SUMMARY
We previously reported that Streptococcus cristatus, an oral commensal, was able to downregulate the interleukin-8 (IL-8) response to Fusobacterium nucleatum, a putative oral pathogen in oral epithelial cells. The aim of this study was to extend the understanding of how S. cristatus regulates cytokine expression in oral epithelial cells on a broad basis, and investigate whether the modulation of a Toll-like receptor (TLR) pathway was involved in this process. KB and TERT-2 cells were co-cultured with F. nucleatum and S. cristatus, either alone or in combination. Total RNA was extracted and pathway-specific focused microarrays were used to profile the transcriptional responses of various cytokine genes and those related to TLR-mediated signal transduction. Reverse transcription–polymerase chain reactions (RT-PCR) and protein assays were performed to confirm the microarray results for selected genes. We found that exposure to either S. cristatus or F. nucleatum alone led to distinct changes in cytokine expression patterns. Fusobacterium nucleatum induced a greater number of gene expression changes than S. cristatus (15% vs 4%, respectively). The presence of S. cristatus with F. nucleatum attenuated the expression of a number of inflammatory cytokines, and upregulated several anti-inflammatory mediators. The RT-PCR confirmed the messenger RNA attenuation of IL-1α, tumor necrosis factor-α and IL-6 by S. cristatus. Profiling of TLR-signaling-related genes revealed that S. cristatus most significantly impacted the downstream pathways, especially nuclear factor-κB, rather than altering TLRs and their adaptors and interacting proteins. Our data suggest that S. cristatus may attenuate the epithelial proinflammatory cytokine response to F. nucleatum by influencing pathways converging on nuclear factor-κB.
Keywords: cytokines, epithelial cells, Fusobacterium nucleatum, inflammatory response, nuclear factor-κB, Streptococcus cristatus
INTRODUCTION
Epithelial cells are the first line of host defense against physical, microbial and chemical insults that may cause local injury. Studies have shown that epithelial cells play an integral role in mucosal immune defense by sensing signals from the external environment, generating various molecules to affect growth, development and function of other cells, and maintaining the balance between health and disease (Kagnoff & Eckmann, 1997). A characteristic response of epithelial cells to bacterial stimuli is synthesis and release of cytokines, chemokines and other inflammatory mediators (Kagnoff & Eckmann, 1997). Putative oral pathogens, such as Fusobacterium nucleatum and Porphyromonas gingivalis, have been shown to induce a wide array of proinflammatory cytokines such as interleukin-8 (IL-8), monocyte chemoattractant protein-1, IL-1β, IL-6 and tumor necrosis factor-α (TNF-α) from epithelial cells (Han et al., 2000, 2003; Sandros et al., 2000; Kusumoto et al., 2004). There is emerging evidence that certain commensal oral streptococci behave in a manner different from that exhibited by the pathogenic species, in that they induce very low levels of proinflammatory cytokines (Cosseau et al., 2008; Zhang et al., 2008; Sliepen et al., 2009). However, the cytokine expression patterns following contact with oral commensal micro-organisms are less studied, and conflicting results have been reported (Vernier et al., 1996; Cosseau et al., 2008; Zhang et al., 2008).
The C-X-C family of cytokines, represented by IL-8, is generally produced at high levels by infected epithelial cells. They can initiate the mucosal influx of polymorphonuclear leukocytes to orchestrate innate mucosal inflammatory responses. The inflammatory process is critical to host defense, but deregulation of inflammatory cytokine production can cause epithelial damage. Excessive recruitment of polymorphonuclear leukocytes to the periodontium has been considered to contribute to the pathogenesis of periodontal disease (Van Dyke & Serhan, 2003). It therefore seems necessary to have regulatory mechanisms controlling innate immunity operating at the level of epithelial cells to prevent persistent inflammation. Recently, two strains of oral commensal streptococci, Streptococcus cristatus and Streptococcus salivarius, were found not to elicit proinflammatory responses by themselves, and were able to inhibit the IL-8 secretion induced by periodontal and respiratory pathogens, indicating a potential role of commensal bacteria in the maintenance of host–microbe homeostasis (Cosseau et al., 2008; Zhang et al., 2008).
Epithelial cells sense bacterial products through a variety of pattern recognition receptors, the best understood of which are Toll-like receptors (TLRs). Each TLR recognizes specific microbial components present on diverse microbes. For example, TLR2 recognizes peptidoglycan, in addition to the lipoproteins and lipopeptides of gram-positive bacteria, whereas TLR4 recognizes lipopolysaccharide from most gram-negative species (Takeuchi et al., 1999). It is known that TLR2 and TLR4 are present on or in oral epithelial cells (Uehara et al., 2001). In general, the cascade of events occurring following ligation of the different TLRs involves the activation of a common set of adapter proteins and protein kinases that results in the activation of nuclear factor-κB (NF-κB) to express cytokine genes relevant to inflammation (Kawai & Akira, 2005). Downregulation of TLR expression (Sweet et al., 2001) or upregulation of inhibitory regulators of TLR signaling (Burns et al., 2000) have been shown to be active in epithelial cells to mediate tolerance.
Given the amount of tissue or cells required for the variety of tests, traditional host–microbe interaction studies generally only focus on one or limited numbers of cytokines in a single assay. Transcriptional profiling using microarrays provides a way to monitor host responses on a broad scale (Mans et al., 2006). Studies of the host transcriptional responses to oral bacteria have begun but the focus in these few studies is mostly on putative oral pathogens (Handfield et al., 2005; Milward et al., 2007). Using pathway-specific microarray technology in a dual infection model, here we demonstrated that the oral commensal species S. cristatus induces distinct cytokine expression patterns as opposed to the putative oral pathogen F. nucleatum, and dampens F. nucleatum-induced proinflammatory epithelial responses. Our data did not show altered expression of TLRs or their adaptors and interacting proteins as a mechanism for attenuation by S. cristatus. In contrast to the strong activation of elements downstream of the NF-κB pathway by F. nucleatum, the minimal NF-κB activation by S. cristatus raises the possibility that the mechanism of cytokine inhibition in the presence of S. cristatus may be controlled at the level of the NF-κB transcription factor.
METHODS
Bacterial strains and culture conditions
Fusobacterium nucleatum (ATCC 10953) and S. cristatus CC5A (ATCC 49999) were routinely grown under anaerobic conditions (85% N2, 10% H2, 5% CO2) at 37°C. The F. nucleatum was cultured in trypticase soy broth (BBL, Becton Dickinson, Sparks, MD) supplemented with 1 g yeast extract, 5 mg hemin and 1 mg menadione per liter; S. cristatus was grown in Todd–Hewitt broth (BBL).
Epithelial cell cultures
The epithelial carcinoma cell line KB was kindly provided by Dr Mark Herzberg (University of Minnesota). The immortalized normal oral epithelial cell line OKF6/TERT-2 was obtained under a materials transfer agreement from Dr James Rheinwald (Brigham and Women’s Hospital, Boston, MA). All cell lines were maintained in 75-cm2 flasks (Corning, Corning, NY) in a humidified atmosphere of 5% CO2 at 37°C. KB cells were grown in minimal essential medium (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum, whereas OKF6/TERT-2 was cultured in keratinocyte serum-free medium (Invitrogen) supplemented with CaCl2 (0.4 mm), bovine pituitary extract (25 µg ml−1) and epidermal growth factor (0.2 ng ml−1). Twenty-four hours before infection, cells were trypsinized, centrifuged and seeded into triplicate wells in duplicate cell culture plates (Corning).
Bacterial infection procedure
Overnight broth cultures of bacteria were harvested by centrifugation at 5000 g for 20 min, washed twice with 1 × Dulbecco’s phosphate-buffered saline, and then re-suspended in serum-free cell culture media. Bacterial concentrations were adjusted to 108 colony-forming units per ml by measuring optical density at 620 nm for each experiment, and confirmed by colony counting on agar plates. Then, F. nucleatum and/or S. cristatus was added to cell monolayers at a multiplicity of infection of 100 for each species, and incubated for 2 h at 37°C in 5% CO2. Wells containing no bacteria served as negative controls. After stimulation, cell supernatants were collected for cytokine assays and cell monolayers were harvested for RNA extraction (see below). All assays were carried out in triplicate, and three independent experiments were performed. For all the experiments, the viability of infected cells was examined by trypan blue exclusion.
RNA extraction and array hybridization
Total RNA was extracted and purified from infected and control cells using the RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The integrity, purity and quantification of RNA were evaluated on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Pathway-specific, non-radioactive GEArray Q Series Human Inflammatory Cytokines & Receptors Gene Arrays (HS-015.2; SuperArray Inc., Bethesda, MD) containing 96 cytokine and receptor genes associated with the inflammatory response were used according to the manufacturer’s instructions. Briefly, 3 µg total RNA from KB cells was used to synthesize biotin-16-dUTP-labeled (Roche, Indianapolis, IN) complementary DNA (cDNA) probes with an AmpoLabeling-LPR kit (L-03; SuperArray). Labeled cDNA probes were denatured and hybridized to GEArray membranes. After washing and blocking, the membranes were incubated with alkaline phosphatase-conjugated streptavidin and CDP-Star substrate. Images of the arrays were recorded by exposure to X-ray films (Kodak, Rochester, NY), and scanned by an imaging densitometer (Bio-Rad Laboratories, Hercules, CA). Scanned images were digitized using Scanalyze software (free download from http://rana.stanford.edu/software), and data were analyzed on the GEArray analyzer program (SuperArray). The cDNA microarray experiments were carried out twice, using RNA isolated from two independent experiments.
Shortly after we finished the Inflammatory Cytokines & Receptors Array assays, SuperArray switched to the Oligo GEArrays® microarray system. To confirm the major cytokine responses obtained from KB cells and further analyse related signal transduction pathways, the Oligo GEArray® Human Toll-Like Receptor Signaling Pathway Microarray (EHS-018.2, SuperArray) was employed to assess the TLR-signaling-related gene expression in our dual infection model. OKF6/TERT2 cells were used because they had shown attenuation of the IL-8 response to F. nucleatum in a previous study (Zhang et al., 2008). The Oligo GEArray® Human Toll-Like Receptor Signaling Microarray profiles the expression of 113 genes related to TLR-mediated signal transduction, including members of the TLR family, key mediators including adaptors and proteins that interact with TLR and effectors, and members of the NF-κB, c-JUN N-terminal kinase (JNK)/p38, nuclear factor-IL-6 (NF/IL6), and interferon regulatory factor (IRF) signaling pathways downstream of TLR signaling, including major proinflammatory cytokines that were also components of the Inflammatory Cytokines & Receptors Array. According to the manufacturer’s instructions, 3 µg total RNA from infected and control TERT2 cells was used to synthesize biotin-16-UTP-labeled cRNA via an in vitro transcription labeling kit (TrueLabeling-AMP kit, GA-030, SuperArray). Labeled cRNA was then quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and hybridized to GEArray membranes. After washing and blocking, the membranes were incubated with alkaline phosphatase-conjugated streptavidin and CDP-Star substrate. Images of the arrays were recorded by exposure to X-ray films (Kodak), and scanned with an imaging densitometer (Bio-Rad). Scanned images were analysed using the specially designed web-based and completely integrated GEArray Expression Analysis Suite software (SuperArray). Oligo microarray assays were repeated three times, using RNA isolated from three independent experiments.
Analysis of microarray data
We first eliminated spots where the signals were so strong that they ‘bled’ into the background area, resulting in regions of high background values or non-specific background. According to the SuperArray software, a transcript’s spot is considered ‘absent’ if the average density of the spot is less than 1.5 × the mean value of the local backgrounds of the lower 75th percentile of all non-bleeding spots. Consequently all transcripts that were ‘absent’ on any array under any experimental condition were removed from further analysis. The hybridization signal of each gene present was normalized to that of the internal positive control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) observed on the same membrane for cytokine arrays, and to β2-microglobulin for TLR arrays. The expression levels of different genes after bacterial exposure were compared with those from uninfected controls, and expressed as relative ratios. Results are presented as the average of the number of replicates noted above for each array. A more than two-fold increase or less than 0.5-fold decrease in signal intensity between experimental groups (bacteria-infected) and the uninfected control group was considered biologically significant. Statistical comparison between conditions was performed by comparing fold differences averaged over all genes expressed for each condition, by using repeated measures analysis of variance (anova) followed by Duncan’s multiple range test. For TLR array data, relative gene expression levels were converted to logs for statistical analysis.
Qualitative reverse transcriptase–polymerase chain reaction
A 3-µg sample of RNA from the same batch as that used for microarray assays was reverse transcribed using a random primer (Promega, Madison, WI) and Moloney monkey leukemia virus reverse transcriptase (Promega) using conditions described by the manufacturer. One microliter of each cDNA sample was used for routine polymerase chain reaction (PCR). The primers for IL-1α, IL-6, IL-8, TNF-α and GAPDH are listed in Table 1. Standard PCR was performed in a total volume of 25 µl with the following components: 1 × PCR buffer, 0.25 mm dNTPs, 2 mm MgCl2, 0.5 µm of primers, and 1.5 U of platinum Taq polymerase (Invitrogen). The PCR amplicons were visualized in 1.5% agarose gels stained by ethidium bromide.
Table 1.
The sequences of primers used for reverse transcription–polymerase chain reaction
| Gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| IL1α | CAC TCC ATG AAG GCT GCA TGG | ACC CAG TAG TCT TGC TTT GTG G |
| IL6 | GTG TGA AAG CAG CAA AGA GG | TGG ACT GCA GGA ACT CCT T |
| IL8 | GAG ACA GCA GAG CAC ACA AGC | TTC TCA GCC CTC TTC AAA AAC T |
| TNF-α | CACCAGCTGGTTATCTCTCAGCTC | GGGACGTGGAGCTGGCCGAGGAG |
| GAPDH | GAC CCC TTC ATT GAC CTC AAC TAC | AGC CTT CTC CAT GGT GGT GAA GAC |
Luminex multiplex cytokine assays
Frozen cell supernatants were sent to the Cytokine Reference Laboratory at the University of Minnesota. Bead-based Fluorokine® MultiAnalyte Profiling Kits (R&D Systems, Minneapolis, MN) run on the Luminex® 100™ platform were used to measure the levels of selected cytokines including IL-1α, IL-8, interferon-γ (IFN-γ) and TNF-α simultaneously in a single sample. Cytokine concentrations were normalized to total protein levels as determined using the bicinchoninic acid protein assay (Pierce, Rockford, IL), and expressed as ng cytokine per mg total protein.
RESULTS
Differential cytokine expression profiles induced by F. nucleatum and S. cristatus
We first examined the effects of F. nucleatum and S. cristatus on 96 cytokine and receptor genes associated with an inflammatory response. After 2 h of exposure, F. nucleatum induced a potent proinflammatory cytokine response with 15% of all tested genes upregulated. No downregulated gene expression was found (Table 2). Among the 14 genes with high expression levels, IL1α was upregulated about 10-fold relative to the control. The relative messenger RNA (mRNA) expression of other upregulated genes varied from two- to approximately five-fold. Unfortunately, the manufacturer did not include the gene for IL-8 in this array. However, our previous work has shown IL8 to be significantly upregulated by F. nucleatum (Zhang et al., 2008).
Table 2.
Gene expression profile in KB cells induced by Fusobacterium nucleatum and/or Streptococcus cristatus, as determined by cytokine arrays1
| Gene2 | Description | Expression ratio3 | Fn/Fn+Sc ratio4 | ||
|---|---|---|---|---|---|
| Fn | Sc | Fn+Sc | |||
| IL1α | Interleukin 1, alpha | 9.92 | 1.41 | 2.53 | 3.92 |
| IL6 | Interleukin 6 | 1.16 | 0.65 | 0.38 | 3.08 |
| IL18 | Interleukin 18 | 0.77 | 0.44 | 0.40 | 1.90 |
| CCL16 | Chemokine ligand 16 | 3.84 | 1.31 | 2.65 | 1.45 |
| LTB | Lymphotoxin beta | 3.56 | 1.61 | 2.51 | 1.41 |
| TNFα | Tumor necrosis factor, | 2.32 | 0.85 | 1.64 | 1.41 |
| XCL2 | Chemokine ligand 2 | 2.56 | 1.06 | 1.84 | 1.40 |
| CCL19 | Chemokine ligand 19 | 2.44 | 1.28 | 1.84 | 1.32 |
| IL11 | Interleukin 11 | 2.17 | 1.21 | 1.65 | 1.31 |
| CXCL11 | Chemokine ligand 11 | 2.31 | 0.92 | 1.77 | 1.30 |
| CCL2 | Chemokine ligand 2 | 2.50 | 1.31 | 2.11 | 1.19 |
| CCL13 | Chemokine ligand 13 | 2.68 | 1.49 | 2.32 | 1.15 |
| CCL22 | Chemokine ligand 22 | 2.93 | 1.89 | 2.84 | 1.03 |
| CCL15 | Chemokine ligand 15 | 2.12 | 1.48 | 2.08 | 1.02 |
| CCL7 | Chemokine ligand 7 | 1.93 | 1.35 | 2.06 | 0.94 |
| IL10Rα | Interleukin 10 receptor, alpha | 4.82 | 2.06 | 6.64 | 0.73 |
| CCR9 | Chemokine receptor 9 | 1.47 | 1.59 | 2.04 | 0.72 |
| CCL1 | Chemokine ligand 1 | 1.50 | 2.79 | 2.52 | 0.60 |
| IL4 | Interleukin 4 | 2.75 | 4.89 | 5.86 | 0.47 |
| Overall mean5 | 2.83 ± 0.86 | 1.56 ± 0.97 | 2.40 ± 1.46 | ||
After background subtraction, the signal intensity of each gene present was normalized to that of GAPDH. The expression levels of different genes after bacterial exposure were compared with those from uninfected controls and expressed as relative ratios. Only the genes that had a fold change > 2-fold or < 0.5-fold in any condition (shown in bold) were included for analysis.
Genes whose expression levels were significantly modulated by any experimental treatment. They were sorted in a descending order according to the Fn/Fn+Sc ratio.
Values represent mean relative ratios from two independent experiments.
Fold changes in expression levels when comparing F. nucleatum and F. nucleatum plus S. cristatus. Fn, F. nucleatum; Sc, S. cristatus; Fn+Sc, F. nucleatum plus S. cristatus.
Values represent the mean ± SD for each ratio column. Statistical comparison showed that the overall expression level induced by S. cristatus was significantly lower than F. nucleatum (P = 0.0046), as well as lower than F. nucleatum plus S. cristatus (P = 0.04). However, the difference between F. nucleatum and F. nucleatum plus S. cristatus was not significant.
A different gene expression pattern was found in cells treated with S. cristatus alone (Table 2). Only 4% (four genes) were modulated, with upregulation of three genes and downregulation of one. Among those, three genes exhibited more than a two-fold increase compared with the control, including IL4, CCL1 and IL-10 receptor α (IL-10RA). One downregulated gene, IL18, was identified with a 2.3-fold decrease in mRNA expression. Duncan’s multiple range test confirmed that the overall expression patterns between F. nucleatum-infected versus S. cristatus-treated cells were significantly distinct (P = 0.0046), with significantly higher expression levels in F. nucleatum-infected cells.
Streptococcus cristatus attenuated epithelial cytokine production induced by F. nucleatum
Next, the cytokine expression pattern induced by the co-infection of F. nucleatum and S. cristatus was analysed. We observed at least two-fold changes in transcription of 14 genes following 2 h of combined infection, with upregulation of 12 genes and downregulation of two genes (Table 2). According to their immunoregulatory effects in inflammation, those 14 genes were classified into five groups. Genes in the first group included those for IL-1α, CCL16, lymphotoxin-β, TNF-α and XCL2, which are recognized as proinflammatory cytokines. Their upregulation in response to F. nucleatum was attenuated at least 1.4-fold by the presence of S. cristatus. Exposure to the two organisms together also downregulated the expression of IL18 and IL-6 signal transducer gp130 (IL6), which were not changed in the F. nucleatum-infected cells. The third group comprised anti-inflammatory genes including IL4 and IL10RA, whose expression remained upregulated in co-infection of KB cells with F. nucleatum and S. cristatus in an additive fashion, compared with the individual infection by either organism. The fourth group contained the chemokine genes CCL1 and CCL7 and chemokine receptor gene CCR9. In contrast to their response to F. nucleatum alone, their expression was upregulated with the addition of S. cristatus. The fifth group included chemokines CCL2, CCL13, CCL15 and CCL22, which had similar levels of upregulation compared with F. nucleatum alone.
The anova indicated that the overall cytokine expression patterns were significantly different between treatments (P = 0.01). Duncan’s multiple range test further revealed that F. nucleatum and co-infection induced significantly higher gene expression levels than S. cristatus alone (P = 0.0046 and P = 0.04, respectively). However, the overall difference between F. nucleatum alone and co-infection was not significant (P = 0.3).
Modulation of TLR signaling pathway by F. nucleatum and S. cristatus
The previously reported IL-8 attenuation (Zhang et al., 2008) and the cytokine array patterns pointed strongly toward S. cristatus having anti-inflammatory activities. Cytokine induction in response to micro-organisms is a recognized consequence of TLR activation in host cells (Akira & Takeda, 2004). To confirm the array results obtained from the KB cell model, and further explore the impacts of F. nucleatum and S. cristatus on TLR-signaling-related gene expression, we then profiled the expression of 113 genes related to TLR-mediated signal transduction in TERT2 cells. This array incorporated several cytokine genes, including those for IL-8 and IL-1β, which were not included in the cytokine array.
Among the 113 genes represented by the array, 7% (eight genes) were significantly modulated by S. cristatus alone. By contrast, exposure to F. nucleatum alone led to differential expression of 16% of genes (18 genes), and under combined infection this proportion was 12% (14 genes). These differentially expressed genes are listed in Table 3. As reported previously, no TLR expression was detected on the gene array, in contrast with their easy amplification by PCR (Durand et al., 2006; Decanis et al., 2009; Li & Dongari-Bagtzoglou, 2009). Such negative results have been attributed to the lower sensitivity resulting from the generation of the probes for the gene array (Luo et al., 2002; Fuke et al., 2004).
Table 3.
Gene expression profile in TERT2 cells induced by Fusobacterium nucleatum and/or Streptococcus cristatus, as determined by Toll-like receptor (TLR) arrays
| Gene1 | Pathway | Expression ratio2 | Fn/Fn+Sc ratio3 | ||
|---|---|---|---|---|---|
| Fn | Sc | Fn+Sc | |||
| GPC1 | Adaptors & TLR Interacting | 2.41 | 0.67 | 0.87 | 3.06 |
| HMGB1 | Adaptors & TLR Interacting | 2.25 | 1.40 | 1.18 | 3.04 |
| HSPA1A | Adaptors & TLR Interacting | 2.45 | 2.49 | 1.24 | 3.03 |
| HSPA4 | Adaptors & TLR Interacting | 0.72 | 0.84 | 0.36 | 3.00 |
| HSPD1 | Adaptors & TLR Interacting | 2.10 | 0.60 | 0.82 | 2.97 |
| IL1A | NF-κB Pathway | 11.16 | 2.20 | 3.83 | 2.95 |
| IL1B | NF-κB Pathway | 5.12 | 1.01 | 1.73 | 2.92 |
| IL8 | NF-κB Pathway | 77.36 | 1.61 | 38.46 | 2.91 |
| CSF2 | NF-κB Pathway | 17.96 | 1.77 | 8.54 | 2.89 |
| CSF3 | NF-κB Pathway | 4.13 | 0.79 | 1.36 | 2.81 |
| IRAK1 | Effectors | 2.20 | 0.61 | 0.73 | 2.76 |
| IRF1 | IRF Pathway | 19.87 | 2.26 | 6.62 | 2.56 |
| IRF3 | IRF Pathway | 2.59 | 1.08 | 1.08 | 2.44 |
| FADD | Effectors | 0.70 | 1.35 | 0.46 | 2.41 |
| JUN | JNK/p38 Pathway | 0.68 | 0.93 | 0.46 | 2.39 |
| MAPK9 | JNK/p38 Pathway | 2.50 | 2.02 | 0.86 | 2.25 |
| MYD88 | Adaptors & TLR Interacting | 1.14 | 1.35 | 0.50 | 2.10 |
| NFKB1 | NF-κB Pathway | 3.99 | 2.23 | 2.27 | 2.04 |
| NFKBIA | NF-κB Pathway | 20.71 | 4.24 | 6.97 | 2.04 |
| NFKBIB | NF-κB Pathway | 2.67 | 4.10 | 2.85 | 2.01 |
| RELA | NF-κB Pathway | 0.58 | 1.07 | 0.31 | 1.90 |
| RELB | NF-κB Pathway | 2.34 | 1.10 | 0.76 | 1.86 |
| FOS | JNK/p38 Pathway | 0.89 | 2.39 | 0.72 | 1.76 |
| TOLLIP | Adaptors & TLR Interacting | 0.20 | 0.56 | 0.10 | 1.54 |
| UBE2V1 | Effectors | 0.81 | 0.77 | 0.33 | 1.48 |
| TNFRSF | NF-κB Pathway | 1.32 | 0.78 | 0.47 | 1.23 |
| Overall mean4 | 7.26 ± 15.49 | 1.55 ± 0.98 | 3.23 ± 7.53 | ||
After background subtraction, the expression level of each gene present was normalized to β2-microglobulin. The expression levels of different genes after bacterial exposure were compared with those from uninfected controls and expressed as relative ratios. Only the genes which had a fold change > 2-fold or < 0.5-fold in any condition (shown in bold) were included for analysis.
Genes whose expression levels were significantly modulated by any experimental treatment. They were sorted in a descending order according to the Fn/Fn+Sc ratio.
Values represented mean relative ratios from three independent experiments. Statistical comparison was performed for log-transformed data, and showed that the overall expression level induced by F. nucleatum was significantly higher than S. cristatus (P = 0.00042), as well as F. nucleatum plus S. cristatus (P = 0.00007). However, the difference between S. cristatus and F. nucleatum plus S. cristatus was not significant.
Fold changes in expression levels when comparing F. nucleatum and F. nucleatum plus S. cristatus. Fn, F. nucleatum; Sc, S. cristatus; Fn+Sc, F. nucleatum plus S. cristatus.
Values represented mean ± SD for each ratio column. Statistical comparison was performed for log-transformed data, and showed that the overall expression level induced by F. nucleatum was significantly higher than S. cristatus (P = 0.00042), as well as F. nucleatum plus S. cristatus (P = 0.00007). However, the difference between S. cristatus and F. nucleatum plus S. cristatus was not significant.
Compared with control cells, S. cristatus-stimulated cells never showed any notable changes in gene expression. Among the eight upregulated genes, modest increases were only seen in the expression of two genes: NFKBIA (NF-κB inhibitor α, 4.24-fold) and NFKBIB (4.10-fold), and the remaining six genes only had small increases in expression (from 2.02-fold to 2.49-fold). In contrast, F. nucleatum-exposed cells demonstrated a significant and dramatic increase in the expression of five genes: CSF2 (colony stimulating factor 2, 17.96-fold), IL1α (11.16-fold), IL8 (77.36-fold), IRF1 (interferon regulatory factor 1, 19.87-fold) and NFKBIA (20.71-fold). Modest increases were seen in three genes: CSF3 (4.13-fold), IL1β (5.12-fold) and NFKB1 (3.99-fold), whereas the nine remaining genes only showed small increases (from 2.10-fold to 2.67-fold). Interestingly, we found that the addition of S. cristatus attenuated, to varying degrees (from 1.8-fold to 3-fold), the expression of almost every gene upregulated by F. nucleatum with the exception of NFKBIB, and ~ 60% of those genes were no longer upregulated.
Downregulation of TLR-related genes was not commonly seen with infection by either F. nucleatum or S. cristatus alone. Specifically, only one gene, TOLLIP (Toll interacting protein, 0.2-fold) was significantly downregulated in the presence of F. nucleatum. No gene was significantly downregulated by S. cristatus. In contrast, the combined infection of F. nucleatum and S. cristatus led to significant downregulation of seven genes, the same number as were upregulated.
When overall gene expression levels were compared by repeat-measures anova, there was a highly significant difference between groups (P = 0.00002). The F. nucleatum alone was significantly higher than S. cristatus alone (P = 0.0004), as well as than F. nucleatum plus S. cristatus (P = 0.00007). There was no significant difference found between S. cristatus alone and F. nucleatum plus S. cristatus (P = 0.29).
Immune function and pathway analysis of differentially expressed genes
The differentially expressed genes from TLR signaling arrays were further examined in the context of molecular function and pathway membership. In TERT2 cells, the majority of F. nucleatum upregulated genes were members of pathways downstream of TLR signaling (Table 3). Specifically, 10 genes were in the NF-κB pathway, two genes were in the IRF pathway and one gene was in the JNK/p38 pathway. The remaining five upregulated genes belong to the family of effectors, adaptors and TLR interacting proteins. TOLLIP, the only gene downregulated by F. nucleatum, is a negative regulator of TLR signaling.
Similar results were also found with S. cristatus alone, and the combined infection. Among the eight genes upregulated by S. cristatus, four were involved in the NF-κB pathway, two genes were in the JNK/p38 pathway, one was in the IRF pathway and one was in the family of adaptors and TLR-interacting proteins. On the other hand, the combined infection of F. nucleatum and S. cristatus modulated 10 genes involved in the NF-κB, IRF and JNK/p38 pathways, and four genes related to effectors, adaptors and TLR-interacting proteins.
Upon closer examination, we found that upregulation of key proinflammatory genes related to the NF-κB pathway (CSF, IL1α, IL1β and IL8) appeared as a major outcome following F. nucleatum stimulation of TERT2 cells, supporting the results obtained from inflammatory cytokine array analysis in KB cells. In the S. cristatus experiment, however, the expression levels of those genes did not change as much compared with control infected cells, except for a slight increase in IL1α level. The presence of S. cristatus clearly attenuated the degree of F. nucleatum-induced proinflammatory gene upregulation, which also confirmed the previous results.
Validation of array data of selected cytokine genes by RT-PCR and protein assays
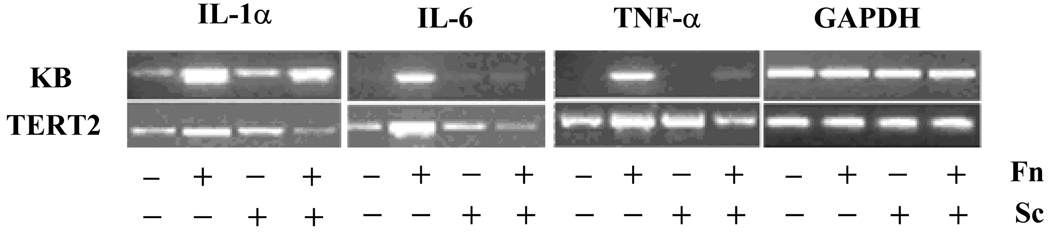
To confirm the results of the array gene expression analysis, we performed qualitative RT-PCR to examine the mRNA expression changes of selected proinflammatory genes (IL1α, IL6 and TNFα). As extensive assays had been performed in our previous study (Zhang et al., 2008), IL-8 was not included in the validation. The RT-PCR was performed on the same set of RNA extracts used for arrays and the lowest possible number of PCR cycles was used to detect small differences. The RT-PCR results are shown in Fig. 1.
Figure 1.
Qualitative reverse transcription–polymerase chain reaction (RT-PCR) analysis of selected cytokine gene expression in infected KB and TERT2 cells. KB and TERT2 cells were co-cultured for 2 h with medium; Fusobacterium nucleatum (Fn); Streptococcus cristatus (Sc); or S. cristatus and F. nucleatum. The same set of total RNA as that used for array analysis was analysed by RT-PCR. Representative gel images of triplicate analysis are shown.
In KB cells, we found that S. cristatus alone barely stimulated IL6 and TNFα expression, but was able to attenuate the robust expression of both cytokine genes induced by F. nucleatum. Constitutive expression of IL1α was detected in unstimulated KB cells and S. cristatus only induced a subtle increase of IL1α expression as opposed to the significant upregulation caused by F. nucleatum. In agreement with the array results, F. nucleatum-induced IL1α upregulation was inhibited in the presence of S. cristatus. The same expression patterns and S. cristatus-elicited attenuation effects were also observed in TERT2 cells for all three cytokines, although control TERT2 cells demonstrated a baseline constitutive expression of these genes.
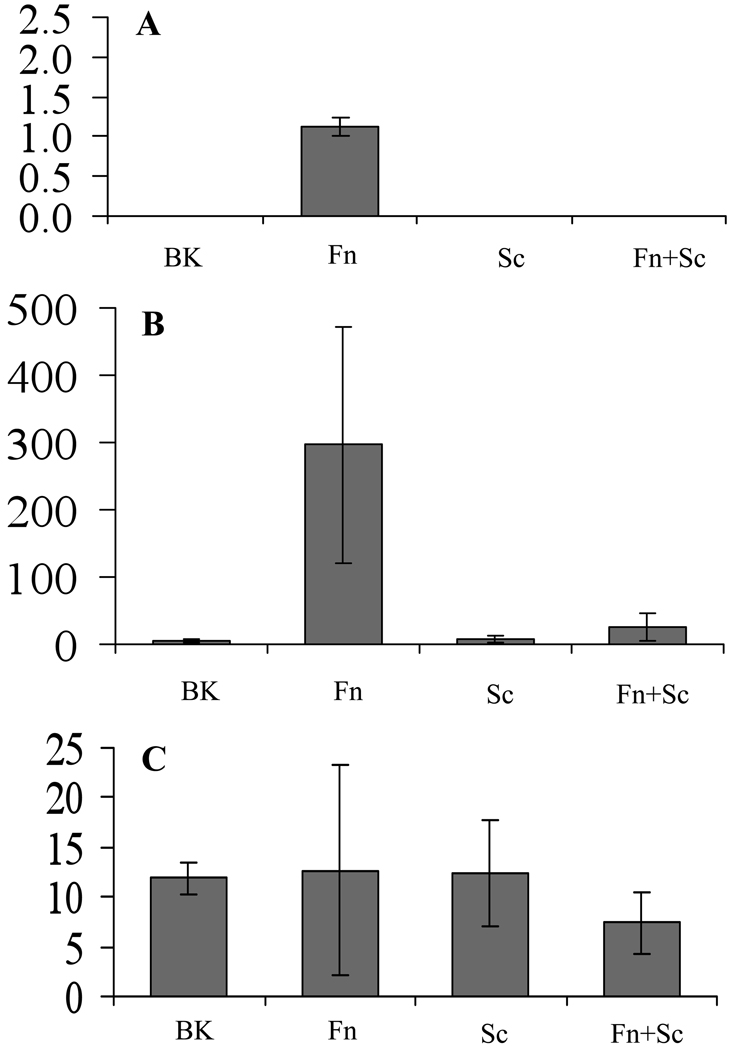
To validate and extend the results of mRNA analysis, we further determined the secreted protein levels of four selected genes from KB cells using the Luminex multiplex cytokine assay (Fig. 2). Those included modulated genes such as those for IL-1α and TNF-α, a non-modulated gene for IFN-γ, and IL-8 as a positive control. No TNF-α protein was detected in any sample, levels were below the limit of detection. Interleukin-1-α was only detected in F. nucleatum-infected cell supernatants, and fell below the limit of detection in all the other three conditions. Interleukin-8 was significantly elevated following stimulation with F. nucleatum, and this elevation was attenuated in the presence of S. cristatus. Secretion of IFN-γ was detected in all samples, and as with the array, no difference was found among the conditions. Results with the proteins selected for analysis were consistent with the data obtained by microarrays.
Figure 2.
Selected cytokine protein levels in KB cells following bacterial challenge. KB cells were co-cultured for 2 h with medium (Blank); Fusobacterium nucleatum (Fn); Streptococcus cristatus (Sc); or F. nucleatum plus S. cristatus (Fn + Sc). Cell supernatants were then collected, and levels of the indicated cytokines were analysed by multiplexed Luminex assay. Cytokine concentrations were normalized to total protein levels, and expressed as ng cytokine per mg total protein. (A) Interleukin-1α (IL-1α); (B) IL-8; (C) interferon-γ (IFN-γ). Statistical analysis confirmed that the significant upregulation of IL-8 by F. nucleatum (P < 0.01) was significantly attenuated to background level by the presence of S. cristatus (P < 0.01).
DISCUSSION
In an effort to characterize the repertoire of cytokine responses of oral epithelia, in this study we have applied pathway-specific microarrays to the mRNA expression analysis of 96 cytokine genes in epithelial cells exposed to two common organisms, S. cristatus, a commensal oral viridans streptocococcus, and F. nucleatum, a putative oral pathogen. We demonstrated that F. nucleatum and S. cristatus induce differential cytokine expression patterns in epithelial cells, and S. cristatus is able to inhibit inflammatory responses provoked by F. nucleatum. By further profiling the expression of 113 genes related to TLR-mediated signal transduction, we confirmed the cytokine-modulating effect of S. cristatus, and our results suggest that S. cristatus exerts its anti-inflammatory activities through influencing pathways converging on NF-κB.
It is well recognized that F. nucleatum can stimulate a wide array of cytokines from oral epithelial cells (Ji et al., 2007; Milward et al., 2007; Peyret-Lacombe et al., 2009). Our microarray data confirmed the upregulation of the genes for IL-1α, IL-1β, IL-8 and TNF-α, and identified a number of upregulated genes whose mRNA expression after F. nucleatum infection had not previously been investigated, including various CC, CXC and C chemokines such as CCL16, CCL2, CXCL11 and XCL2. These chemokines are chemoattractants of monocytes/macrophages, eosinophils and subpopulations of T cells. It is speculated that the expression of various chemokines by activated epithelial cells plays a role in orchestrating the initiation of mucosal inflammatory and immune responses in which a number of different cell types participate (Kagnoff & Eckmann, 1997). Other than the proinflammatory cytokines and chemokines, it is noteworthy that IL4 and IL10RA were also found to be up-regulated to various degrees in F. nucleatum-infected KB cells. Interleukin-4 is known to be a potent anti-inflammatory cytokine that acts by inhibiting the synthesis of proinflammatory cytokines such as IL-1, TNF-α, IL-6 and IL-8 by hematopoietic cells (D'Andrea et al., 1995) and keratinocytes (Raingeaud & Pierre, 2005). Although mainly produced by T helper type 2 lymphocytes and mast cells, expression of IL-4 was also observed in bile duct epithelial cells (Chedid et al., 1999), indicating that IL-4 might be able to exert its activity directly at epithelial surfaces. Interleukin-10 is another potent immunosuppressive cytokine that initiates a wide variety of anti-inflammatory activities in cells of different lineages when it binds to its cellular receptor complex. The IL-10 receptor complex is a heterodimer consisting of the specific IL-10R1 (alias of IL10RA) and the common IL-10R2, but only the IL-10R1 is essential for initial ligand binding and receptor signaling (Moore et al., 2001). Primarily expressed by hematopoietic cells such as B cells, T cells, natural killer cells, monocytes and macrophages, IL10R1 expression had also been described in keratinocytes (Michel et al., 1997) and colonic epithelium (Denning et al., 2000). The presence of IL-10RA in KB cells is an addition to this short list of non-hematopoietic cells. Upregulation of IL4 and IL10RA induced by F. nucleatum indicated that pathogenic bacteria not only activate a proinflammatory state of acute-phase response but also trigger a potentially compensatory antagonistic mechanism involving anti-inflammatory mediators such as IL-4 or IL-10 receptor.
The cytokine response to oral streptococci is less studied, and the results have been conflicting (Vernier et al., 1996; Scannapieco et al., 2001; Ji et al., 2007; Peyret-Lacombe et al., 2009). In contrast to F. nucleatum, here we demonstrated that S. cristatus did not provoke a strong host response in either KB or TERT2 cells, which was in agreement with most previous studies (Scannapieco et al., 2001; Hasegawa et al., 2007; Ji et al., 2007; Cosseau et al., 2008; Peyret-Lacombe et al., 2009). Only four genes were altered following S. cristatus infection. Among those was a proinflammatory cytokine, IL-18, which was down-regulated. It has been reported that oral epithelial cells can produce IL-18 (Rouabhia et al., 2002). Accumulated IL-18 is positively correlated to unresolved periodontal inflammation (Johnson & Serio, 2005). Therefore, downregulation of IL-18 may indicate an anti-inflammatory mechanism exerted by S. cristatus. Two of the three genes upregulated after exposure to S. cristatus were again those for IL-4 and IL-10R, which further suggested that S. cristatus induces an anti-inflammatory response in epithelial cells. The distinct transcriptional responses to F. nucleatum and S. cristatus are relevant to their biological behaviors, and provide experimental support for their distinct roles in health and disease. It appears that S. cristatus is a ‘true’ commensal that has developed a balanced relationship with the epithelial cells, whereas F. nucleatum is a potential threat to the host, and has to be monitored closely.
Communities in the oral cavity are polymicrobial, and host immune responses to complex microbial communities may differ greatly from those to single species. Oral bacteria implicated in periodontal diseases appear to have the capacity to downregulate the cytokine response of the host to other pathogenic species through various components, which has been related to periodontal pathogenesis (Huang et al., 2004a; Asai et al., 2005). Little is known about the interactions between oral commensal bacteria and pathogens in the host cytokine response. In this study we found that the presence of S. cristatus decreased the levels of epithelial proinflammatory cytokine production induced by F. nucleatum, and upregulated anti-inflammatory mediators such as IL-4 and IL-10RA. Such community-based responses are of clinical significance. In the oral cavity, oral mucosa coexists in intimate contact with a diverse bacterial flora including putative periodontal pathogens but dominated by commensal streptococci (Rudney et al., 2005). We previously demonstrated that S. cristatus could be transported into KB and TERT-2 epithelial cells by invasive F. nucleatum through a combination of coaggregation and invasion mechanisms (Edwards et al., 2006); however, the inhibitory effect of S. cristatus on F. nucleatum-induced IL-8 was independent of interspecies’ coaggregation and appeared to require bacterial contact with host cells (Zhang et al., 2008). The inhibition of an acute inflammatory response demonstrated by S. cristatus might represent an effective anti-inflammatory mechanism for polymicrobial flora to live in harmony with the host on epithelial surfaces. On the other hand, the mucosal hyporesponsiveness to invasive bacteria may also provide a protected environment for these microbes to later colonize the gingival crevice, and even remote locations.
Cytokine synthesis in response to microorganisms is largely a consequence of TLR activation (Kawai & Akira, 2009). Activation of TLR is essential for provoking the innate response and enhancing adaptive immunity against pathogens, but if it is left unchecked, the host would be overwhelmed by immune activation. Mechanisms exist to tightly regulate TLR signaling and functions at multiple levels (Liew et al., 2005). As no TLR genes were detected on our arrays, no conclusions could be drawn regarding the differential effects of bacteria on TLR expression in our model system. Further analysis of the TLR array data revealed that most of the F. nucleatum up-regulated genes identified in our study are NF-κB-dependent proinflammatory genes, which was generally in agreement with previous reports (Huang et al., 2004b; Milward et al., 2007). Typically pathogenic bacteria can prevent or limit the inflammatory response by targeting NF-κB (Schesser et al., 1998). Here we observed that almost all of F. nucleatum upregulated NF-κB-dependent genes were attenuated by the presence of S. cristatus (some of those genes were also upregulated by S. cristatus alone, but at much lower levels). In support of this observation, it was recently reported that nonpathogenic/commensal gut flora are also able to act on the NF-κB activation pathway, either inducing its precocious nuclear clearance (Kelly et al., 2004) or preventing the nuclear translocation of its active dimer (Neish et al., 2000), to attenuate proinflammatory cytokine expression induced by various inflammatory stimuli. Given the limited number of NF-κB pathway-related genes activated by S. cristatus alone, and its attenuation of most NF-κB-targeted genes induced by F. nucleatum, we speculate that S. cristatus may also exert its attenuating activity through interfering with the signal transduction pathways of the host cells.
Other pathways stimulated by either species alone and attenuated in combination were mitogen-activated protein kinases (MAPK) and interferon-regulatory factor (IRF) signal transduction. We have not been able to locate any previous reports of IRF pathway induction by F. nucleatum or S. cristatus, so the role of the IRF pathway remains elusive. Considerably more information is available for the MAPK pathway. For example, MAPK–JNK/p38 pathways, rather than NF-κB, were involved in the induction of an antimicrobial peptide human β-defensin-2 in epithelial cells by F. nucleatum (Krisanaprakornkit et al., 2000; Chung & Dale, 2008). Largely mediated by NF-κB activation, IL-8 upregulation by F. nucleatum also involved MAPK p38 and MAPK kinase/extracellular regulated kinase pathways (Huang et al., 2004b). As cross-talk between various pathways in mammalian cells can regulate the amplitude of cytokine responses in inflammation, differences in the cytokine expression profiles between F. nucleatum alone and F. nucleatum with the addition of S. cristatus may be related to differential regulation of individual components of various pathways. The upregulation of c-Fos mRNA by S. cristatus, but not by F. nucleatum, was of interest. c-Fos is a transcription factor that, together with c-Jun, composes activator protein-1 (AP-1). It can be induced by a variety of stimuli in diverse cell types including keratinocytes. It was reported that c-Fos may function as an anti-inflammatory transcription factor to suppress systemic inflammatory response to endotoxin through inhibiting NF-κB activity (Ray et al., 2006). In Drosophila, the lipopolysaccharide response of NF-κB target genes can be downregulated by AP-1 (Kim et al., 2005). Interestingly, MAPK signaling pathways regulate AP-1 activity by increasing transcription and by phosphorylation of AP-1. Based on these findings, it is tempting to speculate that the addition of S. cristatus induces a negative cross-talk between the c-Fos/AP-1 and NF-κB signaling modules, and consequently, the amplitude of cytokine responses in inflammation is dampened. Further study is warranted for confirmation.
In conclusion, the data presented here extend our previous study on the effects of S. cristatus on epithelial responses to F. nucleatum, and highlight many previously unreported findings. It is clear from our results that oral pathogens and commensals can induce distinct cytokine expression patterns in oral epithelial cells, and S. cristatus is able to attenuate the acute inflammatory responses to F. nucleatum, probably by influencing pathways converging on NF-κB.
ACKNOWLEDGEMENTS
This work was supported by Public Health Service grant 1 R01 DE 14214 from the National Institute for Dental and Craniofacial Research, Bethesda, MD, USA.
REFERENCES
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Asai Y, Ohyama Y, Taiji Y, et al. Treponema medium glycoconjugate inhibits activation of human gingival fibroblasts stimulated with phenol-water extracts of periodontopathic bacteria. J Dent Res. 2005;84:456–461. doi: 10.1177/154405910508400511. [DOI] [PubMed] [Google Scholar]
- Burns K, Clatworthy J, Martin L, et al. Tollip, a new component of the IL-1RI pathway, links IRAK to the IL-1 receptor. Nat Cell Biol. 2000;2:346–351. doi: 10.1038/35014038. [DOI] [PubMed] [Google Scholar]
- Chedid A, Mendenhall C, Moritz T. The antigenic heterogeneity of the bile duct epithelium in alcoholic liver disease. VA Cooperative Study Group 275. Arch Pathol Lab Med. 1999;123:411–414. doi: 10.5858/1999-123-0411-TAHOTB. [DOI] [PubMed] [Google Scholar]
- Chung WO, Dale BA. Differential utilization of nuclear factor-kappaB signaling pathways for gingival epithelial cell responses to oral commensal and pathogenic bacteria. Oral Microbiol Immunol. 2008;23:119–126. doi: 10.1111/j.1399-302X.2007.00398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosseau C, Devine DA, Dullaghan E, et al. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect Immun. 2008;76:4163–4175. doi: 10.1128/IAI.00188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea A, Ma X, Aste-Amezaga M, Paganin C, Trinchieri G. Stimulatory and inhibitory effects of interleukin (IL)-4 and IL-13 on the production of cytokines by human peripheral blood mononuclear cells: priming for IL-12 and tumor necrosis factor alpha production. J Exp Med. 1995;181:537–546. doi: 10.1084/jem.181.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decanis N, Savignac K, Rouabhia M. Farnesol promotes epithelial cell defense against Candida albicans through Toll-like receptor 2 expression, interleukin-6 and human beta-defensin 2 production. Cytokine. 2009;45:132–140. doi: 10.1016/j.cyto.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Denning TL, Campbell NA, Song F, et al. Expression of IL-10 receptors on epithelial cells from the murine small and large intestine. Int Immunol. 2000;12:133–139. doi: 10.1093/intimm/12.2.133. [DOI] [PubMed] [Google Scholar]
- Durand SH, Flacher V, Romeas A, et al. Lipoteichoic acid increases TLR and functional chemokine expression while reducing dentin formation in in vitro differentiated human odontoblasts. J Immunol. 2006;176:2880–2887. doi: 10.4049/jimmunol.176.5.2880. [DOI] [PubMed] [Google Scholar]
- Edwards AM, Grossman TJ, Rudney JD. Fusobacterium nucleatum transports noninvasive Streptococcus cristatus into human epithelial cells. Infect Immun. 2006;74(1):654–662. doi: 10.1128/IAI.74.1.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieling J, Mulder J, Hendriks T, Curfs J, van der Linden C, Sauerwein R. Differential induction of pro- and anti-inflammatory cytokines in whole blood by bacteria: effects of antibiotic treatment. Antimicrob Agents Chemother. 1997;41:1439–1443. doi: 10.1128/aac.41.7.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuke S, Betsuyaku T, Nasuhara Y, Morikawa T, Katoh H, Nishimura M. Chemokines in bronchiolar epithelium in the development of chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2004;31:405–412. doi: 10.1165/rcmb.2004-0131OC. [DOI] [PubMed] [Google Scholar]
- Han D, Huang G, Lin L, Warner N, Gim J, Jewett A. Expression of MHC Class II, CD70, CD80, CD86 and pro-inflammatory cytokines is differentially regulated in oral epithelial cells following bacterial challenge. Oral Microbiol Immunol. 2003;18:350–358. doi: 10.1046/j.0902-0055.2003.00094.x. [DOI] [PubMed] [Google Scholar]
- Han Y, Shi W, Huang G, et al. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68:3140–3146. doi: 10.1128/iai.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handfield M, Mans JJ, Zheng G, et al. Distinct transcriptional profiles characterize oral epithelium–microbiota interactions. Cell Microbiol. 2005;7:811–823. doi: 10.1111/j.1462-5822.2005.00513.x. [DOI] [PubMed] [Google Scholar]
- Huang G, Zhang H, Dang H, Haake S. Differential regulation of cytokine genes in gingival epithelial cells challenged by Fusobacterium nucleatum and Porphyromonas gingivalis. Microb Pathog. 2004a;37:303–312. doi: 10.1016/j.micpath.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Huang GT, Zhang HB, Dang HN, Haake SK. Differential regulation of cytokine genes in gingival epithelial cells challenged by Fusobacterium nucleatum and Porphyromonas gingivalis. Microb Pathog. 2004b;37:303–312. doi: 10.1016/j.micpath.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Ji S, Kim Y, Min BM, Han SH, Choi Y. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria. J Periodontal Res. 2007;42:503–510. doi: 10.1111/j.1600-0765.2007.00974.x. [DOI] [PubMed] [Google Scholar]
- Johnson R, Serio F. Interleukin-18 concentrations and the pathogenesis of periodontal disease. J Periodontol. 2005;76:785–790. doi: 10.1902/jop.2005.76.5.785. [DOI] [PubMed] [Google Scholar]
- Kagnoff MF, Eckmann L. Epithelial cells as sensors for microbial infection. J Clin Invest. 1997;100:6–10. doi: 10.1172/JCI119522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Pathogen recognition with Toll-like receptors. Curr Opin Immunol. 2005;17:338–344. doi: 10.1016/j.coi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D, Campbell J, King T, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- Kim T, Yoon J, Cho H, et al. Downregulation of lipopolysaccharide response in Drosophila by negative crosstalk between the AP1 and NF-kappaB signaling modules. Nat Immunol. 2005;6:211–218. doi: 10.1038/ni1159. [DOI] [PubMed] [Google Scholar]
- Krisanaprakornkit S, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun. 2000;68:2907–2915. doi: 10.1128/iai.68.5.2907-2915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto Y, Hirano H, Saitoh K, et al. Human gingival epithelial cells produce chemotactic factors interleukin-8 and monocyte chemoattractant protein-1 after stimulation with Porphyromonas gingivalis via toll-like receptor 2. J Periodontol. 2004;75:370–379. doi: 10.1902/jop.2004.75.3.370. [DOI] [PubMed] [Google Scholar]
- Li L, Dongari-Bagtzoglou A. Epithelial GM-CSF induction by Candida glabrata. J Dent Res. 2009;88:746–751. doi: 10.1177/0022034509341266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- Luo Y, Cai J, Liu Y, et al. Microarray analysis of selected genes in neural stem and progenitor cells. J Neurochem. 2002;83:1481–1497. doi: 10.1046/j.1471-4159.2002.01260.x. [DOI] [PubMed] [Google Scholar]
- Mans JJ, Lamont RJ, Handfield M. Microarray analysis of human epithelial cell responses to bacterial interaction. Infect Disord Drug Targets. 2006;6:299–309. doi: 10.2174/187152606778249926. [DOI] [PubMed] [Google Scholar]
- Michel G, Mirmohammadsadegh A, Olasz E, et al. Demonstration and functional analysis of IL-10 receptors in human epidermal cells: decreased expression in psoriatic skin, down-modulation by IL-8, and up-regulation by an antipsoriatic glucocorticosteroid in normal cultured keratinocytes. J Immunol. 1997;159:6291–6297. [PubMed] [Google Scholar]
- Milward MR, Chapple IL, Wright HJ, Millard JL, Matthews JB, Cooper PR. Differential activation of NF-kappaB and gene expression in oral epithelial cells by periodontal pathogens. Clin Exp Immunol. 2007;148:307–324. doi: 10.1111/j.1365-2249.2007.03342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KW, de Waal Malefyt R, Coffman R, O'Garra A. Interleukin-10 andtheinterleukin-10receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- Neish A, Gewirtz A, Zeng H, et al. Prokaryotic regulation of epithelial responses by inhibition of IkappaB-alphaubiquitination. Science. 2000;289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- Peyret-Lacombe A, Brunel G, Watts M, Charveron M, Duplan H. TLR2 sensing of F. nucleatum and S. sanguinis distinctly triggered gingival innate response. Cytokine. 2009;46:201–210. doi: 10.1016/j.cyto.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Raingeaud J, Pierre J. Interleukin-4 downregulates TNFalpha-induced IL-8 production in keratinocytes. FEBS Lett. 2005;579:3953–3959. doi: 10.1016/j.febslet.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Ray N, Kuwahara M, Takada Y, et al. c-Fos suppresses systemic inflammatory response to endotoxin. Int Immunol. 2006;18:671–677. doi: 10.1093/intimm/dxl004. [DOI] [PubMed] [Google Scholar]
- Rouabhia M, Ross G, Page N, Chakir J. Interleukin-18 and gamma interferon production by oral epithelial cells in response to exposure to Candida albicans or lipopolysaccharide stimulation. Infect Immun. 2002;70:7073–7080. doi: 10.1128/IAI.70.12.7073-7080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudney JD, Chen R, Zhang G. Streptococci dominate the diverse flora within buccal cells. J Dent Res. 2005;84:1165–1171. doi: 10.1177/154405910508401214. [DOI] [PubMed] [Google Scholar]
- Sandros J, Karlsson C, Lappin D, Madianos P, Kinane D, Papapanou P. Cytokine responses of oral epithelial cells to Porphyromonas gingivalis infection. J Dent Res. 2000;79:1808–1814. doi: 10.1177/00220345000790101301. [DOI] [PubMed] [Google Scholar]
- Scannapieco F, Wang B, Shiau H. Oral bacteria and respiratory infection: effects on respiratory pathogen adhesion and epithelial cell proinflammatory cytokine production. Ann Periodontol. 2001;6:78–86. doi: 10.1902/annals.2001.6.1.78. [DOI] [PubMed] [Google Scholar]
- Schesser K, Spiik A, Dukuzumuremyi J, Neurath M, Pettersson S, Wolf-Watz H. The yopJ locus is required for Yersinia-mediated inhibition of NF-kappaB activation and cytokine expression: YopJ contains a eukaryotic SH2-like domain that is essential for its repressive activity. Mol Microbiol. 1998;28:1067–1079. doi: 10.1046/j.1365-2958.1998.00851.x. [DOI] [PubMed] [Google Scholar]
- Sliepen I, Van Damme J, Van Essche M, Loozen G, Quirynen M, Teughels W. Microbial interactions influence inflammatory host cell responses. J Dent Res. 2009;88:1026–1030. doi: 10.1177/0022034509347296. [DOI] [PubMed] [Google Scholar]
- Sweet MJ, Leung BP, Kang D, et al. A novel pathway regulating lipopolysaccharide-induced shock by ST2/T1 via inhibition of Toll-like receptor 4 expression. J Immunol. 2001;166:6633–6639. doi: 10.4049/jimmunol.166.11.6633. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- Uehara A, Sugawara S, Tamai R, Takada H. Contrasting responses of human gingival and colonic epithelial cells to lipopolysaccharides, lipoteichoic acids and peptidoglycans in the presence of soluble CD14. Med Microbiol Immunol (Berl) 2001;189:185–192. doi: 10.1007/s004300100063. [DOI] [PubMed] [Google Scholar]
- Van Dyke TE, Serhan CN. Resolution of inflammation: a new paradigm for the pathogenesis of periodontal diseases. J Dent Res. 2003;82:82–90. doi: 10.1177/154405910308200202. [DOI] [PubMed] [Google Scholar]
- Vernier A, Diab M, Soell M, et al. Cytokine production by human epithelial and endothelial cells following exposure to oral viridans streptococci involves lectin interactions between bacteria and cell surface receptors. Infect Immun. 1996;64:3016–3022. doi: 10.1128/iai.64.8.3016-3022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Chen R, Rudney JD. Streptococcus cristatus attenuates Fusobacterium nucleatum-induced interleukin-8 expression in oral epithelial cells. J Periodontal Res. 2008;43:408–416. doi: 10.1111/j.1600-0765.2007.01057.x. [DOI] [PubMed] [Google Scholar]