Abstract
mTOR has emerged as an important regulator of T helper cell differentiation. Here we demonstrate that TH1 and TH17 differentiation is selectively regulated by Rheb-dependent mTOR complex 1 (mTORC1) signaling. Rheb-deficient T cells fail to generate TH1 and TH17 responses in vitro and in vivo and cannot induce classical experimental autoimmune encephalomyelitis (EAE). However, they retain their ability to become TH2 cells. Alternatively, when mTORC2 signaling is deleted in T cells, they fail to generate TH2 cells in vitro and in vivo but preserve their ability to become TH1 and TH17 cells. Our data provide mechanisms by which the two distinct signaling pathways downstream of mTOR differentially regulate helper cell fate. These findings define a novel paradigm linking T cell differentiation with selective metabolic signaling pathways.
The mammalian Target of Rapamycin (mTOR) is an evolutionarily conserved member of the phosphatidylinositol-3-OH (PI-3)-kinase-related kinase (PI3KK) family that plays a central role in the regulation of metabolism, protein synthesis, energy balance, proliferation and survival 1. mTOR senses environmental cues such as amino acids, insulin, and growth factors and then integrates these signals in order to regulate cellular metabolism 2. mTOR forms the core of two distinct signaling complexes whose activation is differentially regulated 3, 4. mTOR complex 1 (mTORC1) contains the scaffolding protein regulatory-associated protein of mTOR (Raptor) as well as mlST8 (mammalian lethal with Sec13 protein 8), PRAS40 (the proline-rich Akt substrate 40 kDa) and DEP-domain-containing mTOR-interacting protein (Deptor) 5. Activation of mTORC1 is achieved through the PI-3K, PDK1, Akt signaling. mTORC1 promotes the phosphorylation of the translational regulators S6K1 and 4E-BP1 and is believed to play a central role in regulating cellular growth and proliferation by modulating metabolism 6. The second mTOR signaling complex (mTORC2) consists of mlST8, the scaffolding protein raptor-independent companion of TOR (Rictor), mSIN1 proteins, and the protein observed with Rictor (Protor) 5. Rictor is a critical adaptor protein for mTORC2 and Rictor-deficient mice and MEFs have been shown to lack mTORC2 activity 4. mTORC2 activity can be measured by the hydrophobic motif phosphorylation of Akt on serine 473 and other AGC kinases (SGK1 and PKCα) 7. The exact upstream activators of mTORC2 signaling have yet to be fully elucidated. Despite ongoing studies delineating both upstream activators and downstream effectors of the two respective signaling pathways, distinct physiological consequences mediated by mTORC1 and mTORC2 have yet to be defined in biologic systems.
A central role for mTOR in sensing the immune microenvironment and dictating immune function and differentiation has begun to emerge8. CD4+ T cells lacking mTOR fail to differentiate into effector cells under appropriate skewing conditions 9. Instead, upon activation, the mTOR-deficient T cells become Foxp3+ regulatory cells. This inability to differentiate into effector cells in the absence of mTOR was associated with a decrease in the activation of the STAT4, STAT6 and STAT3 transcription factors in response to the skewing cytokines interleukin 12 (IL-12), IL-4 and IL-6 respectively. mTOR controls CD8+ memory T cell development in part by regulating the expression of the transcription factors T-bet and Eomesodermin10, 11. In addition, the PI-3K-mTOR axis regulates lymphocyte trafficking12
The Ras Homologue Enriched in Brain (Rheb) is a small GTPase isolated from the central nervous system, and originally thought to be a mediator of the Ras-MAPK pathway13, 14. The mammalian Rheb family consists of two proteins, Rheb1 and Rheb2, both of which are expressed in T cells but their function has yet to be elucidated13, 15. Rheb is a crucial regulator of mTORC1 signaling16. The tuberous sclerosis complex (TSC, which is comprised of TSC1 and TSC2) functions as a GTPase-activating protein (GAP) for Rheb. When TSC is inhibited, the active, GTP-bound form of Rheb interacts with mTORC1 to stimulate its activity15.
Despite the emerging appreciation for the central role of mTOR in regulating T cell effector, memory and regulatory cell differentiation, the downstream signaling pathways involved in regulating these processes have yet to be elucidated. By selectively knocking out Rheb in T cells we demonstrate that both TH1 and TH17 effector differentiation require mTORC1 signaling, while TH2 differentiation is preserved. Mice which harbor T cell-specific deletions of Rheb cannot mount TH1 and TH17 responses in vivo and are resistant to the development of classical experimental autoimmune encephalomyelitis (EAE). Since T cells lacking mTORC1 activity were still able to become TH2 cells, we further investigated the role of mTORC2 signaling on TH2 development. By deleting mTORC2 signaling in T cells we found that they were unable to become TH2 cells but maintained their ability to differentiate into TH1 and TH17 cells. Our findings define distinct downstream signaling pathways as the mechanism by which mTOR regulates T helper cell differentiation.
RESULTS
Rheb regulates mTORC1 activity in T cells
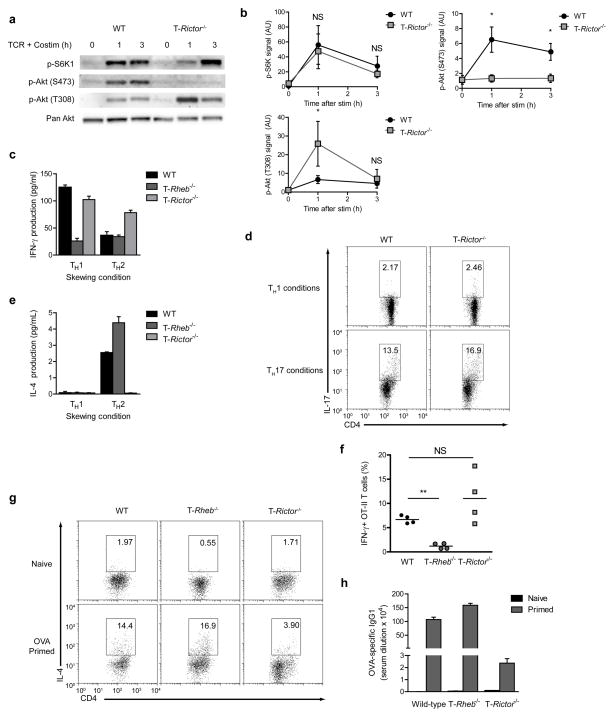
We hypothesized that genetic deletion of Rheb in T cells would inhibit mTORC1 activation. As such, Rheb floxed mice were bred to CD4-Cre mice leading to the deletion of Rheb from T cells (Supplementary Fig. 1). CD4+ T cells from wild-type and T-Rheb−/− mice were stimulated with anti-CD3+ anti-CD28 and interrogated biochemically for mTORC1 and mTORC2 activity. Upon stimulation, wild-type T cells markedly phosphorylate S6K1, indicative of mTORC1 activity (Fig. 1a, b). In contrast, S6K1 phosphorylation was virtually absent from the activated T-Rheb−/− T cells. Phosphorylation of Akt at threonine 308 (which is PDK1-dependent and upstream of mTORC1) remained intact in the T-Rheb−/− T cells (Fig. 1a, b). Likewise, phosphorylation of Akt at serine 473 (which is downstream of mTORC2) remained intact.
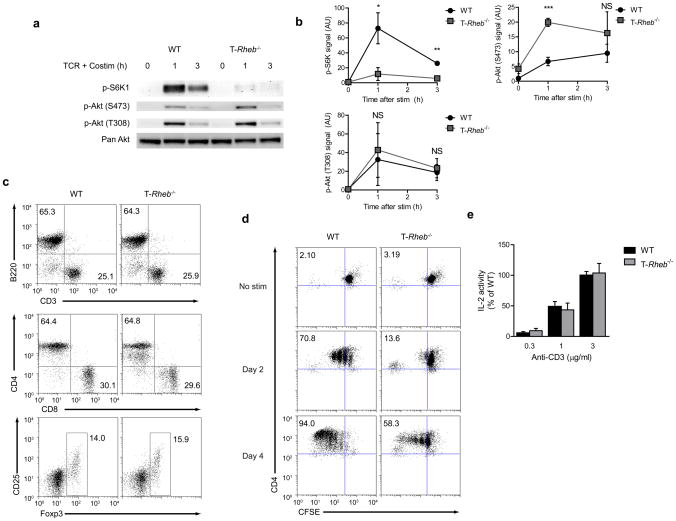
Figure 1.
Rheb controls mTORC1 activity in T cells. (a) Immunoblot (IB) of mTOR activation in lysates from wild-type and T-Rheb−/− CD4+ T cells stimulated with anti-CD3 and anti-CD28 in serum free media for 1 or 3 h. (b) Densitometry analysis across 4 experiments for mTORC1 (p-S6K1), mTORC2 (p-Akt (S473)), and PI3K (p-Akt (T308)) activity (n=4, * p <.05, ** p <.01, *** p <.001 by Student’s t test). (c) Flow cytometric analysis of splenocytes harvested from wild-type or T-Rheb−/− mice. CD4 vs CD8 plots are gated on CD3+, Treg plots are gated CD3+CD4+. (d) Dilution of CFSE in wild- type and T-Rheb−/− CD4+ T cells stimulated with anti-CD3 and irradiated autologous APCs for 48 or 96h. (e) IL-2 production in wild-type and T-Rheb−/− CD4+ T cells stimulated with anti-CD3 and anti-CD28 overnight Data are representative of at least 3 independent experiments.
Next we sought to determine the functional consequences of deleting Rheb in T cells. The percentages of CD8+, CD4+ T cells and B cells were similar between the wild-type and T-Rheb−/− mice (Fig. 1c and Supplementary Fig. 2). In addition, in spite of the lack of mTORC1 activity, we did not see any differences in the development of Foxp3+ T cells. Similar to T cells lacking mTOR9, T cells from the T-Rheb−/− mice did not demonstrate a block in proliferation but rather proliferated slower than their wild-type counterparts (Fig. 1d). Likewise, IL-2 production was similar between the wild-type and T-Rheb−/− T cells indicating that TCR-induced activation and subsequent expression of IL-2 was not impacted by the absence of Rheb (Fig. 1e). In some systems, mTOR signaling can affect apoptosis. Upon in vitro activation, there was no evidence for increased apoptosis by Annexin V and 7-AAD staining in T cells from the T-Rheb−/− mice (Supplementary Fig. 3). Thus, deleting Rheb in T cells specifically eliminates mTORC1 activity while preserving mTORC2 activity. Furthermore, elimination of Rheb in T cells still permits TCR-induced cytokine expression and proliferation.
TH1 and TH17 but not TH2 differentiation require mTORC1
In light of the fact that mTOR-deficient T cells fail to differentiate into TH1, TH2 or TH17 effector cells, we wanted to determine the effect of Rheb deletion on T cell differentiation. There was no difference in naïve and memory T cell compartments from wild-type and T-Rheb−/− littermates, as assayed using freshly-isolated lymphocytes (Supplementary Fig. 4). Similar to T cells lacking mTOR, T-Rheb−/− T cells failed to differentiate into TH1 cells under appropriate skewing conditions in vitro (Fig. 2a). T-Rheb−/− T cells also failed to differentiate into TH17 cells (Fig. 2b). These observations identify mTORC1 signaling as being the downstream mediator of mTOR-dependent TH1 and TH17 differentiation. In marked contrast to mTOR-deficient T cells, the T-Rheb−/− T cells were capable of differentiating into TH2 cells (Fig. 2c). Therefore, the inhibition of mTORC1 activity by the deletion of Rheb selectively inhibited TH1 and TH17 differentiation but preserved TH2 differentiation.
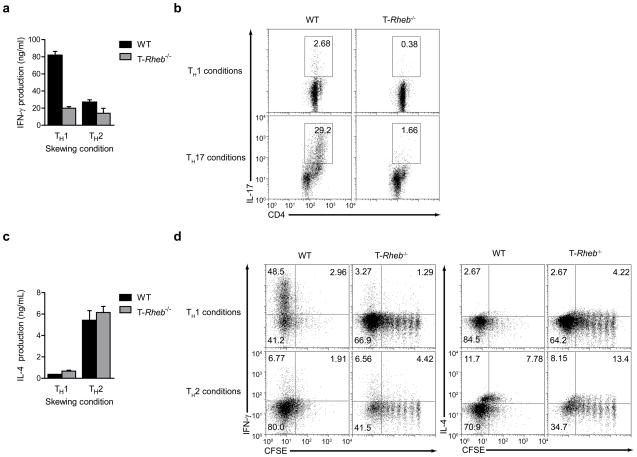
Figure 2.
mTORC1 is required for TH1 and TH17 differentiation in vitro, but is dispensable for TH2 differentiation. (a) IFN-γ production in wild-type and T-Rheb−/− CD4+ T cells stimulated with anti-CD3 and APCs in TH1 or TH2 skewing conditions, expanded 5 d with IL-2, and restimulated in vitro with anti-CD3 and anti-CD28. (b) IL-17 production in wild-type and T-Rheb−/− CD4+ T cells stimulated with anti-CD3 and APCs in TH1 or TH17 skewing conditions, expanded 5 d, then restimulated with anti-CD3 and APCs in the presence of a protein transport inhibitor. (c) As in a, but for IL-4. (d) CFSE dilution and cytokine production of wild-type and T-Rheb−/− CD4+ T cells labeled and skewed as in a and restimulated with anti-CD3 and anti-CD28 in the presence of a protein transport inhibitor. Data are representative of at least 3 independent experiments.
To examine if the defect in TH1 skewing was linked to the proliferative defect in the Rheb-deficient T cells, we labeled wild-type and Rheb-deficient T cells with CFSE, cultured them in TH1 and TH2 skewing conditions and interrogated them for cytokine production upon rechallenge. Rheb-deficient T cells did proliferate in vitro, although at slower rates. Furthermore, Rheb-deficient T cells that divided several times under TH1 skewing conditions still could not make interferon (IFN)-γ in amounts comparable to wild-type cells (Fig. 2d). Further, the slightly increased IL-4 production observed did not directly correlate with the number of cell divisions. Thus, the inability of Rheb-deficient T cells to differentiate into TH1 cells was not secondary to their slower rate of proliferation.
Next we sought to confirm these results in vivo. Vaccinia virus infection leads to a robust TH1 response. T cells from wild-type and T-Rheb−/− mice crossed onto the OT-II TCR transgenic background specific for ovalbumin (OVA) were adoptively transferred into wild-type C57/BL6 mice17. Recipient mice were infected with Vaccinia virus expressing ovalbumin (Vaccinia-OVA) and clonotypic T cells were evaluated 5 days later for the expression of IFN-γ and IL-4. As expected, the wild-type OT-II cells differentiated into TH1 cells producing robust levels of IFN-γ IL- 2 and TNF-α but little IL-4 (Fig. 3a, b). In marked contrast, T-Rheb−/− clonotypic T cells failed to produce robust IFN-γ, but produced significant levels of IL-4. To assess the role of proliferation in this in vivo system, OT-II cells were labeled with CFSE. Cells transferred into non-virus-infected host mice failed to proliferate or produce cytokines upon rechallenge, while wild-type T cells transferred into immunized mice proliferated in vivo and produced IFN-γ (Fig. 3c). However, Rheb-deficient T cells, even in the case of cells undergoing multiple rounds of proliferation fail to produce IFN-γ. Addition of an IL-4 neutralizing antibody to the culture conditions failed to rescue IFN-γ production (data not shown). These data demonstrate that selective deletion of mTORC1 signaling leads to enhanced TH2 differentiation. Thus, selective inactivation of mTORC1 in T cells did not lead to T cell tolerance, but rather promoted differentiation down an alternative effector pathway.
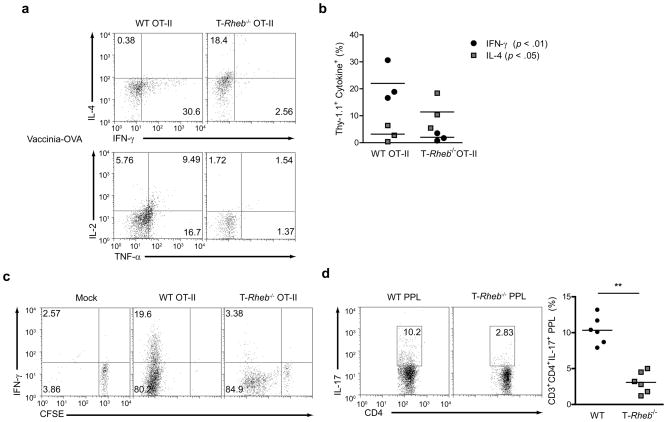
Figure 3.
mTORC1-deficient T cells cannot skew to TH1 or TH17 in vivo. (a) Cytokine production in C57/BL6 (Thy-1.2+) mice that were immunized with Vaccinia-OVA and received an adoptive transfer of 1 × 106 wild-type or T-Rheb−/− CD4+ OT-II T cells (Thy-1.1+). Splenocytes were harvested 4 days after transfer and rechallenged overnight in vitro with OVA peptide in the presence of a protein transport inhibitor. Plots are gated on Thy-1.1+ cells. (b) Tabulated results from one independent experiment as in a. (n=3, p < .01 for IFN-γ, p < .05 for IL-4, by Student’s t test). (c) As in a, but some mice received a mock immunization, and adoptively transferred cells were CFSE-labeled. (d) Cytokine production from Peyer’s Patch Lymphocytes (PPL) isolated from wild-type and T-Rheb−/− mice and stimulated with PMA and ionomycin (n = 6, ** p <.01). Data are representative of at least 3 independent experiments.
The ability of Rheb-deficient T cells to differentiate into TH17 cells in vivo was also determined. TH17 cells develop in vivo in the gut in response to pathogens. Following a brief PMA and Ionomycin stimulation in vitro, there was a marked decrease in in vivo differentiated CD3+CD4+ TH17 in the Peyer’s Patch in T-Rheb−/− mice compared to wild-type (Fig. 3d). Overall, these data show that TH1 and TH17 but not TH2 differentiation is specifically regulated by mTORC1 signaling in vivo and in vitro.
T-Rheb−/− mice are resistant to EAE
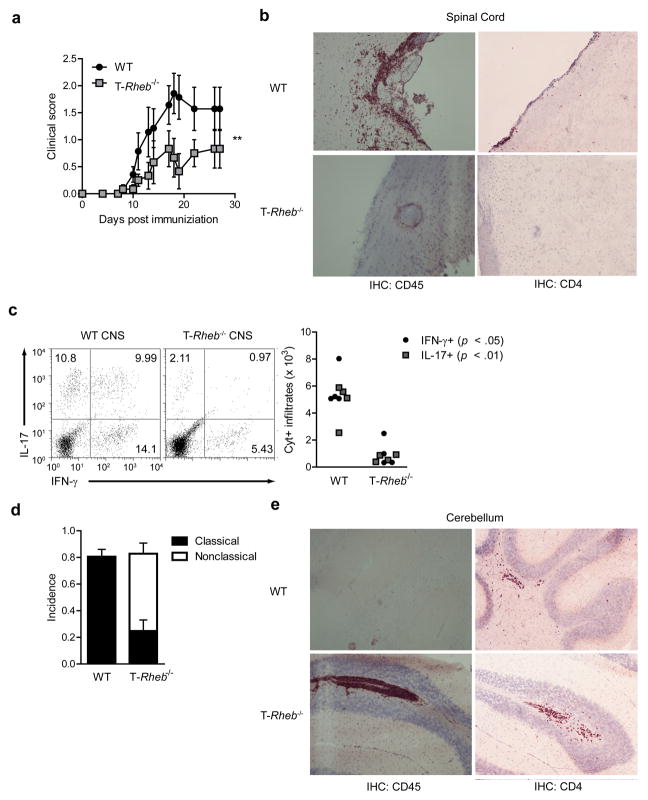
Because both TH1 and TH17 T cells are implicated in promoting autoimmune diseases and IFN-γ and IL-17 participate in promoting the pathology seen in EAE, we determined the susceptibility of the T-Rheb−/− mice to this experimental model 18. Following MOG35–55 peptide immunization, wild-type mice developed clinical EAE (Fig. 4a) while T-Rheb−/− mice had decreased clinical scores, associated with decreased leukocytic infiltration in the spinal cord and the decreased expansion of TH1 and TH17 cells (Fig. 4b, c). However, nearly 60% (on average) of MOG-immunized T-Rheb−/− mice developed “non-classical EAE” (Supplementary Video 1), which is characteristic of TH2 responses to MOG (Fig. 4d). MOG-specific TH2 cells or IFN-γ−/− cells can induce a neurologic disorder characterized by ataxia (not paralysis) and infiltration of the cerebellum with immune cells 19. T-Rheb−/− mice displayed increased leukocytic infiltration, including CD4 cells, in the cerebellum, (Fig. 4e) and T cells isolated from these mice produced increased levels of IL-4 and IL-5 in response to MOG peptide in vitro (data not shown). Thus, the mTORC1-deficient T cells were neither “immunodeficient” nor incapable of trafficking to the CNS. Thus, the selective deletion of mTORC1 signaling leads to enhanced TH2 differentiation in vivo, even under strongly TH1/TH17 promoting conditions.
Figure 4.
T-Rheb−/− mice do not develop EAE, but instead develop an alternative autoimmune disease. (a) Disease progression of wild-type and T-Rheb−/− mice immunized with MOG and CFA to induce EAE (n=7, **, p < 0.01 by one-way ANOVA). (b) Immunohistochemistry (IHC) for CD45 and CD4 of CNS sections frozen at the height of disease in wild-type mice. (c) Cytokine production of CNS samples rested in unsupplemented media for 16 h and stimulated with PMA and ionomycin in the presence of a protein transport inhibitor (n = 4, p values indicated are from Student’s t test). Plots are gated by CD4 expression and side scatter. (d) Incidence of classical or nonclassical EAE in our system. (e) As in b, but sections of cerebellum are shown. Data are representative of 3 independent experiments.
TH2 but not TH1 or TH17 differentiation requires mTORC2 signaling
We further bred Rictor floxed mice to CD4-Cre mice to determine the effect of selectively eliminating mTORC2 in T cells (T-Rictor−/−, Supplementary Fig. 5)20. Following anti-CD3 + anti-CD28 stimulation, T-Rictor−/− T cells manifested a decrease in mTORC2 activity as determined by Akt hydrophobic motif (serine 473) phosphorylation. mTORC1 activity in these cells, as determined by the phosphorylation of S6K1, was normal (Fig. 5a, b). In agreement with observations made in Rictor-deficient mice7, proximal (PI3K-PDK1) signaling, assayed by Akt phosphorylation at threonine 308, was intact and consistently higher. T-Rictor−/− CD4 T cells developed normally (a slight decrease in the percentage of peripheral CD8s was observed) (Supplementary Fig. 6) and produced equivalent amounts of IL-2 (Supplemental Fig. 7) as the wild-type T cells indicating that TCR-induced signaling was intact. Rictor−/− T cells displayed slower proliferation upon stimulation, but had a less severe defect than Rheb−/− T cells (Supplementary Fig. 7). In addition, T-Rictor−/− mice did not demonstrate an increase in activation-induced apoptosis (Supplementary Fig. 3) and have normal proportions of CD4+ T cells in the naïve compartment (Supplementary Fig. 8).
Figure 5.
mTORC2-deficient T cells cannot skew to TH2, but retain TH1 and TH17 skewing. (a) IB of mTOR activation in lysates from wild-type and T-Rictor−/− CD4+ T cells stimulated with anti-CD3 and anti-CD28 in serum free media for 1 or 3 h. (b) Densitometry analysis across 4 experiments for mTORC1 (p-S6K1), mTORC2 (p-Akt (S473)), and PI3K (p-Akt (T308)) activity (n=4, * <.05, ** <.01, *** <.001 by Student’s t test). (c) IFN-γ production in wild-type, T-Rheb−/−, and T-Rictor−/− CD4+ T cells stimulated with anti-CD3 and APCs overnight in TH1 or TH2 skewing conditions, expanded 5 d with IL-2, and restimulated in vitro with anti-CD3 and anti-CD28. (d) IL-17 production of wild-type and T-Rictor−/− CD4+ T cells stimulated with anti-CD3 and APCs in TH1 or TH17 skewing conditions, expanded 5 d, and restimulated with anti-CD3 and APCs in the presence of a protein transport inhibitor. (e) As in c, but for IL-4. (f) Cytokine production as in 3a, but some host mice received an adoptive transfer of T-Rictor−/− CD4+ T cells. (g) IL-4 production in CD4+ T cells from wild-type, T-Rheb−/−, and T-Rictor−/− mice immunized with alum or ovalbumin with alum and boosted after 14 days. Splenocytes were stimulated 48 h with 100 μg/ml OVA protein (protein transport inhibitor present for the last 8 h). (h) OVA-specific serum antibody titers from mice in g. Data are representative of at least 3 independent experiments, except for f, which was performed twice.
T-Rictor−/− T cells were able to develop into TH1 and TH17 T cells under appropriate skewing conditions in vitro (Fig. 5c, d), but failed to differentiate down a TH2 pathway (Fig. 5E).
To exclude that the presence of previously activated cells may alter skewing conditions in our system, we sorted naïve (CD62LhiCD44lo) CD4+ cells from wild-type, T-Rheb−/−, and T-Rictor−/− mice and skewed them using plate-bound anti-CD3 and soluble anti-CD28. As seen with bulk CD4+ T cells, Rheb-deficient cells fail to differentiate into TH1 or TH17 cells, and Rictor-deficient cells fail to differentiate into TH2 cells (Supplementary Fig. 9). TCR signaling strength can affect TH1 and TH2 responses 21. Specifically, low concentrations of peptide can impair IFN-γ production, and create a bias toward a TH2 response To determine the impact of strength of peptide signaling in the differentiation of Rheb- and Rictor-deficient T cells, wild-type, Rheb-, and Rictor-deficient OT-II T cells were stimulated under conditions of widely varying doses of OVA peptide as previously described22. However, even at a dose of 100μM of peptide, there was no evidence of increased IFN-γ production from Rheb-deficient T cells (Supplementary Fig. 10). Alternatively, Rictor-deficient T cells produced TH1 cytokines even at exceedingly low doses of peptide.
Consistent with the in vitro data, T-Rictor−/− T cells produced robust levels of IFN-γ in response to Vaccinia infection (Fig. 5f). T-Rictor−/− mice had normal susceptibility to EAE (Supplementary Fig. 11), associated with equivalent numbers of IFN-γ and IL-17 producing cells in response to MOG peptide as the wild-type. To assess TH2 responses in vivo, wild-type (C57/BL6), T-Rheb−/− and T-Rictor−/− mice were primed and boosted with OVA using alum as an adjuvant to elicit a TH2 response. Compared with wild-type and T-Rheb−/− mice, T-Rictor−/− mice had reduced induction of IL-4-secreting OVA specific T cells (Fig. 5g) and failed to generate antigen-specific IgG1 antibodies (that are dependent upon TH2 cell help) (Fig. 5h). Thus, mTORC2 regulates the ability of helper T cell to differentiate toward TH2 cells.
Distinct STAT activation by mTORC1 and mTORC2
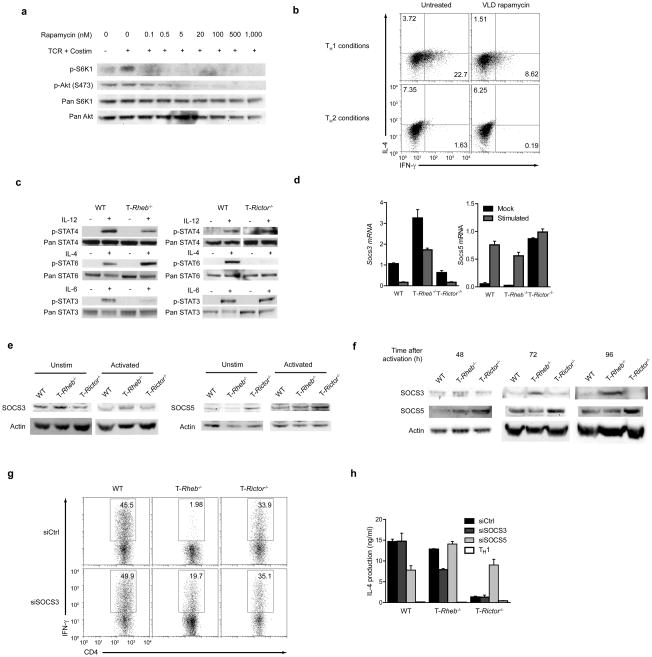
Next we determined if we could reproduce these results in wild-type primary T cells using pharmacological inhibition of mTOR activity. Biochemical analysis of T cells revealed that rapamycin can inhibit mTORC2 activity (Fig. 6a)23. However, at very low doses (100–500pM, referred to as VLD) rapamycin selectively inhibits mTORC1 in T cells. VLD of rapamycin specifically inhibited TH1 differentiation in naïve wild-type T cells activated under either TH1 or TH2 skewing conditions (Fig. 6B), consistent with the findings in Rheb-deficient T cells.
Figure 6.
mTORC1 and mTORC2 influence cytokine signaling through differential inhibition of SOCS proteins. (a) IB of mTOR activation in lysates from wild-type, naive T cells stimulated with anti-CD3 and anti-CD28 in varying doses of rapamycin for 8 h in serum-containing media. (b) Cytokine production of wild-type, 5C.C7 T cells stimulated with PCC peptide for 48 h in TH1 or TH2 skewing conditions in the presence or absence of very low dose (VLD, 100pM) rapamycin, washed and expanded in fresh drug and IL-2 for 5 days, then restimulated with anti-CD3 and anti-CD28. (c) IB of STAT phosphorylation in lysates from freshly isolated wild-type, T-Rheb−/− and T-Rictor−/− CD4+ T cells resuspended in serum-free media for 1 h and stimulated with IL-12, IL-4, or IL-6 for 30 min. (d) SOCS3 and SOCS5 mRNA levels from wild-type, T-Rheb−/− and T-Rictor−/− CD4+ T cells stimulated with mock or anti-CD3 and anti-CD28 simulation overnight. Values are normalized to 18s rRNA and scaled to wild-type unstimulated (for SOCS3) or stimulated (for SOCS5). (e) IB of SOCS3 and SOCS5 of cells treated as in d. (f) Kinetic analysis of SOCS3 and SOCS5 protein expression in lysates from wild-type, T-Rheb−/− and T-Rictor−/− CD4+ T stimulated with anti-CD3 and anti-CD28 for the indicated times. Actin is included as a loading control. (g) IFN-γ production in wild-type, T-Rheb−/− and T-Rictor−/− CD4+ T cells transfected with control, SOCS3, and SOCS5 siRNA by nucleofection, rested 16 h, stimulated in TH1 and TH2 skewing conditions 48 h, then expanded in IL-2 for 48 h and restimulated with anti-CD3 and anti-CD28. (h) IL-4 production from cells in g. All data are representative of at least three independent experiments, except for g and h, which were performed twice.
It is known that T cells lacking mTOR cannot differentiate into effector cells, due in part to diminished responses to T helper skewing cytokines9. Freshly isolated T cells from wild-type, T-Rheb−/− and T Rictor−/− mice were exposed to skewing cytokines and STAT activation was assessed. Consistent with their inability to become TH1 and TH17 cells, T-Rheb−/− T cells showed decreased STAT4 and STAT3 phosphorylation in response to IL-12 and IL-6 respectively (Fig. 6c, left panel). However, T-Rheb−/− T cells demonstrated enhanced STAT6 phosphorylation in response to IL-4 (a TH2-promoting cytokine). T-Rictor−/− T cells showed intact IL-12 and IL-6-induced STAT4 and STAT3 activation, while IL-4-induced STAT6 activation was diminished (Fig. 6C, right panel). These findings indicate that the ability of mTORC1 and mTORC2 to differentially regulate T cell effector differentiation is due in part to their ability to differentially regulate STAT activation. In response to different cytokines, Rheb-deficient T cells showed decreased IL-23- and IFN-β–induced STAT4 activation and decreased IL-10- and IL-21-dependent STAT3 activation (data not shown). Rictor-deficient T cells showed decreased STAT6 activation in response to IL-13 (data not shown).
mTOR was previously shown to influence STAT activation9, 24, 25. To determine how distinct STATs are differentially regulated in T cells, we examined cytokine receptor expression on Rheb- and Rictor-deficient T cells. IL-12Rβ1, IL-4R and IL-6R were all expressed at equivalent levels in the wild-type, Rheb and Rictor-deficient T cells, indicating that mTOR did not regulate cytokine receptor expression (Supplementary Fig. 12).
STAT activation is partially regulated by the expression of SOCS (Suppressor Of Cytokine Signaling) proteins, which can differentially dephosphorylate JAK-dependent residues on STAT proteins. We determined the expression pattern of SOCS3 and SOCS5 in the Rheb and Rictor-deficient T cells. Consistent with previously published findings, wild-type T cells expressed SOCS3 (mRNA and protein) at baseline and downregulated its expression upon activation26–28 (Fig. 6d, e) while SOCS5 expression was low in naïve T cells and increased with activation29, 30. In resting Rheb-deficient T cells SOCS3 mRNA and protein expression was markedly increased, and retained higher expression upon activation (Fig. 6d, e), while SOCS5 expression was similar to wild-type cells. Rictor-deficient T cells displayed SOCS3 at baseline (albeit lower amounts than the wild-type cells) and this expression decreased upon activation, while SOCS5 expression was markedly upregulated in the resting cells and remained elevated upon activation (Fig. 6d, e). Thus, at baseline and upon activation, the levels of SOCS3 and SOCS5 are differentially regulated in the Rheb-deficient and Rictor-deficient T cells.
SOCS protein expression is dynamically regulated over time. To examine the expression patterns of SOCS proteins at later time points, T cells were activated for 48, 72, and 96 h. At these time points, SOCS3 protein remained elevated in Rheb-deficient T cells, while wild-type and Rictor-deficient cells downregulated SOCS3 (Fig. 6f). Further, while wild-type and Rheb-deficient cells consistently upregulated SOCS5 upon activation, Rictor-deficient T cells displayed significantly elevated levels of SOCS5 at later time points.
To prove that the increased levels of SOCS3 in the Rheb-deficient T cells and the increased levels of SOCS5 in the Rictor-deficient T cells had functional significance, we transfected freshly isolated wild-type, Rheb-, and Rictor-deficient CD4+ T cells with siRNA against SOCS3 and SOCS5. This resulted in significant knockdown of SOCS3 and SOCS5 mRNA (Supplementary Fig. 13). In T cells stimulated in TH1 and TH2 differentiation conditions knockdown of SOCS3 mRNA resulted in increased IFN-γ production in Rheb-deficient T cells (Fig. 6g), while knock down of SOCS5 mRNA in Rictor-deficient T cells resulted in increased ability to produce IL-4 (Fig. 6h). Such observations demonstrate that SOCS proteins regulated TH1, TH17 and TH2 differentiation in the Rheb- and Rictor-deficient T cells.
Transcription factor expression in T-Rheb−/− and T-Rictor−/− T cells
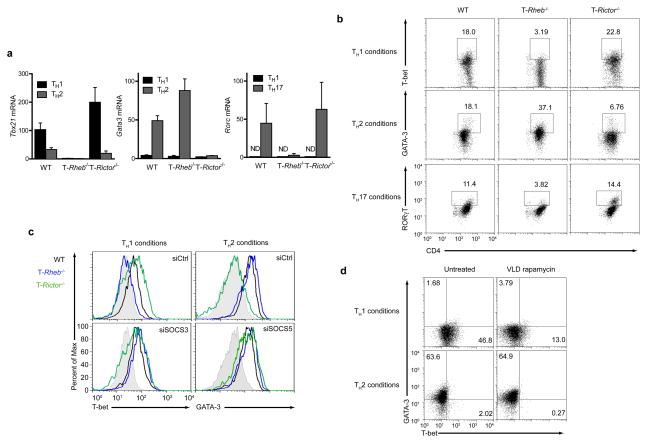
Next we sought to determine if the transcription factors T-bet, RORγT and GATA-3, which play critical roles in the development of TH1, TH17 and TH2 cells are selectively affected by Rheb or Rictor deletion. Rheb-deficient T cells failed to fully upregulate T-bet message and protein under TH1 conditions (Fig. 7a, b) and RORγT under TH17 conditions, while strongly expressing GATA-3 under TH2 conditions. In fact, GATA-3 was expressed at higher levels in Rheb-deficient cells than in wild-type cells, consistent with their enhanced ability to phosphorylate STAT6 and produce IL-4 in vivo. In Rictor−/− T cells, both T-bet and RORγT were transcribed and translated under TH1 and TH17 conditions respectively, but GATA-3 failed to be expressed under TH2 conditions (Fig. 7a and b). In T cells stimulated in TH1 and TH2 differentiation conditions, knockdown of SOCS3 mRNA resulted in increased in T-bet expression in Rheb-deficient T cells (Fig. 7c), while knock down of SOCS5 mRNA in Rictor-deficient T cells resulted in increased GATA-3 expression (Fig. 7c). Furthermore, wild-type T cells exposed to VLD rapamycin (which inhibits TH1 but not TH2 development) demonstrated decreased T-bet expression but not decreased GATA3 expression in response to the appropriately differentiating conditions (Fig. 7d). Thus, in the absence of the nutrient sensitive signaling pathways mediated by mTOR, the ability to respond to cytokines is diminished, and T cells fail to properly upregulate lineage-specific transcription factors.
Figure 7.
Control of transcription factor induction by mTORC1 and mTORC2. (a) Lineage-specific transcription factor mRNA expression in wild-type, T-Rheb−/− and T-Rictor−/− CD4+ T cells skewed to TH1, TH2, or TH17 as before. ND = not detected. Units are fold over 18S rRNA (for Tbx21 and Gata3) or ddCT arbitrary units (for Rorc). (b) Flow cytometric analysis of wild-type, T-Rheb−/− and T-Rictor−/− CD4+ T cells skewed to TH1, TH2, or TH17 and stained for T-bet, GATA-3, and RORγT. (c) Flow cytometric analysis of cells nucleofected as in 6g and skewed to TH1 and TH2. The shaded histogram represents wild-type TH2 cells (for T-bet) and wild-type TH1 cells (for GATA-3). (d) Flow cytometric analysis of naive 5C.C7 T cells skewed to TH1 or TH2 in the presence or absence of VLD rapamycin. All data are representative of at least three independent experiments, except for c, which was performed twice.
Treg development in the absence of mTORC1 and mTORC2
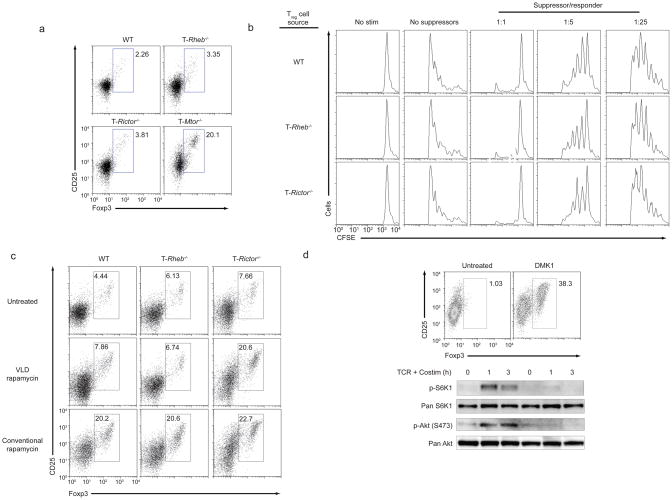
It is known that mTOR deficient T cells differentiate down a Foxp3+ T regulatory pathway and that inhibition of mTORC1 alone is not sufficient for the generation of Tregs 9. Rapamycin can enhance the generation of inducible regulatory T cells 31–33. To determine the specific effect of mTORC1 and mTORC2 on regulatory T cell differentiation, wild-type, T-Rheb−/−, T-Rictor−/− and T-Mtor−/− T cells were cultured in vitro under activating conditions (in the absence of exogenous TGF-β) and then assessed for the generation of Foxp3+ T cells. Neither the elimination of mTORC1 nor mTORC2 signaling alone led to an increase in Foxp3+ cells (Fig. 8a). Notably however, inducible regulatory T cells (made with TGF-β and IL-2) from wild-type, T-Rheb−/−, T-Rictor−/− mice all appeared to inhibit T cell function equally (Fig. 8b).
Figure 8.
Inhibition of both mTORC1 and mTORC2 are required for spontaneous Treg induction. (a) Foxp3 and CD25 expression in CD4+ T cells from wild-type, T-Rheb−/−, T-Rictor−/− and T-Mtor−/− mice stimulated with anti-CD3 APCs overnight and rested in IL-7 supplemented media for 5 d. (b) CFSE-dilution of wild-type T cells stimulated in vitro with anti-CD3 APCs, and varying numbers of Foxp3+ Treg generated from wild-type, T-Rheb−/−, and T-Rictor−/− CD4+ T cells (using TGF-β and IL- ). Treg were enriched by CD25 magnetic separation, and Foxp3 expression was confirmed by intracellular staining. (c) Foxp3 and CD25 expression in wild-type, T-Rheb−/−, and T-Rictor−/− CD4+ T cells stimulated with anti-CD3 and APCs in the presence of VLD or conventional dose rapamycin and rested in IL-7 supplemented media (with drug) for 5 days. (d) Foxp3 and CD25 expression of wild-type T cells stimulated with anti-CD3 and APCs in the presence or absence of DMK1, a kinase inhibitor of mTOR, and expanded for 5 days in the presence of IL-2 and drug (top panel). IB of mTOR activation in lysates from wild-type T cells stimulated with anti-CD3 and anti-CD28 for 1 or 3 h in serum free media with or without DMK1 (bottom panel). All data are representative of at least 3 independent experiments.
To further test the hypothesis that both mTORC1 and mTORC2 must be inhibited in order to generate regulatory T cells, wild-type, T-Rheb−/− and T-Rictor−/− T cells were activated in the presence of a conventional dose (500nM, which inhibits mTORC1 and mTORC2) or VLD (500pM, which only inhibits mTORC1) of rapamycin. Wild-type T cells differentiated into Foxp3+ T cells at the conventional 500 nM dose that inhibits mTORC1 and mTORC2 (Fig. 8c). T-Rheb−/− T cells required mTORC2 inhibition induced by the conventional dose. In contrast, T-Rictor−/− T cells require only VLD rapamycin to differentiate into Tregs. Finally, a mTOR-specific kinase inhibitor DMK1 (which acutely inhibits both mTOR downstream signaling pathways) 34 promoted Treg generation from naïve wild-type T cells, consistent with our data from mTOR-, mTORC1- and mTORC2-deficient T cells (Fig. 8d). Thus, both mTORC1 and mTORC2 inhibition is required for the generation of regulatory T cells in the absence of exogenous TGF-β.
DISCUSSION
In this report, we define precise and reciprocal roles for mTORC1 and mTORC2 signaling in regulating CD4+ T helper cell differentiation. Our data ascribe tissue distinct physiologic functions of mTORC1 and mTORC2. Thus, mTORC1 signaling promotes TH1 and TH17 differentiation, mTORC2 signaling promotes TH2 differentiation and mTOR inhibition leads to regulatory T cells. Preliminary studies from our lab have failed to demonstrate the differential activation of mTORC1 and mTORC2 under TH1 and TH2 skewing conditions. As such we hypothesize that it is the differential expression and activation of downstream substrates and signaling pathways that confer the ability of mTORC1 to regulate TH1 and TH17 differentiation and mTORC2 to regulate TH2 differentiation.
Our studies provide a critical link between metabolism and T cell differentiation. mTORC1 signaling integrates growth factors, energy, oxygen and amino acids to promote processes necessary for cell growth 2, 5. In addition, in T cells, the PI3K-Akt-mTORC1 axis is activated by CD28 engagement and IL-2 receptor signaling35. IL-7 promotes mTOR activity, helping to prevent atrophy in T cells, 36 while IL-4-induced mTOR activity prevents apoptosis37. IL-12 and IFN-γ promote the sustained activation of mTORC1 activity in CD8+ T cells.11, 38 Relatively little is known about the precise role of mTORC2 signaling in regulating cellular function 5. The ability of mTORC2 to regulate TH2 differentiation provides potential clues for determining the specific role of mTORC2 in other cell types.
The small GTPase Rheb was recently identified as an activator of mTORC115. Our data demonstrate that Rheb signaling plays an important role in regulating T cell differentiation. mTOR-deficient T cells, Rheb-deficient T cells and T cells treated with very low doses of rapamycin, which only inhibits mTORC1, all inhibit CD4+ effector cell differentiation, supporting the hypothesis that the ability of Rheb to direct differentiation is through the activation of mTORC1. Furthermore, mTORC1 signaling appears to be both necessary and sufficient for TH1 and TH17 differentiation because Rictor-deficient T cells still maintain their ability to become TH1 and TH17 effectors.
A recent study has examined Rictor-deficient T cells generated by crossing Lck-Cre and floxed Rictor mice 39. Consistent with our study, Rictor deletion inhibited TH2 differentiation. However, a decrease in the generation of TH1 cells (albeit modest compared to TH2 inhibition) was also reported. The cause of these divergent observations remains unclear. The use of Lck-Cre can lead to subtle differences in T cell development and affect TH differentiation. Additionally, we found that under TH1 conditions, Rictor-deficient T cells can generate greater TH17 cells than normal. In addition, there appear to be differences in signaling between the Rictor deficient T cells employed in each study. Notably, unlike our mice, which demonstrate robust phosphorylation of Akt threonine 308 and subsequent S6K1 activation, T cells in this other study show decreased signaling upstream of mTOR39, which could also impair mTORC1 signaling. We suggest that when the signaling defects are restricted downstream of mTORC2 signaling, there is a selective block in TH2 but not TH1 development. Thus, it is possible that Rictor-deficient T cells in the abovementioned report behave more like mTOR-deficient T cells.
Rapamycin promotes the generation of regulatory T cells in the absence of exogenous TGF-β40–42. PD-L1 was shown to promote inducible regulatory T cells by inhibiting mTOR activation43. In addition, local depletion of essential amino acids can promote the generation of regulatory T cells by inhibiting mTOR activity44. We show here that both mTORC1 and mTORC2 signaling must be inhibited in order to skew cells down the regulatory T cell pathway. Our genetic studies were complemented by a pharmacologic approach. The simultaneous inhibition of mTORC1 and mTORC2 by an mTOR kinase inhibitor was a potent inducer of regulatory T cells in the absence of exogenous TGF-β indicating that this class of drugs can act as potent immunosuppressive/tolerance-inducing agents.
mTORC1 and mTORC2 can promote cell differentiation in other tissues. Cerebral cortical development is mTORC1 dependent while terminal differentiation of myoblasts is mTORC2 mediated45, 46. In this model, mTORC2 signaling inactivates Rho-associated kinase ROCK1, which in turn promotes the activation of the transcription factors FoxO1 and FoxO3a. We demonstrate a similar role for mTORC1 and mTORC2 signaling in regulating distinct tissue specific transcription factors. It was recently shown that rapamycin promotes CD8+ memory cell differentiation10. Thus, by inhibiting T-bet expression, rapamycin not only inhibits effector functions (such as IFN-γ production) but promotes the expression of Eomesodermin which in turn promotes the generation of memory T cells11. The decrease in TH specific transcription factors is due to a decrease in specific STAT activation in response to cytokines. While the IL-12-, IL-6- and IL-4-induced STAT4, STAT3 and STAT6 phosphorylation is not completely lost in the mTORC1-and mTORC2-deficient T cells, the decreased response contributes to the observed differentiation failure. The ability of mTOR to regulate STAT3 phosphorylation was described in cancer and neuron activation however, the precise mechanism for this regulation is unclear 24, 25, 47. In T cells we show that differences in SOCS3 and SOCS5 expression contribute to the STAT activation defects. Along these lines, mTORC2 inhibits the activity of Foxo transcription factors7, 48. Foxos control KLF2 expression, which in turn promotes SOCS5 expression49. Preliminary studies show an increase in KLF2 in Rictor-deficient T cells, consistent with the observed increase in SOCS5 in these cells (data not shown). We propose a model whereby mTORC1 and mTORC2 activate distinct signaling programs the net sum of which is to promote specific effector differentiation. Future studies will seek to identify the mTORC1 and mTORC2 specific targets that control differentiation.
Supplementary Material
Acknowledgments
We thank members of the Powell laboratory, as well as Drs. C. Drake and D. Pardoll for discussion and reagent contributions. This work was supported by NIH grant R01AI077610-01A2.
Footnotes
AUTHOR CONTRIBUTIONS
G.M.D. performed the research, helped design experiments, and wrote the paper. K.N.P. assisted with in vivo experiments and biochemistry. A.T.W. assisted with EAE induction, CNS isolation, and performed immunohistochemistry. E.H. assisted with VLD rapamycin experiments. D.J.M. synthesized the mTOR kinase inhibitor. M.R.H. helped design experiments and contributed reagents. B.X. and P.F.W. generated the original Rheb floxed mouse line. J.D.P. designed experiments, oversaw research, and wrote the paper.
References
- 1.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 3.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 4.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 5.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holz MK, Blenis J. Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J Biol Chem. 2005;280:26089–26093. doi: 10.1074/jbc.M504045200. [DOI] [PubMed] [Google Scholar]
- 7.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Delgoffe GM, Powell JD. mTOR: taking cues from the immune microenvironment. Immunology. 2009;127:459–465. doi: 10.1111/j.1365-2567.2009.03125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delgoffe GM, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinclair LV, et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat Immunol. 2008;9:513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamagata K, et al. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269:16333–16339. [PubMed] [Google Scholar]
- 14.Yee WM, Worley PF. Rheb interacts with Raf-1 kinase and may function to integrate growth factor- and protein kinase A-dependent signals. Mol Cell Biol. 1997;17:921–933. doi: 10.1128/mcb.17.2.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. Trends Biochem Sci. 2003;28:573–576. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Saucedo LJ, et al. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 17.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 18.Park H, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wensky AK, et al. IFN-gamma determines distinct clinical outcomes in autoimmune encephalomyelitis. J Immunol. 2005;174:1416–1423. doi: 10.4049/jimmunol.174.3.1416. [DOI] [PubMed] [Google Scholar]
- 20.Kumar A, et al. Muscle-specific deletion of rictor impairs insulin-stimulated glucose transport and enhances Basal glycogen synthase activity. Mol Cell Biol. 2008;28:61–70. doi: 10.1128/MCB.01405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul WE. What determines Th2 differentiation, in vitro and in vivo? Immunol Cell Biol. 2010;88:236–239. doi: 10.1038/icb.2010.2. [DOI] [PubMed] [Google Scholar]
- 22.Yamane H, Zhu J, Paul WE. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J Exp Med. 2005;202:793–804. doi: 10.1084/jem.20051304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 24.Wang B, et al. Nogo-66 promotes the differentiation of neural progenitors into astroglial lineage cells through mTOR-STAT3 pathway. PLoS One. 2008;3:e1856. doi: 10.1371/journal.pone.0001856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J, et al. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci U S A. 2007;104:16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto K, Yamaguchi M, Miyasaka N, Miura O. SOCS-3 inhibits IL-12-induced STAT4 activation by binding through its SH2 domain to the STAT4 docking site in the IL-12 receptor beta2 subunit. Biochem Biophys Res Commun. 2003;310:1188–1193. doi: 10.1016/j.bbrc.2003.09.140. [DOI] [PubMed] [Google Scholar]
- 27.Yu CR, et al. Suppressor of cytokine signaling 3 regulates proliferation and activation of T-helper cells. J Biol Chem. 2003;278:29752–29759. doi: 10.1074/jbc.M300489200. [DOI] [PubMed] [Google Scholar]
- 28.Palmer DC, Restifo NP. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol. 2009;30:592–602. doi: 10.1016/j.it.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozaki A, Seki Y, Fukushima A, Kubo M. The control of allergic conjunctivitis by suppressor of cytokine signaling (SOCS)3 and SOCS5 in a murine model. J Immunol. 2005;175:5489–5497. doi: 10.4049/jimmunol.175.8.5489. [DOI] [PubMed] [Google Scholar]
- 30.Seki Y, et al. Expression of the suppressor of cytokine signaling-5 (SOCS5) negatively regulates IL-4-dependent STAT6 activation and Th2 differentiation. Proc Natl Acad Sci U S A. 2002;99:13003–13008. doi: 10.1073/pnas.202477099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battaglia M, Stabilini A, Roncarolo MG. Rapamycin selectively expands CD4+CD25+FoxP3+ regulatory T cells. Blood. 2005;105:4743–4748. doi: 10.1182/blood-2004-10-3932. [DOI] [PubMed] [Google Scholar]
- 32.Sauer S, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ballou LM, Selinger ES, Choi JY, Drueckhammer DG, Lin RZ. Inhibition of mammalian target of rapamycin signaling by 2-(morpholin-1-yl)pyrimido[2,1-alpha]isoquinolin-4-one. J Biol Chem. 2007;282:24463–24470. doi: 10.1074/jbc.M704741200. [DOI] [PubMed] [Google Scholar]
- 35.Colombetti S, Basso V, Mueller DL, Mondino A. Prolonged TCR/CD28 engagement drives IL-2-independent T cell clonal expansion through signaling mediated by the mammalian target of rapamycin. J Immunol. 2006;176:2730–2738. doi: 10.4049/jimmunol.176.5.2730. [DOI] [PubMed] [Google Scholar]
- 36.Rathmell JC, Farkash EA, Gao W, Thompson CB. IL-7 enhances the survival and maintains the size of naive T cells. J Immunol. 2001;167:6869–6876. doi: 10.4049/jimmunol.167.12.6869. [DOI] [PubMed] [Google Scholar]
- 37.Stephenson LM, Park DS, Mora AL, Goenka S, Boothby M. Sequence motifs in IL-4R alpha mediating cell-cycle progression of primary lymphocytes. J Immunol. 2005;175:5178–5185. doi: 10.4049/jimmunol.175.8.5178. [DOI] [PubMed] [Google Scholar]
- 38.Lekmine F, et al. Interferon-gamma engages the p70 S6 kinase to regulate phosphorylation of the 40S S6 ribosomal protein. Exp Cell Res. 2004;295:173–182. doi: 10.1016/j.yexcr.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 39.Lee K, et al. Mammalian Target of Rapamycin Protein Complex 2 Regulates Differentiation of Th1 and Th2 Cell Subsets via Distinct Signaling Pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang J, Huddleston SJ, Fraser JM, Khoruts A. De novo induction of antigen-specific CD4+CD25+Foxp3+ regulatory T cells in vivo following systemic antigen administration accompanied by blockade of mTOR. J Leukoc Biol. 2008;83:1230–1239. doi: 10.1189/jlb.1207851. [DOI] [PubMed] [Google Scholar]
- 41.Kopf H, de la Rosa GM, Howard OM, Chen X. Rapamycin inhibits differentiation of Th17 cells and promotes generation of FoxP3+ T regulatory cells. Int Immunopharmacol. 2007;7:1819–1824. doi: 10.1016/j.intimp.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valmori D, et al. Rapamycin-mediated enrichment of T cells with regulatory activity in stimulated CD4+ T cell cultures is not due to the selective expansion of naturally occurring regulatory T cells but to the induction of regulatory functions in conventional CD4+ T cells. J Immunol. 2006;177:944–949. doi: 10.4049/jimmunol.177.2.944. [DOI] [PubMed] [Google Scholar]
- 43.Francisco LM, et al. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cobbold SP, et al. Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc Natl Acad Sci U S A. 2009;106:12055–12060. doi: 10.1073/pnas.0903919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S, et al. The apical complex couples cell fate and cell survival to cerebral cortical development. Neuron. 2010;66:69–84. doi: 10.1016/j.neuron.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shu L, Houghton PJ. The mTORC2 complex regulates terminal differentiation of C2C12 myoblasts. Mol Cell Biol. 2009;29:4691–4700. doi: 10.1128/MCB.00764-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson MD, O’Connell M, Vito F, Bakos RS. Increased STAT-3 and synchronous activation of Raf-1-MEK-1-MAPK, and phosphatidylinositol 3-Kinase-Akt-mTOR pathways in atypical and anaplastic meningiomas. J Neurooncol. 2009;92:129–136. doi: 10.1007/s11060-008-9746-7. [DOI] [PubMed] [Google Scholar]
- 48.Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 49.Dekker RJ, et al. KLF2 provokes a gene expression pattern that establishes functional quiescent differentiation of the endothelium. Blood. 2006;107:4354–4363. doi: 10.1182/blood-2005-08-3465. [DOI] [PubMed] [Google Scholar]
- 50.Delgoffe GM, Kole TP, Cotter RJ, Powell JD. Enhanced interaction between Hsp90 and raptor regulates mTOR signaling upon T cell activation. Mol Immunol. 2009;46:2694–2698. doi: 10.1016/j.molimm.2009.05.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.