Abstract
The highly conserved let-7 microRNA (miRNA) regulates developmental pathways across animal phyla. Mis-expression of let-7 causes lethality in Caenorhabditis elegans and has been associated with several human diseases. We show that timing of let-7 expression in developing worms is under complex transcriptional and post-transcriptional control. Expression of let-7 primary transcripts oscillates during each larval stage but precursor and mature let-7 miRNAs do not accumulate until later in development after lin-28 activity has diminished. We demonstrate that LIN-28 binds endogenous primary let-7 transcripts co-transcriptionally. We further show that LIN-28 binds endogenous primary let-7 transcripts in the nuclear compartment of human ES cells, suggesting that this LIN-28 activity is conserved across species. We conclude that co-transcriptional interaction of LIN-28 with let-7 primary transcripts blocks Drosha processing and, thus, precocious expression of mature let-7 during early development.
MicroRNAs (miRNAs) function as ∼22 nucleotide guide RNAs in the RNA induced silencing complex (RISC) by binding to partially complementary sites in target mRNAs, causing inhibition of translation or destabilization1. Typically, mature miRNAs originate from long, capped and polyadenylated primary miRNAs (pri-miRNAs) that are transcribed by RNA polymerase II1. Endonucleolytic cleavage of the pri-miRNA by the RNase-III enzyme Drosha in cooperation with the RNA-binding protein Pasha (also known as DGCR8) releases the ∼70 nucleotide hairpin precursor miRNA (pre-miRNA)1. Exportin-5 translocates the pre-miRNA to the cytoplasm, where subsequent endonucleolytic cleavage by the RNase III enzyme Dicer produces the mature miRNA that functions in the RISC complex1,2.
Originally discovered in C. elegans, the let-7 miRNA is conserved across species in both sequence and temporal expression3,4. In C. elegans, let-7 regulates developmental timing and promotes cellular differentiation pathways5,6. The human let-7 miRNAs also have anti-proliferative functions and down-regulation of let-7 levels is associated with many cancers, including lung, breast and colon5,6. Over-expression of let-7 early in worm development causes premature adoption of adult fates, while cells in let-7 under-expression mutants fail to terminally differentiate at the larval to adult transition4. Thus, the level and timing of mature miRNA expression are paramount in determining organismal development.
The worm let-7 gene encodes two nascent and one trans-spliced primary transcripts (Fig. 1a)7. Deletion of the 3′ splice site sequence, required for trans-splicing, abolishes let-7 rescue activity, indicating that the splicing event or the sequence/structural changes produced by it are important for let-7 biogenesis7. Mature let-7 accumulation is first observed during the third larval stage (L3) and is maintained into adulthood4. Recently, lin-28 activity was shown to prevent premature accumulation of let-7 in the second larval stage (L2)8. The lin-28 gene encodes a nucleo-cytoplasmic localized cold shock domain and zinc finger containing protein that is conserved across animal species9-13. The LIN-28 protein is expressed early in worm development but is down-regulated by more than a factor of 10 from L1 to L3 through the action of lin-4 miRNA and other pathways9,14,15. Decreases in LIN-28 protein levels coincide with mature let-7 accumulation during the L3 stage4,14. Likewise, opposite expression patterns for LIN-28 protein and mature let-7 miRNA have been documented in several mammalian cell types12,16-19. Moreover, LIN-28 has been shown to regulate the accumulation of mature let-7 miRNA in mammalian systems through multiple mechanisms, including blocked Drosha or Dicer processing and destabilization of let-7 precursor RNAs16-22. What determines the utilization of one mechanism versus another to regulate accumulation of mature let-7 in vivo is yet to be resolved.
Figure 1.
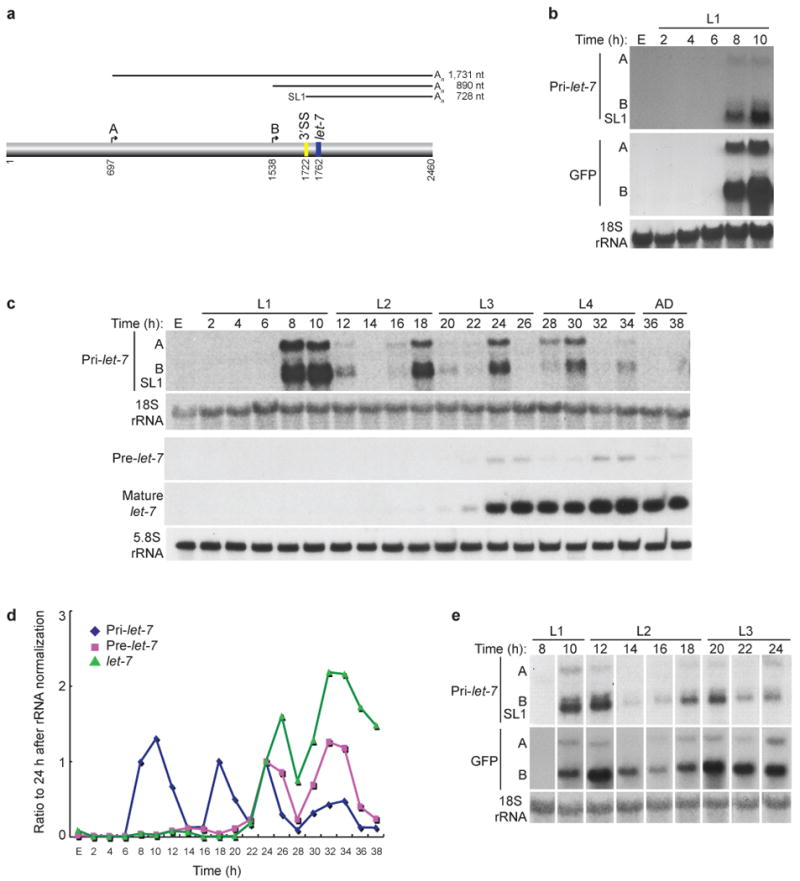
Expression of let-7 is transcriptionally and post-transcriptionally regulated. (a) Depiction of the 2460 nt long let-7 rescue construct with the positions of the mature let-7 sequence (shaded blue), 3′ splice site (SS; shaded yellow), two start sites (A and B) and approximate sizes of the spliced and unspliced transcripts indicated4,7. (b) Total RNA was isolated from embryos (E) or synchronized plet-7B∷GFP transgenic worms and analyzed by northern blotting. The similar sized B and SL1 transcripts often do not clearly resolve. (c) Total RNA was isolated from embryos (E) or synchronized WT N2 worms and analyzed by agarose or PAGE northern blotting. Representative blots from four independent experiments are shown. (d) Average pri-, pre- and mature let-7 levels after normalization to 18s or 5.8s rRNA from four independent experiments. (e) Total RNA was isolated from synchronized plet-7B∷GFP transgenic worms and analyzed as in Figure 1b. The entire blot is shown in Supplementary Figure 1c.
In this study we examine the role of lin-28 in regulating endogenous let-7 expression in a whole organism throughout development. We find that let-7 primary transcript expression is dynamic and accumulation of primary transcripts is uncoupled from pre-and mature let-7 in wild-type (WT) but not lin-28 mutant animals. We further show that LIN-28 binds endogenous pri-let-7 in both C. elegans and human embryonic stem cells and that this interaction is co-transcriptional in C. elegans. Altogether our results suggest that LIN-28 acts co-transcriptionally at the Drosha processing step to inhibit precocious expression of let-7 during animal development. The ability of LIN-28 to interact with primary let-7 transcripts as they are being synthesized provides an efficient mechanism for blocking production of this essential miRNA in multiple organisms.
Results
Uncoupling of primary and mature let-7 miRNA expression
Mature let-7 miRNA accumulates during the third larval stage (L3) of development in C. elegans4,7,23,24. Previous studies also found that the two unspliced (A and B) and one trans-spliced (SL1) pri-let-7 transcripts were first detected during the L3 stage, suggesting that mature let-7 production is transcriptionally regulated7. However, reporter constructs consisting of GFP fused to sequences upstream of mature let-7 revealed potential transcriptional activity earlier than the L3 stage 23,25,26. In agreement, we observed fluorescence at the end of the L1 stage in transgenic worms that express GFP fused to the pri-let-7B start site (data not shown). Detection of GFP mRNA, driven by both let-7 promoter A and B sequences in the transgenic worms, mirrored that of endogenous let-7 primary transcripts, indicating that expression of let-7 is repressed largely at the transcriptional level from embryogenesis until the late first larval stage (Fig. 1b and Supplementary Fig. 1a).
To further investigate the possibility of uncoupled expression of let-7 primary and mature RNAs, we used northern blotting and qRT-PCR to analyze the endogenous expression patterns of all three pri-let-7 isoforms as well as pre- and mature let-7 in RNA collected from embryos and every two hours of larval development to adulthood (Fig. 1c,d and Supplementary Fig. 1b and Supplementary Fig. 2). Consistent with our reporter analysis, pri-let-7 was first observed during the late L1 stage (Fig. 1c,d). All three pri-let-7 isoforms were detected, and coordinate expression of these isoforms oscillated throughout development (Fig. 1c,d and Supplementary Fig. 2). This cycling pattern of expression was specific to pri-let-7 since other endogenous mRNAs, like act-1, maintained steady levels throughout the time course (Supplementary Fig. 1b). The low levels of pri-let-7 at most mid larval time points and the slight shifts in the timing of pri-let-7 expression between experiments indicate that expression of endogenous pri-let-7 transcripts is dynamic, and that even slight changes in culture conditions can affect the rate of development and thus pri-let-7 expression (Fig. 1 and Supplementary Fig. 1a,b). Therefore, the failure of prior studies to detect expression of let-7 primary transcripts in L1 and L2 was likely due to the analysis of single time points at each stage7. GFP mRNA levels of our let-7 promoter reporter oscillated with a frequency identical to endogenous pri-let-7 expression, suggesting that transcriptional mechanisms largely control the cycling pattern of pri-let-7 expression (Fig. 1e and Supplementary Fig. 1c).
Consistent with previous reports, pre- and mature let-7 RNAs were undetectable until the L3 stage and mRNA levels of its target lin-41 decreased concordantly with let-7 appearance (Fig. 1c,d and Supplementary Fig. 1b)4,24. In the L3 and L4 stages, pre-let-7 levels oscillated in parallel to pri-let-7, while mature let-7 accumulated to a relatively constant level (Fig. 1c,d and Supplementary Fig. 1b). Taken together, our results indicate that expression of let-7 is regulated by transcriptional and post-transcriptional control mechanisms during development in C. elegans.
Primary let-7 processing is developmentally regulated
The detection of primary but not precursor or mature let-7 in the first two larval stages could be due to blocked Drosha processing of pri-let-7 or destabilization of pre-or mature let-7 RNAs. To distinguish between these possibilities, we used a sensitive cloning strategy to detect potential Drosha cleavage products and/or degradation intermediates. Drosha processing is expected to release the let-7 miRNA hairpin precursor from primary transcripts, leaving specific 5′ and 3′ products comprised of flanking sequences. We assayed for these cleavage products by performing 5′ or 3′ RNA oligo ligation reactions using total RNA isolated from 10 hour (h) (L1) and 24 h (L3) time points, and then conducted standard RACE (rapid amplification of cDNA ends) cloning experiments to detect the ligation junctions. Drosha cleavage products were evident in the 24 h RNA sample, as the majority of 3′ RACE results mapped to the 3′ end of the let-7 precursor and almost half of the 5′ RACE results mapped to the expected cleavage site between the precursor and 3′ product (Fig. 2a and Supplementary Fig. 3). In contrast, no 5′ RACE products from the 10 h time point mapped to canonical Drosha cleavage sites; instead these clones may represent general degradation intermediates (Fig. 2a and Supplementary Fig. 3). The 3′ RACE of 10 h RNA samples, which was performed in parallel with RNA from the 24 h time point, yielded no products that could be cloned (Fig 2a). Since we purposefully selected clones from the 5′ RACE with different sized inserts, the identification of 8/19 clones that mapped to the Drosha cleavage site from the 24 h RNA sample is not a quantitative measure of frequency. Indeed, another 5′ RACE clone from the 24 h RNA sample mapped to the Drosha cleavage position at the 5′ end of the let-7 hairpin, likely representing a molecule where 3′ cleavage had not yet been accomplished (Supplementary Fig. 3). To further assess the presence of Drosha cleavage products within the primary transcript population of N2 (wild-type, WT) worms at the 10 versus 24 h time points, the 3′ cleavage products were analyzed by PCR. 5′ RACE cDNA samples were amplified with primers corresponding to the 5′ RNA oligo linker (P1), pri-let-7 sequence upstream of the let-7 hairpin (P2), or pri-let-7 sequence downstream of the cleavage site (P3) and a common reverse primer (P4) (Fig. 2b). No amplification of the P1+P4 PCR product was detected at the 10 h time point from two independent samples, while consistent amplification was seen from 24 h samples (Fig. 2b). The P2+P4 PCR product was detected at a slightly higher level at 10 versus 24 hours, while the P3+P4 PCR product was readily detected from all samples at both time points at similar levels (Fig. 2b). These differences in detection of Drosha cleavage products at 10 and 24 hours indicate that processing of let-7 primary transcripts is inhibited during the first larval stages of development.
Figure 2.
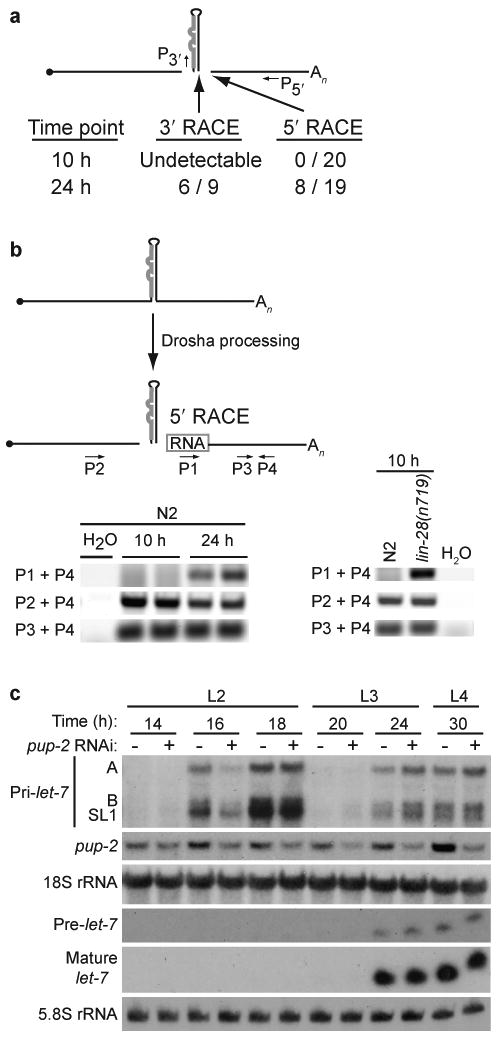
Developmentally regulated processing of let-7 pri-miRNA transcripts. (a) Depiction of expected Drosha cleavage products: 5′ flanking, let-7 hairpin precursor, and 3′ flanking. The number of sequenced RACE clones that mapped to the precise 3′ and 5′ Drosha cleavage products at each time point from two independent experiments is shown. The sequences of all Drosha cleavage products are shown in Supplementary Figure 3. (b) RT-PCR was performed on two independent 5′ RACE samples from N2 (left panel) or N2 and lin-28(n719) worms (right panel). (c) Total RNA was isolated from synchronized eri-1(mg366) RNAi hypersensitive worms at the indicated time points after vector control (-) or pup-2 (+) RNAi treatment, and analyzed by agarose and PAGE northern blotting. Representative blots from three independent experiments are shown.
A recent study reported that RNAi inactivation of the pup-2 poly(U) polymerase results in increased levels of a precursor let-7 miRNA processed from transcripts encoded by a transgene with truncated let-7 sequences driven by a heterologous promoter8. Using similar RNAi conditions, we also achieved an approximately fifty percent decrease in pup-2 mRNA levels but did not detect substantial effects on the accumulation of let-7 RNAs (Fig. 2c). The strong pulse of endogenous let-7 primary transcript expression during L2 did not give rise to detectable precursor in vector control or pup-2 (RNAi) samples (Fig. 2c). No appreciable difference in accumulation of precursor or mature let-7 miRNA during the L3 and L4 stages was observed in worms depleted of pup-2 compared to control (Fig. 2c). Similar results were also observed in the pup-2(tm4344) deletion strain (Supplementary Fig. 4). Thus, regulation of endogenous let-7 miRNA expression is independent of pup-2 activity. All together our results indicate that regulation of let-7 processing occurs at a step prior to precursor formation in developing worms.
lin-28 blocks early accumulation of mature let-7 miRNA
lin-28 acts upstream of let-7 in the C. elegans developmental timing pathway6, and multiple mechanisms by which LIN-28 inhibits let-7 expression have been proposed8,16-22. Thus, we next tested if lin-28 mediates post-transcriptional regulation of endogenous let-7 expression in C. elegans. In contrast to N2 worms, we observed accumulation of mature let-7 concordant with expression of pri-let-7 in lin-28(n719) putative null mutant worms (Fig. 3a). In RNA samples from N2 and lin-28(n719) worms, primary let-7 transcripts were undetectable in embryos and early L1, but by the 10 h L1 time point unspliced pri-let-7 RNAs were apparent in both strains (Fig. 3a). Because lin-28(n719) worms develop precociously and skip the L2 stage of development9, pri-let-7 levels at the 24 h time point in lin-28(n719) resemble the decreased levels observed in N2 at the later L4 stage (Fig. 3a, and Supplementary Figs. 2 and 5). Notably, precursor and mature let-7 accumulated while the SL1 trans-spliced primary transcript was underrepresented in lin-28 mutants at the 10 h time point (Fig. 3a). Consistent with these results, we detected 3′ Drosha cleavage products of pri-let-7 in lin-28(n719) worms at the 10 h time point (Fig. 2b). Thus, maturation of let-7 occurs two stages earlier in lin-28(n719) compared to WT worms (Fig. 3a). Expression of mature lin-4 and mir-58 miRNAs was unaffected in lin-28(n719) worms (Fig. 3a and Supplementary Fig. 6), indicating a specific role for lin-28 in regulation of let-7 as opposed to a general role in miRNA biogenesis.
Figure 3.
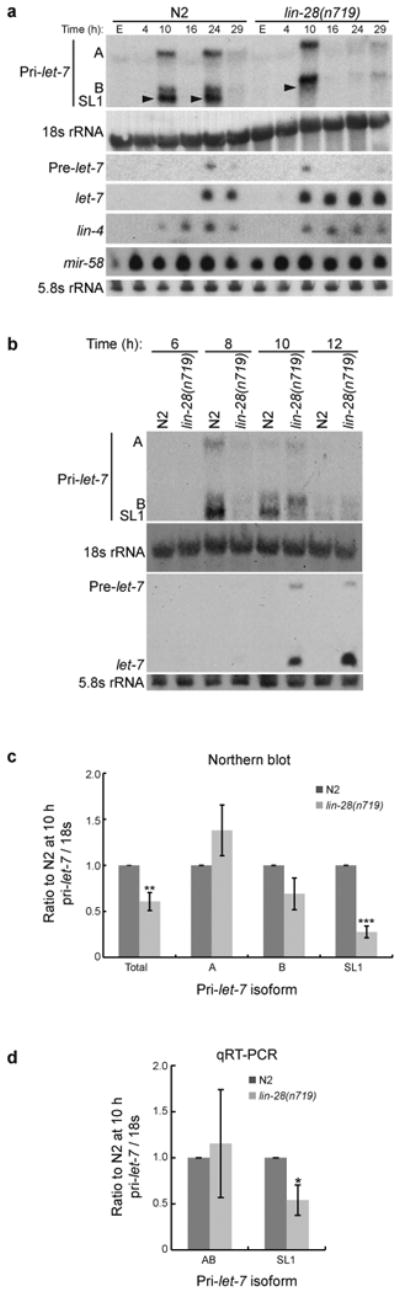
Regulation of let-7 processing by lin-28. (a) Total RNA was isolated from WT N2 or lin-28(n719) embryos (E) and synchronized worms at the indicated time points, and analyzed by agarose and PAGE northern blotting. Representative blots from three independent experiments are shown. The arrowheads mark the location of the SL1 pri-let-7 transcript. (b) Total RNA was isolated and analyzed as in Figure 3a. Representative blots from three independent experiments are shown. (c-d) Levels of each pri-let-7 isoform at the 10 h time point in lin-28(n719) relative to WT N2 worms after 18s rRNA normalization were calculated from six independent northern blot experiments (c) or 3 independent qRT-PCR experiments (d) and analyzed by Student's t-tests (*, p<0.05**, p<0.005; ***, p<0.0005). Error bars show s.e.m.
Closer analysis of pri-let-7 levels during the late L1 and early L2 stages, revealed significantly reduced levels of total pri-let-7 during the initial peak of expression at 10 hours in lin-28(n719) compared to WT worms (Fig. 3b-c). Furthermore, this reduction is largely accounted for by under-representation of the SL1 trans-spliced primary transcript isoform as seen by both northern blotting and qRT-PCR analyses (Fig. 3b-d). The correlation between decreased pri-let-7 levels and increased pre- and mature let-7 levels in lin-28 mutant worms suggests that LIN-28 normally functions to block primary to precursor let-7 processing during development in C. elegans.
LIN-28 interacts with endogenous primary let-7 transcripts
Expression of LIN-28 protein is developmentally regulated with strongly reduced levels by the mid L3 stage when mature let-7 begins to accumulate4,9,14. Decreased LIN-28 in mammalian cells and tissues has also been linked to up-regulation of mature let-712,16-19. Furthermore, association of LIN-28 with let-7 primary or precursor RNAs expressed from transgenes in cell culture or synthesized in vitro has been shown to block processing or promote degradation of these substrates, respectively16-22. Since our results suggest that LIN-28 inhibits the pri- to pre-let-7 processing step, we tested if LIN-28 binds endogenous let-7 primary transcripts in C. elegans by RNA immunoprecipitation (RIP). We utilized a strain that expresses LIN-28 tagged with GFP in the lin-28(n719) mutant background (PQ272); the integrated transgene fully rescues lin-28 mutant phenotypes and is developmentally regulated like the endogenous protein with a gradual reduction from late L1 to L3 (Fig. 4a,b)9,14. Extracts from 10 h late L1 transgenic worms were used for RIP experiments to test for specific association of let-7 and control RNAs with LIN-28:GFP (Fig. 4c and Supplementary Fig. 7a). Primers designed to amplify all 3 isoforms of pri-let-7 produced a robust signal from the anti-GFP precipitate. The unspliced A and B transcripts and the SL1 trans-spliced isoform were co-immunoprecipitated with LIN-28:GFP, indicating that LIN-28 does not substantially discriminate among these let-7 primary transcripts (Fig. 4c and Supplementary Fig. 7a). Sequences upstream of the A start site in the let-7 gene could not be amplified, verifying that the PCR signals are dependent on RNA transcripts (Fig. 4c and Supplementary Fig. 7a). Additionally, signals for the abundant actin or other primary miRNAs transcripts, like pri-mir-58, were not enriched in the LIN-28:GFP immunoprecipitates (Fig. 4c and Supplementary Fig. 7a), indicating that LIN-28 specifically binds let-7 primary transcripts in C. elegans.
Figure 4.
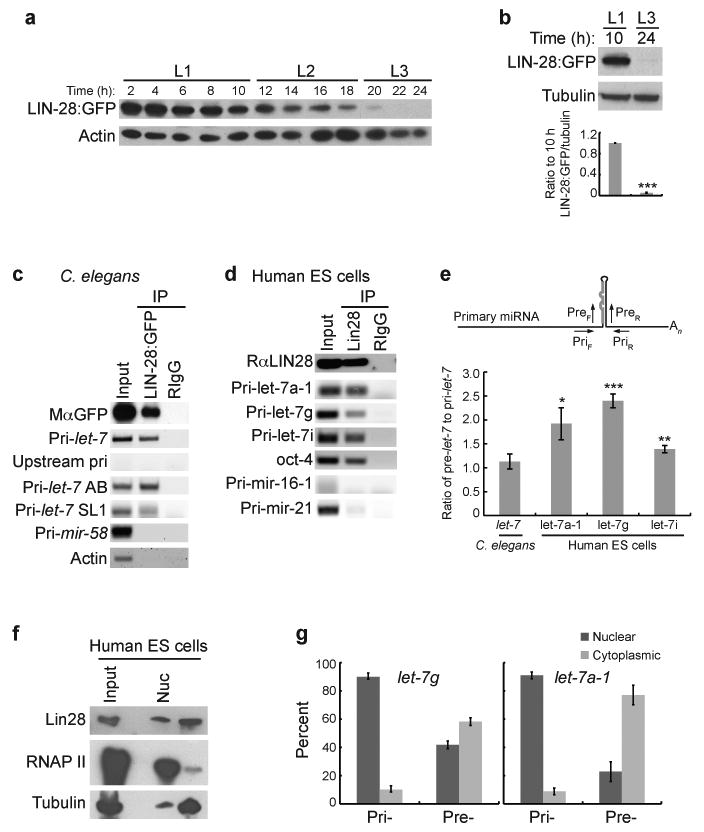
LIN-28 binds endogenous let-7 primary transcripts in C. elegans and human ES cells. (a-b) Total protein was isolated from PQ272 (LIN-28:GFP) worms and analyzed by western blotting. (b) Ratios of LIN-28:GFP levels to the 10 h time point after tubulin normalization were calculated from three independent experiments and analyzed by Student's t-tests (***, p<0.0005). Error bars show s.e.m. (c) Synchronized PQ272 worms were collected at 10 h and analyzed by RNA immunoprecipitation (RIP). Input, and LIN-28:GFP and IgG immunoprecipitates were analyzed by western blotting or RT-PCR. (d) Undifferentiated HUES6 cells were analyzed by RIP. Input, Lin28 and IgG immunoprecipitates were analyzed by western blotting or RT-PCR. (e) The worm and human cell samples from Figure 4c-d were analyzed by qRT-PCR to determine the levels of input or LIN-28 immunoprecipitated pri- or pre-let-7 RNAs using primers specific for pri-let-7 (priF and priR) or pre-let-7 and pri-let-7 transcripts containing the precursor hairpin (preF and preR). The ratio of precursor containing let-7 transcripts to pri-let-7 transcripts for immunoprecipitated samples after normalization to input samples for at least three independent experiments is shown, and was analyzed by Student's t-tests (*, p<0.05; **, p<0.005; ***, p<0.0005). Error bars show s.e.m. (f-g) Undifferentiated HUES6 cells were fractionationated into nuclear and cytoplasmic extracts before analysis by RIP and western blotting (f) or qRT-PCR as in Figure 4e (g). Results from three independent experiments are shown.
Although LIN-28 has been reported to regulate Drosha processing in mammalian embryonic stem (ES) cells, association of LIN-28 with endogenous let-7 primary transcripts has not yet been demonstrated16,17. To determine if LIN-28 also binds human pri-let-7 transcripts in vivo, we performed RIP in the human embryonic stem cell line HUES6. As a positive control, oct-4 was specifically detected in the LIN-28 immunoprecipitate (Fig. 4d and Supplementary Fig. 7b)27. Primary transcript sequences for human let-7a-1, let-7g, or let-7i also were present in the anti-LIN-28 immunoprecipitation samples (Fig. 4d and Supplementary Fig. 7b). In contrast, other primary miRNA transcripts expressed in ES cells, like pri-mir-21 and pri-mir-16-128, were not enriched in the LIN-28 immunoprecipitates (Fig. 4d and Supplementary Fig. 7b). Thus, LIN-28 binds endogenous let-7 primary transcripts in worm and human cells.
To determine if LIN-28 bound pre-let-7 in addition to pri-let-7, we performed quantitative PCR (qPCR) after RIP with primers specific for pri-let-7 (priF and priR) or primers residing within the precursor sequence (preF and preR), which would amplify cDNA representing precursor and the hairpin–containing primary let-7 transcripts (Fig. 4e). Comparison of the LIN-28 immunoprecipitated pre- to pri-let-7 signal showed no significant increase in precursor compared to primary let-7 levels in C. elegans (Fig. 4e). However, though the amount of increase differed among the let-7 genes, the ratio of precursor to primary for each human let-7 isoform was significantly higher than one (Fig. 4e). Thus, in C. elegans LIN-28 predominately interacts with endogenous let-7 primary transcripts, while in human ES cells LIN-28 interacts with both endogenous primary and precursor let-7 transcripts.
To determine the cellular location of LIN-28 interaction with endogenous pri- and pre-let-7, we performed RIP on fractionated HUES6 cells (Fig. 4f,g). Consistent with prior studies in C. elegans and human cells9,11,13,18, we detected endogenous LIN-28 in both nuclear and cytoplasmic fractions with a greater relative distribution in the cytoplasm (Fig. 4f). qRT-PCR analysis of immunoprecipitated LIN-28 showed that the majority of pri-let-7g and pri-let-7a-1 was nuclear localized (Fig. 4g). In contrast, immunoprecipitated pre-let-7g and pre-let-7a-1 were predominantly cytoplasmic (Fig. 4g). Thus, in human ES cells LIN-28 interacts with pri- and pre-let-7 in cellular fractions consistent with the sites of Drosha and Dicer processing, respectively.
LIN-28 co-transcriptionally binds endogenous primary let-7
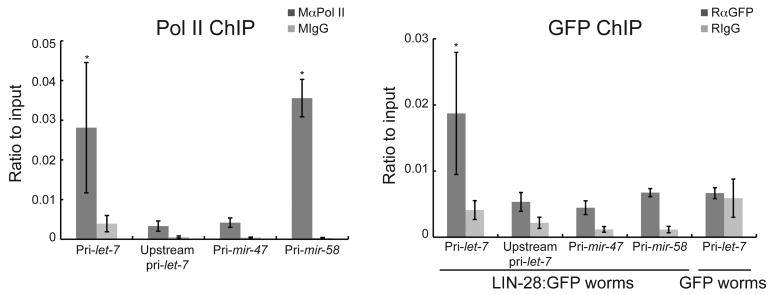
Our results suggest that LIN-28 negatively regulates let-7 expression at the Drosha processing step. Since Drosha processing can be co-transcriptional, we asked if the association of LIN-28 with pri-let-7 also acts at this step29-31. To test if LIN-28 binds the endogenous let-7 gene in C. elegans, we performed chromatin immunoprecipitation (ChIP) experiments. Worms expressing LIN-28:GFP or GFP alone were collected at the 10 h time point in late L1 and processed to detect association of RNA Polymerase II (RNAP II), GFP, or a control IgG antibody with specific DNA sequences. qPCR was used to analyze the immunoprecipitated genomic DNA levels for multiple primary miRNAs relative to the amount of genomic DNA in the input sample. Unlike sequences for pri-mir-47 and an untranscribed region ∼20 kb upstream of pri-let-732, sequences for pri-let-7 and pri-mir-58 were significantly enriched for association with RNAP II relative to IgG (Fig. 5). The let-7 gene also showed significant association with LIN-28:GFP relative to IgG in LIN-28:GFP worms (Fig. 5). In contrast, no significant increase in GFP versus IgG was detected for the untranscribed region upstream of pri-let-7, pri-mir-47, or pri-mir-58 in LIN-28:GFP worms or pri-let-7 in GFP only worms (Fig. 5). Taken together, we conclude that LIN-28 associates with endogenous let-7 transcripts co-transcriptionally in C. elegans.
Figure 5.
LIN-28 binds endogenous let-7 genomic DNA. Synchronized PQ272 (LIN-28:GFP) or pD4792 (GFP) worms were collected at 10 h and analyzed by chromatin immunoprecipitation (ChIP) with polyclonal antibodies against RNA pol II, GFP or IgG. The ratio of the indicated genomic DNA in the immunoprecipitated sample to the input sample for at least three independent experiments is shown and was analyzed by Student's t-tests (*, p<0.05). Error bars show s.e.m.
Discussion
The levels and timing of mature let-7 expression are critical for animal development and viability. In C. elegans, under-expression of let-7 late in development or over-expression of let-7 early in development causes abnormal cell fates that ultimately result in lethality4. In humans, inappropriate let-7 levels are found in multiple types of tumors and, in some cases, mis-expression of let-7 has been shown to have a causal role in disease progression5. Accordingly, multiple genes have been found that negatively, like hnRNP A1, or positively, like KSRP, regulate let-7 expression in mammalian cells33,34. Here we demonstrate that both transcriptional and post-transcriptional mechanisms contribute to let-7 miRNA expression during the development of C. elegans. Our results indicate that early in development LIN-28 binds and prevents processing of endogenous pri-let-7 transcripts as they are being synthesized. Down-regulation of LIN-28 levels by late larval stages permits efficient processing of pri-let-7 to the precursor and mature forms.
Pri-let-7 is first detected during the late L1 stage, and its levels cycle throughout development with peak expression coinciding with each molt early in development (Fig. 1 and Supplementary Fig. 1 and 2). Identical patterns of timing and oscillation of GFP mRNA and pri-let-7 RNAs in let-7 reporter worms indicate that transcriptional control mechanisms largely regulate the pulses of pri-let-7 expression during development (Fig. 1e and Supplementary Fig. 1c). The cycling of pri-let-7 accumulation warrants caution when choosing time points to analyze pri-let-7 levels, since less than two hours is sufficient for dramatically different expression levels (Fig. 1 and Supplementary Fig 1). Furthermore, synchronization and the rates of worm development within a population are sensitive to slight changes in culture conditions, such as temperature and food availability, and this is reflected in shifts in the timing of let-7 transcription (Fig. 1 and Supplementary Fig 1). Indeed, previous studies of pri-let-7 levels in staged worm samples showed varying or no pri-let-7 expression, likely because of the limited time points that were chosen for analysis7,26.
The LIN-28 RNA binding protein is an important regulator of let-7 biogenesis across species5,6,35. Originally discovered as a gene that regulates developmental timing in C. elegans9, LIN-28 has been shown to promote stem cell fates in mammalian cells35. Developmental abnormalities in lin-28 mutant worms are partially rescued by loss of let-7 expression4. Recent work from the Miska lab demonstrated that let-7 miRNA is expressed prematurely in the absence of lin-28 activity in C. elegans8. We show that, in contrast to WT worms, the initial pulse of primary let-7 expression at the end of the first larval stage coincides with accumulation of mature let-7 miRNA in lin-28 mutant worms (Fig. 3). Thus, lin-28 uncouples primary from mature let-7 expression early in development, and loss of this control results in premature engagement of let-7 miRNA regulatory pathways and abnormal development.
Our studies indicate that lin-28 blocks processing of endogenous primary let-7 transcripts. In the presence of lin-28, neither precursor nor flanking Drosha cleavage products were detected, loss of pup-2 activity did not affect regulation of let-7, and levels of let-7 primary transcripts diminished as precursor and mature let-7 accumulated in lin-28 mutant worms. Additionally, LIN-28 specifically bound let-7 primary transcripts in vivo and LIN-28 associated with the let-7 gene co-transcriptionally. In contrast, the Lehrbach et al., 2009 study concluded that LIN-28, in conjunction with PUP-2, inhibits the processing and stability of let-7 precursor RNAs in C. elegans8. This model was based largely on the analysis of transgenic let-7 expression under the control of a heterologous promoter8. This construct also lacked the 3′ splice site required for generation of the SL1 isoform previously shown to be important for let-7 rescue activity7. Notably, endogenous primary transcript significantly decreased as mature let-7 increased in lin-28 mutants but this correlation was not detected in the transgenic strain8. Since depletion of pup-2 by RNAi was only shown to result in let-7 precursor up-regulation in the transgenic strain8, and we detected no effect on regulation of endogenous let-7 miRNA expression after RNAi treatment or in a pup-2 mutant strain (Fig. 2c and Supplementary Fig. 4), it is possible PUP-2 helps cull excess precursor RNAs that escape the LIN-28-mediated block in primary transcript processing. In the endogenous context, there may be sufficient lin-28 activity to fully prevent the first step of let-7 processing, but this mechanism may become limiting in cells over-expressing let-7 transcripts, resulting in the detection of additional pathways that can repress maturation of let-7 miRNA. Additionally, our findings that LIN-28 associates with let-7 co-transcriptionally and that the spliced primary transcript is particularly sensitive to lin-28 activity suggest that natural regulation of let-7 expression may not be fully recapitulated by some transgenes.
A function for LIN-28 in repressing let-7 expression was first discovered in mammalian systems16-19,35. Consistent with our findings in C. elegans, some studies concluded that LIN-28 blocks the processing of let-7 primary transcripts in human and mouse embryonic cells16,17. Other reports proposed that LIN-28 binds let-7 precursors and inhibits Dicer processing and/or recruits TUT4/Zcchc11/PUP-2 poly(U) polymerase to catalyze 3′ end tailing, which results in destabilization of pre-let-7 RNAs18-20,22. We found that LIN-28 binds both primary and precursor endogenous let-7 RNAs in human ES cells, indicating that LIN-28 regulates let-7 biogenesis at multiple steps in this cell type. This ability could be required for regulation of the multiple, highly similar let-7 genes expressed in mammalian cells. In contrast, lin-28 appears to primarily block the first step of let-7 processing during normal worm development.
Association of LIN-28 with the let-7 gene provides an efficient mechanism for preventing processing of primary transcripts. In mammalian cells, Drosha can bind and cleave primary miRNA transcripts co-transcriptionally29-31. Thus, recognition of let-7 transcripts as they are being synthesized would allow LIN-28 to effectively compete with Drosha and prevent processing. A rescuing LIN-28:GFP protein exhibits fluorescence in the cytoplasm and occasionally in the nucleus and nucleoli of most worm cell types early in development9. Endogenous mammalian LIN-28 protein also displays a nucleo-cytoplasmic distribution that fluctuates with the cell cycle11,13. Exit from the nucleus may be dependent on association with RNA as mutation of both RNA binding domains renders LIN-28 entirely nuclear in mouse P19 cells13. We also detected LIN-28 in both the nucleus and the cytoplasm of human ES cells, and found that LIN-28 predominantly interacted with endogenous pri-let-7 in the nucleus and pre-let-7 in both the nucleus and cytoplasm (Fig. 4f,g). Taken together, the pulses of endogenous let-7 primary transcript expression may coincide with sufficient accumulation of LIN-28 in the nucleus to bind newly synthesized let-7 primary transcripts and block processing in C. elegans. Association of LIN-28 with let-7 RNAs may then facilitate export of the complex to the cytoplasm where the primary transcripts are subject to general mRNA decay pathways. Recent evidence suggests that C. elegans let-7 primary transcripts may also undergo processing in the cytoplasm36. Thus, the nucleo-cytoplasmic distribution of LIN-28 could be poised to regulate processing of let-7 primary transcripts in either cellular compartment.
Methods
Nematode culture and strains
C. elegans were grown under standard conditions37, and synchronized by hypochlorite treatment. Starvation arrested L1 worms were plated on OP50 bacteria, cultured at 25°C and collected at the desired time points. The wild-type (WT) strain was N2 Bristol. The pD4792(mIs11 IV) strain expresses myo-2∷GFP, pes-10∷GFP, and gut∷GFP. plet-7B∷GFP [plet-7B∷GFP;pha-1(+)] expresses plet-7B∷GFP and a pha-1(+) rescue construct as transgenes in a pha-1(e2123) background. PQ272 [lin-28(n719); plin-28∷LIN-28:GFP; pRF4 (rol-6 marker)] was made by crossing lin-28(n719) with a strain containing stably integrated copies of rescuing LIN-28-GFP, flanked by the lin-28 promoter and 3′ UTR9.
RNAi Treatment
Two generation feeding RNAi experiments used the eri-1(mg366) RNAi hypersensitive strain as described8.
ES Culture
The hESC line HUES6 was cultured as described (http://www.mcb.harvard.edu/melton/HUES/)38. Briefly, cells were grown to 80% confluency on growth factor–reduced (GFR) matrigel–coated plates (BD) in StemPro® hESC serum free medium (Invitrogen) before collection for RIP.
DNA constructs
plet-7B∷GFP was made by PCR amplifying the let-7 promoter (Supplementary Table 1) and fusing it upstream of three NLS repeats and GFP sequence.
Northern blotting
PAGE and agarose northern blotting analysis for small (<200 nt) and larger RNA species respectively was performed as described7, with probe templates listed in Supplementary Table 2, and analyzed with ImageQuant software (Molecular Dynamics).
RNA Ligase-mediated Rapid Amplification of cDNA Ends (RACE)
RACE was completed with the GENERACER kit (Invitrogen) and primers listed in Supplementary Table 37. For 5′ RACE, total RNA was ligated to the kit 5′ linker and reverse transcribed with Superscript III (Invitrogen) and a pri-let-7 primer downstream of pre-let-7. PCR and nested PCR used 5′ linker and pri-let-7 sequence primers. For 3′ RACE, gel-purified, 50-100 nt, dephosphorylated RNA was ligated to a RNA linker with a 5′ phosphate group and a 3′ puromycin tag. cDNA was made as above with a primer complementary to the 3′ linker. PCR used mature let-7 and the 3′ linker primers. Nested 5′ and 3′ RACE PCR products were analyzed by gel electrophoresis (Supplementary Table 3) or sequenced after TOPO cloning (Invitrogen).
Western Blotting
Western blotting was performed as described with mouse monoclonal antibodies against GFP (Santa Cruz), actin (MP Biomedicals), tubulin (Sigma), and RNA Pol II (Santa Cruz), or a rabbit polyclonal antibody against LIN-28 (Abcam)24. The Rabbit IgG TrueBlot secondary antibody (eBioscience) was used for LIN-28 western blots.
C. elegans RNA Immunoprecipitation (RIP)
PQ272 worms were crosslinked by UV treatment. Equal lysate amounts were precleared before immunoprecipitation with the appropriate antibody and Protein G Dynabeads (Invitrogen). Immunoprecipitated material associated with the beads was subjected to protein degradation and RNA extraction before RT-PCR with the primers listed in Supplementary Table 4. For further details see Supplementary Methods.
ES cell RNA Immunoprecipitation (RIP)
Equal amounts of pre-cleared lysates from un-crosslinked HUES6 cells were immunoprecipitated and treated as described above. For further details see Supplementary Methods.
ES cell fractionation
Cell fractionation was performed as previously described39. For further details see Supplementary Methods.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as described previously24,40, with some modifications. PQ272 or pD4792 worms were crosslinked with formaldehyde. Equal amounts of sonicated worm lysates were precleared before immunoprecipitation with the appropriate antibody and Protein G Dynabeads (Invitrogen). Immunoprecipitated material was eluted from the beads, reverse crosslinked, subjected to protein degradation and DNA extracted. qPCR was performed with primers listed in Supplementary Table 4. For further details see Supplementary Methods.
qPCR
qPCR was performed with SYBR green (Applied Biosystems) and 6.25 pmol of each primer (Supplementary Table 4) on an ABI Prism 7000 real time PCR machine.
Supplementary Material
Acknowledgments
We thank J. Lykke-Andersen (UCSD) and members of the Pasquinelli lab for their suggestions and critical reading of this manuscript. We thank E. Moss (Univ. of Medicine and Dentistry of NJ) for providing the plin-28:LIN-28:GFP strain, R. Gassmann and A. Desai (UCSD) for providing the GFP polyclonal antibody, M. Li and M. David (UCSD) for sharing their real time PCR machine, and the Caenorhabditis Genetics Center for worm strains. P.M.V. was supported by a Ruth L. Kirschstein National Research Service Award (F32GM087004) from NIGMS. Z.S.K. was supported by NIH CMG and NIH/NCI T32 CA009523 Graduate Student Training Grants. V.H.B. was supported by a University of California – San Diego Chancellor's Undergraduate Research Scholarship. This work was funded by the US National Institutes of Health (GM071654), Keck, Searle, V, Emerald and Peter Gruber Foundations (A.E.P.) and the California Institute of Regenerative Medicine (RB1-01413) and the Stem Cell Program at the University of California, San Diego (G.W.Y.).
Footnotes
Author Contributions: A.E.P and P.M.V designed the project and wrote the paper; P.M.V. (all Figs.), Z.S.K (Fig. 1 and Supplementary Fig. 1), K.B.M. (Fig. 4), V.H.B. (Fig. 4) performed the experiments; A.E.P. and G.W.Y. supervised the studies.
References
- 1.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–9. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–94. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehrbach NJ, et al. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:1016–20. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–46. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 10.Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432–42. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- 11.Guo Y, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137:891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- 13.Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- 14.Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–25. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- 15.Morita K, Han M. Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. Embo J. 2006;25:5794–804. doi: 10.1038/sj.emboj.7601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–93. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 20.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–5. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piskounova E, et al. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–4. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 22.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev Biol. 2003;259:364–79. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- 24.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 25.Esquela-Kerscher A, et al. Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev Dyn. 2005;234:868–77. doi: 10.1002/dvdy.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez NJ, et al. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 2008;18:2005–15. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–8. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suh MR, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 29.Ballarino M, et al. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol Cell Biol. 2009;29:5632–8. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morlando M, et al. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–9. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pawlicki JM, Steitz JA. Subnuclear compartmentalization of transiently expressed polyadenylated pri-microRNAs: processing at transcription sites or accumulation in SC35 foci. Cell Cycle. 2009;8:345–56. doi: 10.4161/cc.8.3.7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Celniker SE, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–30. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–8. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–4. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–9. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Bussing I, Yang JS, Lai EC, Grosshans H. The nuclear export receptor XPO-1 supports primary miRNA processing in C. elegans and Drosophila. Embo J. 2010 doi: 10.1038/emboj.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood W. The Nematode Caenorhabditis elegans. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1988. [Google Scholar]
- 38.Cowan CA, et al. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–6. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 39.Gondran P, Amiot F, Weil D, Dautry F. Accumulation of mature mRNA in the nuclear fraction of mammalian cells. FEBS Lett. 1999;458:324–8. doi: 10.1016/s0014-5793(99)01175-8. [DOI] [PubMed] [Google Scholar]
- 40.Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc. 2008;3:698–709. doi: 10.1038/nprot.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.