tetramethylbenzidine (Pierce) by H2O2 (10 mM). The reaction was monitored by absorbance at 650 nm. As controls, TSPO without hemin or hemin alone was assessed in parallel.
Total ROS/Reactive Nitrogen Species Staining and Quantification
Intracellular production of ROS and/or reactive nitrogen species was measured according to Maxwell et al. (1999). Briefly, 15 μM 5-(and-6)-chloromethyl-2',7'- dichlorodihydrofluorescein diacetate (CM-H2DCFDA; Molecular Probes), dissolved in ethanol, was added to the cell suspension. This nonpolar compound is actively taken up by cells and converted by esterases into 2′,7′-dichlorodihydrofluorescein, a nonfluorescent molecule, which is rapidly oxidized to the highly fluorescent 2′,7′-dichlorofluorescein by intracellular peroxides. The cells were incubated for 15 min in the dark, and then excess dye was removed by extensive washing of the cells with fresh culture medium, and the cells were observed within 30 min of treatment under a Zeiss 710 confocal microscope. The 2′,7′-dichlorofluorescein was imaged as described by Maxwell et al. (1999). These experiments were repeated at least two times.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries and in the Arabidopsis Genome Initiative and Carica papaya genome databases under the following accession numbers: At2g47770 (At-TSPO), AT4G34450 (At-SEC21), AT2G45170 (At-ATG8e), At5g17290 (At-ATG5), At1g09570 (Arabidopsis PhyA), CAA89140 (S. cerevisiae Gas1p), XP_002531881 (Ricinus communis TSPO1), EEF30518 (R. communis TSPO2), EEF07520 (Populus trichocarpa TSPO1), XP_002301472 (P. trichocarpa TSPO2), XP_002334319 (P. trichocarpa TSPO3), EEE80745 (P. trichocarpa TSPO4), XP_002263702 (Vitis vinifera TSPO), CAH10765 (Solanum tuberosum TSPO), XP_002439293 (Sorghum bicolor TSPO), CF069942.1 (Medicago truncatula EST translated, TSPO), evm.model.supercontig_9.210 (C. papaya, predicted gene translated, TSPO), CAQ09717 (Homo sapiens TSPO1), and XP_167072 (H. sapiens TSPO2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transcriptional Regulation of At-TSPO Expression.
Supplemental Figure 2. ABA Transiently Upregulates Unbound Heme Levels in Plant Cells.
Supplemental Figure 3. At-TSPO Degradation Is Sensitive to Wortmannin.
Supplemental Figure 4. YFP-AtTSPO Degradation in Vivo Is Regulated by Tetrapyrrole Metabolism.
Supplemental Figure 5. YFP-AtTSPO Degradation in Vivo Is Affected by Senescence.
Supplemental Figure 6. The Intrinsic Peroxidase Activity of Hemin Is Enhanced after Interaction with At-TSPO.
Supplemental Figure 7. ABA and At-TSPO Increases Reactive Oxygen Species Levels in the Plant Cell.
Supplemental Figure 8. Quantification of ABA- and At-TSPO–Increased Reactive Oxygen Species in Plant Cells.
Acknowledgments
We thank D. Bassham, P. Morsomme, O. Park, RIKEN, ABRC, and the Nottingham Arabidopsis Stock Centre for providing reagents or plant materials and A. Errachid of the imaging platform IMABIOL for assistance. This work was partly funded by the Interuniversity Attraction Poles Programme–Belgian Science Policy, the Communauté Française de Belgique-Actions de Recherches Concertées (Grant ARC-0510-329), the Belgian Funds for Scientific Research (FNRS), and Université Catholique de Louvain (FSR). H.B. is a Research Associate of the FRS-FNRS.
References
- Ahuja U., Thöny-Meyer L. (2006). The membrane anchors of the heme chaperone CcmE and the periplasmic thioredoxin CcmG are functionally important. FEBS Lett. 580: 216–222 [DOI] [PubMed] [Google Scholar]
- Awai K., Xu C., Lu B., Benning C. (2006). Lipid trafficking between the endoplasmic reticulum and the chloroplast. Biochem. Soc. Trans. 34: 395–398 [DOI] [PubMed] [Google Scholar]
- Balestrasse K.B., Noriega G.O., Batlle A., Tomaro M.L. (2005). Involvement of heme oxygenase as antioxidant defense in soybean nodules. Free Radic. Res. 39: 145–151 [DOI] [PubMed] [Google Scholar]
- Balla J., Balla G., Jeney V., Kakuk G., Jacob H.S., Vercellotti G.M. (2000). Ferriporphyrins and endothelium: A 2-edged sword-promotion of oxidation and induction of cytoprotectants. Blood 95: 3442–3450 [PubMed] [Google Scholar]
- Bassham D.C. (2007). Plant autophagy—More than a starvation response. Curr. Opin. Plant Biol. 10: 587–593 [DOI] [PubMed] [Google Scholar]
- Batoko H., Zheng H.Q., Hawes C., Moore I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart E.F., Krause U., Schellens J.P., Vreeling-Sindelárová H., Meijer A.J. (1997). The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 243: 240–246 [DOI] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Braestrup C., Albrechtsen R., Squires R.F. (1977). High densities of benzodiazepine receptors in human cortical areas. Nature 269: 702–704 [DOI] [PubMed] [Google Scholar]
- Callis J. (1995). Regulation of protein degradation. Plant Cell 7: 845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R., Ouyang J., Abreu I.A., Hu Y., Seo H., Zhang X., Chua N.H. (2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento A.L., Xiong Y., Bassham D.C. (2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 42: 598–608 [DOI] [PubMed] [Google Scholar]
- Corsi L., Avallone R., Geminiani E., Cosenza F., Venturini I., Baraldi M. (2004). Peripheral benzodiazepine receptors in potatoes (Solanum tuberosum). Biochem. Biophys. Res. Commun. 313: 62–66 [DOI] [PubMed] [Google Scholar]
- Dinneny J.R., Long T.A., Wang J.Y., Jung J.W., Mace D., Pointer S., Barron C., Brady S.M., Schiefelbein J., Benfey P.N. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Dixon D.P., Lapthorn A., Madesis P., Mudd E.A., Day A., Edwards R. (2008). Binding and glutathione conjugation of porphyrinogens by plant glutathione transferases. J. Biol. Chem. 283: 20268–20276 [DOI] [PubMed] [Google Scholar]
- Fan J.J., Rone M.B., Papadopoulos V. (2009). Translocator protein 2 is involved in cholesterol redistribution during erythropoiesis. J. Biol. Chem. 284: 30484–30497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Gampala S.S.L., Rock C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl): S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank W., Baar K.M., Qudeimat E., Woriedh M., Alawady A., Ratnadewi D., Gremillon L., Grimm B., Reski R. (2007). A mitochondrial protein homologous to the mammalian peripheral-type benzodiazepine receptor is essential for stress adaptation in plants. Plant J. 51: 1004–1018 [DOI] [PubMed] [Google Scholar]
- Fujii H., Zhu J.K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavish M., Bachman I., Shoukrun R., Katz Y., Veenman L., Weisinger G., Weizman A. (1999). Enigma of the peripheral benzodiazepine receptor. Pharmacol. Rev. 51: 629–650 [PubMed] [Google Scholar]
- Geelen D., Leyman B., Batoko H., Di Sansebastiano G.P., Moore I., Blatt M.R. (2002). The abscisic acid-related SNARE homolog NtSyr1 contributes to secretion and growth: Evidence from competition with its cytosolic domain. Plant Cell 14: 387–406 Erratum. Plant Cell 14: 963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M., Dohmen R.J., Lévy F., Varshavsky A. (1996). Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15: 4884–4899 [PMC free article] [PubMed] [Google Scholar]
- Ghislain M., Udvardy A., Mann C. (1993). S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature 366: 358–362 [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A. (2002). Transformation of yeast by the Liac/ss carrier DNA/PEG method. Methods Enzym. 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Guillaumot D., Guillon S., Déplanque T., Vanhee C., Gumy C., Masquelier D., Morsomme P., Batoko H. (2009a). The Arabidopsis TSPO-related protein is a stress and abscisic acid-regulated, endoplasmic reticulum-Golgi-localized membrane protein. Plant J. 60: 242–256 [DOI] [PubMed] [Google Scholar]
- Guillaumot D., Guillon S., Morsomme P., Batoko H. (2009b). ABA, porphyrins and plant TSPO-related protein. Plant Signal. Behav. 4: 1087–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K., Lippincott-Schwartz J. (2010). Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141: 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. (2009). A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11: 1433–1437 [DOI] [PubMed] [Google Scholar]
- Hermans C., Vuylsteke M., Coppens F., Craciun A., Inzé D., Verbruggen N. (2010). Early transcriptomic changes induced by magnesium deficiency in Arabidopsis thaliana reveal the alteration of circadian clock gene expression in roots and the triggering of abscisic acid-responsive genes. New Phytol. 187: 119–131 [DOI] [PubMed] [Google Scholar]
- Hoffman M., Góra M., Rytka J. (2003). Identification of rate-limiting steps in yeast heme biosynthesis. Biochem. Biophys. Res. Commun. 310: 1247–1253 [DOI] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K., Izumi M., Reisen D., Yano Y., Makino A., Ohsumi Y., Hanson M.R., Mae T. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N. (2008). Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19: 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanczewska J., Marco S., Vandermeeren C., Maudoux O., Rigaud J.L., Boutry M. (2005). Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimer into a hexamer. Proc. Natl. Acad. Sci. USA 102: 11675–11680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant P., Gordon M., Kant S., Zolla G., Davydov O., Heimer Y.M., Chalifa-Caspi V., Shaked R., Barak S. (2008). Functional-genomics-based identification of genes that regulate Arabidopsis responses to multiple abiotic stresses. Plant Cell Environ. 31: 697–714 [DOI] [PubMed] [Google Scholar]
- Kitajima A., Asatsuma S., Okada H., Hamada Y., Kaneko K., Nanjo Y., Kawagoe Y., Toyooka K., Matsuoka K., Takeuchi M., Nakano A., Mitsui T. (2009). The rice alpha-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell 21: 2844–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T., Kindgren P., Benedict C., Hendrickson L., Strand A. (2007). Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., et al. (2008). Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4: 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkhov V.M., Sachse C., Short J.M., Tate C.G. (2010). Three-dimensional structure of TspO by electron cryomicroscopy of helical crystals. Structure 18: 677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., Walter P. (2009). An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325: 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T.C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Kreps J.A., Wu Y.J., Chang H.S., Zhu T., Wang X., Harper J.F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy P.C., Du G.Q., Fukuda Y., Sun D.X., Sampath J., Mercer K.E., Wang J.F., Sosa-Pineda B., Murti K.G., Schuetz J.D. (2006). Identification of a mammalian mitochondrial porphyrin transporter. Nature 443: 586–589 [DOI] [PubMed] [Google Scholar]
- Kwok E.Y., Hanson M.R. (2004). Plastids and stromules interact with the nucleus and cell membrane in vascular plants. Plant Cell Rep. 23: 188–195 [DOI] [PubMed] [Google Scholar]
- Kwon S.I., Cho H.J., Jung J.H., Yoshimoto K., Shirasu K., Park O.K. (2010). The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 64: 151–164 [DOI] [PubMed] [Google Scholar]
- Lacapère J.J., Papadopoulos V. (2003). Peripheral-type benzodiazepine receptor: Structure and function of a cholesterol-binding protein in steroid and bile acid biosynthesis. Steroids 68: 569–585 [DOI] [PubMed] [Google Scholar]
- Lara F.A., Lins U., Bechara G.H., Oliveira P.L. (2005). Tracing heme in a living cell: Hemoglobin degradation and heme traffic in digest cells of the cattle tick Boophilus microplus. J. Exp. Biol. 208: 3093–3101 [DOI] [PubMed] [Google Scholar]
- Lee K.S., et al. (2007). Hemin interactions and alterations of the subcellular localization of prion protein. J. Biol. Chem. 282: 36525–36533 [DOI] [PubMed] [Google Scholar]
- Levine B., Kroemer G. (2008). Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann P., Koch A., Degenhardt B., Hause G., Grimm B., Papadopoulos V. (2004). A novel Arabidopsis thaliana protein is a functional peripheral-type benzodiazepine receptor. Plant Cell Physiol. 45: 723–733 [DOI] [PubMed] [Google Scholar]
- Lucana D.O.D., Schaa T., Schrempf H. (2004). The novel extracellular Streptomyces reticuli haem-binding protein HbpS influences the production of the catalase-peroxidase CpeB. Microbiology 150: 2575–2585 [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Matsunaga K., et al. (2009). Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11: 385–396 [DOI] [PubMed] [Google Scholar]
- Matsuoka K., Higuchi T., Maeshima M., Nakamura K. (1997). A vacuolar-type H+-ATPase in a nonvacuolar organelle is required for the sorting of soluble vacuolar protein precursors in tobacco cells. Plant Cell 9: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauzerall D., Granick S. (1956). The occurrence and determination of δ-amino-levulinic acid and porphobilinogen in urine. J. Biol. Chem. 219: 435–446 [PubMed] [Google Scholar]
- Maxwell D.P., Wang Y., MacIntosh L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.N., Klionsky D.J. (2008). Autophagy fights disease through cellular self-eating. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Levine B. (2010). Methods in mammalian autophagy research. Cell 140: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Tanaka R., Grimm B., Masuda T., Moulin M., Smith A.G., Tanaka A., Terry M.J. (2010). The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 15: 488–498 [DOI] [PubMed] [Google Scholar]
- Morsomme P., Prescianotto-Baschong C., Riezman H. (2003). The ER v-SNAREs are required for GPI-anchored protein sorting from other secretory proteins upon exit from the ER. J. Cell Biol. 162: 403–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Müller R., Funk M. (1994). Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22: 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa F., Alev C., Shin M., Nakaya Y., Jakt L.M., Sheng G.J. (2009). PBRL, a putative peripheral benzodiazepine receptor, in primitive erythropoiesis. Gene Expr. Patterns 9: 114–121 [DOI] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Noda N.N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. (2008). Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13: 1211–1218 [DOI] [PubMed] [Google Scholar]
- Noda N.N., Ohsumi Y., Inagaki F. (2010). Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584: 1379–1385 [DOI] [PubMed] [Google Scholar]
- Nott A., Jung H.S., Koussevitzky S., Chory J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Otterbein L.E., Soares M.P., Yamashita K., Bach F.H. (2003). Heme oxygenase-1: Unleashing the protective properties of heme. Trends Immunol. 24: 449–455 [DOI] [PubMed] [Google Scholar]
- Ozaki H., Zoghbi S.S., Hong J., Verma A., Pike V.W., Innis R.B., Fujita M. (2010). In vivo binding of protoporphyrin IX to rat translocator protein imaged with positron emission tomography. Synapse 64: 649–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos V., Baraldi M., Guilarte T.R., Knudsen T.B., Lacapère J.J., Lindemann P., Norenberg M.D., Nutt D., Weizman A., Zhang M.R., Gavish M. (2006). Translocator protein (18kDa): New nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol. Sci. 27: 402–409 [DOI] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampon C., Bouzaffour M., Ostuni M.A., Dufourcq P., Girard C., Freyssinet J.M., Lacapere J.J., Schweizer-Groyer G., Vriz S. (2009). Translocator protein (18 kDa) is involved in primitive erythropoiesis in zebrafish. FASEB J. 23: 4181–4192 [DOI] [PubMed] [Google Scholar]
- Rubinsztein D.C. (2006). The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443: 780–786 [DOI] [PubMed] [Google Scholar]
- Rupprecht R., et al. (2009). Translocator protein (18 kD) as target for anxiolytics without benzodiazepine-like side effects. Science 325: 490–493 Erratum. Science 325: 1072 [DOI] [PubMed] [Google Scholar]
- Schulz H., Hennecke H., Thöny-Meyer L. (1998). Prototype of a heme chaperone essential for cytochrome c maturation. Science 281: 1197–1200 [DOI] [PubMed] [Google Scholar]
- Seglen P.O., Gordon P.B. (1982). 3-Methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA 79: 1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Severance S., Hamza I. (2009). Trafficking of heme and porphyrins in metazoa. Chem. Rev. 109: 4596–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J., Jabben M., Vierstra R.D. (1987). Red light-induced formation of ubiquitin-phytochrome conjugates: Identification of possible intermediates of phytochrome degradation. Proc. Natl. Acad. Sci. USA 84: 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhawat G.S., Verma K. (2010). Haem oxygenase (HO): An overlooked enzyme of plant metabolism and defence. J. Exp. Bot. 61: 2255–2270 [DOI] [PubMed] [Google Scholar]
- Shen Y.Y., et al. (2006). The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443: 823–826 [DOI] [PubMed] [Google Scholar]
- Sláviková S., Shy G., Yao Y.L., Glozman R., Levanony H., Pietrokovski S., Elazar Z., Galili G. (2005). The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56: 2839–2849 [DOI] [PubMed] [Google Scholar]
- Spielewoy N., Schulz H., Grienenberger J.M., Thony-Meyer L., Bonnard G. (2001). CCME, a nuclear-encoded heme-binding protein involved in cytochrome c maturation in plant mitochondria. J. Biol. Chem. 276: 5491–5497 [DOI] [PubMed] [Google Scholar]
- Stevens J.M., Daltrop O., Higham C.W., Ferguson S.J. (2003). Interaction of heme with variants of the heme chaperone CcmE carrying active site mutations and a cleavable N-terminal His tag. J. Biol. Chem. 278: 20500–20506 [DOI] [PubMed] [Google Scholar]
- Takahashi S., Ogawa T., Inoue K., Masuda T. (2008). Characterization of cytosolic tetrapyrrole-binding proteins in Arabidopsis thaliana. Photochem. Photobiol. Sci. 7: 1216–1224 [DOI] [PubMed] [Google Scholar]
- Takatsuka C., Inoue Y., Matsuoka K., Moriyasu Y. (2004). 3-Methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol. 45: 265–274 [DOI] [PubMed] [Google Scholar]
- Taketani S. (2005). Aquisition, mobilization and utilization of cellular iron and heme: Endless findings and growing evidence of tight regulation. Tohoku J. Exp. Med. 205: 297–318 [DOI] [PubMed] [Google Scholar]
- Taketani S., Kohno H., Furukawa T., Tokunaga R. (1995). Involvement of peripheral-type benzodiazepine receptors in the intracellular transport of heme and porphyrins. J. Biochem. 117: 875–880 [DOI] [PubMed] [Google Scholar]
- Thomas J., Weinstein J.D. (1990). Measurement of heme efflux and heme content in isolated developing chloroplasts. Plant Physiol. 94: 1414–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues P.E., Zhu J.K. (2007). New developments in abscisic acid perception and metabolism. Curr. Opin. Plant Biol. 10: 447–452 [DOI] [PubMed] [Google Scholar]
- Villarejo A., et al. (2005). Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7: 1224–1231 [DOI] [PubMed] [Google Scholar]
- Wagner D., Przybyla D., Op den Camp R., Kim C., Landgraf F., Lee K.P., Würsch M., Laloi C., Nater M., Hideg E., Apel K. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]
- Weinstein J.D., Beale S.I. (1983). Separate physiological roles and subcellular compartments for two tetrapyrrole biosynthetic pathways in Euglena gracilis. J. Biol. Chem. 258: 6799–6807 [PubMed] [Google Scholar]
- Wendler G., Lindemann P., Lacapère J.J., Papadopoulos V. (2003). Protoporphyrin IX binding and transport by recombinant mouse PBR. Biochem. Biophys. Res. Commun. 311: 847–852 [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.Q., et al. (2009). The magnesium-chelatase H subunit binds abscisic acid and functions in abscisic acid signaling: New evidence in Arabidopsis. Plant Physiol. 150: 1940–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2007a). Disruption of autophagy results in constitutive oxidative stress in Arabidopsis. Autophagy 3: 257–258 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Nguyen P.Q., Bassham D.C. (2007b). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.H., Kim K.D., Lucas A., Drahos K.E., Santos C.S., Mury S.P., Capelluto D.G.S., Finkielstein C.V. (2008). A novel heme-regulatory motif mediates heme-dependent degradation of the circadian factor period 2. Mol. Cell. Biol. 28: 4697–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.F., Klionsky D.J. (2010). Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeliseev A.A., Kaplan S. (1995). A sensory transducer homologous to the mammalian peripheral-type benzodiazepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 270: 21167–21175 [DOI] [PubMed] [Google Scholar]
- Yeliseev A.A., Kaplan S. (1999). A study of the expression and intracellular localization of the protein TspO in the phototrophic bacterium Rhodobacter sphaeroides. Appl. Biochem. Microbiol. 35: 284–290 [Google Scholar]
- Yeliseev A.A., Kaplan S. (2000). TspO of rhodobacter sphaeroides. A structural and functional model for the mammalian peripheral benzodiazepine receptor. J. Biol. Chem. 275: 5657–5667 [DOI] [PubMed] [Google Scholar]
- Yeliseev A.A., Krueger K.E., Kaplan S. (1997). A mammalian mitochondrial drug receptor functions as a bacterial “oxygen” sensor. Proc. Natl. Acad. Sci. USA 94: 5101–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W.L., Shintani T., Nair U., Cao Y., Richardson B.C., Li Z.J., Hughson F.M., Baba M., Klionsky D.J. (2010). The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J. Cell Biol. 188: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.L. (2009). Electron microscopic tomography reveals novel features of the relationship between phagophore membranes and endoplasmic reticulum. Autophagy 5: 897–898 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]