Abstract
Many abbreviations are used in the literature especially in the life sciences, and polysemous abbreviations appear frequently, making it difficult to read and understand scientific papers that are outside of a reader’s expertise. Thus, we have developed Allie, a database and a search service of abbreviations and their long forms (a.k.a. full forms or definitions). Allie searches for abbreviations and their corresponding long forms in a database that we have generated based on all titles and abstracts in MEDLINE. When a user query matches an abbreviation, Allie returns all potential long forms of the query along with their bibliographic data (i.e. title and publication year). In addition, for each candidate, co-occurring abbreviations and a research field in which it frequently appears in the MEDLINE data are displayed. This function helps users learn about the context in which an abbreviation appears. To deal with synonymous long forms, we use a dictionary called GENA that contains domain-specific terms such as gene, protein or disease names along with their synonymic information. Conceptually identical domain-specific terms are regarded as one term, and then conceptually identical abbreviation-long form pairs are grouped taking into account their appearance in MEDLINE. To keep up with new abbreviations that are continuously introduced, Allie has an automatic update system. In addition, the database of abbreviations and their long forms with their corresponding PubMed IDs is constructed and updated weekly.
Database URL: The Allie service is available at http://allie.dbcls.jp/.
Introduction
With the fast pace of progress in the life sciences and the increase of accompanying literature, new domain-specific terms such as gene, protein, chemical compound or disease names are routinely introduced. These terms often consist of multiple words, and many researchers create or use abbreviations for them in their articles. Chang et al. (1) reported on average one new abbreviation appears in every five to ten abstracts, and our survey showed that MEDLINE entries have increased by about 650 000 per year on average from 2004 to 2009. Existing dictionaries cannot keep up with this situation. As a result, the clarity of articles decreases (2) and polysemy or synonymy issues arise. Another study (3) reported that 81.2% of abbreviations are ambiguous and have an average of 16.6 meanings. For example, the abbreviation SPF may stand for any one of ‘specific pathogen-free’, ‘S-phase fraction’, ‘sun protection factor’ and more. Here, we call these terms that have abbreviations ‘long forms’. In addition, several long forms have lexical variants. For example, ‘acute myeloid leukemia’ and ‘acute myeloid leukaemia’ share identical concepts, and both are abbreviated as AML. Both of these long forms frequently appear (5652 and 1270) in the MEDLINE data.
A significant problem is that not all abbreviations in the MEDLINE data appear with their corresponding long forms (4). This situation can make it difficult for researchers to understand articles, especially when these are outside of their fields of expertise. This circumstance often happens with the emergence of new high-throughput technologies such as microarrays. Moreover, document search systems such as PubMed would return many non-relevant entries when a polysemous abbreviation is used as a query.
To help researchers learn domain-specific abbreviations easily, we have developed a system called Allie that looks up abbreviation-long form pairs from the entire MEDLINE database. Allie displays either long forms or abbreviations that correspond to a query consisting of either an abbreviation or a long form, respectively. Thus, if ‘SPF’ is given as a query, Allie displays a list of its corresponding long forms mentioned in the above example (i.e. ‘specific pathogen-free’, etc.).
In addition, for each hit pair, Allie returns the research field in which it frequently appears along with other abbreviations that co-occur with it to help users quickly learn about the context of its appearance. For example, ‘Dermatology’ is the research field of ‘sun protection factor’, which is usually abbreviated as SPF, and UV, UVR or MED are some of the abbreviations that co-occur with the pair. This novel functionality thus provides users with a way of disambiguating polysemous abbreviations in addition to indicating PubMed/MEDLINE data in which a target pair appears. This information can also be used to narrow down a set of hit pairs or PubMed/MEDLINE entries; thus Allie can be used to find articles that contain a particular target pair in a contextual manner. Moreover, for those who want to use Allie from their own programs or web servers, Allie also implements Simple Object Access Protocol (SOAP) and Representational State Transfer (REST) interfaces.
As mentioned above, new abbreviations are introduced rapidly, and we take this issue seriously since Allie’s target users are actively working researchers including database annotators and curators in life sciences. To update Allie periodically, we built an automatic update system that extracts pairs from newly added MEDLINE data and reflects them in Allie. Although there have already been several abbreviation search systems (1, 4–9), some do not exist any more or have not been updated for a long time (more than a year). Thus, to our knowledge, we can claim that Allie is the only system of its kind that is updated periodically.
Methods
Database construction
The database used by Allie is constructed in advance. The construction process consists of the following six consecutive tasks: (i) splitting MEDLINE data into sentences, (ii) extracting abbreviation-long form pairs from the sentences, (iii) merging lexical variants, (iv) applying a domain-specific dictionary to identify conceptually identical terms, (v) forming groups of conceptually identical pairs considering their appearances in MEDLINE and (vi) for each group, choosing representatives of the abbreviations and their corresponding long forms.
MEDLINE titles and abstracts are split into sentences by the tool sptoolkit, and pairs are extracted from the sentences by ALICE (10). ALICE achieved a recall of 95% and a precision of 97% on randomly selected MEDLINE data, and so Allie inherits this performance.
After obtaining a list of pairs, Allie merges some lexical variants in the long forms using UMLS SPECIALIST Lexicon (11). More precisely, Allie uses the ‘Agreement and Inflection’ file to map a term to its basic form. If there is a basic form whose inflectional form exactly matches a long form, it is replaced with the basic form only if the basic form is used as a long form elsewhere and it appears more frequently than the original one. If it appears less, all the long forms that exactly match the basic form are replaced with that original one (i.e. the inflectional form). In addition, if a subset of a long form that includes its last word exactly matches an inflectional form, Allie processes it similarly to cope with those long forms which consist of adjectives and an inflectional form such as ‘Acute lymphatic leukaemia’ (Acute + lymphatic leukaemia). In more detail, when a long form consists of n words, we express it as ‘w1 w2 … wn’, where w1 is the first word and the wn is the last. In this situation, if a term ‘wi … wn’ (1 < i < n) or ‘wn’ exactly matches an inflectional form, Allie processes it in the same way.
Next, Allie normalizes those terms that are conceptually identical but that have different expressions using GENA (12) by applying an identifier to terms having identical concepts. This normalization is at a more conceptual level than the previous process is. For example, GENA returns the same concept ID to both ‘premature atrial contractions’ and ‘premature atrial complexes’, both of which are abbreviated as ‘PAC’, but these are not in the file mentioned above. In developing our database, we used a customized version of GENA, developed by its creator, which can identify not only gene names, but also chemical compounds or disease names. The method of identifying named entities and normalizing them is described in (12), which states that a trie-based algorithm with several heuristics is used to recognize entities in text. In addition, UMLS Metathesaurus (13) is used to identify and normalize chemical compounds and disease names.
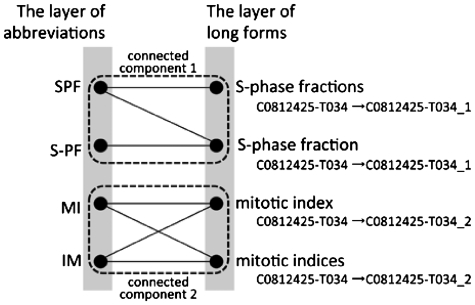
For each concept ID, taking all pairs of abbreviations and long forms, if those pairs whose long forms share the same ID are treated as one pair, the synonymy problem can be solved to some extent. However, it introduces another problem. Here, we take two terms ‘mitotic index’ and ‘S-phase fraction’ as an example. In MEDLINE, ‘S-phase fraction’ is usually abbreviated as ‘SPF’, and ‘mitotic index’ is always abbreviated as either ‘MI’ or ‘IM’. Therefore, Allie should not display ‘mitotic index’ when a user searches for long forms that correspond to ‘SPF’. However, it is displayed if Allie identifies pairs by only applying concept IDs added by GENA because GENA does not consider relationships between abbreviations and their long forms. In this example, GENA would apply the same concept ID to ‘mitotic index’ and ‘S-phase fraction’.
To take care of this problem, Allie changes the concept IDs of long forms after the application of GENA as follows. Allie constructs a bipartite graph with one layer representing a set of abbreviations and the other layer representing a set of long forms. Each edge denotes the existence of a pair in MEDLINE, and each long form is labeled by its concept ID given by GENA. Next, Allie computes the connected components of the graph. Then, Allie changes the concept ID given by GENA to a new one by concatenating a connected component ID as a suffix to the original one. For example, as shown in Figure 1, GENA gives the same concept ID ‘C0812425-T034’ to ‘S-phase fractions’, ‘S-phase fraction’, ‘mitotic index’ and ‘mitotic indices’. Since the connected component that includes ‘S-phase fraction’ is different from the one that includes ‘mitotic index’, Allie appends the connected component ID to each concept ID, such as ‘C0812425-T034_1’ or ‘C0812425-T034_2’. In other words, Allie divides each group of pairs with a same concept ID into subgroups by generating intersections of groups and connected components. Allie thus obtains the final groups of conceptually identical pairs by using the new concept IDs. Finally, for each group, the pair appearing most frequently in MEDLINE is selected as the representative of the group.
Figure 1.
A part of a bipartite graph used in Allie. GENA gives the concept ID ‘C0812425-T034’ to ‘S-phase fractions’, ‘S-phase fraction’, ‘mitotic index’ and ‘mitotic indices’. Allie changes the ID to one that corresponds to each cluster using connected components of the graph.
To obtain the research field of each pair, we use Journal Subject Terms, which are assigned by National Library of Medicine (NLM) to MEDLINE journals to describe the journals’ overall scope. When multiple research fields are added to a pair, the most frequently added one is chosen.
Database update
The database update mainly consists of two parts. The first part performs tasks (1) and (2) (i.e. splitting MEDLINE data into sentences, extracting abbreviation-long form pairs from the sentences) and the second part consists of the remaining tasks (3) through (6). Since MEDLINE is usually updated every weekday, and the first part takes relatively a short amount of time to complete the task, it is run weekday. Since the second part takes more time, it is run once a month. Therefore, this update is reflected in the Allie search service monthly. We make notice here that the daily MEDLINE update data are obtained from NLM under a license agreement between NLM and DBCLS.
Search system description
Allie has four main pages, described below.
Top page
On the top page, Allie accepts a user query with advanced search options. A query can be an abbreviation, a long form, or a substring, and it must contain at least two ASCII characters. If a query matches either an abbreviation or a long form, Allie returns the corresponding long form or abbreviation clusters, respectively. A cluster is a group of conceptually identical abbreviations or long forms. In other words, if a query matches an abbreviation, the abbreviation becomes the search key and the list of its corresponding long form clusters is displayed. When a query matches both, the user may choose either one. We use the term ‘item’ to denote either abbreviation or long form, depending on the search key.
The user is provided options for the search method (i.e. exact match or partial match), the sorting order of the results (by hit clusters in a result page and by PubMed/MEDLINE information in each cluster), and the number of hit clusters shown per result page. The hit clusters can be sorted in ascending or descending order for each of the following:
alphabetical order of cluster-representative items;
appearance frequency (the number of the pairs appearing in MEDLINE);
publication year of papers that contain the pairs in their titles or abstracts.
The sort order of PubMed/MEDLINE information for each cluster can be in ascending or descending order by publication year. The default values for the user options are exact match, descending frequency of appearance for the order of hit clusters, ascending publication year for PubMed/MEDLINE information, and 30 clusters per result page. Using these default values, a user can quickly find the most frequently used long form for an abbreviation and when the pair was first used in the literature along with its research field and co-occurring abbreviations.
Hit cluster-list page
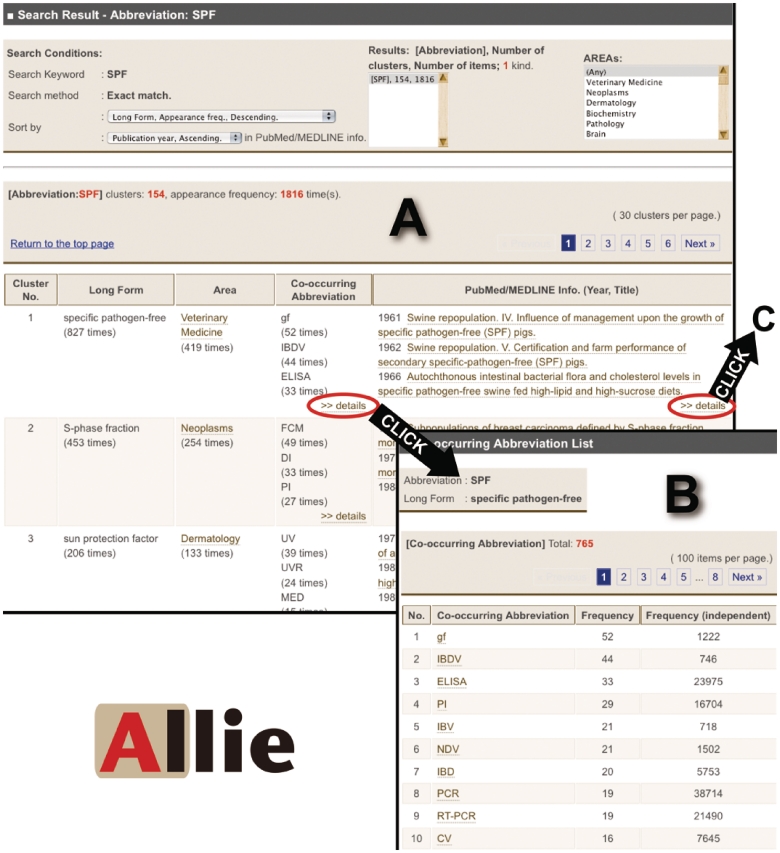
When a query hits either abbreviations or long forms, Allie shows a hit cluster-list page (Figure 2A). Allie displays buttons for the user to choose if there are both or a message if it hits neither.
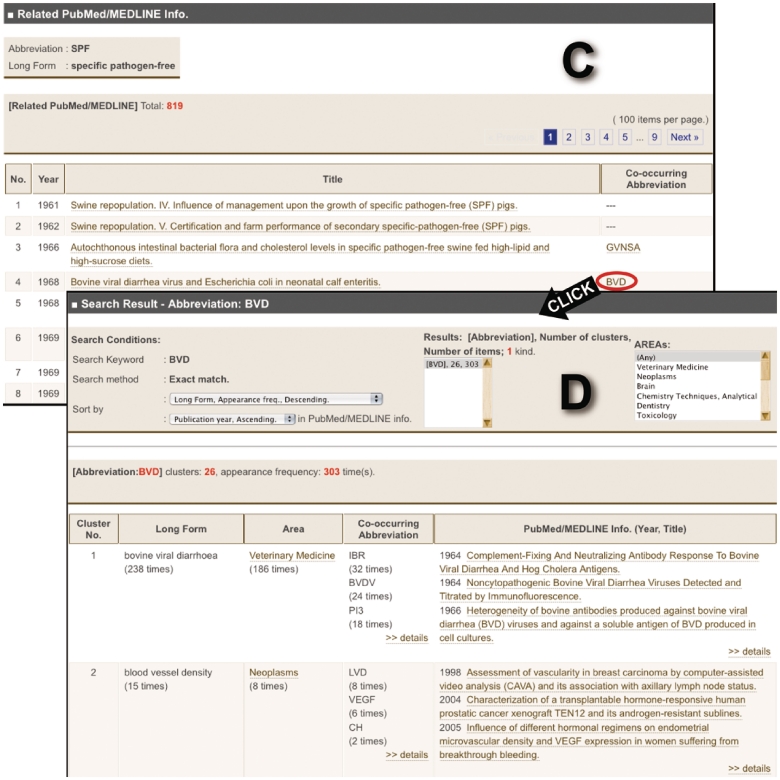
Figure 2.
Images of Allie’s outputs. (A) Hit cluster-list page for the abbreviation ‘SPF’. By clicking links in the ‘Co-occurring Abbreviation’ or the ‘PubMed/MEDLINE Info.’ cells, the user can access these corresponding pages (A to B or A to C, respectively). (B) Co-occurring abbreviation page. Here, the user is provided with all the co-occurring abbreviations, and by clicking one of the listed abbreviations, one can access the hit cluster-list page. (C) PubMed/MEDLINE Information page. Here, the user is provided with all publication years, titles, and co-occurring abbreviations that appear in the titles or abstracts with the pair. Each title is anchor text that links to the corresponding PubMed page. By clicking one of the co-occurring abbreviations, the user can access the hit cluster-list page (C to D). (D) Hit cluster-list page for the abbreviation ‘BVD’.
A hit cluster-list page is vertically divided into three parts. The top section shows the search conditions, where users can rearrange the order or change the cluster-list displayed if partial match is chosen for the search method and if the query hits multiple items. In addition, this section shows a menu by which the user can filter out those clusters that do not frequently appear in the articles of the chosen research field. The middle section shows the meta-data of the list shown, including the item that the query matches and the numbers of clusters and pairs. The lower section is a table with each cluster’s information on a separate row. It contains the representative item (long form or abbreviation), research field, co-occurring abbreviations, and PubMed/MEDLINE information (publication years and titles of articles in which the pair appears in the titles or the abstracts). In cells of co-occurring abbreviations and PubMed/MEDLINE information, there are links to pages where users can find detailed information.
Co-occurring abbreviation page
On this page, there is a table where each row shows a co-occurring abbreviation, the frequency of co-occurrence with the hit pair, and its total appearance frequency (Figure 2B). Each abbreviation is anchor text that links to the hit cluster-list page for the abbreviation by exact match.
PubMed/MEDLINE information page
On this page, there is a table which lists the publication year, title, and co-occurring abbreviations that appear in the title or the abstract with the pair (Figure 2C). Each title is anchor text that links to the corresponding PubMed page. In addition, each co-occurring abbreviation is anchor text, similar to the co-occurring abbreviation pages.
Database download
While the Allie database for the search service is updated monthly, raw data extracted by ALICE are updated daily and are published weekly, which are freely downloadable from our FTP site (ftp://ftp.dbcls.jp/allie/). Since these are data that ALICE extracts without any post-processing such as clustering, these data may not reflect the same results as would be obtained from the Allie search system. These data are provided such that users can develop their own applications using them. These are tab-delimited text, where each line consists of a pair of an abbreviation and its corresponding long form with their unique IDs (i.e. an abbreviation ID and a long form ID), the PubMed ID of the title or the abstract where the pair appears, and its publication year.
Implementation
Allie consists of two main parts: an updating part (updater) and a search system part (searcher). The updater is a set of scripts that process MEDLINE data to generate a list of pairs and to update the database. The searcher was designed using Ruby on Rails and MySQL. Since some of the datasets needed for Allie are large (about one gigabyte) and since data retrieval takes time, the datasets are cached in the main memory of the Allie server.
As for the SOAP/REST interfaces, there are four types of searches, consisting of the combination of search methods (exact or partial) and search keys (abbreviation or long form). In addition, by using a pair ID obtained by a search, the co-occurring abbreviations and the PubMed/MEDLINE information can be obtained, respectively.
Results and an example usage
Table 1 shows examples of Allie’s outputs. There are three abbreviations and their corresponding long forms with their research fields and the co-occurring abbreviations. It also shows the year of each long form’s first appearance. At the time of writing of this manuscript, the total number of non-redundant pairs is 1 564 399, and the total numbers of the abbreviation and long form clusters are 406 372 and 1 341 981, respectively. The history of the updates shows that around 9000 new pairs are added monthly.
Table 1.
Examples of Allie’s outputs The long forms are sorted by descending frequency of appearance
| Abbreviation | Long form | Research field | Co-occurring abbreviation | Year |
|---|---|---|---|---|
| SPF | Specific pathogen-free | Veterinary medicine | GF/IBDV/ELISA … | 1961 |
| S-phase fraction | Neoplasms | FCM/DI/PI … | 1978 | |
| Sun protection factor | Dermatology | UV/UVR/MED … | 1978 | |
| MAP | Mean arterial pressure | Physiology | HR/CO/CI … | 1974 |
| Mitogen-activated protein | Biochemistry | ERK/JNK/PKC … | 1991 | |
| Mean arterial blood pressure | Physiology | HR/NO/CO … | 1975 | |
| Microtubule-associated protein | Neurology | AD/GFAP/NGF … | 1979 | |
| BAC | Bacterial artificial chromosome | Genetics | FISH/YAC/PCR … | 1994 |
| Blood alcohol concentration | Substance-Related Disorders | DUI/DWI/BrAC … | 1994 | |
| Bronchioloalveolar carcinoma | Neoplasms | AAH/NSCLC/EGFR … | 1983 | |
| Benzalkonium chloride | Ophthalmology | EDTA/CPC/CMC … | 1979 |
The following is an example usage of Allie. A researcher wants to know about the abbreviation ‘SPF’ that appears in a document without its long form nearby. The document describes a vaccine and that enzyme-linked immunosorbent assay (ELISA) was used in the experiment. Using Allie, he can find out that ‘SPF’ is a polysemous abbreviation, and that many articles are published in the journals pertaining to the research field of ‘Veterinary Medicine’ where it was used as an abbreviation of ‘specific pathogen-free’. In addition, ‘GF’, ‘IBDV’ and ‘ELISA’ often co-occur with it as an abbreviation of ‘specific pathogen-free’ in MEDLINE. Since ‘ELISA’ appears in the document, he can speculate that the ‘SPF’ stands for ‘specific pathogen-free’. By clicking the details link below the ‘ELISA’, and then clicking some of the listed abbreviations, he can find out that the pairs GF—‘germ-free’ and ‘IBDV’—‘infectious bursal disease virus’ also co-occur with ‘SPF’. Consequently, he can verify that the pair is correct.
We assume that the document in which an abbreviation in question appears contains several domain specific terms, and in many cases, he/she can find some clues to identify the proper long form by checking the research field, the co-occurring abbreviations, and the PubMed/MEDLINE information. Allie does not guess or predict the right long form for a given abbreviation, but instead it provides various related evidence for the user to quickly identify it.
Conclusion
Allie is a search system that returns not only pairs of abbreviations and their long forms appearing in the MEDLINE data, but also their relevant research fields and abbreviations. Providing the relevant information is a unique feature and makes it much easier for researchers in the life sciences to find the pair that they are looking for as demonstrated in the example usage. In addition, it is useful for users to utilize the database used in Allie in their environment since the entire database updated periodically is freely downloadable.
Discussions and future plans
The granularity of relevant research fields used to disambiguate polysemous abbreviations may be considered to be too coarse. To help users disambiguate easily, we considered that several granularity levels of the contexts where each pair appears should be provided. For example, if a user wants to know the correct long form for ‘SPF’ appearing in a document, as described above, providing several contexts assumed to be helpful. Enumerating from the finest to coarser levels, articles in which each pair appears are at the finest level of the granularity. At a coarser level there are co-occurring abbreviations. MeSH terms frequently annotated to articles in which each pair appears could be at a next coarser level. However, we considered MeSH terms at even coarser granularity would be better to be provided because no single user grasps the whole MeSH vocabulary. As for our pair database, a frequently appearing MeSH term would be chosen from about the 20 000 terms for each pair if we take the most frequently annotated MeSH term as a research field of each pair. The set of Journal Subject Terms is a subset of the MeSH terms, and the total number of these is 123. Therefore, these would be familiar enough to users to determine which research field is the right one even if they search for a pair outside of their areas of expertise. To our knowledge, there is no other vocabulary of its kind available now. Moreover, there is a delay (about three months) in the times of registration into MEDLINE and annotation of MeSH terms per each article since MeSH terms are manually annotated. Consequently, some newly emerged pairs do not have any MeSH terms. We also assume that abbreviation has one meaning within a research field, and we believe that the granularity is suitable. After providing relevant research fields, we have received positive feedbacks from users. Nevertheless, there may be a better way for users to disambiguate polysemous abbreviations. We will continue to survey user experiences and reflect its outcomes in Allie promptly.
Since the process of generating the data needed for Allie is fully automatic, the displayed results may contain incorrect pairs. These are caused by errors in extracting pairs, tagging terms, or grouping pairs. While ALICE exhibits good performance, a few new tools to extract pairs of abbreviations and long forms from text have been proposed such as BIOADI (8), Ab3P (14) and NatLab (15). To compare the extraction accuracy, we are evaluating ALICE on BIOADI and Ab3P corpora used in these studies and considering how we can increase the extraction performance. In addition, neither GENA nor SPECIALIST Lexicon have a complete list of conceptually identical terms, and we will research another way to complement it. Concerning handling of long forms that are lexically similar to each other but that are conceptually different, we have emphasized manually crafted dictionaries unless there is a special method. For example, chemical compound names are hard to determine whether two lexically similar names are conceptually identical or not (‘albendazole sulphoxide’ and ‘albendazole sulfoxide’ are conceptually identical to each other, but ‘glutamic acid’ and ‘glutaric acid’ are not). Therefore, we are developing an improved clustering method for grouping conceptually identical pairs based on graph algorithms and dynamic programming techniques. In this way, we are improving the performance as much as possible.
In addition, the downloadable database currently contains raw data generated by ALICE, but we are now planning to release a database used by the search system in which clustering results and the main research field and the co-occurrence abbreviations of each pair are included.
Funding
Integrated Database Project, Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest. None declared.
Acknowledgements
We thank Dr Shin Kawano for checking Allie and reporting an issue, Mr Toyofumi Fujiwara for helping with the development of Allie, and Mr Sebastian R. Riedel and Dr Kiyoko F. Aoki-Kinoshita for their comments on English writing.
References
- 1.Chang JT, Schutze H, Altman RB. Creating an online dictionary of abbreviations from MEDLINE. J. Am. Med. Inform. Assoc. 2002;9:612–620. doi: 10.1197/jamia.M1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloom DA. Acronyms, abbreviations and initialisms. BJU Int. 2000;86:1–6. doi: 10.1046/j.1464-410x.2000.00717.x. [DOI] [PubMed] [Google Scholar]
- 3.Liu H, Lussier YA, Friedman C. Proceedings of AMIA Symposium. Philadelphia, PA, USA: Hanley & Belfus, Inc.; 2001. A study of abbreviations in the UMLS; pp. 393–397. [PMC free article] [PubMed] [Google Scholar]
- 4.Okazaki N, Ananiadou S. Building an abbreviation dictionary using a term recognition approach. Bioinformatics. 2006;22:3089–3095. doi: 10.1093/bioinformatics/btl534. [DOI] [PubMed] [Google Scholar]
- 5.Rimer M, O’Connell M. BioABACUS: a database of abbreviations and acronyms in biotechnology and computer science. Bioinformatics. 1998;14:888–889. doi: 10.1093/bioinformatics/14.10.888. [DOI] [PubMed] [Google Scholar]
- 6.Wren JD, Garner HR. Heuristics for identification of acronym definition patterns within text: towards an automated construction of comprehensive acronym-definition dictionaries. Methods Inform. Med. 2002;41:426–434. [PubMed] [Google Scholar]
- 7.Zhou W, Torvik VI, Smalheiser NR. ADAM: another database of abbreviations in MEDLINE. Bioinformatics. 2006;22:2813–2818. doi: 10.1093/bioinformatics/btl480. [DOI] [PubMed] [Google Scholar]
- 8.Kuo C-JJ, Ling MH, Lin K-TT, Hsu C-NN. BIOADI: a machine learning approach to identifying abbreviations and definitions in biological literature. BMC Bioinformatics. 2009;10(Suppl. 15):S7. doi: 10.1186/1471-2105-10-S15-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okazaki N, Ananiadou S, Tsujii J. Building a high-quality sense inventory for improved abbreviation disambiguation. Bioinformatics. 2010;26:1246–1253. doi: 10.1093/bioinformatics/btq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ao H, Takagi T. ALICE: An algorithm to extract abbreviations from medline. J. Am. Med. Inform. Assoc. 2005;12:576–586. doi: 10.1197/jamia.M1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Library of Medicine (US) 2009 doi: 10.1080/15360280801989377. UMLS® Reference Manual [Internet] http://www.ncbi.nlm.nih.gov/books/NBK9676/ (9 March 2011, date last accessed) [DOI] [PubMed] [Google Scholar]
- 12.Koike A, Takagi T. Gene/protein/family name recognition in biomedical literature HLT-NAACL 2004 Workshop: BioLINK 2004, Linking Biological Literature, Ontologies and Databases. Boston, Massachusetts, USA: Association for Computational Linguistics; 2004. pp. 9–16. [Google Scholar]
- 13.Schuyler PL, Hole WT, Tuttle MS, Sherertz DD. The UMLS Metathesaurus: representing different views of biomedical concepts. Bull. Med. Libr. Assoc. 1983;81:217–222. [PMC free article] [PubMed] [Google Scholar]
- 14.Sohn S, Comeau DC, Kim W, Wilbur WJ. Abbreviation definition identification based on automatic precision estimates. BMC Bioinformatics. 2008;9:402. doi: 10.1186/1471-2105-9-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeganova L, Comeau DC, Wilbur WJ. 2010 Ninth International Conference on Machine Learning and Applications. Los Alamitos, CA, USA: IEEE Computer Society; 2010. Identifying Abbreviation Definitions—Machine Learning with Naturally Labeled Data; pp. 499–505. [Google Scholar]