Abstract
Purpose
The importance of genetic and epigenetic alterations maybe in their aggregate role in altering core pathways in tumorigenesis.
Experimental Design
Merging genome-wide genomic and epigenomic alterations, we identify key genes and pathways altered in colorectal cancers (CRC). DNA Methylation analysis was tested for predicting survival in CRC patients using Cox proportional hazard model.
Results
We identified 29 low frequency mutated genes that are also inactivated by epigenetic mechanisms in CRC. Pathway analysis showed the extracellular matrix (ECM) remodeling pathway is silenced in CRC. 6 ECM pathway genes were tested for their prognostic potential in large CRC cohorts (n=777). DNA Methylation of IGFBP3 and EVL predicted for poor survival (IGFBP3: HR=2.58, 95%CI:1.37-4.87, p=0.004; EVL: HR=2.48, 95%CI:1.07-5.74, p=0.034) and simultaneous methylation of multiple genes predicted significantly worse survival (HR=8.61, 95%CI:2.16-34.36, p<0.001 for methylation of IGFBP3, EVL, CD109 and FLNC). DNA Methylation of IGFBP3 and EVL was validated as a prognostic marker in an independent contemporary matched cohort (IGFBP3 HR=2.06, 95% CI:1.04-4.09, p=0.038; EVL HR=2.23, 95%CI:1.00-5.0, p=0.05) and EVL DNA methylation remained significant in a secondary historical validation cohort (HR=1.41, 95%CI:1.05-1.89, p=0.022). Moreover, DNA methylation of selected ECM genes helps to stratify the high-risk Stage 2 colon cancers patients who would benefit from adjuvant chemotherapy (HR: 5.85, 95%CI:2.03-16.83, p=0.001 for simultaneous methylation of IGFBP3, EVL and CD109).
Conclusions
CRC that have silenced in ECM pathway components show worse survival suggesting that our finding provides novel prognostic biomarkers for CRC and reflects the high importance of integrative analyses linking genetic and epigenetic abnormalities with pathway disruption in cancer.
Keywords: DNA Methylation, Extracellular Matrix Pathway, Prognostic Biomarker, Colorectal cancer
INTRODUCTION
Recent efforts in genome wide sequencing of multiple human cancers has identified a large number of mutations which cluster within a small number of intracellular pathways such as cellular senescence bypass, invasion, and metastasis(1, 2). Epigenetic abnormalities can also operate with genetic alterations to effect aberrant gene function in cancer (3). DNA hypermethylation of promoter- associated CpG islands is a frequent and early mechanism of tumor-suppressor gene inactivation in many cancers and may have great promise as biomarkers (4-6). We have previously emphasized that DNA hypermethylation can affect many of the new genes found to be mutated in both colorectal (CRC) and breast cancer cells (7, 8) However, an understanding of such genome wide alterations has not led to clinically applicable biomarkers in predicting tumor behaviour. We hypothesized that integration of the genome and hypermethylome may provide insights into the key pathways altered in human cancers and lead to insights into relevant biomarkers. In the present study, we have continued to globally search for epigenetic alterations of genes also mutated in colon cancers to narrow down the key pathways altered in colorectal cancers.
We now find that many genes in the extracellular matrix (ECM) maintenance and remodeling pathway, critical for many normal developmental processes and for tumorigenesis (9), are epigenetically altered in nearly every human colon cancer. ECM can be remodeled by many processes, including changes in synthesis, contraction, and proteolytic degradation (9). Recently, misregulated ECM environments have also been implicated in the capacity of tumors to undergo epithelial mesenchymal transition (EMT) and in the acquisition of metastatic potential in tumors as cells lose cell-cell and cell-extracellular matrix adherence along with profound changes in their cystoskeleton architecture (10-12). We now show that ECM pathways alterations are frequent in all colon cancers and DNA methylation induced silencing of these genes can occur at multiple genes in the ECM pathway.
Importantly, we also now show that the simultaneous DNA hypermethylation of a subset of these ECM genes is also associated significantly with poor survival in adjusted analyses of a large cohort of colon cancer patients. Our data highlights the importance of integrative approaches in obtaining a better understanding of carcinogenesis as well as identifying novel biomarkers.
MATERIALS AND METHODS
Patients
Johns Hopkins (JHU) Training and Validation cohort samples for the colorectal cancer studies were prepared from formalin fixed and paraffin embedded (FFPE) colon cancer tissue samples from the pathology archives of the Johns Hopkins Hospital in accordance the Institutional Review Board (IRB) and as per HIPAA compliance. The JHU training cohort consisted of 147 tissue samples from stages 1-4 colon cancer patients who underwent primary surgery from 1995 to 2005 (Median follow up of 6.4 years). The JHU validation cohort consisted of 72 tissue samples from stages 1-4 colon cancer patients who underwent primary surgery from 1995 through 2005 (Median follow up of 7.5 years). Patients with neoadjuvant chemotherapy, including all rectal cancers, were excluded from the current study. Patients in both JHU cohorts were similar with respect to age, sex, race, proportion of cases with lymphovascular invasion, pathologic grade, location, and proportion of cases with recurrence (Table 1).
Table 1.
Demographics of training and validation cohorts in this study
| Training Cohort | Validation Cohorts |
||
|---|---|---|---|
| JHU | JHU | NLCS Set (Netherlands) | |
| Time Period | 1998-2005 | 1998-2005 | 1986-1991 |
| N | n=147 | n=72 | n=558 |
| Median Age (years) | 66 | 70 | 64 |
| 5 year survival | 67.4% | 65.3% | 54.8% |
| Stage I | 83.3% | 83.3% | 78.4% |
| Stage II | 67.4% | 70.4% | 70.2% |
| Stage III | 75.7% | 60.0% | 42.0% |
| Stage IV | 17.0% | 14.3% | 1.6% |
| Gender | |||
| Male | 84 (57%) | 30 (42%) | 296 (53%) |
| Female | 63( 43%) | 42 (58%) | 262 (47%) |
| Location | |||
| Right Colon | 79 (54%) | 40 (56%) | 239 (43%) |
| Left Colon | 66 (45%) | 32 (44%) | 310 (56%) |
| Unknown | 9 (1%) | ||
| Stage | |||
| Stage I | 36 (24.4%) | 18 (25.0%) | 111 (19%) |
| Stage II | 49 (33.3%) | 27 (37.5%) | 201 (36%) |
| Stage III | 37 (25.2%) | 20 (27.8%) | 143 (25%) |
| Stage IV | 25 (17.1%) | 7 (9.7%) | 62 (11%) |
| Unknown | 41 (9%) | ||
Netherlands Cohort study on Diet and Cancer (NLCS) Validation cohort were prepared from 558 formalin-fixed, paraffin-embedded primary colon cancers from stages 1-4 (median follow-up: 7.2 years). The NLCS has been previously described in detail (13). 925 incident colorectal cancer cases were identified from 1989-1994, and DNA was available for analyses on our current subset (Table 1).
DNA methylation and gene expression analyses
Primer pairs were preferentially designed near the putative transcriptional start site (TSS) in the 5′ CpG islands of the genes. Primer sequences for Methylation Specific PCR (MSP) analysis were designed using MSPPrimer (7). All primer sequences are listed in supplementary Table S5. For expression studies using RT-PCR, primers were designed using the open access program Primer3.
For MSP analysis, DNA was extracted using the standard phenol-chloroform extraction method. Bisulfite modification of genomic DNA was carried out using the EZ DNA methylation Kit (Zymo Research). DNA Methylation analysis was performed using MSP primer pairs located close to the putative transcriptional start site in the 5′ CpG island with 2μl of bisulfite-treated DNA as template and JumpStart Red Taq DNA Polymerase (SIGMA) for amplification as previously described (7).
Total RNA was extracted from cell lines using the RNeasy Mini Kit (QIAGEN), treated with DNase (QIAGEN). For reverse transcription (RT) reaction, 1μg of total RNA was subjected to the first strand cDNA synthesis using Superscript III first strand cDNA synthesis kit (INVITROGEN) according to the manufacturer’s instructions. Expression analysis was performed by RT-PCR using 1μl of cDNA as template and JumpStart Red Taq DNA Polymerase (SIGMA) for amplification.
Quantitative methylation (qMSP) analysis
Quantitative MSP (qMSP) was performed after sodium bisulfite modification for selected genes including IGFBP3, EVL, CD109, FLNC and FBN2 on a small series of normal colons and colon cancers (n=5, each) (supplementary Fig. S2). qMSP analysis helped us to confirm whether our candidate genes showed cancer specific DNA methylation patterns consistent with the qualitative MSP analysis. For quantitative real-time analyses, the Power SYBR Green PCR kit (Applied Biosystems) was used and the amplification conditions consisted of an initial 10-min denaturation step at 95 °C, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing and extension for 30 s and 60 s, respectively. An ABI StepOnePlusReal-Time PCR System was used (Applied Biosystems), and for quantitation the comparative cycle threshold (Ct) method was used, normalizing the Ct values for the indicated gene to the Ct values of unmethylated reaction relative to a methylated reaction sample.
Cell culture and treatment
Cancer cell lines (Colorectal cancer cell lines; HCT116, SW480, RKO, HT29, Caco-2, Lovo, COLO 320, COLO 205, DLD1, SW48, and SW620; breast cancer cell lines; MCF7, T47D, MDA-MB-231 and MDA-MB-468) were obtained from ATCC and cultured in appropriate media and under conditions described by ATCC, with media obtained from INVITROGEN, supplemented with 10% fetal bovine serum (Gemini Bio-Products) and 1% penicillin/streptomycin (INVITROGEN). DKO cells (HCT116 cells with genetic disruption of DNMT1 and DNMT3b) were cultured as described previously(7).
Signaling pathway analysis of methylated genes
We used a highly curated database (Metacore) that includes human protein-protein interactions, signal transduction and metabolic pathways, and a variety of cellular functions and processes for signaling pathway analysis. Functional ontology enrichment selected the significantly altered canonical pathways, networks and gene ontology terms. Analysis was run to detect deregulated interaction networks in colorectal cancer, using a subset of genes that are both mutated (1, 2) and hypermethylated in colorectal cancer tissue. Significant pathways were selected and intersected with those found by Wood et al (2). While the paper by Wood et al, (2007) was focusing on those networks targeted by mutations alone, the combination of both approaches was designed to zoom in on those pathways that are important to colorectal cancer as a whole through alterations of these molecular interaction networks, be it by gene mutations or promoter hypermethylation.
Survival and statistical analysis
Univariate and multivariate Cox regression was used to identify genes which could significantly predict poor overall survival. All ties were handled by the Breslow method and the proportional hazards assumption was verified by examination of residual plots. Adjusted Kaplan–Meier survival plots were created to demonstrate the ability of the genes to predict poor overall survival. Statistical significance in this study was set at 0.050. To control for multiple testing, Sidak adjustment method was used to correct for family-wise error rate. Results of all models are reported as odds ratios with 95% confidence intervals. All data were analyzed using StataTM v9.2 statistical analyses software (Stata Corporation).
RESULTS
DNA Methylation analysis of mutated genes
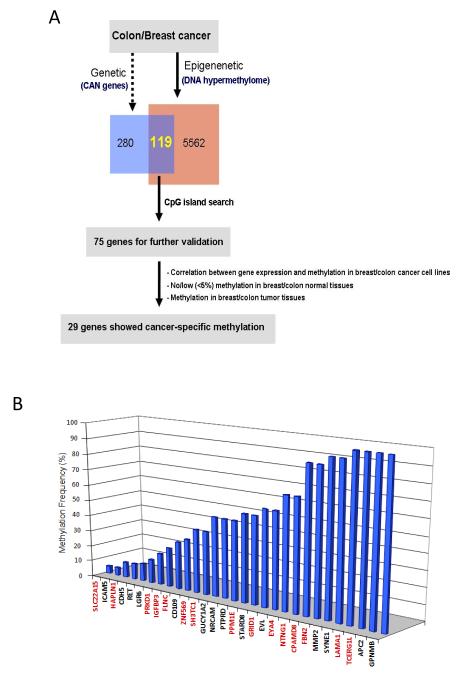
Using the recently compiled database of human cancer mutations in CRC and breast cancers, termed the candidate cancer genes (or CAN genes) (1, 2), we have now identified 119 genes which, in colon and breast cancer cell cultures, are also identified by our recently published gene discovery approaches (7, 8) as candidate genes for promoter DNA hypermethylation. Since the majority of these genes were found to be hypermethylated in colon tumors, we focused the remainder of our studies on colon cancers. These 119 genes then represent the intersection of the consensus mutated and DNA hypermethylated genes in colon cancer. Seventy-five of these 119 genes contained promoter CpG islands (14) and and were then first experimentally validated for 1) expression in normal colon tissues by RT-PCR (n=65 genes); Of these, 45 genes showed 2) correlation between loss of gene expression by RT-PCR and promoter CpG-island DNA methylation by MSP analysis in colon cancer cell lines (HCT116, RKO, SW480, and Colo320) (Fig. 1A, supplementary Table S1). In order to filter down to genes that only showed cancer specific DNA methylation, we next looked at DNA methylation frequencies of these 45 genes in a series of primary colon cancers (n=21, Stages 2 and 3) as well as normal colon from cancer-free individuals (n=20). 3) 34 genes showed >5% frequency of DNA methylation in a series of primary colon cancers and 4) 29 of these genes showed cancer-specific DNA methylation with absence of DNA methylation in normal colon (supplementary Table S1 for schema and genes; supplementary Table S2 for primary colon cancer sample information). Cancer specific DNA methylation was also confirmed using quantitative MSP for a subset of genes and showed identical results to the gel-based MSP assay (supplementary Fig. S2).
Figure 1.
Identification of mutated and DNA hypermethylated genes in colon cancers. (A) Schematic representation of the intersection of consensus mutated and DNA hypermethylated genes in colon cancer (see also supplementary Table S1 for schema). (B) DNA methylation frequencies of the 29 low frequency mutated genes in CRC patients (n=20). A subset of genes (Black letters) have been noted in our previous studies (7, 8) and newly identified genes in this study are indicated with red letters.
A subset of these genes had been previously identified by us during genome-wide screens (7, 8) and were included in the current integrative approaches to provide a more comprehensive view of frequent epigenetic alterations in colon cancer. Among the 29 genes, 15 genes are newly identified by this integrative approach as potential targets of both mutation and DNA methylation (FLNC, SH3TC1, ZNF569, SLC22A15, HAPLN1, PRKD1, IGFBP3, TCERG1L, LAMA1, FBN2, CPAMD8, NTNG1, EYA4, GRID1, and PPM1E) in this study with 5-100% DNA methylation frequencies in CRC (Fig. 1B) whereas the remaining 14 genes were also previously identified (7). A direct comparison of DNA methylation and known mutation frequencies of these 29 genes reveals that they are more often epigenetically than genetically altered in colon cancer (supplementary Fig. S1). DNA Methylation frequencies of the 29 genes show variable frequencies in the primary colon cancers ranging from 5-100% (supplementary Fig. S1). 17 out of 29 genes (GUCY1A2, NRCAM, PTPRD, PPM1E, STARD8, GRID1, EVL, EYA4, NTNG1, CPAMD8, FBN2, MMP2, SYNE1, LAMA1, TCERG1L, APC2, and GPNMB) displayed high level (>=50%) hypermethylation in colon cancers while 12 genes (SLC22A15, ICAM5, HAPLN1, CHD5, RET, LGR6, PRKD1, IGFBP3, FLNC, CD109, ZNF569, and SH3TC1) showed lower frequency of DNA methylation (<50%) in colon cancers (Fig. 1B).
Pathway analysis identifies global disruption of the ECM pathway in CRC
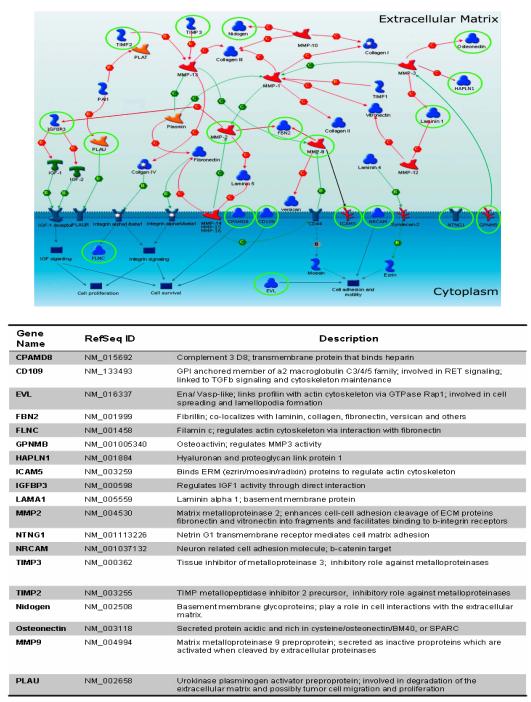
A growing body of research is defining that, particularly with respect to new genes being discovered in cancer re-sequencing efforts, and those genes which are genetically altered with low frequency, that alterations of pathways rather than of individual genes may be the important issue for affecting the course of tumorigenesis (1, 2). These mutated genes, then, belong to a handful of important signalling pathways in breast, colorectal, and other cancer types (2, 15). We queried an extensive gene ontology set (MetaCore; GeneGo) and nearly half of the genes that we identified to be both mutated and hypermethylated (13 of 29, 44.8%) mapped to the ECM pathway in CRC (Fig. 2). ECM remodeling is critical for many developmental processes, and recently, abnormally remodeled ECM has been shown to contribute to the neoplastic process (9).
Figure 2.
Widespread extracellular matrix (ECM) pathway silencing by DNA hypermethylation in colon cancer. (A) Hypermethylated genes in colon cancer (green circled) are highlighted on a map of the ECM pathway. The network shows interaction of the genes with each other within the extracellular matrix. Circled (green) genes include 13 hypermethylated genes in CRC derived from our gene discovery approach (LAMA1, MMP2, FBN2, CPAMD8, NTNG1, NRCAM, EVL and GPNMB, FLNC, CD109, ICAM5, HAPLN1, and IGFBP3). Arrows indicate incoming (←) or outcomming (→) connector between proteins. Colors (Green/Red/Black) of line describe positive, negative, and unspecified effect of functional interaction (www.genego.com). An additional 6 genes including TIMP2, TIMP3, MMP9, Nidogen, Osteonectin, and PLAU were previously identified to be down-regulated in association with DNA methylation in other cancer types, and now are shown to be similarly altered in colon cancers in the present study. The functional gene ontology analysis was performed using the MetaCore database (GeneGo). The Table below the schematic figure shows the putative role of all of these 19 genes in the ECM pathway.
We, therefore, focused the remainder of our studies to understanding the functional implications of the dysregulation of the ECM pathway in CRC. In our analyses of these ECM pathway genes, 8 of the genes (NRCAM, EVL, NTNG1, CPAMD8, FBN2, MMP2, LAMA1, and GPNMB) show high frequency (>50%) of DNA methylation in CRC are amongst those mapping to the ECM pathway(10, 16, 17) (Fig. 2), and the remainder show lower frequency of DNA methylation in CRC (ICAM5, HAPLN1, IGFBP3, FLNC, and CD109) (18-20).
In addition, we also noted that genes such as TIMP2 and TIMP3 that have previously been identified by ourselves and others as being hypermethylated in colorectal and other cancers also belong to the ECM pathways.(21, 22) Based on this observation, we also investigated, for potential epigenetic silencing in CRC, other genes in the ECM remodeling pathway which have previously been identified to show hypermethylation in other cancers by us and others. Six other genes, in addition to TIMP2 and TIMP3, were identified, including PLAU, Nidogen (NID1), Osteonectin, CD44, MMP9 and Laminin 5 (LAMA5)(23-26) and these 8 genes were then investigated for potential DNA methylation in our current set of primary CRC. Six of these 8 ECM genes, including TIMP2, TIMP3, MMP9, PLAU, Nidogen, and Osteonectin were found to be methylated in CRC with variable DNA methylation frequencies, ranging from 5 to 95% (supplementary Table S3). All 8 additional ECM genes also showed no evidence of DNA methylation in normal colon (data not shown; n=5). The TIMPs (TIMP2 and TIMP3) and MMPs (MMP9) have long been thought to be critical components of the ECM pathway and their expression has been critical in primary tumor growth, angiogenesis, invasion, and metastasis in different type of cancers (18). Nidogen forms a complex with laminins and collagen IV and helps to stabilize the basement membrane structure as well as being important for cell adhesion by establishing contact with integrins(24). Osteonectin (also known as SPARC) and PLAU are involved in wound repair (25), cell migration and differentiation but their role in tumorigenesis has not yet been defined.
Together all our data shows that at least 19 genes in the ECM pathway are hypermethylated in CRC (Fig. 2 and supplementary Table S3), and strongly suggests that the ECM pathway is globally and universally altered in CRC by mutation or epigenetic silencing but much more commonly by the latter abnormality.
CRC with ECM dysregulation demonstrate worse survival
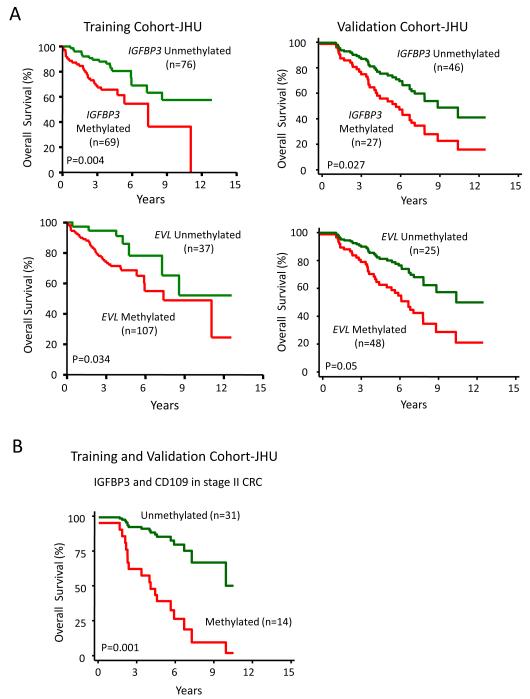
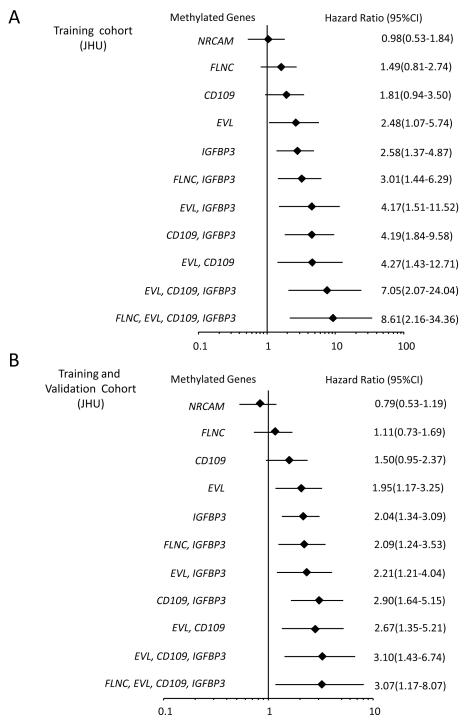
We next wanted to study the clinical correlates of our gene silencing data in terms of the previously observed juxtaposition of embryonic gene expression patterns and EMT with such tumors having the most primitive and aggressive phenotypes(27). In this respect, an exploratory clinical outcome analysis in our initial small cohort of colon cancer patients (n=21 patients) was performed for hypothesis-generation and showed that DNA methylation of selected ECM genes (IGFBP3 and EVL) was associated with significantly worse outcomes on unadjusted analysis (p=0.02 and p=0.03, respectively) while DNA methylation of other genes (CD109, NRCAM, NTNG, and FLNC) showed a trend towards poor survival which was not statistically significant. To explore the potential clinical significance of these data, we tested these above 6 ECM pathway genes for their prognostic potential in a large training cohort of patients with colon cancers and well-annotated clinical data which could be correlated with survival and gene methylation status. Strikingly, in an analyses of tumors from 147 patients with Stages 1-4 colon cancers (Training cohort JHU; Table 1), we observed a statistically significant increased risk for mortality, when either individual genes or combinations of genes were methylated after adjusting for other prognostic variables as age and stage of cancers (Fig. 3A and Fig. 4A). For example, adjusted Kaplan Meier survival curves for two of the individual ECM genes, IGFBP3 and EVL, show that DNA methylation of each is associated with decreased survival (adjusted for age and stage) (Fig. 3A). Multivariate hazard ratios (Fig. 4A) for survival depicted in a forest plot show the hazard ratios (HR) for DNA methylation of these two genes predicted risk for poor survival (IGFBP3 HR=2.58, 95% CI, 1.37- 4.87, p=0.004; EVL HR=2.48, 95% CI, 1.07- 5.74, p=0.034) while another gene CD109 shows a trend towards statistical significance (HR=1.81, 95% CI, 0.94- 3.50, p=NS). Moreover, in addition, in patients with simultaneous DNA methylation of multiple genes in the ECM pathway, significantly worse survival is predicted in adjusted analysis. For example, for DNA methylation of IGFBP3, EVL, CD109 and FLNC, the odds of dying are increased to greater than eight-fold after adjusting for age and stage of cancer compared to patients who lack DNA methylation of these genes (HR: 8.61, 95% CI:2.16-34.36, p<0.001) (Fig. 4A).
Figure 3.
Kaplan-Meier estimates of overall survival according to IGFBP3 and EVL methylation status (adjusted for age and stage). (A) IGFBP3 and EVL methylation was tested in 147 colon cancer patients (Stages 1-4; JHU training cohort) and (B) in 72 CRC patients (stages 1-4; JHU validation cohort)
Figure 4.
Odds ratios for ECM genes in CRC patients. (A) Forest plot depicting Multivariate Hazard Ratios and corresponding 95% confidence intervals for overall mortality risk associated with DNA methylation of individual, or combinations of genes (n = 147 colon cancer samples; JHU training cohort) and (B) combined JHU training and validation cohorts (n=219 colon cancer samples). Selected statistically significant gene combinations are shown. Multivariate Cox regression analysis was performed for six ECM genes (NRCAM, FLNC, CD109, EVL and IGFBP3) which showed either significant, or trends towards significance, for hazard ratios on bivariate analyses. The prognostic value of each gene was adjusted for stage and age and then graphed on the Forest plot.
We further examined DNA methylation status of IGFBP3, EVL, CD109, NRCAM, and FLNC in an independent validation cohort of 72 patients with stages 1-4 colon cancers (Validation cohort JHU) (Table 1 and supplementary Table S4). In multivariate adjusted analysis, both IGFBP3 (HR: 2.06, 95% CI: 1.04-4.09, p=.0.038) and EVL (HR: 2.23, 95% CI: 1.00-5.06, p=0.050) remained statistically significant as markers associated with worse survival. CD109 showed a trend towards worse survival outcome which was not statistically significant (HR: 1.49, 95%CI: 0.72-3.08, p=NS) while FLNC and NRCAM had no prognostic value. Adjusted Kaplan Meier survival curves (adjusted for age and stage) for two of the individual ECM genes, IGFBP3 and EVL, again shows that DNA methylation of each is associated with decreased survival in our validation cohort (Fig. 3A). Multivariate hazard ratios (Fig. 4B) for survival depicted in a forest plot of both the training and validation cohort shows that simultaneous DNA methylation of multiple genes in the ECM pathway is associated with significantly worse survival. These findings support our hypothesis that DNA methylation of ECM pathway genes may have an important role during cancer progression and metastasis.
We also looked at the prognostic significance of these ECM genes (IGFBP3, EVL, CD109, NRCAM, and FLNC) in a secondary validation cohort NLCS (13) of 558 patients with Stages 1-4 colon cancers (Table 1). Of note, the NLCS cohort differs significantly from the JHU training and validation in being a historical cohort (study period 1986-1991) with marked differences in survival of colon cancers, a preponderance of distal colon cancers and markedly worse outcomes (see Table 1). EVL (HR: 1.41, 95% CI: 1.05-1.89, p=0.022) again is statistically significant as a marker associated with worse survival and CD109 again shows a trend towards statistical significance (HR: 1.19; 95%CI: 0.95-1.48, p=NS) while IGFBP3 methylation was not significant in this historical cohort.
ECM Biomarkers in Stage 2 colon cancers
In the clinical management of colon cancer patients, there is an urgent need for identification of biomarkers that may stratify the high risk stage 2 colon cancers patients who are at the highest risk of recurrence. Previous large randomized trials have not shown a consistent benefit of adjuvant chemotherapy (28) but approximately 30-40% of these patients will nevertheless recur/metastasize (29). Identification of this high risk cohort of Stage 2 colon cancer patients would be useful since this subset may derive a benefit from adjuvant chemotherapy. Therefore, we investigated the biomarker potential of the ECM gene methylation in the Stage 2 colon cancers in the combined JHU training and validation cohort (n=219) (supplementary Table S6). Both IGFBP3 and CD109 are associated with worse survival in these Stage 2 colon cancers (IGFBP3 HR: 3.02; 95%CI: 1.42-6.4; p=0.004 and CD109 HR: 2.41; 95% CI: 1.14-5.10; p=0.021 respectively). Similarly, simultaneous DNA methylation of multiple genes in the ECM pathway is again associated with significantly worse survival for the Stage 2 colon cancers (HR: 5.85, 95% CI: 2.03-16.83, p=0.001 for DNA methylation of both IGFBP3 and CD109) (Fig. 3B).
DISCUSSION
Our data highlights the important role that integrative approaches may have in understanding cancer biology and implications of such in the translational arena. Our current approach analyzing the genomic and epigenomic changes in colon cancer identified the role of the ECM pathway in colorectal carcinogenesis.
These findings also are potentially useful in finding novel prognostic biomarkers as highlighted by our finding of DNA methylation of IGFBP3, EVL and CD109 showing a modest effect in being associated with worse survival. As mentioned in the results, this may have direct clinical implications in stratifying the high-risk Stage 2 colon cancers patients who have worse outcomes and who may benefit from adjuvant chemotherapy; although our findings would need to be validated in other retrospective studies of carefully clinically annotated tissues or a well-designed prospective study. Our findings were validated in an internal validation cohort (JHU validation) where DNA methylation of both IGFBP3 and EVL again showed the association with worse survival while CD109 showed a trend towards worse survival. We then validated our findings in another large external NLCS for DNA methylation of EVL and CD109 associated with worse outcomes. However, IGFBP3 methylation was not associated with worse survival in this cohort. Although at present it is unclear why IGFBP3 methylation was not validated in the NLCS cohort, there are multiple possible explanations for this. This may simply reflect the different biology in the two cohorts. In fact, epigenetic alterations have been shown to vary in different populations and races and potentially affect outcomes (30, 31). Netherlands also has a lack of colon cancer screening program and this is reflected in the markedly different 5-yr survival of this cohort (54.8% NLCS vs. 67.4%-JHU Training cohort or 65.3% JHU validation cohort) and the biases associated with screening (32). Moreover, the NLCS cohort has a preponderance of left-sided cancers while the JHU cohorts reflect a distribution and survival similar to what is seen in the Surveillance, Epidemiology and End Results database (a large national cancer database in the United States) (33). The location of colon cancer has been shown to affect both genetic and epigenetic alterations (34) (35, 36) and survival (33), (37). In addition, adjuvant chemotherapy was not offered routinely for high risk patients in the NLCS cohort. Thus lack of validation of IGFBP3 may also reflect the fact that this is a predictive rather than a prognostic marker. Further validation studies in comparable modern-day cohorts similar to the Johns Hopkins cohort may clarify this. In particular, our data on IGFBP3 and survival is intriguing since previous studies have shown that IGFBP3 induces apoptosis and inhibits DNA synthesis in breast, prostate and non-small cell lung cancer (38). Moreover, prior studies have also reported that high levels of circulating IGFBP3 decrease the risk of recurrent colorectal adenoma formation (39). Further studies are needed to confirm these findings particularly the risk stratification in stage 2 colon cancers.
Previous studies have also alluded to the potential of epigenetic changes, especially DNA hypermethylation of promoter-associated CpG islands, in serving as prognostic biomarkers. Brock et al. showed that hypermethylation of two genes, p16 and H-cad, in the primary lung tumor and the mediastinal lymph nodes could predict recurrence for early stage lung cancers (5). In another study, DNA methylation profiles of multiple methylated-in tumor (MINT) loci predicted distal recurrence in a small cohort of rectal cancer patients (40) while ID4 methylation has also been associated with worse outcomes (41). However most of these studies used candidate genes from the literature and tested them for prognostic significance in a cohort of patients. In our study, we have not only identified a unique pathway, the ECM pathway, that is altered by DNA methylation of many genes in CRC, but then show that dysregulation of this core pathway has prognostic significance as well. However, our current studies do not clearly distinguish between the DNA hypermethylation arising from the tumorigenic cells or from the non-tumorigenic stromal cells. This will need to be investigated in future studies.
What are the functional consequences of loss of function for the above ECM genes in CRC? We also considered, from several perspectives, our findings in another context – namely, that they might represent an example of properties which reflect the primitive cell status increasingly attributed to multiple tumor types(42). First, multiple previous studies(3) have stressed that some 50% of genes that become DNA hypermethylated in colon cancers are marked by a chromatin pattern, termed bivalent chromatin, that keeps these same genes in a low expression pattern in ESC and embryonic mesenchymal (EM) progenitor cells(43). Indeed, a key chromatin component of bivalent chromatin, the presence of polycomb group (PcG) constituents around gene start sites, is present in ESC and EM cells in the promoter regions of several of the ECM genes (IGFBP3, NRCAM, CPAMD8, FBN2, MMP2, TIMP3, MMP9), based on our database analyses (unpublished data).
In summary, our results suggest that CRC may harbor defects in key ECM genes, induced infrequently by gene mutations, and even more frequently by gene promoter DNA hypermethylation. This loss of function for such genes appears to correlate with loss of cellular differentiation and to be associated with aspects of primitive, stem/progenitor cell, features of CRC (supplementary Fig. S3). Loss of ECM gene function could foster metastatic behavior by altering events which require ECM remodelling such as allowing cells to invade through the basement membrane, migrate into the lymphovascular space, and establish metastatic foci in a distant organ. Our study suggests that this ECM alteration by hypermethylated genes may contribute to carcinogenesis, to some degree in virtually every CRC, through silencing of selected ECM pathway genes by both genetic and epigenetic alterations. Finally, our data indicate that detection of DNA hypermethylation of selected of the genes we have studied may provide a biomarker strategy for predicting the clinical behaviour of CRC. Our data emphasize the importance of integrating cancer gene mutations and DNA hypermethylation changes to uncover the molecular events disrupting cell signaling in CRC and other cancers.
TRANSLATIONAL RELEVANCE.
We have used an exciting integrative approach of multiple whole genome analysis (genetic and epigenetic) to identify an important pathway, the extracellular matrix (ECM) maintenance and remodeling pathway, which is silenced in all colon cancers. Colon cancers that have silenced multiple genes in this pathway have a poor prognosis based on analyses of large cohort of patients. DNA Methylation of these genes is particularly useful in stratifying the low- vs. high- risk Stage 2 colon cancer patients. Colorectal cancers that have silenced multiple genes in the ECM pathway appear to show a significantly worse survival in adjusted multivariate analysis of a large cohort of colon cancer patients. Our study has important ramifications in the clinical management of colon cancer patients since 30-40% of early stage colon cancers will present with recurrence/metastases but current strategies are not helpful in stratifying this high-risk cohort. We show that our prognostic markers are the most efficacious in stratifying the high risk subset of Stage 2 colon cancers who may then benefit from adjuvant chemotherapy.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. Steven Goodman for Biostatistical discussion. We would like to thank Sharon Metzger-Gaud and the Johns Hopkins Cancer Registry for assistance with the primary cancer databases. We would like to thank Iris van Vlodrop for technical assistance and Kathy Bender for administrative support.
GRANT SUPPORT The study was supported by the National Institute of Environmental Health Sciences (grant number: ES011858), the National Institutes of Health (grant number: K23CA127141), the Jeannik M. Littlefield-AACR Grant in Metastatic Colon Cancer, the Mary Kay Ash Foundation, the Susan G. Komen Foundation, and the Ross Clinician Scientist Award.
Footnotes
Disclosure of Potential Conflicts of Interest: SBB and JGH is a consultant of OncoMethylome Sciences (OMS) and research support from OMS.
REFERENCES
- 1.Sjoblom T, Jones S, Wood LD, et al. The Consensus Coding Sequences of Human Breast and Colorectal Cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 2.Wood LD, Parsons DW, Jones S, et al. The Genomic Landscapes of Human Breast and Colorectal Cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The Epigenomics of Cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 5.Brock MV, Hooker CM, Ota-Machida E, et al. DNA Methylation Markers and Early Recurrence in Stage I Lung Cancer. The New England Journal of Medicine. 2008;358:1118–28. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 6.Kim MSLJ, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Review. 2010;29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 7.Schuebel KE, Chen W, Cope L, Glöckner SC, Suzuki H, Yi JM, Chan TA, Neste LV, Criekinge WV, Bosch SV, van Engeland M, Ting AH, Jair K, Yu W, Toyota M, Imai K, Ahuja N, Herman JG, Baylin SB. Comparing the DNA Hypermethylome with Gene Mutations in Human Colorectal Cancer. PLoS Genetics. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan TAGS, Yi JM, Chen W, Van Neste L, Cope L, Herman JG, Velculescu V, Schuebel KE, Ahuja N, Baylin SB. Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Medicine. 2008;5:0823–38. doi: 10.1371/journal.pmed.0050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 10.DeClerck YA, Mercurio AM, Stack MS, et al. Proteases, Extracellular Matrix, and Cancer: A Workshop of the Path B Study Section. American Journal of Pathology. 2004;164:1131–9. doi: 10.1016/S0002-9440(10)63200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Migrating cancer stem cells [mdash] an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–9. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Weinberg RA. Epithelial-Mesenchymal Transition: At the Crossroads of Development and Tumor Metastasis. Developmental Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 13.van den Brandt PA, Goldbohm RA, Van ’T Veer P, Volovics A, Hermus RJJ, Sturmans F. A large-scale prospective cohort study on diet and cancer in the Netherlands. Journal of Clinical Epidemiology. 1990;43:285–95. doi: 10.1016/0895-4356(90)90009-e. [DOI] [PubMed] [Google Scholar]
- 14.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3740–5. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parsons DW, Jones S, Zhang X, et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane RP, Chen X-N, Yamakawa K, Vielmetter J, Korenberg JR, Dreyer WJ. Characterization of a Highly Conserved Human Homolog to the Chicken Neural Cell Surface Protein Bravo/Nr-CAM That Maps to Chromosome Band 7q31. Genomics. 1996;35:456–65. doi: 10.1006/geno.1996.0385. [DOI] [PubMed] [Google Scholar]
- 17.Beck K, Hunter I, Engel J. Structure and function of laminin: anatomy of a multidomain glycoprotein. The FASEB Journal. 1990;4:148–60. doi: 10.1096/fasebj.4.2.2404817. [DOI] [PubMed] [Google Scholar]
- 18.Chambers AF, Matrisian LM. Changing views of the role of matrix metalloproteinases in metastasis. JNCI Journal of the National Cancer Institute. 1997;89:1260–70. doi: 10.1093/jnci/89.17.1260. [DOI] [PubMed] [Google Scholar]
- 19.Finnson KWTB, Liu K, Marcoux A, Lepage P, Roy S, Bizet AA, Philip A. Identification of CD109 as part of the TGF-beta receptor system in human keratinocytes. FASEB Journal. 2006;20:1525–7. doi: 10.1096/fj.05-5229fje. [DOI] [PubMed] [Google Scholar]
- 20.Tian LSM, Ning L, Van Lint P, Nyman-Huttunen H, Libert C, Itohara S, Mishina M, Rauvala H, Gahmberg CG. Activation of NMDA receptors promotes dendritic spine development through MMP-mediated ICAM-5 cleavage. Journal of cell Biology. 2007;178:687–700. doi: 10.1083/jcb.200612097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachman KE, Herman JG, Corn PG, et al. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 22.Galm O, Suzuki H, Akiyama Y, et al. Inactivation of the tissue inhibitor of metalloproteinases-2 gene by promoter hypermethylation in lymphoid malignancies. Oncogene. 2005;24:4799–805. doi: 10.1038/sj.onc.1208599. [DOI] [PubMed] [Google Scholar]
- 23.de Caceres I Ibanez, Dulaimi E, Hoffman AM, Al-Saleem T, Uzzo RG, Cairns P. Identification of Novel Target Genes by an Epigenetic Reactivation Screen of Renal Cancer. Cancer Research. 2006;66:5021–8. doi: 10.1158/0008-5472.CAN-05-3365. [DOI] [PubMed] [Google Scholar]
- 24.Ulazzi LSS, Miotto E, Veronese A, Angusti A, Gafà R, Manfredini S, Farinati F, Sasaki T, Lanza G, Negrini M. Nidogen 1 and 2 gene promoters are aberrantly methylated in human gastrointestinal cancer. Mol Cancer. 2007;28:17. doi: 10.1186/1476-4598-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheetham S, Tang MJ, Mesak F, Kennecke H, Owen D, Tai IT. SPARC promoter hypermethylation in colorectal cancers can be reversed by 5-Aza-2[prime]deoxycytidine to increase SPARC expression and improve therapy response. Br J Cancer. 2008;98:1810–9. doi: 10.1038/sj.bjc.6604377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chicoine E, Esteve P-O, Robledo O, Van Themsche Ci, Potworowski EF, St-Pierre Y. Evidence for the role of promoter methylation in the regulation of MMP-9 gene expression. Biochemical and Biophysical Research Communications. 2002;297:765–72. doi: 10.1016/s0006-291x(02)02283-0. [DOI] [PubMed] [Google Scholar]
- 27.Mani SA, Guo W, Liao M-J, et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andre T, Boni C, Mounedji-Boudiaf L, et al. Oxaliplatin, Fluorouracil, and Leucovorin as Adjuvant Treatment for Colon Cancer. The New England Journal of Medicine. 2004;350:2343–51. doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- 29.Kuebler JP, Wieand HS, O’Connell MJ, et al. Oxaliplatin Combined With Weekly Bolus Fluorouracil and Leucovorin As Surgical Adjuvant Chemotherapy for Stage II and III Colon Cancer: Results From NSABP C-07. Journal of Clinical Oncology. 2007;25:2198–204. doi: 10.1200/JCO.2006.08.2974. [DOI] [PubMed] [Google Scholar]
- 30.Mehrotra J, Ganpat MM, Kanaan Y, et al. Estrogen Receptor/Progesterone Receptor-Negative Breast Cancers of Young African-American Women Have a Higher Frequency of Methylation of Multiple Genes than Those of Caucasian Women1. Clinical Cancer Research. 2004;10:2052–7. doi: 10.1158/1078-0432.ccr-03-0514. [DOI] [PubMed] [Google Scholar]
- 31.Chan AO, Soliman AS, Zhang Q, et al. Differing DNA methylation patterns and gene mutation frequencies in colorectal carcinomas from Middle Eastern countries. Clin Cancer Res. 2005;11:8281–7. doi: 10.1158/1078-0432.CCR-05-1000. [DOI] [PubMed] [Google Scholar]
- 32.Pelikan S, Moskowifz M. Effects of lead time, length bias, and false-negative assurance on screening for breast cancer. Cancer. 1998;71:1998–2005. doi: 10.1002/1097-0142(19930315)71:6<1998::aid-cncr2820710613>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 33.Meguid RASM, Wolfgang CL, Chang DC, Ahuja N. Is there a difference in survival between right- versus left-sided colon cancers? Annals of Surgical Oncology. 2008;15:2388–94. doi: 10.1245/s10434-008-0015-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barry I. Are there two sides to colorectal cancer? Internaltional Journal of Cancer. 2002;101:403–8. doi: 10.1002/ijc.10635. [DOI] [PubMed] [Google Scholar]
- 35.Annika L. Different mechanisms in the tumorigenesis of proximal and distal colon cancers. Curr Opin Oncol. 2001;13:63–9. doi: 10.1097/00001622-200101000-00013. [DOI] [PubMed] [Google Scholar]
- 36.Sugai T, Habano W, Jiao Y-F, et al. Analysis of Molecular Alterations in Left- and Right-Sided Colorectal Carcinomas Reveals Distinct Pathways of Carcinogenesis: Proposal for New Molecular Profile of Colorectal Carcinomas. Journal of Molecular Diagnostics. 2006;8:193–201. doi: 10.2353/jmoldx.2006.050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benedix FKR, Meyer F, Schmidt U, Gastinger I, Lippert H. Colon/Rectum Carcinomas (Primary Tumor) Study Group. Comparison of 17,641 patients with right- and left-sided colon cancer: differences in epidemiology, perioperative course, histology, and survival. Diseases of the Colon and Rectum. 2010;53:57–64. doi: 10.1007/DCR.0b013e3181c703a4. [DOI] [PubMed] [Google Scholar]
- 38.Yamada PM, Lee K-W. Perspectives in Mammalian IGFBP-3 Biology: Local vs. Systemic Action. AJP - Cell Physiology. 2009 doi: 10.1152/ajpcell.00598.2008. 00598.2008. [DOI] [PubMed] [Google Scholar]
- 39.Flood AMV, Pfeiffer R, Kahle L, Rosen CJ, Lanza E, Schatzkin A. Serum concentrations of insulin-like growth factor and insulin-like growth factor binding protein 3 and recurrent colorectal adenomas. Cancer Epidemiol Biomarkers Prev. 2008;17:1493–8. doi: 10.1158/1055-9965.EPI-08-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Maat MFG, van de Velde CJH, van der Werff MPJ, et al. Quantitative Analysis of Methylation of Genomic Loci in Early-Stage Rectal Cancer Predicts Distant Recurrence. Journal of Clinical Oncology. 2008;26:2327–35. doi: 10.1200/JCO.2007.14.0723. [DOI] [PubMed] [Google Scholar]
- 41.Umetani N, Takeuchi H, Fujimoto A, Shinozaki M, Bilchik AJ, Hoon DSB. Epigenetic Inactivation of ID4 in Colorectal Carcinomas Correlates with Poor Differentiation and Unfavorable Prognosis. Clinical Cancer Research. 2004;10:7475–83. doi: 10.1158/1078-0432.CCR-04-0689. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernstein BE, Meissner A, Lander ES. The Mammalian Epigenome. Cell. 2007;128:669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.