Abstract
Laccases and their homologues form the protein superfamily of multicopper oxidases (MCO). They catalyze the oxidation of many, particularly phenolic substances, and, besides playing an important role in many cellular activities, are of interest in biotechnological applications. The Laccase Engineering Database (LccED, http://www.lcced.uni-stuttgart.de) was designed to serve as a tool for a systematic sequence-based classification and analysis of the diverse multicopper oxidase protein family. More than 2200 proteins were classified into 11 superfamilies and 56 homologous families. For each family, the LccED provides multiple sequence alignments, phylogenetic trees and family-specific HMM profiles. The integration of structures for 14 different proteins allows a comprehensive comparison of sequences and structures to derive biochemical properties. Among the families, the distribution of the proteins regarding different kingdoms was investigated. The database was applied to perform a comprehensive analysis by MCO- and laccase-specific patterns.
The LccED combines information of sequences and structures of MCOs. It serves as a classification tool to assign new proteins to a homologous family and can be applied to investigate sequence–structure–function relationship and to guide protein engineering.
Database URL: http://www.lcced.uni-stuttgart.de
Introduction
Multicopper oxidases (MCOs) catalyze the one-electron oxidation of their substrates with a concomitant four-electron reduction of molecular oxygen to water. MCOs consist of four enzyme families: laccases (EC 1.10.3.2), ascorbate oxidases (EC 1.10.3.3), ferroxidases (EC 1.16.3.1) and ceruloplasmin (EC 1.16.3.1). Functional studies have revealed that MCOs have two active sites: one blue type 1 (T1) copper site where the substrate is oxidized, and a trinuclear copper cluster [consisting of three type 2 (T2)/type 3 (T3) coppers] where oxygen is bound, activated and reduced (1). The electrons are transferred from the T1 site to the T2/T3 site via highly conserved amino acids which have previously been described in PROSITE (2, 3) as MCO-specific patterns, further referred to as M2 and M4 (4, 5). In addition, laccase-specific signature sequences, namely L1 and L3, were generated from 100 plant and fungal laccase sequences. L1 and L3 have been suggested to be specific for laccases and were proposed to distinguish laccases from other MCOs (6). While there is only low overall sequence similarity, the structure and catalytic mechanism is conserved (7). Most MCOs consist of three cupredoxin domains, except for ceruloplasmin and some bacterial laccases which contain six or two domains, respectively (8). Depending on the number of domains, MCOs vary in size, from 300 to 1000 residues, and contain up to six copper ions (4).
Laccases, which constitute the largest subfamily of MCOs, are widely distributed among fungi, higher plants (9, 10), bacteria (11) and insects (12). In fungi, they are supposed to be involved in lignin degradation (13), pigment production (14) and plant pathogenesis (15). In plants, their potential function is the biosynthesis of lignin (16). In bacteria, they are suggested to play a role in melanin production, spore coat resistance, morphogenesis and detoxification of copper (17). In particular laccases, which form the largest subgroup of MCOs, also have a high biotechnological potential as versatile catalysts in textile and in pulp and paper industries, as well as in food applications, bioremediation and organic synthesis (18, 19). However, their redox potential often is restricted and they react in a relatively non-specific way (20). Engineered laccases promise to have improved enzymatic properties such as activity, specificity and selectivity (21). It is expected that understanding the relationships between sequence, structure and function would greatly help the engineering of laccases. Therefore, we integrated data on MCO sequences and structures and built up the Laccase Engineering Database (LccED) using the data warehouse system DWARF (22). Previously, 350 MCOs were assigned to 10 superfamilies (23): (A) basidiomycete laccases, (B) ascomycete laccases, (C) insect laccases, (D) fungal pigment MCOs, (E) fungal ferroxidases, (F) fungal and plant ascorbate oxidases, (G) plant laccase-like MCOs, (H) copper resistance proteins (CopA), (I) bilirubin oxidases and (J) copper efflux (CueO) proteins. For the MCOs that lack the second domain, referred to as small laccases (SLAC), a distinct family was established based on SLAC from of Streptomyces coelicolor (24). Homologous MCO sequences were retrieved and assigned to families by sequence similarity. In order to assist comprehensive sequence analysis, reliable multisequence alignments were generated and annotated either by an automated pattern search or by information extracted from GenBank (25). In addition, family-specific HMM profiles (26) and a BLAST (27) interface are provided to allow an assignment of new sequences to families. Thus, the LccED is the first data resource that combines information on sequences, sequence alignments, annotations, and structures of MCOs.
Construction and content
Database construction
The LccED was established within the data warehouse system DWARF, which provides a data model for the integration of sequences and structures in a family-specific protein database, as well as tools for extracting and loading data from various data sources (22). Previously, more than 350 MCO sequences were assigned to 10 superfamilies (23). From this data set 248 sequences, for which a GenBank entry was available, were selected as seed sequences and assigned according to their initial classification by Hoegger et al. to the 10 superfamilies, which were named based on the origin of their seed sequences. An additional superfamily was created for two domain laccases and SLAC from Streptomyces coelicolor was used as seed sequence for this family. Subsequently, for each seed sequence a BLAST search (27) was performed in the non-redundant sequence database at NCBI (http://ncbi.nlm.nih.gov) with an E-value of E = 10−10. The E-value for the BLAST search was determined empirically. Therefore, for the more diverse bacterial families a higher E-value of E = 10−5 was applied. Each BLAST hit was assigned to the superfamily of the respective seed sequence if the sequence identity was higher than 40%. Sequences within a superfamily were classified into homologous families based on multiple sequence alignments and phylogenetic trees, as calculated by CLUSTAL W (28). Information on source organism, sequence annotations and sequence was extracted by the sequence data loader of the DWARF system. The species information was adapted to the NCBI taxonomy. Different names denominating the same organism are listed as synonyms on the organism page. Sequences from the same source organism and sharing >98% identical residues were represented as one single protein entry. This assignment is implemented in an automated script, thus preventing that one protein from the same organism may occur in duplicate within the database and avoiding redundancy even if it may occur in GenBank.
In case of BLAST hits specifying a protein structure, the respective structure was extracted from the PDB (29), stored as structural monomers and secondary structure information was generated for each chain by DSSP (30). For all families, multiple sequence alignments were performed by CLUSTAL W (28) and manually checked to improve consistency and quality. Proteins which were not assignable to any superfamily and are not similar to the class of MCOs were removed from the database. BLAST hits which are proteins fragments or putative sequences which were longer than usual MCOs, or proteins with high sequence similarity to MCOs where chosen to be included if they locally align in the active site regions.
The LccED provides regular yearly updates using the automated update function of the DWARF system which applies a protein ‘blacklist’ of removed entries, keeping pace with the permanently growing GenBank data (25).
Contents
The LccED contains data on 2828 sequences and 2297 proteins. For 21 proteins from 10 different homologous families crystal structures are deposited, which results in a total of 82 structural monomers. The proteins were assigned to 11 superfamilies based on the origin of the seed sequences and to 56 homologous families based on phylogeny (Table 1). For each superfamily and homologous family, an annotated multiple sequence alignment, a phylogenetic tree and a family-specific HMM profile (http://hmmer.janelia.org/) were generated.
Table 1.
LccED families, sequences and structures
| Superfamily | Homologous families | Proteins | Structures |
|---|---|---|---|
| A (Basidiomycete Laccases) | 4 | 201 | 13 |
| B (Ascomycete Laccases) | 6 | 421 | 6 |
| C (Insect Laccases) | 8 | 168 | 0 |
| D (Fungal Pigment MCOs) | 4 | 55 | 0 |
| E (Fungal Ferroxidases) | 6 | 117 | 6 |
| F (Fungal and plant AOs) | 6 | 137 | 8 |
| G (Plant Laccases) | 5 | 333 | 0 |
| H (Bacterial CopA Proteins) | 6 | 383 | 0 |
| I (Bacterial Bilirubin Oxidases) | 5 | 149 | 24 |
| J (Bacterial CueO Proteins) | 5 | 310 | 11 |
| K (SLAC homologs) | 1 | 18 | 3 |
Web interface
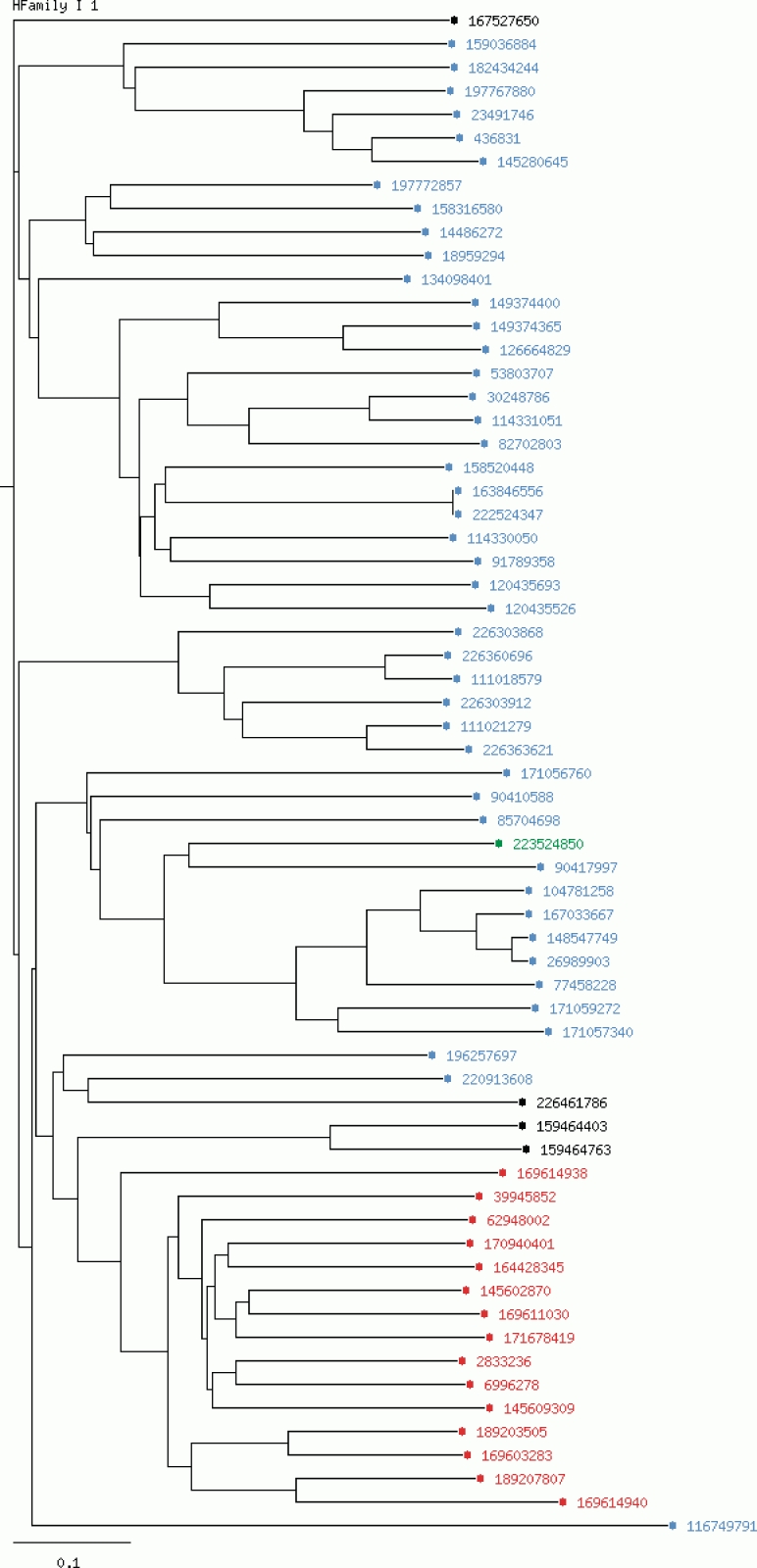
The LccED is publicly available on http://www.LccED.uni-stuttgart.de. It can be browsed by family, organism or structure. For each family, pre-calculated annotated multiple sequence alignments, phylogenetic trees and HMMs are provided. All protein entries in the alignments and trees are linked to their original NCBI entries. Functionally relevant amino acids are color coded, and further information is displayed upon moving the mouse over the respective residue in the multiple sequence alignment. The conservation degree of the alignment was calculated using PLOTCON (31). PFAM (32) links to all protein entries were added as far they were available. Further, the PFAM annotation to each protein can be accessed within the multiple sequence alignments by scrolling the mouse over the respective region. Phylogenetic trees are visualized by an in-house developed tree-visualizer which allows coloring each entry by properties such as homologous family (in superfamily-trees), organism, sequence length and kingdom of the source organism (Figure 1). Via a local BLAST interface, unknown MCO sequences can be classified by sequence similarity to the existing LccED entries. A tar-archive comprising all information on families, sequences, structures, multiple sequence alignments, trees and profiles can be downloaded.
Figure 1.
Phylogenetic tree for the homologous family I1 (Bilirubin oxidases).The chosen coloring option is ‘by kingdom’. Entries of bacterial origin are shown in blue, fungal entries in red, plant proteins in green and non-specified entries are colored in black.
Analysis of organism distribution and patterns
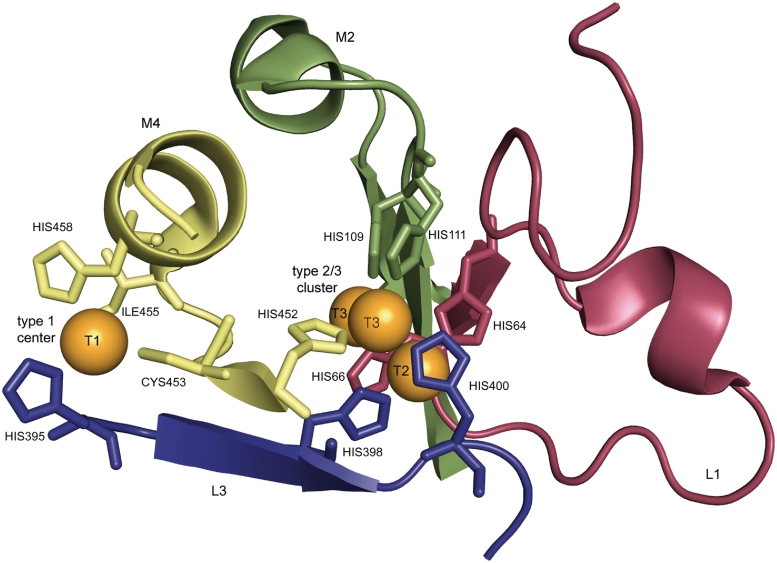
In this study, 2297 MCO proteins from a wide spectrum of source organisms were assigned to superfamilies and homologous families based on sequence similarity and phylogenetic analysis. A comprehensive analysis of the relationships between sequence similarity, source organism and of patterns forming the binding sites of copper was performed in 2274 proteins of families A–K. The proposed patterns L1 (H-W-H-G-x(9)–D-G-x(5)–Q-C-P-I) and L3 (H-P-x-H-L-H-G-H) have been suggested to be specific for laccases, the patterns M2 (G-x-[FYW]-x-[LIVMFYW]-x-[CST]-x-{PR}-{K}-x(2)-{S}-x-{LFH}-G-[LM]-x(3)-[LIVMFYW], PROSITE entry PS00079) and M4 (H-C-H-x(3)-H-x(3)-[AG]-[LM], PROSITE entry PS00079) for MCOs (Figure 2). Pattern L1 includes one histidine which binds the T2 copper and one histidine which binds the T3 copper. Pattern M2 includes two further T3 copper ligands. Pattern L3 includes ligands of the T1, T2 and T3 coppers. Within pattern M4 three of the four ligands of the T1 centre and one T3 copper ligand are located (Figure 2). For annotation and evaluation purposes regular expressions generated from L1, L3, M2 and M4 were applied. Because of the short length of their sequences and the missing domain, proteins of the SLAC family were excluded from the analysis of the appearance of the pattern within the families (Supplementary Table S1) and the resulting numbers for false negatives (Supplementary Table S2).
Figure 2.
Copper binding residues of laccase from Trametes versicolor [PDB entry 1GYC, (39)].The copper centers are shown in orange, the residues that match the defined pattern L1, M2, L3, M4 are colored in red, green, blue, and yellow, respectively [visualization by PyMOL (40)].
Family A (Basidiomycete Laccases) contains exclusively fungal proteins, 91% are from basidiomycetes (homologous families A1–A4), 9% from ascomycetes (homologous family A2). Eighty percent are annotated as laccases in GenBank. Twenty-three percent of the proteins contain the pattern L1, 22% M2, 14% L3 and 46% M4. Seventeen percent of the entries are annotated as putative.
Family B (Ascomycete Laccases) contains 36% from ascomycetes. All of them cluster into the homologous family B1 and 62% are annotated as laccases in GenBank. The other proteins are all of bacterial origin (homologous families B2–B6) and 3% are annotated as laccases in GenBank. Yet, they show a considerable sequence identity of over 40% to ascomycetous laccases. Eighty-eight percent of the proteins contain the pattern L1, 92% M2, 49% L3 and 15% M4. Thirty-three percent of the entries are annotated as putative.
Family C (Insect Laccases) resulted in 78% proteins of insect origin (homologous families C1–C8). The remaining 22% consist of euechinoidea (in homologous family C1), cephalochordata and cnidaria (in homologous family C6). Thirty-eight percent are annotated as laccases in GenBank. Thirty percent of the proteins contain pattern L1, 75% M2, 75% L3 and 3% M4. Thirty-five percent of the entries are annotated as putative.
Family D (Fungal Pigment MCOs) contains exclusively fungal proteins. Thirty-six percent of the proteins are annotated in GenBank as laccases. Ninety percent of the proteins contain pattern L1, 78% M2, 82% L3 and 11% M4. Fifty-eight percent of the entries are annotated as putative.
Family E (Fungal Ferroxidases) contains exclusively fungal proteins. Seventeen percent of the proteins are annotated in GenBank as laccases. Eighty-three percent of the proteins contain pattern L1, 40% M2, 23% L3 and 3% M4. Forty-five percent of the entries are annotated as putative.
Family F (Fungal and Plant Ascorbate Oxidases) mainly contain proteins of plant origin (homologous families F2–F6). Twelve percent are of fungal origin and clustered all to the homologous family F1. Two percent are annotated as laccases in GenBank. In this family, 88% of the proteins contain pattern L1, 56% M2, 66% L3 and 66% M4. Sixty-two percent of the entries are annotated as putative.
Family G (Plant Laccases) exclusively contains proteins of plant origin and 83% are annotated as laccases in GenBank (homologous families G1–G5). Fifteen percent of the proteins contain pattern L1, 88% contain pattern M2, 77% contain pattern L3 and 2% contain pattern M4. Fifty-three percent of the entries are annotated as putative.
Family H (Bacterial CopA Proteins) contains proteins of which 98% were of bacterial origin (homologous families H1–H6). Eighty-three percent are annotated as laccases in GenBank. Fifty percent contain pattern L1, 50% M2, 42% L3 and 3% M4. Six percent of the entries are annotated as putative.
Family I (Bilirubin Oxidases) contains proteins of which 70% were of bacterial origin (homologous families I1–I5), 15% of plant origin (homologous family I3), 10% of fungal origin (homologous family I1) and 5% of unspecified source organism (Figure 1). Three percent are annotated in GenBank as laccases. Seventy percent contain pattern L1, 92% M2, 91% L3 and 65% M4. Twenty-six percent of the entries are annotated as putative.
Family J (Bacterial CueO Proteins) contains proteins of which 90% were of bacterial origin (homologous families J1–J4) and 10% of eukaryotic origin (homologous family J5). Twelve percent are annotated as laccases in GenBank. Seventy-four percent of the proteins contain pattern L1, 75% M2, 76% L3 and 3% M4. Thirty-nine percent of the entries are annotated as putative.
Family K (SLAC) which contains exclusively members of the ‘small laccase family’ which all are of bacterial origin, annotated as MCOs in GenBank and contain only pattern L4. Six percent of the entries are annotated as putative.
Besides the slight variations within the laccase and MCO sequence patterns almost all MCO sequences share the same highly conserved copper binding residues (Figure 2). They could be identified and annotated by a manual validation of each family-specific multisequence alignment. Only within homologous families F5, F6, H3 and J2 these residues could not be detected.
Discussion
As suggested previously (23), the 10 MCO superfamilies were named by combining the name of the prevailing source organism and the putative enzymatic function. The overall distribution of source organisms among the families generally agreed with the initial classification. Since the assignment of a protein to a superfamily was exclusively based on sequence similarity, it was expected to find in the same family proteins from different source organisms, even from different kingdoms of life. Indeed, most families consisted of a majority of proteins belonging to one kingdom with a minority from other kingdoms, despite a sequence similarity as high as 40%. A systematic classification of all proteins by sequence similarity only was prerequisite to a reliable sequence alignment of superfamilies and to identify conserved, functionally relevant sequence patterns. However, a systematic analysis of previously described MCO- and laccase-specific patterns (4–6), which have been derived from a small number of MCOs and laccases, demonstrated their low sensitivity. This is also supported by the high number of false negatives which were retrieved by a manual analysis of the respective regions of the multisequence alignments (Supplementary Table S2). The MCO patterns M2 and M4 were only found in 15 and 65% of all MCOs (Supplementary Table S1), respectively. Nine percent of all MCOs contain both M2 and M4. To differentiate laccases from other MCOs is even more difficult. If we assume that sequence similarity is an indication of function similarities, there are four superfamilies which contain putative laccases. However, for these families the laccase-specific patterns L1 and L3 were only found in 45 and 37% of the sequences, respectively (Supplementary Table S1). Only 8% of all putative laccases contain all four patterns simultaneously. This low percentage of positive hits could either indicate that ‘laccase superfamilies’ contain MCOs without laccases activity, or it might be caused by the too restrictive patterns. As an alternative to patterns, sequence profiles are widely used to specify functionally related protein families (32, 33). Therefore, for each superfamily, a hidden Markov profile is provided, and the four copper binding regions are consistently annotated in the LccED.
To define discriminating rules for laccases, more detailed functional studies are needed. Newly gained information, for example, on redox potential or the effect of mutations, can be added to readily prepared tables and may be transferred on closely related proteins. It has been shown previously that a systematic classification of large protein families based on sequence similarity and comprehensive analysis tools as provided by the LccED serve as a reliable framework for studying sequence–structure–function relationships of enzyme families (34–36) and for the design of mutants or focused mutant libraries with improved biochemical properties (37, 38).
Conclusion
The LccED enables the systematic classification and analysis of MCO sequences and structures from different public sources. The integration of protein data in a relational database system has been used to study the molecular basis of biochemical properties and to investigate sequence–structure–function relationships. The LccED comes with a set of tools for phylogenetic analysis and classification. The annotated multisequence alignments allow the identification of the regions which house the copper atoms and other functionally relevant residues.
Availability
The LccED is available at http://www.LccED.uni-stuttgart.de. Via this web interface all sequences, alignments and trees are accessible and all data are supplied for download.
Supplementary Data
Supplementary data are available at Database online.
Funding
Financial support by the Deutsche Forschungsgemeinschaft (SFB 706) is gratefully acknowledged, as well as funding for open access charge.
Conflict of interest. None declared.
Acknowledgements
We acknowledge Constantin Vogel for the extension of the DWARF workbench to allow structure annotation.
References
- 1.Solomon E, Sundaram U, Machonkin T. Multicopper oxidases and oxygenases. Chem. Rev. 1996;96:2563–2606. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- 2.Hulo N, Bairoch A, Bulliard V, et al. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigrist CJ, Cerutti L, Hulo N, et al. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 2002;3:265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- 4.Messerschmidt A, Huber R. The blue oxidases, ascorbate oxidase, laccase and ceruloplasmin. Modelling and structural relationships. Eur. J. Biochem. 1990;187:341–352. doi: 10.1111/j.1432-1033.1990.tb15311.x. [DOI] [PubMed] [Google Scholar]
- 5.Ouzounis C, Sander C. A structure-derived sequence pattern for the detection of type I copper binding domains in distantly related proteins. FEBS Lett. 1991;279:73–78. doi: 10.1016/0014-5793(91)80254-z. [DOI] [PubMed] [Google Scholar]
- 6.Kumar SVS, Phale PS, Durani S, et al. Combined sequence and structure analysis of the fungal laccase family. Biotechnol. Bioeng. 2003;83:386–394. doi: 10.1002/bit.10681. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura K, Go N. Function and molecular evolution of multicopper blue proteins. Cell. Mol. Life Sci. 2005;62:2050–2066. doi: 10.1007/s00018-004-5076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy ME, Lindley PF, Adman ET. Structural comparison of cupredoxin domains: domain recycling to construct proteins with novel functions. Protein Sci. 1997;6:761–770. doi: 10.1002/pro.5560060402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Messerschmidt A. Multi-Copper Oxidases. Singapore: World Scientific; 1997. [Google Scholar]
- 10.Mayer AM, Staples RC. Laccase: new functions for an old enzyme. Phytochemistry. 2002;60:551–565. doi: 10.1016/s0031-9422(02)00171-1. [DOI] [PubMed] [Google Scholar]
- 11.Alexandre G, Zhulin IB. Laccases are widespread in bacteria. Trends Biotechnol. 2000;18:41–42. doi: 10.1016/s0167-7799(99)01406-7. [DOI] [PubMed] [Google Scholar]
- 12.Dittmer NT, Suderman RJ, Jiang H, et al. Characterization of cDNAs encoding putative laccase-like multicopper oxidases and developmental expression in the tobacco hornworm, Manduca sexta, and the malaria mosquito, Anopheles gambiae. Insect. Biochem. Mol. Biol. 2004;34:29–41. doi: 10.1016/j.ibmb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Bourbonnais R, Paice MG. Oxidation of non-phenolic substrates. An expanded role for laccase in lignin biodegradation. FEBS Lett. 1990;267:99–102. doi: 10.1016/0014-5793(90)80298-w. [DOI] [PubMed] [Google Scholar]
- 14.Clutterbuck AJ. Absence of laccase from yellow-spored mutants of Aspergillus nidulans. J. Gen. Microbiol. 1972;70:423–435. doi: 10.1099/00221287-70-3-423. [DOI] [PubMed] [Google Scholar]
- 15.Geiger JP, Nicole M, Nandris D, Rio B. Root-rot diseases of Hevea brasiliensis. 1. Physiological and biochemical aspects of host aggression. Eur. J. Forest Pathol. 1986;16:22–37. [Google Scholar]
- 16.O'Malley DM, Whetten R, Bao W, et al. The role of of laccase in lignification. Plant J. 1993;4:751–757. [Google Scholar]
- 17.Sharma P, Goel R, Capalash N. Bacterial laccases. World J. Microbiol. Biotechn. 2007;23:823–832. [Google Scholar]
- 18.Rodriguez Couto S, Toca Herrera JL. Industrial and biotechnological applications of laccases: a review. Biotechnol. Adv. 2006;24:500–513. doi: 10.1016/j.biotechadv.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Riva S. Laccases: blue enzymes for green chemistry. Trends Biotechnol. 2006;24:219–226. doi: 10.1016/j.tibtech.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Burton SG. Oxidizing enzymes as biocatalysts. Trends Biotechnol. 2003;21:543–549. doi: 10.1016/j.tibtech.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Kunamneni A, Camarero S, Garcia-Burgos C, et al. Engineering and Applications of fungal laccases for organic synthesis. Microb. Cell Fact. 2008;7:32. doi: 10.1186/1475-2859-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer M, Thai QK, Grieb M, et al. DWARF–a data warehouse system for analyzing protein families. BMC Bioinformatics. 2006;7:495. doi: 10.1186/1471-2105-7-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoegger PJ, Kilaru S, James TY, et al. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 2006;273:2308–2326. doi: 10.1111/j.1742-4658.2006.05247.x. [DOI] [PubMed] [Google Scholar]
- 24.Machczynski MC, Vijgenboom E, Samyn B, et al. Characterization of SLAC: a small laccase from Streptomyces coelicolor with unprecedented activity. Protein Sci. 2004;13:2388–2397. doi: 10.1110/ps.04759104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benson DA, Karsch-Mizrachi I, Lipman DJ, et al. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durbin R, Eddy S, Krogh A, Mitchison G. Biological Sequence Analysis. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- 27.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman HM, Battistuz T, Bhat TN, et al. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 31.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 32.Finn RD, Tate J, Mistry J, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Servant F, Bru C, Carrere S, et al. ProDom: automated clustering of homologous domains. Brief Bioinform. 2002;3:246–251. doi: 10.1093/bib/3.3.246. [DOI] [PubMed] [Google Scholar]
- 34.Fischer M, Pleiss J. The Lipase Engineering Database: a navigation and analysis tool for protein families. Nucleic Acids Res. 2003;31:319–321. doi: 10.1093/nar/gkg015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knoll M, Hamm TM, Wagner F, Martinez V, Pleiss J. The PHA Depolymerase Engineering Database: a systematic analysis tool for the diverse family of polyhydroxyalkanoate (PHA) depolymerases. BMC Bioinformatics. 2009;10:89. doi: 10.1186/1471-2105-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sirim D, Wagner F, Lisitsa A, Pleiss J. The Cytochrome P450 Engineering Database: integration of biochemical properties. BMC Biochem. 2009;10:27. doi: 10.1186/1471-2091-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seifert A, Pleiss J. Identification of selectivity-determining residues in cytochrome P450 monooxygenases: a systematic analysis of the substrate recognition site 5. Proteins. 2009;74:1028–1035. doi: 10.1002/prot.22242. [DOI] [PubMed] [Google Scholar]
- 38.Seifert A, Vomund S, Grohmann K, et al. Rational design of a minimal and highly enriched CYP102A1 mutant library with improved regio-, stereo- and chemoselectivity. Chembiochem. 2009;10:853–861. doi: 10.1002/cbic.200800799. [DOI] [PubMed] [Google Scholar]
- 39.Piontek K, Antorini M, Choinowski T. Crystal structure of a laccase from the fungus Trametes versicolor at 1.90-A resolution containing a full complement of coppers. J. Biol. Chem. 2002;277:37663–37669. doi: 10.1074/jbc.M204571200. [DOI] [PubMed] [Google Scholar]
- 40.Delano WL. San Carlos, CA, USA: DeLano Scientific; 2002. The PyMOL Molecular Graphics System. [Google Scholar]