Summary
Acquired resistance to ABL1 tyrosine kinase inhibitors (TKIs) through ABL1 kinase domain mutations, particularly the gatekeeper mutant T315I, is a significant problem for chronic myeloid leukemia (CML) patients. Using structure-based drug design, we developed compounds that bind to residues (Arg386/Glu282) ABL1 uses to switch between inactive and active conformations. The lead “switch-control” inhibitor, DCC-2036, potently inhibits both unphosphorylated and phosphorylated ABL1 by inducing a type II inactive conformation, and retains efficacy against the majority of clinically relevant CML resistance mutants, including T315I. DCC-2036 inhibits BCR-ABL1T315I-expressing cell lines, prolongs survival in mouse models of T315I-mutant CML and B-lymphoblastic leukemia, and inhibits primary patient leukemia cells expressing T315I in vitro and in vivo, supporting its clinical development in TKI-resistant Ph+ leukemia.
Introduction
Chronic myeloid leukemia (CML) and a subset of B-cell acute lymphoblastic leukemia (B-ALL) are characterized by the Philadelphia (Ph) chromosome and its product, the fusion tyrosine kinase BCR-ABL1, which recapitulates CML-like myeloproliferative disease when expressed in hematopoietic stem cells in mice (Daley et al., 1990). Motivated by this, imatinib, a small molecule tyrosine kinase inhibitor (TKI) of ABL1, was developed for the treatment of CML (Deininger and Druker, 2003). Structural studies demonstrate that imatinib binds to the kinase domain of ABL1 in an inactive conformation, referred to as the “DFG-out” or Type II conformation (Nagar et al., 2003; Schindler et al., 2000), in which the ABL1 activation loop tyrosine 393 is unphosphorylated and acts as pseudo-substrate to impair access to the substrate pocket, while the ATP pocket is blocked by the DFG Phe382. Such inactive kinase conformations provide unique binding pockets that are distinct from the corresponding active (Type I) conformations, and targeting of these unique inactive conformations provides a general strategy for designing selective kinase inhibitors (Huse and Kuriyan, 2002) that exploit additional binding sites adjacent to the ATP pocket (Liu and Gray, 2006). The search for robust approaches for the development of Type II inhibitors remains an intense area of research.
Imatinib induces durable hematologic and cytogenetic remissions in the majority of CML patients (Druker et al., 2006), but a significant proportion eventually experience disease progression, frequently as a consequence of mutations in the BCR-ABL1 kinase domain that render the enzyme resistant to the drug (Gorre et al., 2001). To date, more than 50 different point mutations in the ABL1 kinase domain have been detected in imatinib-resistant patients (Shah et al., 2002), some of which confer resistance by impairing the “induced fit” binding of imatinib to the kinase domain (Roumiantsev et al., 2002). The second generation BCR-ABL1 TKIs dasatinib (Shah et al., 2004), nilotinib (Kantarjian et al., 2006), and bosutinib (Puttini et al., 2006) inhibit many of these BCR-ABL1 mutants and provide clinical responses in imatinib-resistant CML. However, mutation of the gatekeeper residue, threonine 315, to isoleucine (T315I) causes virtually absolute resistance to all four TKIs, in part through steric interference with drug binding (Gorre et al., 2001). ABL1 mutations may also confer imatinib resistance by driving ABL1 towards the active conformation to which imatinib cannot bind (Azam et al., 2003), which we refer to as conformational escape. Interestingly, the T315I mutation has been shown to activate c-ABL1 (Azam et al., 2008) by conformational escape through stabilization of a “hydrophobic spine” that is a structural feature shared by many activated kinases (Kornev et al., 2006). Conformational escape may also underlie the resistance of secondary mutants of the c-KIT kinase to imatinib and sunitinib in patients with gastrointestinal stromal tumors (Gajiwala et al., 2009).
In CML, T315I accounts for ~15% of the mutations recovered from patients with imatinib resistance, but represents the predominant mechanism of acquired resistance to multiple TKIs (Shah et al., 2007). Mutation of gatekeeper residues in epidermal growth factor receptor and c-KIT also leads to resistance to TKIs, including gefitinib and erlotinib in lung cancer (Pao et al., 2005), and imatinib in gastrointestinal stromal tumors (Wardelmann et al., 2005). Hence, the development of TKIs that retain potency for gatekeeper mutants is of major clinical importance. While there are ongoing efforts to develop agents to treat CML with the T315I BCR-ABL1 mutation, no drugs have yet won approval for this indication. The aurora kinase inhibitors MK-0457 and PHA-739358 inhibit T315I mutant BCR-ABL1 in vitro (Giles et al., 2007; Gontarewicz et al., 2008), but hematologic responses observed in clinical trials of these agents may be due predominantly to inhibition of Aurora kinase rather than BCR-ABL1 (Donato et al., 2010). AP-24534, a TKI that inhibits T315I BCR-ABL1, has been described recently (O’Hare et al., 2009), and is currently in clinical trials for the treatment of refractory CML.
We have approached the general problem of inhibitor resistance, and specifically the issue of conformational escape resistance, by using the concept of ‘switch control pocket’ inhibition to guide drug design (Flynn and Petillo, 2004). When a tyrosine kinase adopts the active Type I conformation, the transition is promoted by specific “switch control” amino acids that interact with and stabilize the phosphorylated activation loop tyrosine. These switch control residues adopt alternative orientations in the inactive, unphosphorylated Type II state. A switch control inhibitor is designed to interact with these residues in the inactive conformation, providing a thermodynamic bias for stabilizing the inhibitor-bound Type II conformation, even in the face of phosphorylation or mutations that otherwise predispose escape of the kinase to the Type I active conformation. We refer to this type of durable Type II inhibition as conformational control inhibition. Here, we report on structure-based design activities that have culminated in the discovery and development of DCC-2036, a conformational control inhibitor of ABL1, including the BCR-ABL1T315I mutant.
Results
Structure-based design of ABL1 kinase inhibitors that engage the Type II conformation Glu282/Arg386 salt bridge
Although there is currently no reported structure of apo-ABL1 in its active, phospho-tyrosine 393 (pY393) Type I conformation, the structure of pY393-ABL1 in complex with dasatinib (Tokarski et al., 2006) has been reported and shows that pY393 makes binding interactions with both the catalytic loop arginine 362 (R362) and an activation loop arginine 386 (R386) (Figure S1A). The same R362 and R386 residues make electrostatic interactions with Y393 in the active conformation co-crystal structure of H396P ABL1 with VX-680 ((Young et al., 2006); Figure S1B), while in the structure of activated LCK (Yamaguchi and Hendrickson, 1996), the homologous R363 and R387 residues also interact with the activation loop pY394. Thus in both ABL1 and LCK kinases, R386 or its homolog R387 functions as a crucial conformational switch residue to stabilize the activation loop phospho-tyrosine in the active Type I state. When ABL1 is dephosphorylated and assumes the Type II inactive conformation, as inferred from the co-crystal structure with imatinib (Nagar et al., 2003), the interaction between Y393 and R386 is disrupted: Y393 moves to occupy the substrate pocket, while R386 moves to a unique inhibitor-accessible region under the C-helix in close proximity to E282 (Figure S1C). The E282/R386 amino acid pair therefore provides a design element for inhibitor binding to the inactive Type II conformation that is not available in the Type I conformation.
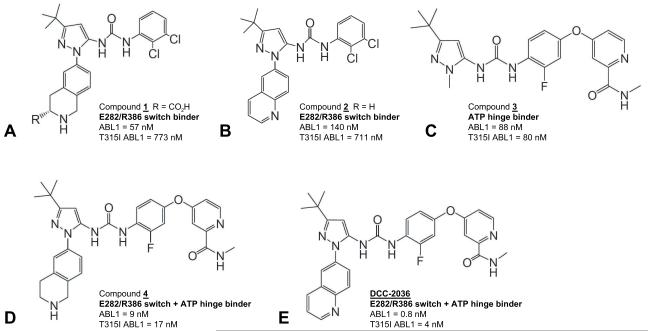
To this end, several prototype diarylurea inhibitors were designed, synthesized, and evaluated. From this effort, compounds 1 and 2 were identified (Figures 1A and 1B). Compound 1 contains a tetrahydro-isoquinoline (THIQ) ring system wherein a basic ring basic nitrogen was designed for hydrogen bonding to the acidic ABL1 E282 residue and a pendant carboxylic acid moiety (R = CO2H) was included for hydrogen bonding to the basic ABL1 R386 residue. Compound 1 also contains a urea moiety to allow a hydrogen bond with the conserved K271-E286 salt bridge of ABL1, a t-butyl moiety to bind into the hydrophobic spine at the third pocket position (Kornev et al., 2006), and a 2,3-dichlorophenyl ring to stabilize the DFG-phenylalanine F382 in the Type II-out conformation. Compound 1 exhibited an IC50 of 57 nM for ABL1native and an IC50 of 773 nM for ABL1T315I. Compound 2 replaced the tetrahydro-isoquinoline ring with a quinoline ring, designed to interact with E282 through an electrostatic interaction. Electrostatic potential calculations revealed that the C2 carbon of the quinoline ring exhibits the highest partial positive charge (Figure S1D), focusing and orienting the positive charge toward E282. Compound 2 exhibited an IC50 of 140 nM for ABL1native and an IC50 of 711 nM for ABL1T315I. Analogs of compounds 1 and 2 lacking the THIQ or the most distal nitrogen-containing portion of the quinoline ring system were essentially inactive against ABL1 (IC50 >5 μM, Figure S1E), emphasizing the significance of the E282/R386 switch control residues as an inhibitor-anchoring site.
Figure 1. Structures of switch-control inhibitors of ABL1 kinase.
(A) Compound 1 contains a tetrahydro-isoquinoline (THIQ) ring nitrogen that interacts with switch control amino acid E282; a pendant carboxylic acid moiety that interacts with switch control amino acid R386; a t-butyl moiety to bind into the hydrophobic spine position #3 pocket; and a 2,3-dichlorophenyl ring to bind into the extended hydrophobic pocket of ABL1. (B) Compound 2 replaces the THIQ ring with a quinoline ring. (C) Compound 3 contains a pyridine ring nitrogen and a pendant carboxamide-NH moiety to form hydrogen bonds with M318 at the ATP hinge. (D) Compound 4 incorporates the THIQ ring of compound 1 and the carboxamide containing pyridine ring of compound 3. (E) DCC-2036 incorporates the quinoline ring of compound 2 and the carboxamide containing pyridine ring of compound 3. The IC50 of each compound for inhibition of the kinase domains of purified ABL1native and ABL1T315I (see Experimental Procedures) are indicated. See also Figure S1 and Table S1.
To confirm the mode of binding of compound 1, an X-ray crystal structure was obtained in complex with the ABL1native kinase domain (Figure S1F). This structure revealed that the THIQ ring system of 1 does indeed bind to the E282/R386 switch control residues, with the THIQ ring basic nitrogen and carboxylic acid moiety making hydrogen bond interactions with the acidic side chain of E282 (3.26 Å) and the guanidinyl side chain of R386 (3.32 Å), respectively. Other features of inhibitor binding include hydrogen bond formation between the urea moiety and the K271-E286 salt bridge, binding of the t-butyl moiety into the third pocket of the hydrophobic spine, and orientation of the 2,3-dichlorophenyl ring to stabilize F382 in an edge-face π-interaction. The key bonding interactions between compound 1 and ABL1native are summarized in Table S1. It is noteworthy that this binding mode affords a 57 nM inhibitor of ABL1 kinase without relying on binding into the ATP pocket hinge region, a typical requirement of classical Type II and ATP-competitive Type I inhibitors.
Design of dual-anchoring inhibitors of ABL1 that bind both the Glu282/Arg386 switch pair and the ATP pocket hinge Met318
To improve potency, we next designed inhibitors that provided dual docking sites to both the switch control pocket E282/R386 pair and also to the ATP hinge, reasoning that such a dual-anchoring inhibitor might provide the increased binding energy required to stabilize oncogenic and resistant forms of ABL1 in the Type II conformation. To probe docking of inhibitor only to the ATP hinge region, we prepared compound 3 (Figure 1C), which contains a carboxamide-substituted pyridine ring to form hydrogen bonds with the backbone of the ATP hinge residue M318. However, compound 3 does not extend in the direction of the activation loop to engage the switch control E282/R386 residues. Compound 3 exhibited an IC50 of 88 nM vs. ABL1native, similar in potency to compounds 1 and 2, indicating that either inhibitor-anchoring mode affords similar potency.
We then combined both E282/R386 switch control-binding and M318-ATP hinge-binding moieties by synthesizing compound 4 (Figure 1D), and DCC-2036 (Figure 1E). Compound 4 and DCC-2036 exhibited IC50 values of 9 nM and 0.8 nM vs. ABL1native, respectively. Significantly, these dual-anchoring inhibitors also showed potent inhibition of the gatekeeper mutant ABL1T315I, with IC50 values of 17 nM and 4 nM, respectively. Thus, incorporation of the additional switch control pocket-binding quinoline ring in DCC-2036 leads to a 100-fold improvement in potency vs. ABL1native and a 20-fold improvement in potency vs. gatekeeper mutant ABL1T315I, compared to the prototypic Type II compound 3.
Structure of DCC-2036 bound to ABL1native and gatekeeper mutant ABL1T315I
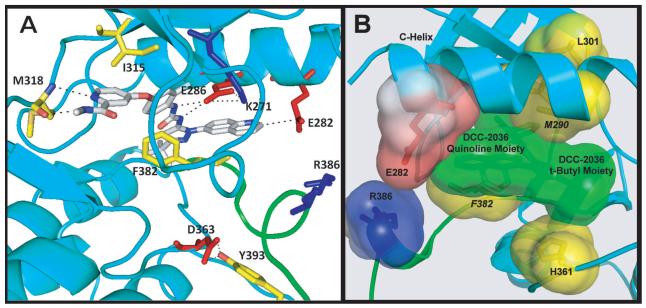
Co-crystal structures of DCC-2036 with ABL1native (Figure S2A) and with ABL1T315I (Figure 2A and Figure S2B) were solved (Table S2). In both structures, DCC-2036 binds to the DFG-out Type II conformation. Similar to compound 1, the quinoline ring of DCC-2036 forms an electrostatic interaction with E282 (d = 4.06 Å in ABL1native, 3.30 Å in ABL1T315I; Table S1). In the ABL1native structure, E282 is held in place to retain a hydrogen bond with R386 (d = 2.76 Å). The urea nitrogens form two hydrogen bonds with the conserved K271-E286 salt bridge and an additional hydrogen bond between the urea carbonyl and the backbone amino group of D381, the t-butyl moiety binds into the hydrophobic spine pocket, thereby forcing the DFG F382 (yellow) out into the mouth of the ATP pocket, and the carboxamide-substituted pyridine ring forms two hydrogen bonds with the ATP hinge residue M318. The fluorine substituent on the central phenyl ring is positioned ortho- to the urea moiety, providing stereoelectronic bias for inducing planarity of the fluoro-phenyl ring and the urea function. Such stereoelectronic restriction optimally orients the phenyl ring for π-interaction with F382.
Figure 2. Co-crystal structure of DCC-2036 bound to ABL1T315I kinase.
(A) Ribbon diagram of the Type II conformation of ABL1T315I in complex with DCC-2036. Note the trajectory of the quinoline C2 carbon oriented towards E282 (d = 3.30 Å). The activation-loop internal switch (green) is in the ‘off state’, with F382 occluding the ATP pocket and Y393 occluding the substrate-binding pocket, making a hydrogen bond with D363. DCC-2036 makes additional hydrogen bonds with E286 (2.80 and 2.90 Å), and with the ATP hinge residue M318 (2.95 and 3.18 Å). Key hydrogen bonds and electrostatic interactions are highlighted as dashed lines. (B) View of the DCC-2036/ABL1T315I complex. The activating hydrophobic spine is disrupted by displacement of F382. DCC-2036 makes stabilizing interactions with spine residues M290, H361, and the displaced residue F382. DCC-2036 (t-Bu moiety) occupies the third position in this ‘inhibitory’ hydrophobic spine. Key interactions of the inhibitor quinoline ring with the E282/R386 switch pair are also shown. See also Figure S2 and Tables S1 and S2.
Recently, an activating hydrophobic spine of ABL1T315I has been described (Azam et al., 2008), wherein a vertical cluster of hydrophobic amino acids stabilizes the Type I active state. This hydrophobic cluster includes L301, M290, F382, H331, and the additional mutant I315 as a fifth reinforcing hydrophobic residue further stabilizing the Type I active state ((Modugno et al., 2007), Figure S2C). In the co-crystal structure of DCC-2036 with ABL1T315I, this activating spine has been disrupted by outward movement of F382 (Figure 2B), which is stabilized by the central 2-fluoro phenyl ring of DCC-2036. The inhibitor also makes productive hydrophobic contact with the mutant gatekeeper I315 and spine residues M290 and H361, while the t-butyl residue of DCC-2036 occupies the third position of the hydrophobic spine as a surrogate of F382, effectively establishing an inhibitory hydrophobic spine in the interior of the kinase, which includes L301, M290, the inhibitor (DCC-2036 t-butyl moiety), and H361 (Figure 2B).
DCC-2036 inhibits unphosphorylated ABL1native, phospho-ABL1native, ABL1H396P, and ABL1T315I in a non-ATP-competitive manner
To investigate the biochemical mechanism of ABL1 inhibition by DCC-2036, we compared the inhibitory activity of DCC-2036 to that of imatinib, dasatinib, and nilotinib (Table 1) against purified native ABL1 in unphosphorylated (u-ABL1native) and phosphorylated (p-ABL1native) forms, unphosphorylated and phosphorylated gatekeeper mutant ABL1T315I, and the activation loop mutant ABL1H396P. Consistent with the structural data in Figure S2A, DCC-2036 potently (IC50 0.82 nM) inhibited u-ABL1native, which is thought to exist predominantly in the inactive type II conformation. In addition, DCC-2036 also strongly inhibited p-ABL1native (IC50 2 nM), which more readily adopts an active, Type I conformation. More significantly, DCC-2036 potently inhibited both u-ABL1T315I (IC50 5 nM) and p-ABL1T315I (IC50 4 nM), both of which exist predominately in the Type I conformation due to stabilization of an activating hydrophobic spine by the T315I mutation (Azam et al., 2008). DCC-2036 also potently inhibited ABL1H396P (IC50 1.4 nM), which, like ABL1T315I, is predisposed to exist predominately in a Type I activated conformation due to the restricted backbone torsional angles imposed by the mutant Pro396 (Young et al., 2006). Together, these results argue that DCC-2036 can effectively inhibit forms of ABL1 predisposed to adopt active, DFG-in conformations by forcing the kinase domains into inhibitor-bound, inactive Type II conformations. By comparison, imatinib had moderate inhibitory activity only for u-ABL1native (IC50 75 nM) while losing significant potency for those forms that are predisposed to exist predominately in the ‘switch on’ activated conformation (p-ABL1native, ABL1H396P, and ABL1T315I). As previously reported (Shah et al., 2004; Weisberg et al., 2006), dasatinib and nilotinib retained potency for p-ABL1native and ABL1H396P, but were essentially inactive against mutant ABL1T315I (IC50 >10,000 and 3,800 nM, respectively).
Table 1.
Inhibition (IC50, nM) of ABL1 kinases by DCC-2036 and other TKIs
| u-ABL1native | p-ABL1native | ABL1H396P | u-ABL1T315I | p-ABL1T315I | |
|---|---|---|---|---|---|
| DCC-2036 | 0.75 ± 0.11 | 2 ± 1 | 1.4 ± 0.1 | 5 ± 1 | 4 ± 1 |
| imatinib | 75 ± 3 | 7,700 ± 1,000 | 3,000 ± 1,000 | >10,000 | >10,000 |
| dasatinib | 1.1 ± 0.1 | 0.7 ± 0.4 | 0.5 ± 0.1 | >10,000 | >10,000 |
| nilotinib | 0.87 ± 0.09 | 2.2 ± 1.2 | 1.1 ± 0.3 | >5,000 | 3,800 ± 1,200 |
DCC-2036 inhibits ABL1 kinase in a non-ATP-competitive manner and exhibits prolonged kinase residency time
For ATP competitive inhibitors such as imatinib, an increase of ATP concentration can result in significant loss of inhibitor potency. However, DCC-2036 lost little activity against AblT315I even in the presence of high (5 mM) ATP concentrations that are typical of the intracellular milieu (Table S3). DCC-2036 exhibited very prolonged off rates (koff) from its complex with all three ABL1 forms (Table S4). The koff value of 0.00172 min−1 for p-ABL1native corresponds to a t1/2 value of 402 minutes, demonstrating that DCC-2036, once bound, exhibits prolonged residency time in complex with ABL1. This off-rate is considerably longer than the respective values obtained for dasatinib (153 min) and nilotinib (180 min).
Kinase inhibition profile of DCC-2036
In addition to ABL1, DCC-2036 also inhibited the SRC family kinases SRC, LYN, FGR, and HCK, and the receptor TKs KDR, FLT3, and TIE2 (Table 2). Notably, DCC-2036 spared c-KIT (IC50 481 nM). DCC-2036 was further evaluated against a broad panel of human kinases, but did not inhibit a significant number of kinases outside this ensemble (Table S5).
Table 2.
Kinase inhibition profile of DCC-2036
| Kinase | IC50, nM |
|---|---|
| ABL1 | 0.75 ± 0.11 |
| SRC | 34 ± 6 |
| LYN | 29 ± 1 |
| FGR | 38 ± 1 |
| HCK | 40 ± 1 |
| KDR | 4 ± 0.3 |
| FLT3 | 2 ± 0.3 |
| TIE2 | 6 ± 0.3 |
| c-KIT | 481 ± 57 |
| PDGFRα | 70 ± 10 |
| PDGFRβ | 113 ± 10 |
| Aurora A | >5,000 |
| CDK2/Cyclin D | >5,000 |
IC50 values (mean ± SD) were determined as described in Experimental Procedures vs. purified kinase domains
See also Table S5
DCC-2036 inhibits cellular proliferation of Ba/F3 cells expressing native or mutant BCR-ABL1
We next tested the ability of DCC-2036 to inhibit proliferation of Ba/F3 cells transformed to interleukin-3-independence by BCR-ABL1native or a series of TKI-resistant BCR-ABL1 mutants arising in the P-loop, the interior N-lobe, the E-helix proximal to the catalytic loop, the catalytic loop, the activation loop, and the hinge/gatekeeper region (Table 3). Similar to imatinib, dasatinib, and nilotinib, DCC-2036 effectively inhibited the proliferation of Ba/F3 cells expressing native BCR-ABL1native (IC50 5.4 nM), but DCC-2036 also retained potency against Ba/F3 cells expressing BCR-ABL1 mutants that are resistant to imatinib, dasatinib (T315A), and nilotinib (L248R, Y253H, E255V, F359C), as well as the gatekeeper mutant BCR-ABL1T315I (IC50 13 nM), where all three FDA-approved TKIs were ineffective. DCC-2036 also inhibited proliferation of the Ph+ cell line K562 (IC50 5.5 nM), and induced apoptosis in both BCR-ABL1-expressing Ba/F3 and K562 cells at nanomolar concentrations (data not shown). Importantly, the growth of parental Ba/F3 cells (in the presence of IL-3) was not appreciably inhibited by DCC-2036 until concentrations exceeded 3 μM, demonstrating that the drug is not generally cytotoxic. By contrast, the dual Aurora A/ABL1 kinase inhibitor MK-0457 did not discriminate between parental and BCR-ABL1-transformed Ba/F3 cells, indicative of global cytostatic activity (Table 3). In addition to the T315I mutant, DCC-2036 also inhibited proliferation of several common TKI-resistant mutants of BCR-ABL1, including G250E, Q252H, Y235F, E255K, V299L, F317L, and M351T, at IC50 values ranging from 6-150 nM (Table 3). Collectively, these mutants mediate the majority of clinical TKI resistance in CML patients (O’Hare et al., 2009; Shah et al., 2002).
Table 3.
Comparison of inhibition of cellular proliferation by ABL1 TKIs (IC50, nM)
| Ba/F3-BCR-ABL1 | DCC-2036 | imatinib | dasatinib | nilotinib | MK-0457 |
|---|---|---|---|---|---|
| Ba/F3 Parent* | 3,500 | >10,000 | 3,800 | 9,500 | 102 |
| Native | 5.4 | 48 | 0.4 | 2.1 | 104 |
| L248R | 51 | >10,000 | 7.3 | 930 | 110 |
| G250E | 98 | 1,900 | 2.8 | 67 | 180 |
| Q252H | 2 | 700 | 0.3 | 5.1 | 130 |
| Y253F | 39 | 2,300 | 2 | 63 | 80 |
| Y253H | 56 | >10,000 | 1 | 120 | 190 |
| E255K | 127 | 6,400 | 8 | 80 | 84 |
| E255V | 150 | >10,000 | 2 | >10,000 | 190 |
| V299L | 6 | 190 | 8 | 10 | 200 |
| T315A | 19 | 536 | 65 | 17 | 88 |
| T315I | 13 | >10,000 | 5,200 | 2,800 | 74 |
| F317L | 36 | 417 | 2.3 | 10 | 84 |
| M351T | 14 | 417 | 1.1 | 4.2 | 65 |
| F359C | 120 | 2,500 | 2.7 | 190 | 130 |
| H396P | 81 | 1,700 | 1.2 | 49 | 160 |
IC50 values were determined as described in Experimental Procedures
Determined in the presence of 10 ng/mL IL-3
DCC-2036 inhibits mutant BCR-ABL1T315I signaling and prolongs survival in a mouse Ba/F3 cell allograft model
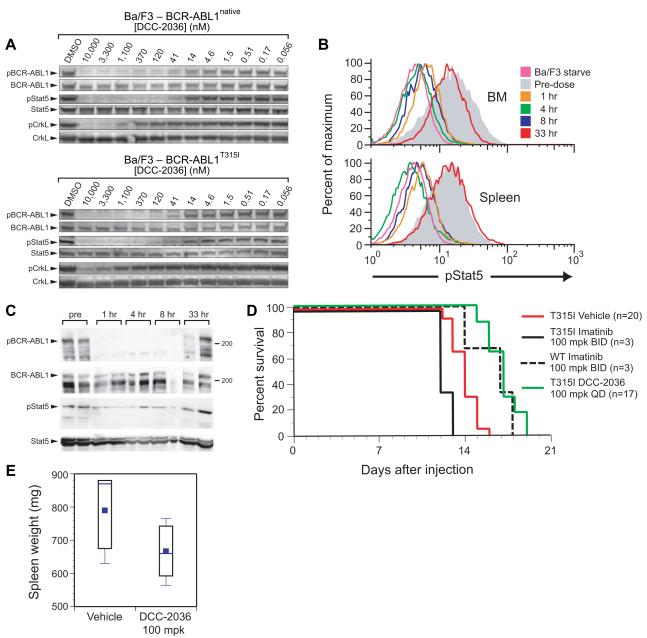
We examined the ability of DCC-2036 to inhibit the phosphorylation of BCR-ABL1 and its downstream targets STAT5 and CrkL in transformed Ba/F3 cells (Figure 3A). At concentrations that correlated with its cytotoxic effects, DCC-2036 effectively inhibited autophosphorylation of BCR-ABL1native (IC50 29 nM) and BCR-ABL1T315I (IC50 18 nM), as well as the phosphorylation of STAT5 in both cell lines (IC50 28 nM and 13 nM, respectively). Phosphorylation of the BCR-ABL1 substrate CrkL was also inhibited, although to a lesser extent, in both cell lines (BCR-ABL1native IC50 495 nM; BCR-ABL1T315I IC50 2,600 nM). Importantly, imatinib, dasatinib, and nilotinib all failed to inhibit phosphorylation of BCR-ABL1, STAT5, and CrkL in Ba/F3 cells expressing BCR-ABL1T315I (IC50 values > 5 μM; data not shown).
Figure 3. DCC-2036 inhibits BCR-ABL1 signaling in Ba/F3 cells in vitro and in a mouse allograft model.
(A) Inhibition of BCR-ABL1 signaling in Ba/F3 cells expressing BCR-ABL1native (top panel) or BCR-ABL1T315I (bottom panel) by different concentrations of DCC-2036.. (B) Complete and sustained inhibition of pSTAT5 in mice bearing Ba/F3-BCR-ABL1T315I leukemia cells following a single oral dose of DCC-2036 at 100 mg/kg. At the indicated time after the dose, mice were sacrificed and the level of phosphorylation of STAT5 assessed in leukemic (GFP+) cells by intracellular staining with anti-pSTAT5 antibody and flow cytometric analysis. The grey histogram represents the level of STAT5 activation prior to the dose, while the pink curve is from parental Ba/F3 cells starved of IL-3 and serum, representing baseline STAT5 activation. (C) Immunoblot analysis of phospho-BCR-ABL1T315I and phospho-STAT5 levels in spleen tissue extracts from the experiment in (B). Note that due to contributions from non-leukemic cells, the extent of pSTAT5 inhibition is not as great as in panel (B). (D) Survival curve of cohorts of mice injected on day 0 with 106 Ba/F3 cells expressing either BCR-ABL1native or BCR-ABL1T315I, and treated beginning on day 3 with either imatinib at 100 mg/kg twice daily or DCC-2036 at 100 mg/kg once daily by oral gavage. The survival of the T315I DCC-2036-treated cohort was significantly longer than either the T315I imatinib-treated or vehicle-treated cohorts (P<0.0001, Wilcoxon test). (E) Box-style plot of spleen weights (mg) from Ba/F3-BCR-ABL1T315I leukemia-bearing mice treated with vehicle or DCC-2036, assessed at the time of morbidity or death. The difference in spleen weight of the DCC-2036-treated and the vehicle-treated cohorts was of borderline significance (p = 0.06, unpaired t-test).
BCR-ABL1-transformed Ba/F3 cells can efficiently engraft syngeneic Balb/c mice following intravenous injection (Ilaria and Van Etten, 1995), proliferating in blood, marrow and spleen and eventually causing morbidity and death of the recipients. A single oral dose of DCC-2036 at 100 mg/kg afforded circulating plasma levels that exceeded 12 μM for up to 24 hours (data not shown), and effectively inhibited BCR-ABL1 signaling for up to 8 hours in Ba/F3-BCR-ABL1T315I leukemia cells isolated from BM and spleen of tumor-bearing mice, as assessed by intracellular flow cytometric staining for phospho-STAT5 (Figure 3B) and immunoblotting of tissue extracts for phospho-BCR-ABL1 and phospho-STAT5 (Figure 3C). Treatment of mice bearing Ba/F3-BCR-ABL1T315I leukemia cells with DCC-2036 at 100 mg/kg once daily by oral gavage significantly prolonged their survival, while imatinib at 100 mg/kg twice daily was ineffective (Figure 3D). In this aggressive allograft model, DCC-2036 was as effective for treatment of BCR-ABLT315I leukemia as imatinib at 100 mg/kg twice daily was for treatment of BCR-ABL1native leukemia, and reduced the leukemia cell burden in the spleens of treated mice (Figure 3E).
DCC-2036 demonstrates efficacy in retroviral transduction/transplantation mouse models of BCR-ABL1T315I CML and Ph+ B-ALL
To extend these results to primary myeloid and lymphoid leukemias induced by BCR-ABL1, we employed a well-characterized retroviral BM transduction/transplantation model system. When BM from 5-fluorouracil (5-FU)-treated donors is transduced with BCR-ABL1 retrovirus and transplanted intravenously into irradiated mice, recipients develop an aggressive and fatal CML-like myeloproliferative neoplasm (MPN), characterized by massive expansion of and extensive organ infiltration by of maturing myeloid cells (Li et al., 1999). The CML-like MPN arises from hematopoietic stem cells (Hu et al., 2006), and is transplantable (Li et al., 1999) and responsive to TKI therapy (Hu et al., 2004). When donors are not treated with 5-FU, recipients of BCR-ABL1-transduced BM instead develop aggressive precursor B-cell acute lymphoblastic leukemia/lymphoma (B-ALL), arising from early lymphoid progenitors (Hu et al., 2006) and characterized by involvement of the BM, spleen, lymphatics, and pleural cavity (Li et al., 1999).
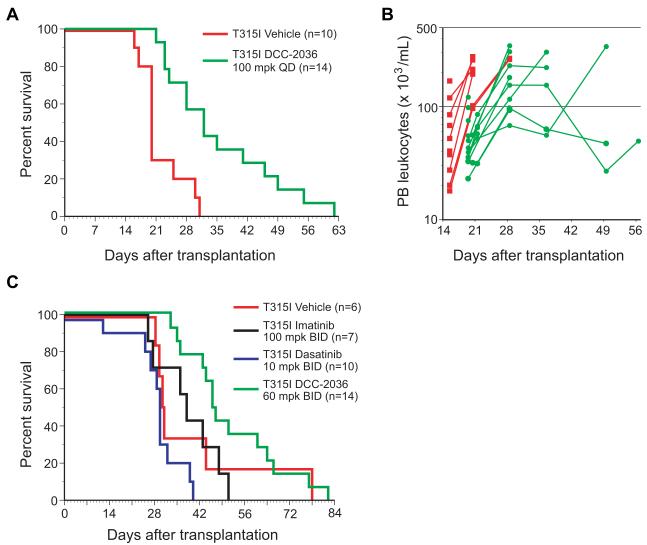
In the CML model, treatment of recipients of BCR-ABL1T315I-transduced BM with DCC-2036 at 100 mg/kg once daily significantly prolonged their survival from a median of 20 days to 32 days (Figure 4A), and was associated with an improvement in the circulating leukocyte counts in treated mice (Figure 4B). In BCR-ABL1-induced B-ALL, genetic studies in mice have implicated SRC family kinases in the disease pathogenesis (Hu et al., 2004), suggesting that the inhibitory activity of DCC-2036 towards LYN, FGR, and HCK (Table 2) might be of therapeutic benefit in this disease. Indeed, a previous report suggested that dasatinib, which also inhibits SRC kinases, has therapeutic activity against B-ALL induced by T315I BCR-ABL1 in mice (Hu et al., 2006). Accordingly, we also compared the activity of DCC-2036 against imatinib and dasatinib in mice with B-ALL induced by BCR-ABL1T315I. DCC-2036 had efficacy that was superior to both imatinib and dasatinib (Figure 4C), significantly prolonging the survival of treated mice (median survival 47 days), although all mice in this cohort eventually succumbed to B-ALL. Interestingly, we noted that the histopathology of the B-lymphoid leukemia/lymphoma appeared to be altered in DCC-2036-treated mice. Rather than developing the malignant hemorrhagic pleural effusion that was the predominant cause of death in the other three cohorts, the majority (10/14) of DCC-2036-treated recipients had absent or minimal (<0.1 mL) pleural effusions, but instead developed hind limb paralysis due to lymphomatous involvement of vertebral spinal cord (data not shown). This suggests that effective treatment of disease outside the central nervous system by DCC-2036 might have altered the natural history of the leukemia and selected for malignant cells residing in a privileged site. Together, these results demonstrate that oral DCC-2036 has efficacy against primary myeloid and lymphoid leukemias induced by BCR-ABL1T315I in mice.
Figure 4. DCC-2036 has efficacy in mouse models of CML and Ph+ B-ALL.
(A) Survival curve for recipients of 5-FU-treated BM transduced with BCR-ABL1T315I retrovirus, treated by oral gavage beginning at day 5 post-transplant with vehicle (red line, n=10) or DCC-2036 at 100 mg/kg once daily (green line, n=14). All recipient mice succumbed to CML-like MPN. The difference in survival between the two cohorts is significant (p = 0.007, Wilcoxon test). (B) Scatter plot of peripheral blood leukocyte counts vs. time from the cohorts in panel (A). Red squares represent individual vehicle-treated mice, while green circles represent individual DCC-2036-treated mice. The difference in mean leukocyte counts on d20 between DCC-2036- and vehicle-treated cohorts was significant (p = 0.0002, unpaired t-test). (C) Survival curve for Balb/c recipients of non-5-FU-treated BM transduced with BCR-ABL1T315I retrovirus, treated by oral gavage beginning at day 10 post-transplant with vehicle (red line, n=6), imatinib at 100 mg/kg once daily (black line, n=7), dasatinib at 10 mg/kg twice daily (blue line, n=10), or DCC-2036 at 60 mg/kg twice daily (green line, n=14). The survival of the DCC-2036-treated cohort was significantly prolonged (Wilcoxon tests) compared to cohorts treated with vehicle (p = 0.0184), imatinib (p = 0.0474), and dasatinib (p < 0.0001).
DCC-2036 inhibits BCR-ABL1, including the T315I mutant, in primary patient cells in vitro and in vivo
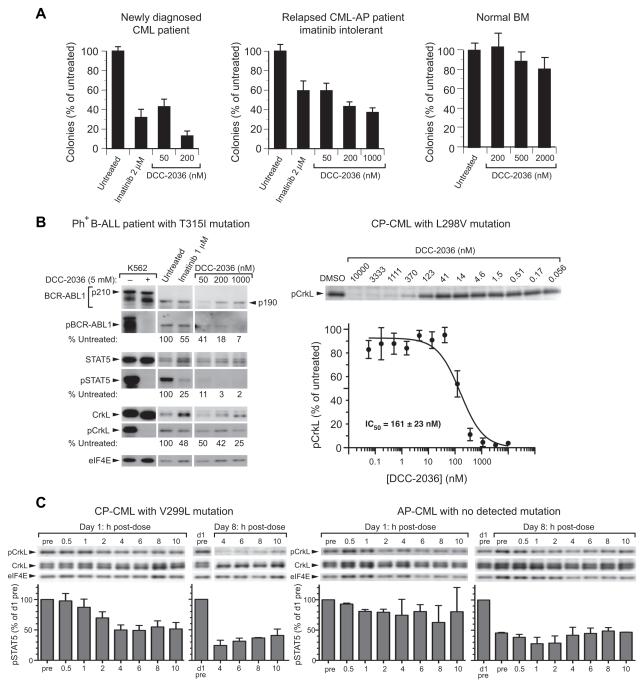
We tested the ability of DCC-2036 to inhibit myeloid colony formation from BM mononuclear cells from a patient with newly diagnosed chronic phase CML, and a patient with relapsed CML in accelerated phase who was intolerant of imatinib. DCC-2036 inhibited the formation of myeloid colonies in a concentration-dependent manner that was equivalent to clinically relevant concentrations of imatinib (Figure 5A). Importantly, DCC-2036 had minimal toxicity to normal hematopoietic colony-forming cells at concentrations up to 2000 nM (Figure 5A and data not shown). In leukemic blasts from a patient with relapsed/refractory Ph+ B-ALL with the T315I mutation, DCC-2036 inhibited phosphorylation of BCR-ABL1, STAT5, and CrkL (Figure 5B) and decreased cell viability in vitro (Figure S3A). DCC-2036 also inhibited phosphorylation of CrkL in primary leukemia cells from a patient with chronic phase CML and L298V mutation (Figure 5B).
Figure 5. DCC-2036 inhibits BCR-ABL1 in patient leukemic cells in vitro and in vivo.
(A) Inhibition of BM-derived myeloid colonies (mean + SD) from a newly diagnosed CML chronic phase patient (left panel), a patient with relapsed CML in accelerated phase who was intolerant of imatinib (middle panel), and a normal individual (right panel). In the left and middle panels, the differences between any of the drug-treated cultures and Untreated were significant (p < 0.001, t-test), while none of the differences in the right panel were statistically significant. (B) Inhibition of BCR-ABL1 kinase activity and signaling by DCC-2036 in leukemic blasts from patients in vitro. Left panel: Patient with Ph+ B-ALL and the T315I mutation, which had an allele frequency of 40% estimated from DNA sequencing data. The relative levels of phospho-BCR-ABL1T315I, phospho-STAT5, and phospho-CrkL vs. untreated are shown. Note that this patient expressed the p190 isoform of BCR-ABL1T315I. Right panel: Patient with CP-CML and a L298V mutation. PB leukocytes were incubated for 3h with the indicated concentrations of DCC-2036, and CrkL phosphorylation quantitated by immunoblot analysis of quadruplicate blots. Data (mean normalized pCrkL ± SD) were fitted to a sigmoidal curve (GraphPad Prism) and an IC50 value calculated. (C) Pharmacodynamic analysis of inhibition of CrkL phosphorylation by DCC-2036 (tosylate salt in formulated tablets, 100 mg BID dose level) in circulating CML cells in vivo obtained on days 1 and 8 of cycle 1 of treatment in a Phase 1 clinical trial. Left panel: patient with CML chronic phase and V299L mutation (note that on day 8, samples before and 0.5-2 h after dosing were not obtained). Right panel: patient with CML accelerated phase and no detectable BCR-ABL1 mutation. pCrkL levels (mean + SD) normalized to percent of day 1 pre-dose level are shown below. See also Figure S3.
Based on these preclinical results, DCC-2036 has been advanced to a phase 1 clinical trial in patients with Ph+ leukemia that has relapsed or been refractory to at least two of the FDA-approved TKIs (ClinicalTrials.gov identifier NCT00827138). In a patient with CML chronic phase and the T315I mutation who received DCC-2036 (tosylate salt) at a 300 mg dose level, we observed significant (~65%) suppression of STAT5 phosphorylation in peripheral blood leukemia cells that was maximum about 2 h following the initial day one dose, with more sustained inhibition observed following a week of continuous daily dosing (Figure S3B). In two additional patients, significant inhibition of CrkL phosphorylation in circulating leukemic cells was observed in the first cycle of treatment, with sustained inhibition apparent in the second week of therapy (Figure 5C). Together, these results demonstrate that DCC-2036 is capable of suppressing BCR-ABL1 activity in primary leukemic cells, both in vitro and in vivo, from patients with Ph+ leukemia who are refractory to multiple TKIs, including those with the T315I mutation.
Discussion
Kinases are involved in a broad range of essential cellular functions, and are regulated in vivo by adopting different conformations. Disruption of kinase conformational regulation through protein fusions, deletions, or missense mutations can lead to pathologic conditions, including cancer. We have initiated a broad-based approach to kinase inhibition through identification of key amino acid residues that are critical for the fluxing of kinases between conformational states, and the design of inhibitors that bind to such residues.
In ABL1, R386 is a critical anchoring partner for stabilizing pY393 in the active conformation. In the inactive Type II conformation, R386 and Y393 separate from each other, with Y393 moving to occupy the substrate pocket, and R386 shifting to the interior of the kinase towards E282 (Figure S1C). This alternative disposition of E282/R386 provides a basis for rational inhibitor design. Here, we have demonstrated the utility of this E282/R386 anchor for design of ABL1 inhibitors, culminating in the discovery of DCC-2036. Prototype inhibitor compounds 1 and 2 (Figure 1), which orient a THIQ or quinoline basic ring system toward the E282-R386 inhibitory anchor, exhibited potencies in the 57-140 nM range for ABL1native, whereas close structural analogs lacking functionality for interaction with E282 or R386 were inactive (Figure S1E). It is noteworthy that this level of inhibition, which is equivalent to the potency of imatinib for ABL1native, is achieved without extending inhibitor binding into the ATP hinge region. Additionally, compounds 1 and 2 also exhibit potencies in the 700 nM range for the gatekeeper mutant ABL1T315I. A similar level of potency was realized by compound 3, which anchors into the ATP hinge but does not extend to the switch E282-R386 salt bridge.
Combining both an ATP hinge binding anchor and an E282/R386 binding anchor within a single inhibitor led to compound 4 and DCC-2036, which exhibit much greater potency vs. ABL1native (IC50 of 9 and 0.8 nM, respectively) and against mutant ABL1T315I (17 and 4 nM, respectively). While compound 3 is equipotent against ABL1native and ABL1T315I and must therefore avoid steric clash with substitutions at the gatekeeper residue, the ability to bind both the ATP hinge and the switch control regions is what affords DCC-2036 the potency (100-fold decrease in ABL1native IC50 vs. compound 3) required for a clinical drug candidate. A derivative of DCC-2036 lacking the 2-fluorine substituent on the phenyl ring maintained full potency for inhibiting ABL1native and ABL1T315I (IC50 1 nM and 7 nM, respectively), demonstrating that the orientation of this fluorine is not critical for inhibition of the gatekeeper mutant by DCC-2036 (data not shown). The structure of DCC-2036 with u-ABL1native also revealed an ideal hydrogen bond (2.76 Å) between the carboxylate side chain of E282 and the guanidinium side chain of R386. We speculate that this Type II switch state E282/R386 hydrogen bond induced by DCC-2036 contributes to its high potency vs. u-ABL1native (IC50 0.8 nM).
DCC-2036’s mode of binding provides a durable mode of Type II inhibition that affords a prolonged off-rate, resiliency to high ATP concentrations, and overcomes the clinical problem of conformational escape resistance by making interactions within the Type II state (Figure 2A) that override the effect of point mutations that drive ABL1 towards the active Type I state. We refer to this type of inhibition as conformational control inhibition. DCC-2036 inhibits both ABL1T315I and ABL1H396P (IC50 values of 4 nM and 1.4 nM, respectively), which are known to exist predominantly in the Type I active conformation (Azam et al., 2008; Young et al., 2006). Structural data (Figure 2B) suggest that DCC-2036 may, in part, lead to conformational control of ABL1 by invoking an inhibitor-participating hydrophobic spine (Azam et al., 2008; Kornev et al., 2006) in which a portion of DCC-2036 substitutes for phenylalanine in the third spine position while also making interactions with the second and fourth spine residues, M290 and H361 (Figure 2B).
DCC-2036 exhibits high potency against cell lines expressing mutant forms of BCR-ABL1, including T315I and H396P, that collectively account for more than 85% of TKI-resistant CML patients in whom ABL1 kinase mutations are identified (Shah et al., 2002; Shah et al., 2007). ABL1 mutants involving the P-loop residue E255 (E255V/K) were relatively less sensitive to DCC-2036 (Table 3), and are also refractory to the type II inhibitors imatinib and nilotinib. In the native ABL1 structure, the acidic side chain of E255 makes an electrostatic interaction with a lysine residue on the other arm of the loop, and disruption of this interaction might distort the nearby ATP hinge region into which DCC-2036 binds. A Ba/F3 cell-based mutagenesis screen for resistance to DCC-2036 recovered no BCR-ABL1 mutations at higher drug concentrations, while mutations at Y253 and E255 emerged at lower concentrations (T. O’Hare, personal communication). However, pharmacokinetic data from the phase 1 clinical trial have demonstrated that DCC-2036 can achieve plasma levels well above these concentrations (data not shown). There were no mutations recovered that are known to destabilize the inactive conformation of BCR-ABL1 towards its active, Type I conformation (Azam et al., 2003; Sherbenou et al., 2010), nor were there mutations identified in the switch control amino acids E282/R386, which may be required for assembly of the catalytically active state (Kornev et al., 2006). Together, these observations suggest that acquired resistance to DCC-2036 therapy in CML may be less frequent than for the three ATP-competitive ABL1 inhibitors in current clinical use.
DCC-2036 inhibits BCR-ABL1, STAT5, and CrkL phosphorylation in BCR-ABL1-expressing cells. In a Ba/F3 cell allograft model, oral administration of DCC-2036 resulted in sustained inhibition of phosphorylation of both BCR-ABL1T315I and STAT5, a significant reduction in leukemic burden, and prolonged survival (Figure 3). Oral administration of DCC-2036 at doses of 60-100 mg/kg/d prolonged survival and reduced circulating leukemia cells in physiologically relevant mouse models of CML-like myeloproliferative neoplasia and Ph+ B-cell acute lymphoblastic leukemia induced by BCR-ABL1T315I (Figure 4) but had no effect on hematopoiesis in normal mice (data not shown). Whereas plasma concentrations of DCC-2036 exceeding 50 μM were observed in recipient mice, after accounting for protein binding (97-99%) the active in vivo drug concentrations were in the range of 100-1000 nM (data not shown). Tested against primary patient cells in vitro, DCC-2036 suppressed Ph+ myeloid colony formation and inhibited BCR-ABL1T315I kinase activity and phosphorylation of STAT5 and CrkL at these concentrations but did not significantly inhibit growth of normal BM progenitors at concentrations up to 2 μM (Figure 5A). Together, these results demonstrate a differential inhibitory effect of DCC-2036 against BCR-ABL1-expressing vs. normal hematopoietic cells at clinically relevant concentrations.
Based on its composite properties and these positive preclinical results, DCC-2036 was selected for clinical development. Correlative studies from the Phase 1 clinical trial of DCC-2036 have demonstrated sustained inhibition of phospho-BCR-ABL1, phospho-STAT5, and phospho-CrkL in refractory CML patients. In summary, DCC-2036 represents a potential therapeutic option for patients with Ph+ leukemia who have relapsed on or are refractory to conventional TKIs. Exploiting the diversity of switch control mechanisms among different kinases is a promising strategy to develop molecularly targeted therapies for hematologic neoplasms, solid tumors, and non-malignant diseases.
Experimental Procedures
Tyrosine kinase expression, purification, crystallography, and kinase assays
His-tagged ABL1 kinase domain proteins were overexpressed in Sf9 cells via a baculovirus vector, and unphosphorylated and phosphorylated proteins purified by sequential Hi-Trap Ni and POROS HQ chromatography. Crystals of ABL1-inhibitor complexes were grown by vapor diffusion and diffraction data obtained with the beam source, data collection and refinement statistics provided in Table S2. Purified ABL1 kinases were assayed by quantitation of ADP production by coupling with the pyruvate kinase/lactate dehydrogenase system (Schindler et al., 2000). Detailed protein methods are provided in Supplemental Information.
Cell Proliferation Assays
Ba/F3 cells (3×103 cells/well) or primary Ph+ leukemia cells (5×104 cells/well) were plated in triplicate in 96-well plates containing test compounds. After 72h, viable cells were quantified by resazurin (O’Brien et al., 2000) or MTT (Roumiantsev et al., 2002) assay as described. Results represent an average of at least three independent experiments.
Immunoblotting Assays
2 × 106 Ba/F3 cells expressing native or BCR-ABL1T315I were incubated for 6 h in media containing test compounds, then lysed in RIPA buffer containing protease inhibitors (Pierce, Rockford, IL). In the case of Ba/F3 cells isolated from allografted mice or primary human leukemia samples, mononuclear cells were isolated by centrifugation through Ficoll-Hypaque and cell pellets solubilized by direct boiling in NuPAGE LDS sample buffer (InVitrogen). Protein was quantitated by the Pierce 660 nm Protein Assay Reagent, equal protein amounts loaded onto NuPAGE 5-15% gradient polyacrylamide gels and transferred to nitrocellulose membranes (InVitrogen LC2001) by electroblotting. Blots were probed using phospho-specific antibodies and bands were detected using ECL Plus (GE Healthcare, Piscataway, NJ) and a Molecular Devices Storm 840 phosphorimager in fluorescence detection mode. Band intensities were quantified using ImageQuant software and normalized for loading differences based on eIF4E content. Blots were stripped and reprobed with the corresponding total antibodies. Antibodies against pABL1, pCrkL, ABL1, STAT5, and eIF4E were obtained from Cell Signaling Technology, pSTAT5 (pTyr694) from BD Biosciences, and CrkL from Santa Cruz Biotechnology.
Allograft and retroviral BM transduction/transplantation mouse models of BCR-ABL1-induced leukemia
Retroviral stocks were generated and titered as previously described (Li et al., 1999). Ba/F3 cells (1×106) transformed to interleukin-3 independence by transduction with either BCR-ABL1native or BCR-ABL1T315I retrovirus were injected intravenously into syngeneic Balb/c recipients, as described (Ilaria and Van Etten, 1995). Beginning day 3 post-injection, mice were treated with imatinib (100 mg/kg in water twice daily via oral gavage) or with DCC-2036 (100 mg/kg in 0.5% CMC/1% Tween-80, once daily via oral gavage) or with vehicle (0.5% CMC/1% Tween-80) alone. For induction of CML-like leukemia, bone marrow (BM) from male Balb/c donor mice was harvested 4d after intravenous administration of 150 mg/kg 5-fluorouracil (5-FU), transduced with BCR-ABL1T315I retrovirus, and 5×105 cells injected intravenously into sublethally irradiated (400 cGy) Balb/c recipients. Beginning at d5 post-transplant, cohorts were treated once daily by oral gavage with vehicle alone, or DCC-2036 at 100 mg/kg. For induction of B-cell acute lymphoblastic leukemia, BM from donors not pretreated with 5-FU was transduced once with BCR-ABL1T315I retrovirus and 1×106 cells injected into sublethally irradiated Balb/c recipients. Beginning at d8 post-transplant, cohorts were treated twice daily by oral gavage with vehicle alone, with DCC-2036 at 60 mg/kg, with imatinib at 100 mg/kg (in water), or with dasatinib at 10 mg/kg (in 80 mM citric acid pH 3.1). All mouse experiments were approved by the Institutional Animal Use and Care Committee of Tufts Medical Center.
Pharmacodynamic analysis of BCR-ABL1T315I inhibition in mice by DCC-2036
Balb/c mice were inoculated with 1×106 Ba/F3 cells co-expressing BCR-ABL1T315I and GFP as described above. On day 9 post-injection, leukemia-bearing mice were given a single dose of 100 mg/kg DCC-2036 by oral gavage. At the indicated times post-dose, pairs of mice were sacrificed, BM and spleen harvested, and single cell suspensions prepared. As a positive control, cultured Ba/F3 cells expressing BCR-ABL1T315I were used, while the negative control was parental Ba/F3 cells starved of serum and IL-3 for 4h. For flow cytometric analysis of pSTAT5 inhibition, cells were fixed with 2% paraformaldehyde, permeabilized with ice-cold 95% methanol with vortexing, and stored at −20°C overnight. For analysis, samples were rehydrated with 3 volumes phosphate-buffered saline/0.5% BSA on ice for 1 hour, then incubated in FACS buffer containing Alexa 647-conjugated anti-phospho-STAT5 antibody (Cell Signaling Technology) in the dark for 30 min. Samples were washed once and analyzed on a Cyan flow cytometer, gating on the GFP+ population. Inhibition of pBCR-ABL1T315I was assessed by immunoblot analysis, as described above.
Inhibition of primary CML myeloid colony formation in vitro
Primary BM cells from a patient with newly diagnosed CML and from a normal individual, and PB mononuclear cells from a patient with CML-AP and imatinib intolerance, were plated (5×104 cells/plate) in triplicate in methylcellulose cultures (H5434, Stem Cell Technologies), either without or with kinase inhibitors at the indicated concentrations. Neither patient had detectable ABL1 kinase domain mutations. Colonies (CFU-GM) of >50 cell size were scored at d14.
Collection of patient samples
Clinical samples were obtained with informed consent and under the approval of the Institutional Review Board of Tufts Medical Center. Peripheral blood or bone marrow from patients or healthy subjects were separated by centrifugation in CPT tubes (Becton-Dickinson) for isolation of mononuclear cells.
Inhibition of BCR-ABL1 kinase activity in leukemic cells in vitro
Peripheral blood blasts from a patient with relapsed and refractory Ph+ B-ALL and detectable T315I mutation (allele frequency 40%) were incubated overnight (initial cell viability >90%) in IMDM supplemented with 100 μM 2-mecaptoethanol and 0.5% BSA, and either without drug or with imatinib (1 μM) or DCC-2036 (50, 200, and 1000 nM). After incubation, cells were lysed and protein extracts subjected to immunoblot analysis as described above. Peripheral blood mononuclear cells (7.5 ×105) from a patient with chronic phase CML and L298V mutation were incubated in varying concentrations of DCC-2036 or DMSO for 3h, followed by lysis and immunoblot analysis.
Pharmacodynamic analysis of pSTAT5 inhibition in CML cells in vivo
After obtaining informed consent, peripheral blood leukocytes from patients with relapsed/refractory CML were collected at the indicated times before and after the oral dose of DCC-2036 on day 1 or day 8 of therapy on a phase 1 dose-escalation clinical trial. Extracts from peripheral blood mononuclear cells were analyzed by immunoblotting.
Accession Numbers
Crystallographic coordinates have been deposited at the RCSB Protein Data Bank under accession numbers 3QRK for the compound 1:ABL1native complex (Figure S1F), 3QRI for the DCC-2036:ABL1native complex (Figure S2A), and 3QRJ for the DCC-2036:ABL1T315 complex (Figure 2 and Figure S2B).
Significance.
Kinase inhibitors have ushered in an era of targeted therapeutics in oncology. In chronic myeloid leukemia (CML), BCR-ABL1 tyrosine kinase inhibitors (TKIs) such as imatinib are now standard treatment, but acquired resistance due to kinase domain mutations is a significant problem, particularly the T315I “gatekeeper” mutation that causes resistance to all approved TKIs. DCC-2036 is an orally active TKI developed to exploit evolutionarily conserved amino acids that regulate switching of ABL1 from inactive to active conformations. DCC-2036 has efficacy against BCR-ABL1T315I in preclinical models and in primary patient cells, and represents a treatment option for patients who fail conventional TKIs. Rational design of “switch-control” inhibitors is a promising strategy for targeting the kinome in cancer and other diseases.
Supplementary Material
Acknowledgements
Supported in part from NIH grant CA090576 and a SCOR grant from the Leukemia and Lymphoma Society (R7059) to R.A.V.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat. Struct. Mol. Biol. 2008;15:1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- Deininger MW, Druker BJ. Specific targeted therapy of chronic myelogenous leukemia with imatinib. Pharmacol. Rev. 2003;55:401–423. doi: 10.1124/pr.55.3.4. [DOI] [PubMed] [Google Scholar]
- Donato NJ, Fang D, Sun H, Giannola D, Peterson LF, Talpaz M. Targets and effectors of the cellular response to aurora kinase inhibitor MK-0457 (VX-680) in imatinib sensitive and resistant chronic myelogenous leukemia. Biochem. Pharmacol. 2010;79:688–697. doi: 10.1016/j.bcp.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Guilhot F, O’Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MW, Silver RT, Goldman JM, Stone RM, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- Flynn DL, Petillo P. Modulation of Protein Functionalities. WO patent 2004/061084. 2004
- Gajiwala KS, Wu JC, Christensen J, Deshmukh GD, Diehl W, DiNitto JP, English JM, Greig MJ, He YA, Jacques SL, et al. KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients. Proc. Natl. Acad. Sci. USA. 2009;106:1542–1547. doi: 10.1073/pnas.0812413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles FJ, Cortes J, Jones D, Bergstrom D, Kantarjian H, Freedman SJ. MK-0457, a novel kinase inhibitor, is active in patients with chronic myeloid leukemia or acute lymphocytic leukemia with the T315I BCR-ABL mutation. Blood. 2007;109:500–502. doi: 10.1182/blood-2006-05-025049. [DOI] [PubMed] [Google Scholar]
- Gontarewicz A, Balabanov S, Keller G, Colombo R, Graziano A, Pesenti E, Benten D, Bokemeyer C, Fiedler W, Moll J, et al. Simultaneous targeting of Aurora kinases and Bcr-Abl kinase by the small molecule inhibitor PHA-739358 is effective against imatinib-resistant BCR-ABL mutations including T315I. Blood. 2008;111:4355–4364. doi: 10.1182/blood-2007-09-113175. [DOI] [PubMed] [Google Scholar]
- Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- Hu Y, Liu Y, Pelletier S, Buchdunger E, Warmuth M, Fabbro D, Hallek M, Van Etten RA, Li S. Requirement of Src kinases Lyn, Hck and Fgr for BCR-ABL1-induced B-lymphoblastic leukemia but not chronic myeloid leukemia. Nat. Genet. 2004;36:453–461. doi: 10.1038/ng1343. [DOI] [PubMed] [Google Scholar]
- Hu Y, Swerdlow S, Duffy TM, Weinmann R, Lee FY, Li S. Targeting multiple kinase pathways in leukemic progenitors and stem cells is essential for improved treatment of Ph+ leukemia in mice. Proc. Natl. Acad. Sci. USA. 2006;103:16870–16875. doi: 10.1073/pnas.0606509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Ilaria RL, Van Etten RA. The SH2 domain of P210BCR/ABL is not required for transformation of hematopoietic factor-dependent cells. Blood. 1995;86:3897–3904. [PubMed] [Google Scholar]
- Kantarjian H, Giles F, Wunderle L, Bhalla K, O’Brien S, Wassmann B, Tanaka C, Manley P, Rae P, Mietlowski W, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N. Engl. J. Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ilaria RL, Million RP, Daley GQ, Van Etten RA. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J. Exp. Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- Modugno M, Casale E, Soncini C, Rosettani P, Colombo R, Lupi R, Rusconi L, Fancelli D, Carpinelli P, Cameron AD, et al. Crystal structure of the T315I Abl mutant in complex with the aurora kinases inhibitor PHA-739358. Cancer Res. 2007;67:7987–7990. doi: 10.1158/0008-5472.CAN-07-1825. [DOI] [PubMed] [Google Scholar]
- Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- O’Brien J, Wilson I, Orton T, Pognan F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000;267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, Adrian LT, Zhou T, Huang WS, Xu Q, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttini M, Coluccia AM, Boschelli F, Cleris L, Marchesi E, Donella-Deana A, Ahmed S, Redaelli S, Piazza R, Magistroni V, et al. In vitro and in vivo activity of SKI-606, a novel Src-Abl inhibitor, against imatinib-resistant Bcr-Abl+ neoplastic cells. Cancer Res. 2006;66:11314–11322. doi: 10.1158/0008-5472.CAN-06-1199. [DOI] [PubMed] [Google Scholar]
- Roumiantsev S, Shah NP, Gorre ME, Nicoll J, Brasher BB, Sawyers CL, Van Etten RA. Clinical resistance to the kinase inhibitor STI-571 in CML by mutation of Tyr253 in the Abl kinase domain P-loop. Proc. Natl. Acad. Sci. USA. 2002;99:10700–10705. doi: 10.1073/pnas.162140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of Abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- Shah NP, Skaggs BJ, Branford S, Hughes TP, Nicoll JM, Paquette RL, Sawyers CL. Sequential ABL kinase inhibitor therapy selects for compound drug-resistant BCR-ABL mutations with altered oncogenic potency. J. Clin. Invest. 2007;117:2562–2569. doi: 10.1172/JCI30890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah NP, Tran C, Lee FY, Chen P, Norrris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- Sherbenou DW, Hantschel O, Kaupe I, Willis S, Bumm T, Turaga LP, Lange T, Dao KH, Press RD, Druker BJ, et al. BCR-ABL SH3-SH2 domain mutations in chronic myeloid leukemia patients on imatinib. Blood. 2010;116:3278–3285. doi: 10.1182/blood-2008-10-183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarski JS, Newitt JA, Chang CY, Cheng JD, Wittekind M, Kiefer SE, Kish K, Lee FY, Borzillerri R, Lombardo LJ, et al. The structure of Dasatinib (BMS-354825) bound to activated ABL kinase domain elucidates its inhibitory activity against imatinib-resistant ABL mutants. Cancer Res. 2006;66:5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- Wardelmann E, Thomas N, Merkelbach-Bruse S, Pauls K, Speidel N, Buttner R, Bihl H, Leutner CC, Heinicke T, Hohenberger P. Acquired resistance to imatinib in gastrointestinal stromal tumours caused by multiple KIT mutations. Lancet Oncol. 2005;6:249–251. doi: 10.1016/S1470-2045(05)70097-8. [DOI] [PubMed] [Google Scholar]
- Weisberg E, Manley P, Mestan J, Cowan-Jacob S, Ray A, Griffin JD. AMN107 (nilotinib): a novel and selective inhibitor of BCR-ABL. Br. J. Cancer. 2006;94:1765–1769. doi: 10.1038/sj.bjc.6603170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Hendrickson WA. Structural basis for activation of human lymphocyte kinase Lck upon tyrosine phosphorylation. Nature. 1996;384:484–489. doi: 10.1038/384484a0. [DOI] [PubMed] [Google Scholar]
- Young MA, Shah NP, Chao LH, Seeliger M, Milanov ZV, Biggs WH, 3rd, Treiber DK, Patel HK, Zarrinkar PP, Lockhart DJ, et al. Structure of the kinase domain of an imatinib-resistant Abl mutant in complex with the Aurora kinase inhibitor VX-680. Cancer Res. 2006;66:1007–1014. doi: 10.1158/0008-5472.CAN-05-2788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.