Abstract
Overactive TH17 responses are tightly linked to the development of autoimmunity, yet the factors that negatively regulate differentiation of this lineage remain unknown. Here, we report that T-bet suppresses the development of the TH17 cell lineage by inhibiting the transcription of Rorc. T-bet interacts with the transcription factor Runx1 and this interaction blocks Runx1-mediated transactivation of Rorc. T-bet residue Tyr304 is required for T-bet-Runx1 complex formation, for blocking Runx1 activity and for inhibiting the TH17 differentiation program. These data reinforce the concept of master regulators that shape immune responses by simultaneously activating one genetic program while silencing the activity of competing regulators in a common progenitor cell.
The signals received during an infection trigger a strong adaptive immune response tailored to combat a particular class of pathogen. In the presence of cytokines produced by cells of innate immunity, naive CD4+ T cells differentiate into a TH cell subset with distinct functions and cytokine profile. Until recently two main TH subsets were described: TH1 and TH21. TH1 cells, which predominantly secrete interferon-γ (IFN-γ), are essential for immunity against intracellular microorganisms, while TH2 cells, which secrete IL-4, IL-5 and IL-13, are important for protection against parasites and extracellular pathogens. More recently, a third subset of TH cells, called TH17, has been described2–7. TH17 cells produce IL-17A, IL-17F, IL-21 and IL-22, which protect the host against bacterial and fungal infections encountered at mucosal surfaces8. In mice, TH17 differentiation is initiated by the combination of transforming growth factor-β (TGF-β) and IL-6 or IL-21, which induces the expression of retinoic acid receptor-related orphan nuclear receptor, RORγt, and IL-23R7, 9–12. This acquisition of responsiveness to IL-23 is necessary for the terminal differentiation of the TH17 cell lineage and for the maintenance of TH17 functions in vivo13.
Generally, lineage-specific transcription factors and cytokines can inhibit the differentiation of other TH subsets. T-bet suppresses the generation of TH2 cells by blocking the expression of the TH2 polarizing cytokine IL-4, and by interfering with the activity of the TH2-cell–specific transcription factor, GATA314, 15. Results from several studies have indicated that TH17 responses are stronger in T-bet-deficient animals, although the mechanism underlying this phenomenon was not described16–19. This raises the question of whether, analogous to its role in inhibiting the TH2 pathway, T-bet also actively suppresses TH17 differentiation. Furthermore, several studies have reported that the re-programming of committed TH17 and TH2 cells into effector cells with TH17-TH1 and TH2-TH1 phenotypes is driven by T-bet in response to inflammatory cytokines such as IL-12 and interferons20, 21.
In this study we sought to determine whether T-bet plays a regulatory role in the development of the TH17 lineage. We investigated TH17 differentiationin Tbx21−/− and WT CD4+ T cells in vitro and in vivo during experimental autoimmune encephalomyelitis (EAE). Here we report that T-bet has a negative effect on the expression of the TH17-cell-specific transcription factor RORγt and TH17 cytokine genes. Ectopic T-bet expression in naïve TH precursor cells or in committed TH17 cells was sufficient to repress the expression of RORγt and TH17 signature cytokine genes under TH17 polarizing conditions. Mechanistic studies revealed that interaction of T-bet with Runx1 via the T-bet residue Tyr304 is critical for blocking Runx1-mediated transactivation of the Rorc promoter and for inhibiting TH17 lineage commitment.
RESULTS
T-bet deficiency promotes IL-17A production in vitro
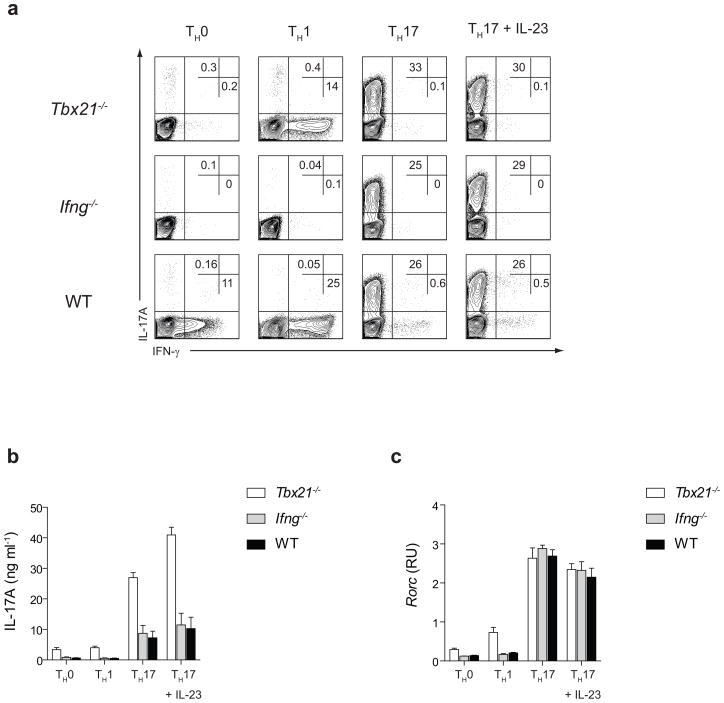
T-bet, encoded by the Tbx21 gene, is a transcriptional activator of IFN-γ and the key regulator of the TH1 differentiation program22. In addition to promoting differentiation of naive CD4+ T cells into the TH1 subset, T-bet actively suppresses the development of the TH2 lineage 14, 15. To investigate whether T-bet expression has a similar antagonistic effect on the development of IL-17A producing TH cells, we cultured Tbx21−/− and wild-type (WT) CD4+ T cells under non-skewing conditions or differentiated them into TH1 cells or TH17 cells which were grown in the absence or presence of IL-23 (TH17 and TH17+IL-23 conditions). Since IFN-γ has a negative effect on the polarization of TH17 cells and Tbx21−/− T cells produce significantly less IFN-γ than WT CD4+ T cells, Ifng−/− TH cells were also tested to delineate T-bet- versus IFN-γ-mediated effects on TH17 development. After five days of in vitro differentiation, Tbx21−/−, Ifng−/− and WT TH0, TH1, TH17 and TH17+IL-23 cells were briefly stimulated with phorbol myristate acetate and ionomycin (PMA+I). We observed a higher percentage of IL-17A producing cells in T-bet-deficient TH0 and TH1 cultures when compared to Ifng−/− and WT cultures (Fig. 1a). Although a similar percentage of IL-17A producing cells was detected under TH17 polarizing conditions, the amount of IL-17A secreted by Tbx21−/− TH cells was higher than that secreted by Ifng−/− and WT TH cells under all differentiating conditions (Fig. 1b).We did not observe substantial differences in the amount of Rorc mRNA expression amongst different TH subsets at 24 hours after activation (data not shown). However, the enhanced IL-17A production by Tbx21−/− TH0 and TH1 cultures correlated with a 2-fold increase in the expression of Rorc mRNA after 5 days of culture. In contrast, Tbx21−/−, Ifng−/− and WT TH17 cells expressed similar levels of Rorc mRNA (Fig. 1c). These results show that T-bet deficiency promotes development of IL-17A producing cells under all polarizing conditions independently of IFN-γ and suggest that T-bet-mediated effects on the generation of IL-17A producing cells in vitro may be through the transcriptional regulation of Rorc and/or Il17a genes in TH0-TH1 and TH17 cells, respectively.
Figure 1.
T-bet deficiency promotes IL-17A production in vitro independently of IFN-γ. (a) Flow cytometry analyzing the IL-17A and IFN-γ production following 4 h stimulation with phorbol ester + ionomycin (PMA+I). Tbx21−/−, Ifng−/− and wild-type CD4+ T cells were cultured in the presence of IL-2 (TH0 cells), IL-2 + IL-12 + anti-IL-4 antibody (TH1 cells), TGF-β + IL-6 + anti-IL-4 + anti-IFN-γ antibodies (TH17 cells) or TGF-β + IL-6 + anti-IL-4 + anti-IFN-γ antibodies followed by IL-23 treatment (TH17+IL-23 cells). Following five days of in vitro differentiation, TH cells were stimulated with PMA+I for 4 hours prior to intracellular cytokine staining with anti-IL-17A and anti-IFN-γ antibodies. (b) The amount of IL-17A in the supernatants of PMA+I stimulated TH0, TH1 and TH17 and TH17+IL-23 cells was determined by ELISA. The data are representative of four independent experiments (a–b). (c) Rorc mRNA expression in Tbx21−/− and WT TH0, TH1 and TH17 and TH17+IL-23 cells was determined by RT-PCR after 4 h stimulation with PMA+I. The data represent mean ± s.e.m. of two independent experiments.
TH17 responses in Tbx21−/− and WT mice during EAE
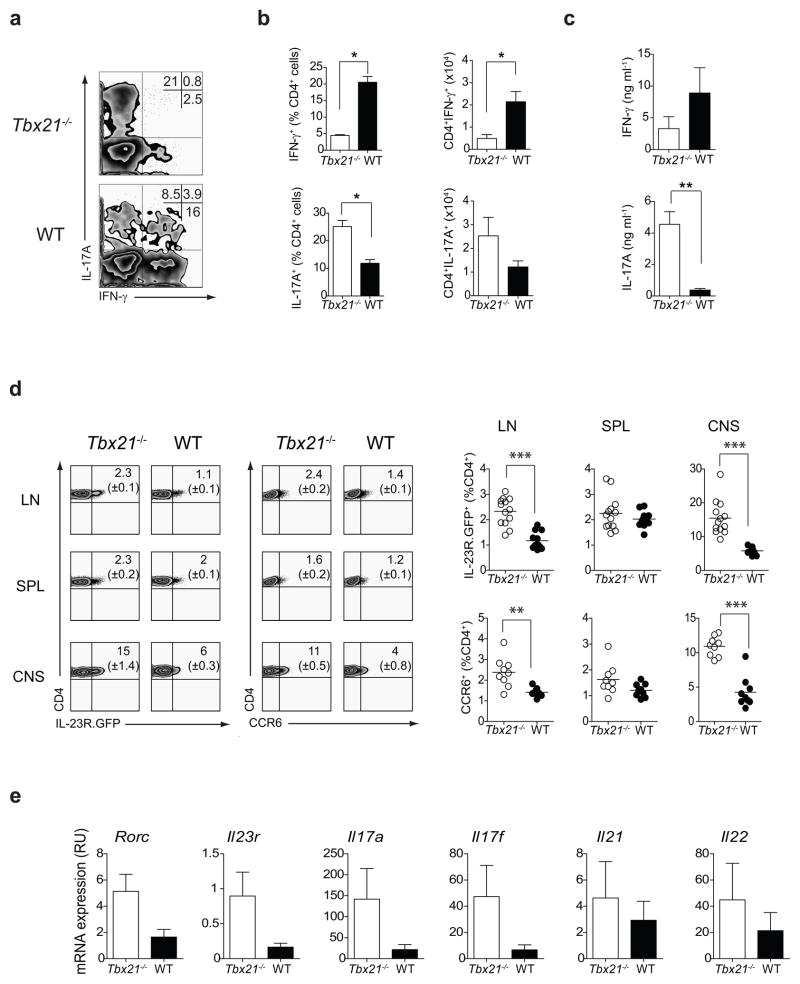
Tbx21−/− mice are protected from developing EAE23. At the time when the results of this study were reported, TH17 cells were yet to be discovered, and the resistance of Tbx21−/− mice to central nervous system (CNS)-specific autoimmune attack was ascribed to the polarization shift of CD4+ T cells from a pathogenic TH1 to a protective TH2 response23. Considering the propensity of T-bet-deficient CD4+ T cells to develop into IL-17A-producing cells in vitro, we investigated whether Tbx21−/− mice generated TH17 responses during EAE, the pathology of which is widely accepted to be dependent on TH17 cells. To determine the types of cytokines produced by CNS-infiltrating CD4+ T cells, we performed intracellular cytokine staining on mononuclear cells isolated from the CNS of Tbx21−/− and WT mice during the peak of disease (day 17 post-immunization). In WT mice, three different cytokine producing populations entered the CNS: those that produced IFN-γ alone (the majority of CD4+ T cells), those that produced only IL-17A, and those that produced both cytokines (Fig. 2a). In contrast, in the CNS of Tbx21−/− mice, IL-17A producing CD4+ T cells represented the majority of cytokine producing cells at day 17 post-immunization (Fig. 2a). Consistent with the role of T-bet in controlling expression of the IFN-γ gene, there was a deficiency in IFN-γ-producing CD4+ T cells in the CNS of Tbx21−/− mice (Fig. 2a). Collectively, there was a shift in the TH1-TH17 balance in the CNS of Tbx21−/− mice during EAE characterized by the preferential recruitment of TH17 cells and significant reduction in the frequency and absolute numbers of IFN-γ-producing CD4+ T cells (Fig. 2b). Moreover, CD4+ T cells isolated from the CNS of Tbx21−/− mice secreted significantly higher levels of IL-17A than WT CD4+ T cells at day 17 post-immunization (Fig. 2c). Thus, there is a strong recruitment of IL-17A-secreting TH17 cells into the CNS of Tbx21−/− mice.
Figure 2.
T-bet deficiency promotes TH17 responses in the CNS during EAE. (a) IL-17A and IFN-γ production by CNS-infiltrating CD4+ lymphocytes on day 17 post-immunization with MOG35–55 plus CFA. Numbers illustrate the percent positive cells in the CD4+ gate. (b) Quantification of IL-17A- and IFN-γ producing CD4+ cells in the CNS of Tbx21−/− and WT mice on day 17 post-immunization. (c) IL-17A and IFN-γ produced by purified CNS-infiltrating CD4+ T cells as determined by ELISA. The concentration of cytokines was normalized to 1×106 cells/ml. The bars represent mean ± s.e.m. of three independent experiments. (d) Cells were prepared from Tbx21−/− IL-23R.GFP and WT-IL-23R.GFP mice on day 17 after EAE induction and analyzed by flow cytometry. Graphs summarize IL-23R and CCR6 expression (percent of the CD4+ gate) in three independent experiments (n=3–4 mice per group in each experiment). (e) Expression of TH17 signature genes in CD4+ T cells purified from the CNS of Tbx21−/− and WT mice on day 17 post-immunization. Cells were pooled from 4 mice per group and were stimulated with PMA+I for 4 h before RNA extraction. The bars represent mean ± s.e.m. of three independent experiments. Statistical analysis was performed using Student-t test; *P<0.05; **P<0.01; ***P<0.0001.
IL-23R signaling drives the pathogenic potential of CNS-infiltrating TH17 cells by promoting the expression of proinflammatory chemokines and by suppressing the expression of IL-104, 24. In addition, IL-23R signaling is essential for the terminal differentiation of TH17 cells and their long term survival13, 25. To address the role of T-bet in controlling IL-23R expression in TH17 cells, we crossed IL-23R.GFP reporter mice onto a T-bet-deficient background. On day 17 after MOG35–55 plus CFA immunization, mononuclear cells were isolated from the draining lymph nodes, spleen and CNS of Tbx21−/−-IL-23R.GFP and WT-IL23R.GFP reporter mice and the percentage of IL-23R positive cells within the CD4+ population was determined by flow cytometry. As shown in Fig. 2d, Tbx21−/− mice displayed a higher percentage of IL-23R+ CD4+ cells in the lymph nodes and the CNS versus controls, while in spleen the frequency of IL-23+ CD4+ T cells was similar. In addition, there was a significantly higher percentage of CD4+ cells expressing the TH17-specific chemokine receptor, CCR6 in the lymph nodes and the CNS of Tbx21−/− mice (Fig. 2d). We looked at the expression of other TH17 signature genes in purified CD4+ T cells isolated from the CNS of Tbx21−/− and WT mice during the disease peak and observed higher levels of Rorc, Il23r, Il17a and Il17f transcripts in Tbx21−/− CD4+ T cells, while the levels of Il21 and Il22 were more variable between Tbx21−/− and WT mice (Fig. 2e). The enhanced Type 17 response in Tbx21−/− mice could be intrinsic to the T cell or reflect differences in cytokine production by innate immune cells which could create a milieu more conducive for the polarization of TH17 cells in vivo. To differentiate between these two possibilities we performed functional analysis of CD4+ T cells in the CNS of Tbx21F/F and Tbx21F/F Cd4cre mice 14 days after EAE induction (Supplementary Fig. 1). Our data demonstrate that T cell-specific deletion of T-bet results in an augmented TH17 response in the CNS during EAE, suggesting that this is a T cell-intrinsic phenomenon. Thus, a dominant TH17 response is generated in Tbx21−/− mice following MOG35–55 plus CFA immunization, suggesting that T-bet expression limits the magnitude of Type 17 responses in the CNS during EAE.
T-bet expression blocks TH17 differentiation
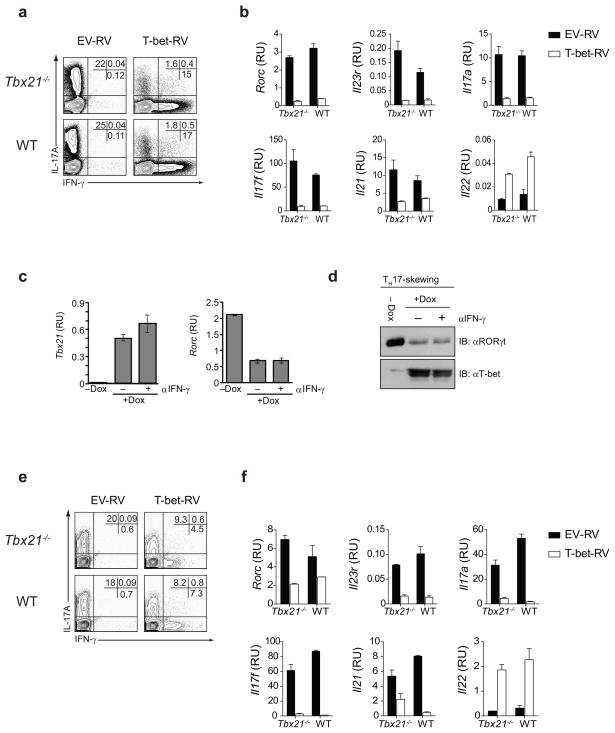
The experiments described above suggest strongly that T-bet negatively regulates TH17 cell lineage commitment. To directly assess whether T-bet plays a negative role in TH17 differentiation, naïve CD4+ T cells (CD62LhiCD25lo) were activated under TH17 polarizing conditions. After 24 h, activated CD4+ cells were transduced with empty retrovirus (EV-RV) or T-bet-RV containing TH17 skewing cytokines + neutralizing IL-4 and IFN-γ antibodies. After 5 days of culture under TH17 polarizing conditions, the cytokine production by sorted GFP+ cells was determined by intracellular cytokine staining (ICS). Transduction of both Tbx21−/− and WT naïve CD4+ T cells with T-bet resulted in a substantial decrease in the percentage of IL-17A producing cells and a marked increase in the frequency of IFN-γ-IL-17A double producers and IFN-γ single producers (Fig. 3a). T-bet expression resulted in a marked reduction of Rorc and Rorc-target genes (Il17a and Il17f), Il21 and Il23r (Fig. 3b). Although we observed no significant difference in the amount of Il22 transcripts between Tbx21−/− and WT TH17 cells, T-bet over-expression augmented Il22 mRNA expression in both Tbx21−/− and WT TH17 cells (Fig. 3b). Ectopic expression of T-bet therefore prevents differentiation of TH precursors into TH17 cells under TH17 polarizing conditions by blocking the expression of the TH17 cell lineage-specific transcription factor, RORγt and consequently, RORγt-target genes.
Figure 3.
T-bet expression in naïve TH precursors and fully differentiated TH17 cells inhibits TH17 response. (a) Flow cytometry of IL-17A and IFN-γ expression by naïve CD4+ T cells transduced with empty retrovirus or T-bet expressing retrovirus under TH17 polarizing conditions. Intracellular cytokine staining was performed on sorted GFP+ cells after a 4 h PMA+I stimulation. (b) Real-time PCR analysis of Rorc, Il23r, Il17a, Il17f, Il21 and Il22 mRNA expression in naïve CD4+ T cells transduced with empty retrovirus or T-bet expressing retrovirus under TH17 polarizing conditions. (c) Tbx21 and Rorc mRNA expression and (d) T-bet and RORγt protein expression in untreated and doxycyline treated Tbx21−/− rtTA+TrTB+ transgenic TH precursor cells activated under TH17 polarizing conditions in the absence or presence of anti- IFN-γ antibody. (e) Flow cytometry of IL-17A and IFN-γ expression by fully differentiated TH17 cells transduced with empty retrovirus or T-bet expressing retrovirus under TH17 polarizing conditions. Intracellular cytokine staining was performed following a 4 h PMA+I stimulation. (f) Real-time PCR analysis of TH17 signature genes in fully differentiated TH17 cells transduced with empty retrovirus or T-bet expressing retrovirus in the presence of TH17 polarizing cytokines. The data in (a–f) are representative of three independent experiments.
In a separate series of experiments, we utilized the transgenic inducible T-bet expression system, where T-bet is induced in naïve TH precursors in response to doxycycline treatment. Naïve CD4+ T cells were activated under TH17 polarizing conditions for 24 h. On the second day, T-bet expression was induced by the administration of 0.5 μg/ml of doxycycline in the absence or presence of neutralizing IFN-γ antibody. The amount of RORγt and T-bet mRNA and protein was determined by real-time PCR and immunoblotting, respectively. Induction of T-bet expression in transgenic TH precursors by doxycycline treatment resulted in marked reduction of both RORγt transcripts and protein under TH17 polarizing conditions (Fig. 3c–d). We observed suppression of RORγt by T-bet even in the presence of neutralizing IFN-γ antibody demonstrating that this mode of RORγt suppression is independent of IFN-γ.
T-bet can redirect fully differentiated TH2 cells into the TH1 pathway22. To determine whether T-bet could similarly reprogram committed TH17 cells, we differentiated naïve CD4+ T cells (CD62LhiCD25lo) under TH17 polarizing conditions for 6 days after which Tbx21−/− and WT TH17 cells were re-activated for 24 h and transduced with either EV-RV or T-bet-RV under TH17 polarizing conditions. Forty-eight hours after retroviral transduction, GFP+ cells were sorted and intracellular cytokines examined. There was a 50% reduction of IL-17A producing cells and an increase in the frequency of IFN-γ-producing cells after transduction with T-bet-RV (Fig. 3e). Similar to the experiments above, enforced T-bet expression in fully differentiated TH17 cells resulted in reduced levels of Rorc, Il17a, Il17f, Il21 and IL23r transcripts and increased expression of Il22 (Fig. 3f). These results indicate that ectopic expression of T-bet is sufficient to suppress expression of the Type 17 signature genes in committed TH17 cells even in the presence of TH17 polarizing cytokines.
T-bet inhibits Runx1 activity
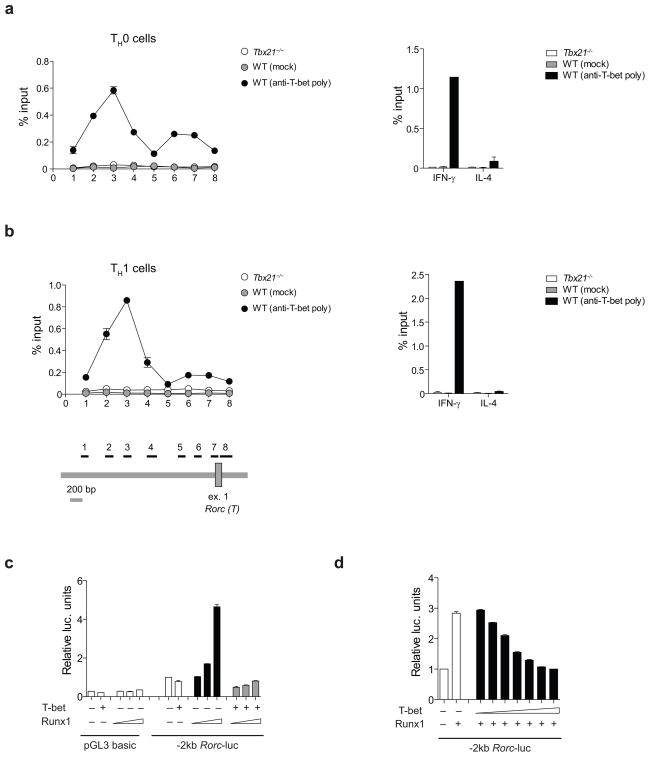
Our results support the idea that the negative effect of T-bet on TH17 differentiation could be mediated by inhibition of the TH17-cell-specific transcription factor RORγt. To determine whether T-bet binds to the Rorc promoter, we performed chromatin immunoprecipitation (ChIP) assays on T-bet-bound chromatin from nuclear lysates of non-polarized TH0 cells and differentiated TH1 and TH17 cells. We detected modest, but reproducible, binding of T-bet to a site located approximately 2kb upstream of the first exon of Rorc (the thymus-specific isoform) in non-skewed TH0 cells, and we found marked enrichment of T-bet bound to the same -2kb site in TH1 cells (Fig. 4a–b). We did not detect binding of T-bet to the Rorc or Ifng promoter in TH17 cells (data not shown). To test whether T-bet directly inhibits Rorc gene expression, we performed reporter luciferase assays with the −2kb Rorc-luc promoter in HEK293 cells. Transfection of T-bet had no effect on Rorc-luc activity indicating that T-bet might not directly suppress Rorc transcription (Fig. 4c). T-bet generally does not act as a direct transcriptional repressor 15, 26, 27. Instead, T-bet exerts its negative effect on gene expression by binding to and sequestering transcriptional activators away from regulatory regions15, 26–29. Runx1 induces RORγt expression30. There are two Runx1 binding sites immediately upstream of the T-bet binding site (2kb upstream of the first exon of the Rorc(T) gene). This finding prompted us to investigate whether T-bet could inhibit RORγt expression by blocking Runx1 transcriptional activity. To analyze the regulation of the Rorc gene, HEK293 cells were transfected with a −2kb Rorc-luc reporter construct in the presence of increasing concentrations of Runx1 plasmid with or without T-bet expression vector. Runx1 expression increased luciferase activity in a dose-dependent manner, which was blocked by co-expression of T-bet (Fig. 4c). Furthermore, T-bet blocked Runx1-mediated transactivation of the Rorc promoter in a dose-dependent manner (Fig. 4d). Several other transcription factors, namely Irf4, Batf and STAT3, control TH17 differentiation by positively regulating Rorc expression31–33. Relative expression of Irf4 and Batf was similar in Tbx21−/− and WT TH cells as determined by RT-PCR (Supplementary Fig. 2). It is currently unknown whether regulation of Rorc expression by Irf4 and Batf is mediated by direct binding of these transcription factors to the Rorc locus, but the binding sites for STAT3 in the Rorc and Il17a loci have been clearly defined33. To determine whether there was a difference in the binding of STAT3 to its target sequences in the Rorc and Il17a loci, we performed ChIP on STAT3-bound chromatin from nuclear lysates of non-polarized TH0 and TH17 cells. The binding of STAT3 to its target sites in the intergenic or intragenic regions of the Rorc locusor Il17a locus was similar in Tbx21−/− and WT TH0 and TH17 cells (Supplementary Fig. 3). These data suggest that interference with Runx1, but not STAT3, transcriptional activity is the likely mechanism by which T-bet blocks the expression of RORγt.
Figure 4.
T-bet blocks Runx1-mediated transactivation of the Rorc promoter. (a–b) T-bet binding to the Rorc promoter in TH0 and differentiated TH1 cells after 6 h PMA+I stimulation, as assessed by ChIP. In all experiments, the Ifng promoter and Il4 promoter were included as positive and negative controls, respectively. As specificity controls, ChIP was performed using pre-immune serum and Tbx21−/− cells. Fold enrichment was calculated by determining the ratios of the amount of immunoprecipitated DNA tothat of the input sample. The ChIP assays were repeated three times. (c) Rorc reporter plasmid was constructed from a 2 kb fragment of the mouse Rorc promoter. HEK293 cells were transfected with empty pGL3 basic plasmid or Rorc-luc reporter construct, pRL-TK, increasing concentration of Runx1 in the absence or presence of T-bet. (d) HEK293 cells were transfected with the Rorc-luc reporter construct, pRL-TK, Runx1 in the absence or presence of increasing concentration of T-bet. Luciferase activity was measured by Dual Luciferase reporter assay and relative luciferase activity was normalized to pRL-TK for transfection efficiency and empty vector control. Bars represent mean ± s.e.m. of duplicate samples. The graphs are representative of three (c) and two (d) independent experiments.
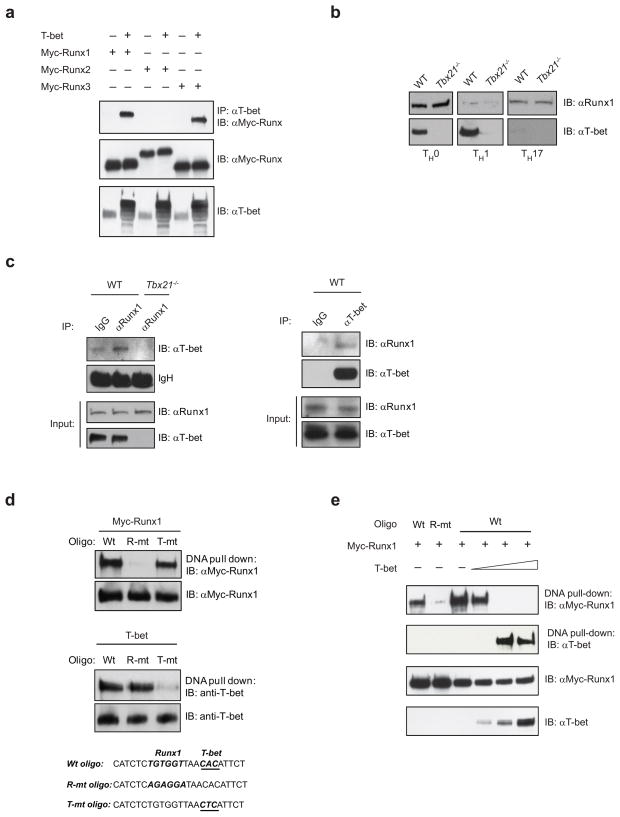
T-bet interacts with Runx1
To examine further the mechanism of T-bet-mediated repression of Rorc expression, we investigated whether T-bet could interact with Runx1. First, we over-expressed T-bet with Myc-tagged Runx1, Runx2 and Runx3 and performed co-immunoprecipitation experiments in HEK293 cells. T-bet interacted with both Runx1 and Runx3, but not Runx2 in HEK293 cells (Fig. 5a). To determine in which TH subset T-bet and Runx1 interact, we performed immunoblot analysis of Runx1 and T-bet protein expression in unskewed TH0 and differentiated TH1 and TH17 cells. We detected T-bet and Runx1 expression in non-polarized TH0 cells (Fig. 5b). There was a marked reduction in the expression of Runx1 protein in in vitro differentiated TH1 cells and conversely, a marked decrease in levels of T-bet protein in in vitro differentiated TH17 cells (Fig. 5b). These data suggest that interaction between T-bet and Runx1 could occur in non-polarized TH0 cells in which both proteins are co-expressed. Indeed, Runx1 immunoprecipitation confirmed the presence of an endogenous T-bet-Runx1 interaction in non-polarized WT TH0 cells, but not in differentiated TH1 and TH17 cells (Fig. 5c and data not shown). T-bet-Runx1 interaction in uncommitted TH0 cells was also confirmed in the reverse co-immunoprecipitation assay (Fig. 5c). These data suggest that a functionally important interaction between T-bet and Runx1 most likely occurs in uncommitted TH cells, but not in fully differentiated TH1 and TH17 cells because of restrictive expression of Runx1 and T-bet, respectively, in these TH cells.
Figure 5.
T-bet interacts with Runx1. (a) HEK293 cells were transfected with empty vector, T-bet and Myc-tagged Runx1, Runx2 and Runx3 expression vectors as indicated. Lysates were immunoprecipitated with anti-T-bet polyclonal antibody followed by immunoblotting with anti-Myc antibody. (b) Runx1 and T-bet protein expression in Tbx21−/− and WT TH0, TH1 and TH17 cells after 6 h stimulation with PMA+I. (c) Endogenous interaction between Runx1 and T-bet in non-polarized TH0 cells. Cell lysates of Tbx21−/− and WT TH0 cells were immunoprecipitated with a control, anti-Runx1 or anti-T-bet antibodies and immunoblotted with anti-T-bet or anti-Runx1 antibody. (d) A DNA pull-down assay with a Wt oligo (containing wild-type Runx1 and T-bet binding sites), R-mt oligo (containing mutated Runx1 binding site) or T-mt oligo (containing mutated T-bet binding site) in the presence of Myc-Runx1 or T-bet. (e) 293HEK cells were transfected with a Wt or R-mt oligo and Runx1 in the absence or presence of increasing doses of T-bet (0.1, 0.5 and 1 μg). Oligo-bound proteins were immunoblotted for over-expressed Myc-Runx1 and T-bet. The data are representative of one - two independent experiments.
Based on the luciferase data and co-immunoprecipitation experiments, one could hypothesize that T-bet interaction with Runx1 could block Runx1 binding to its consensus sites located -2kb in the Rorc(T) promoter . We detected Runx1 binding to the oligo containing the WT Runx1 target sequence, but not to the oligo in which the Runx1 target sequence was mutated by a T to A substitution (Fig. 5d). T-bet bound to the WT oligo containing a T-bet specific half-site, but not to the oligo in which the T-bet half-site was mutated (Fig. 5d). After confirming that Runx1 and T-bet binding to the -2kb Rorc(T) site was sequence-specific, we performed a DNA pull-down assay with the WT oligo and Runx1 in the absence or presence of increasing concentrations of T-bet. In the absence of T-bet, Runx1 bound strongly to the DNA oligo containing the WT Runx1 binding site (Fig. 5e). Increasing concentrations of T-bet ablated the ability of Runx1 to bind to its target sequence in a dose-dependent manner showing that T-bet interaction with Runx1 interferes with Runx1 binding to its -2kb Rorc(T) site (Fig. 5e).
Runx1 reversed T-bet effects on TH17 polarization
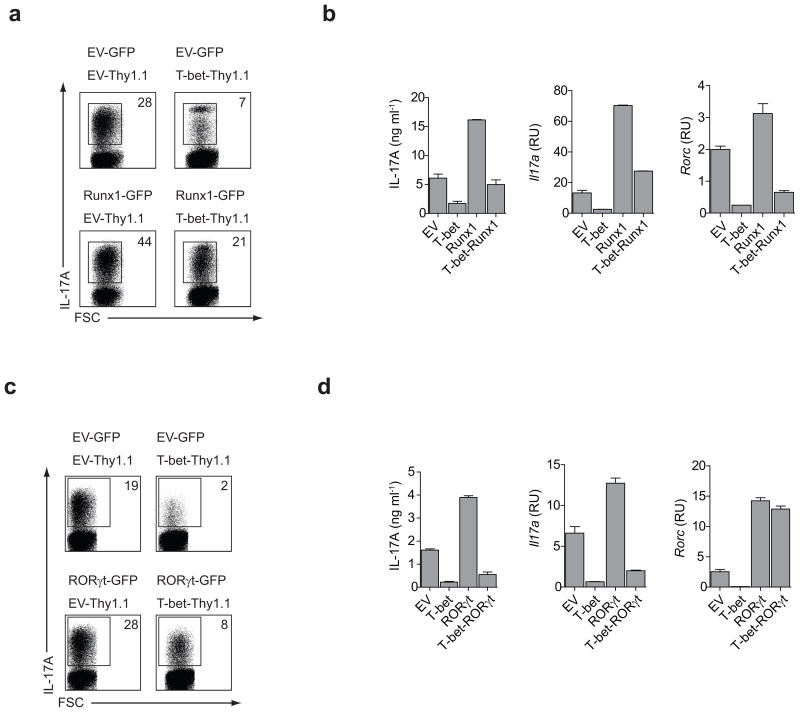
Next we investigated whether Runx1 over-expression could block the inhibitory effects of T-bet on TH17 differentiation. Purified CD4+ T cells were transduced with various combinations of retroviruses expressing GFP alone (EV-GFP), Thy1.1 alone (EV-Thy1.1), Runx1-GFP and/or T-bet-Thy1.1. The transduced cells were grown under TH17 conditions for 5 days after which GFP-Thy1.1 double-positive cells were sorted and examined for TH17 commitment. Retroviral transduction of TH cells with T-bet had a negative effect on TH17 commitment under TH17 polarizing conditions, while transduction of TH cells with Runx1 augmented the differentiation of TH17 cells (Fig. 6a–b). Over-expression of Runx1 reversed the inhibitory effect of T-bet and fully restored TH17 polarization in cells co-expressing Runx1 and T-bet (EV vs. T-bet-Runx1). However, Rorc expression was only partially up-regulated, suggesting that there may be additional mechanism(s) by which T-bet inhibits transcription of the Rorc gene in TH17 cells (Fig. 6b). Conversely, transduction of purified Tbx21−/− CD4+ T cells with Runx1 dominant negative (DN) retrovirus during TH17 differentiation reversed the effects of T-bet deficiency on IL-17A production by TH17 cells (Supplementary Fig. 4).
Figure 6.
Runx1 over-expression restores IL-17A production in T-bet expressing TH17 cells. (a) CD4+ T cells were transduced with retroviruses expressing GFP, Thy1.1, Runx1-GFP and/or T-bet-Thy1.1 within 24 hours of activation. Cells were cultured under TH17 polarizing conditions for 5 days and stimulated with PMA+I for 4 h prior to intracellular cytokine staining for IL-17A. Numbers indicate the percentage of IL-17A producing cells within GFP+/Thy1.1+ gate. (b) CD4+ T cells were transduced with various combinations of retroviruses as described in (a). Following 5 days of in vitro differentiation under TH17 polarizing conditions, GFP+-Thy1.1+ cells were sorted and stimulated with PMA+I for 4 h prior to the analysis of IL-17A production and the Rorc gene expression. (c) Activated CD4+ T cells were transduced with retroviruses expressing GFP, Thy1.1, RORγt-GFP and/or T-bet-Thy1.1. The transduced cells cultured under TH17 conditions for 5 days were stimulated with PMA+I for 4 h prior intracellular cytokine staining for IL-17A. The numbers indicate percentage of IL-17A producing cells within GFP+/Thy1.1+ gate. (d) Cells were transduced under TH17 condition as described in (c). IL-17A production was measured by ELISA and RT-PCR and Rorc mRNA transcripts were analyzed by RT-PCR. The results in (a–d) are representative of two independent experiments.
In addition to directly promoting RORγt expression, Runx1 also acts as a co-activator and together with RORγt induces the expression of the Il17a and Il17f genes30. T-bet suppresses TH17 differentiation by inhibiting the expression of the Rorc gene (Fig. 3). However, it is possible that T-bet’s interaction with Runx1 serves to sequester this transcriptional co-activator and blocks the expression of TH17 signature genes in this manner. To investigate this possibility, we asked whether RORγt was able to restore a TH17 developmental program in TH cells co-expressing T-bet and RORγt under TH17 polarizing conditions. Expression of RORγt independently of the T-bet transcriptional block (i.e. from the retroviral LTR control elements) in developing TH17 cells was unable to fully reverse the T-bet mediated inhibition of TH17 differentiation (Fig. 6c–d). We were unable to co-immunoprecipitate T-bet and RORγt in 293HEK cells, suggesting that the sequestration of RORγt from its target genes by T-bet is unlikely. These data demonstrate that in addition to inhibiting the transcription of the Rorc gene, T-bet interaction with Runx1 depletes the pool of free Runx1 which is available for the formation of transcriptionally active Runx1-RORγt complexes in TH17 cells.
T-bet Tyr304 is crucial for suppression of TH17 cells
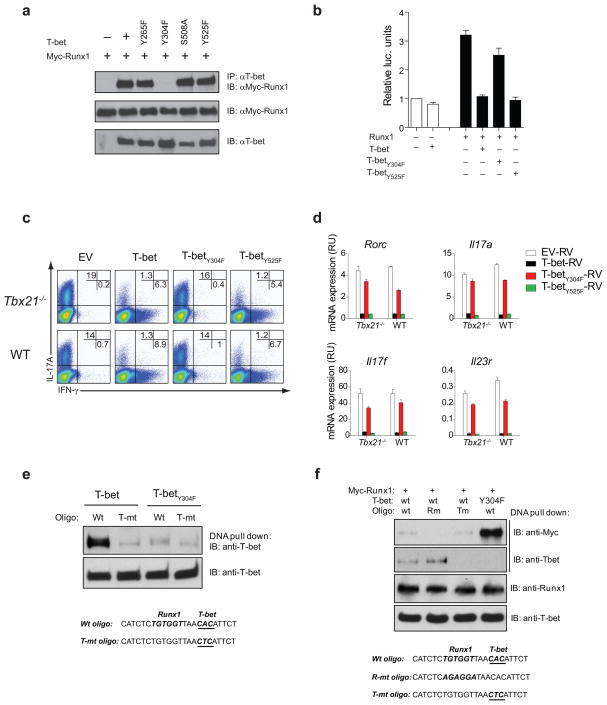
To investigate which amino-acid residue is important for T-bet-Runx1 complex formation, we tested the ability of a series of T-bet mutants to interact with Runx1 in HEK293 cells. Two of these point mutants, T-betS508A and T-betY525F, were shown previously to be functionally important in IL-2 and in TH2 lineage suppression, respectively15, 27. Coimmunoprecipitation studies revealed that Runx1 interacted with the T-betY265F, T-betS508A and T-betY525F mutants, but not with the T-betY304F mutant (Fig. 7a). Furthermore, the T-betY304F mutant was unable to suppress Runx1 transcriptional activity in the -2kb Rorc-luciferase assay (Fig. 7b) suggesting that this T-bet Tyr304 residue may be important for the suppression of TH17lineage commitment. To answer this question, sorted naïve TH cells were activated under TH17 polarizing conditions for 24 h and transduced with control, WT T-bet, T-betY304F and T-betY525F mutant retroviruses. The transduced cells were cultured under TH17 polarizing conditions for an additional 5 days and the percentage of IFN-γ and IL-17 producing cells was determined by ICS. WT T-bet and the T-betY525F control mutant suppressed TH17differentiation under TH17 polarizing conditions. In contrast, mutation of T-bet residue Tyr304 abrogated the ability of T-bet to repress TH17 lineage commitment (Fig. 7c). When compared with WT T-bet and the T-betY525F mutant, the T-betY304F mutant was unable to suppress expression of Rorc, Il17a, Il17f and Il23r in developing TH17 cells (Fig. 7d). These data indicate that T-bet residue Tyr304 is important for T-bet-Runx1 complex formation, for inhibition of Runx1 transcriptional activity and for suppression of TH17 lineage development.
Figure 7.
T-bet Tyr304 is essential for T-bet-Runx1 interaction and for inhibiting the TH17 differentiation program. (a) Interaction of Runx1 with WT T-bet or T-bet point mutants in co-immunoprecipitation experiments in HEK293 cells. (b) HEK293 cells were transfected with Rorc-luc reporter construct, pRL-TK, Runx1 in the absence or presence of WT T-bet or T-bet point mutants as indicated. (c) Flow cytometry analyzing IL-17A and IFN-γ expression by naïve CD4+ T cells transduced with empty vector, WT T-bet, or T-bet point mutants under TH17 polarizing conditions. Numbers in quadrants illustrate the percent positive cells in the CD4+ gate. (d) RT-PCR analysis of TH17 signature genes by CD4+ T cells transduced with empty vector, WT T-bet, or T-bet point mutants under TH17 conditions. (e) 293HEK cells were transfected with WT T-bet or T-betY304F mutant in the presence of a wild-type or T-bet mutant oligo. Oligo-bound proteins were immunoblotted for T-bet. (f) 293HEK cells were transfected with Myc-tagged Runx1, WT T-bet or T-betY304F mutant in the presence of wild-type oligo, Runx1 mutant oligo or T-bet mutant oligo as indicated. Oligo-bound proteins were immunoblotted with anti-Myc and anti-T-bet antibodies. The data are representative of three independent experiments (ad) and one experiment (e–f).
In DNA pull-down assays, the T-betY304F mutant failed to bind to the T-bet binding site in the -2kb Rorc(T) promoter region suggesting that this residue is also important for T-bet binding to DNA (Fig. 7e). To delineate whether T-bet-mediated inhibition of Runx1 activity was dependent on DNA binding or T-bet-Runx1 protein-protein interaction, we investigated whether WT T-bet could inhibit Runx1 binding to its target site using oligos in which the T-bet binding site was mutated. T-bet blocked Runx1 binding to the Runx1-specific sequence independently of T-bet’s ability to bind to DNA (lane 3, Fig. 7f). Collectively, these data suggest that the T-bet-Runx1 protein-protein interaction is mainly responsible for inhibition of Runx1 activity with a T-bet protein-DNA interaction playing a minor if any role.
DISCUSSION
Lineage-specific transcription factors can both activate and repress differentiation programs. T-bet simultaneously promotes TH1 differentiation and represses TH2 differentiation 3, 6. While several studies have reported an enhanced Type 17 response in Tbx21−/− animals in different disease models16–19, no mechanistic explanation for this increase in TH17 responses was provided. Here, we demonstrate that T-bet suppresses TH17 cell lineage commitment by inhibiting the transcription of the TH17-cell-specific transcription factor RORγt and its target genes. T-bet did not directly repress the Rorc promoter. Instead, T-bet interacted with the Runx1 transcription factor and blocked Runx1-mediated transactivation of Rorc. Over-expression of Runx1 was sufficient to reverse the inhibitory effects of T-bet on IL-17A production by TH17 cells. Furthermore, we demonstrate that T-bet residue Tyr304 is crucial not only for T-bet-Runx1 complex formation, but also for blocking Runx1 activity and for inhibiting the TH17 differentiation program. Thus, our data uncover a molecular mechanism to explain the exaggerated TH17 responses observed in T-bet deficient hosts.
In addition to activating a set of genes that promote TH cell differentiation towards a specific lineage, a master regulator can also suppress the developmental program of the opposing T cell lineages34. Among the TH cell-specific transcription factors, T-bet seems to be particularly active in this regard. The ability of T-bet to negatively regulate the differentiation of TH2 cells15, 22, IL-2 production from TH1 cells27 and tumor necrosis factor (TNF) production in dendritic cells26 prompted us to investigate the contribution of T-bet to the regulation of the TH17 response. Our data show that T-bet deficiency results in an augmented Type 17 response in vitro and in vivo during CNS inflammation in the EAE model of multiple sclerosis. Our results differ from a previous publication which reported that mice injected with T-bet-specific siRNA had lower expression of IL-23R and lacked TH17 cells after immunization with myelin basic protein (MBP) Ac1–11 plus CFA or MOG35–55 plus CFA35, 36. We demonstrate instead that TH17 cells are present in the CNS of Tbx21−/− mice in greater numbers and with strong expression of TH17 signature genes after MOG35–55 plus CFA induced EAE. The disparity between the two studies may arise from differences in experimental conditions, such as the use of T-bet-specific siRNA rather than complete genetic deletion in vivo. In support of our results, other studies also detected a higher frequency of myelin-specific TH17 cells in MOG35–55 plus CFA immunized Tbx21−/− mice than WT mice36. Interestingly, despite a strong TH17 response, Tbx21−/− mice are largely protected from the development of EAE. This observation might be explained by postulating that in the inflammatory milieu of the CNS, T-bet controls expression of a novel set of genes that is important for the pathogenicity, but not for the development of TH17 cells. In support of this hypothesis, a recent study reported that T-bet is expressed in IL-23-treated TH17 cells and these T-bet-expressing TH17 cells were pathogenic during CNS inflammation pointing to the important functional role of T-bet in a subset of TH17 cells37. Alternatively, the presence of both TH1 and TH17 cells might be required for CNS pathology. Finally, T-bet expression in other cell types might be important for driving disease development.
Here, we have focused on T-bet’s function in TH17 lineage commitment of CD4+ T cells. In this study, we have demonstrated that T-bet over-expression in naïve TH cell precursors or committed TH17 cells had a negative effect on Rorc transcription and consequently on the expression of Rorc-target genes. In addition, we observed downregulation of Il21 gene expression. T-bet suppressed IL-21 in TH1 cells by interacting with NFATc2, thus preventing NFATc2 from binding to the Il21 promoter29. Since IL-21 promotes IL-23R expression in TH17 cells10, T-bet-mediated suppression of Il21 could also contribute to reduced levels of Il23r in TH17 cells after transduction with a T-bet expressing retrovirus. In contrast to Rorc, Il17a, Il17f, Il23r, which were suppressed by T-bet, the expression of Il22 was augmented by ectopic expression of T-bet. The aryl hydrocarbon receptor (AHR) controls IL-22 production by TH17 cells, as CD4+ TH17 cells from AHR-deficient mice fail to produce IL-22 when exposed to AHR ligands38, 39. Thus, it is plausible that T-bet expression has a synergistic effect on AHR-mediated induction of IL-22.
We detected endogenous T-bet binding in the region located 2kb upstream of the first Rorc exon in non-skewed and TH1 cells, but not TH17 cells. Interestingly, differentiation of TH cells down the TH1 pathway resulted in markedly reduced expression of Runx1. Conversely, culturing TH cells in the presence of TH17-polarizing cytokines resulted in suppression of T-bet protein expression. This is not surprising since TGF-β has a negative effect on T-bet expression, and IFN-γ, the most potent inducer of T-bet expression, is neutralized under in vitro TH17 polarizing conditions. Thus, the lack of T-bet binding to the Rorc promoter in committed TH17 cells could be explained by substantially reduced expression of T-bet in this TH subset.
We did not detect any significant effects of T-bet over-expression on Rorc promoter activity in luciferase assays. Evidence that T-bet acts as a direct transcriptional repressor or can recruit co-repressors to these promoters is currently lacking. However, T-bet can exert a negative regulatory effect on gene expression by blocking the activity of competing transcription factors15,26, 27. Our understanding of the transcriptional regulation of RORγt is still incomplete. It has been reported that the expression of RORγt is substantially reduced in Irf4−/− and Stat3−/− CD4+ cells31, 40–42. Runx1 induces RORγt expression and Batf is important for the maintenance of RORγt expression in stimulated T c ells30, 32. It is currently unknown whether regulation of Rorc expression by Irf4 and Batf is mediated by direct binding of these transcription factors to the Rorc locus, but the binding sites for STAT3 in the Rorc and Il17a loci have been identified33. STAT3 binding to both the Rorc and Il17a loci was similar between Tbx21−/− and WT TH cells. Hence, we focused on the transcriptional activity of Runx1 since we detected two Runx1 consensus sites in the proximity of the T-bet peak binding site located -2kb from the first exon of Rorc as shown by ChIP.
Based on the protein expression data and co-immunoprecipitation experiments, we propose that the functionally important interaction of T-bet and Runx1 occurs in uncommitted TH cells. We mapped the function of T-bet in interacting with Runx1, repressing Runx1 activity, and suppressing TH17 differentiation program to Tyr304. Our preliminary data indicate that this tyrosine is not phosphorylated and that the protein is stably expressed in HEK293 cells and primary T cells. In DNA pull-down assays, the T-betY304F mutant failed to bind to DNA. Although the Tyr304 residue is important for T-bet-DNA interaction, our results indicate that this interaction is not essential for T-bet’s ability to block Runx1. WT T-bet was still able to block Runx1 binding to its target site in a RORγt promoter bearing a mutated T-bet half-site. Thus, the T-bet-Runx1 protein-protein interaction is the major mechanism by which T-bet blocks Runx1 activity by sequestering Runx1 away from the RORγt promoter.
In both infectious and autoimmune diseases, T cell-mediated responses are characterized by the presence of cells co-expressing IL-17A and IFN-γ, the so called IFN-γ+ TH17 cell. T cell clones from MBP-peptide immunized mice were genetically either T-bet+-RORγt− or T-bet+-RORγt+ 28. T-bet+-RORγt+ cells were very responsive to exogenous cytokines such as IL-12 and IL-23, which influenced the relative levels of T-bet and RORγt, and shifted cytokine production towards IFN-γ or IL-17A, respectively28. The unstable phenotype of TH17 cells is not restricted to CNS-specific autoimmunity. IL-17F reporter mice in a transfer model of colitis demonstrated that IFN-γ-producing CD4+ T cells can emerge fromTH17 committed cells during T cell driven inflammation. This transition of TH17 cells into IFN-γ-producing cells was STAT4- and T-bet-dependent21. In the context of these recent findings, it is tempting to propose that this T-bet-mediated transition of TH17 cells into a “TH1-like” subset is controlled partly by T-bet-mediated interference with Runx1 transcriptional activity. Ectopic expression of T-bet in TH17 cells results in the suppression of Rorc. However, in fully differentiated TH17 cells, which already express RORγt, T-bet could still interfere with RORγt transcriptional activity by sequestering its co-activator, Runx1. Thus, over-expression of Runx1 in TH17 cells overcame the inhibitory effect of T-bet and completely restored IL-17A production. Although Runx1 fully restored IL-17A production, the expression of Rorc was only partially increased. These results indicate that there are additional mechanism(s) by which T-bet inhibits Rorc gene expression. One potential mechanism could be via IL-12 signaling inducing repressive epigenetic changes of the Rorc locus, which are dependent on T-bet and STAT443. Thus, T-bet re-expression in TH17 cells turns off RORγt expression through the sequestration of Runx1 and through the introduction of epigenetic changes resulting in expression of TH1 signature genes and acquisition of the “TH1- like” phenotype by TH17 cells.
METHODS
Mice
Tbx21−/−, Tbx21−/− IL-23R.GFP and IL-23R.GFP mice (all C57BL/6 background) were housed at Harvard School of Public Health and were handled in accordance with guidelines from the Center for Animal Resources and Comparative Medicine (ARCM) at Harvard Medical School. Tbx21−/− rtTA+ TrTB+ mice were housed at College of Pharmacy; Ewha Womans University, Soul, S. Korea. Ifng−/− mice were purchased from Jackson Laboratories. Cd4cre and Tbx21F/F mice were provided by Dr. Christopher B. Wilson (The Bill and Melinda Gates Foundation) and Dr. Steven L. Reiner (the University of Pennsylvania), respectively.
Plasmids
Constitutively active STAT3 (pRc/CMV STAT3-C) was obtained from Addgene (submitted by Dr. J.E. Darnell)44. pMCsIg-EV, pMCsIg-Runx1 and pMCsIg-Runx1-DN retroviral plasmids were generously provided by Dr. Warren Strober (NIAID, NIH).
CD4+ T helper differentiation in vitro
Tbx21−/−, Ifng−/− and WT CD4+ T cells were stimulated for 48 h with anti-CD3 antibody (2 μg/ml) in the presence of irradiated splenocytes at a ratio of 5:1. CD4+ T cells were cultured under TH0 conditions (200 U/ml of hIL-2; NCI BRB Preclinical Repository) or differentiated into TH1 cells with hIL-2 (200 U/ml), mIL-12 (10 ng/ml; Peprotech) and anti-mIL-4 antibody (10 μg/ml; clone 11B11; BioXCell) or into TH17 cells by the addition of hTGF-β (2 ng/ml; R&D Systems), mIL-6 (20 ng/ml; R&D Systems), anti-mIL-4 (10 μg/ml) and anti-mIFN-γ (10 μg/ml; clone XMG1.2; BioXCell) antibodies for 5 days. TH17+IL-23 cells were cultured in the presence of hTGF-β, mIL-6, anti-IL-4 and anti-IFN-γ antibodies for the first 48 hours of activation after which TH17 cells were cultured in the presence of IL-23 (10 ng/ml; R&D Systems).
Isolation and functional analysis of CNS mononuclear cells
EAE was induced in 8 – 10 week old mice as described previously23. CNS-infiltrating cells were re-stimulated with PMA+I for 4 h before performing intracellular cytokine staining for IFN-γ (XMG1.2; BD Pharmingen)and IL-17A (TC11-18H10; BD Pharmingen). For ELISA and RT-PCR analysis, CNS-derived CD4+ T cells were purified using CD4 negative selection kit (Stem Cell Technologies). Purified CD4+ T cells (>95% purity) were pooled from 4–5 mice/group and cells were stimulated for 4 h with PMA+I. The RT-PCR primer sequences are listed in the Supplementary Table 1.
Retroviral transduction
Naïve (CD62LhiCD25lo) CD4+ T cells were transduced with indicated retroviruses 24 h after activation. Transduced cells were cultured for 5 days under TH17 conditions, and the sorted GFP+ cells were tested for cytokine production after 4 h of PMA+I stimulation. For transduction of committed TH17 cells, naïve CD4+ T cells were cultured under TH17 conditions for 6 days. TH17 cells were activated on plate-bound anti-CD3/CD28 plates and transduced with retroviruses for 24 h under TH17 conditions. Transduced cells were cultured for additional 48 h under TH17 conditions. The sorted GFP+ were stimulated with PMA+I for 4 h prior to functional analysis.
Doxycycline inducible T-bet transgenic TH17 in vitro cultures
Twenty-four hours after anti-CD3/CD28 activation, T-bet was induced in TH cells by the addition of 0.5 μg/ml of doxycycline to the TH17 culture media. Twenty-fours after T-bet induction with doxycycline, lysates were analyzed for the expression of Tbx21 and Rorc by RT-PCR. In addition, cells were immunoblotted with anti-T-bet and anti-RORγt antibody (clone B2D; eBioscience).
ChIP
Naïve cells were sorted from spleens and lymph nodes of Tbx21−/− and WT mice and differentiated under TH0, TH1 and TH17 polarizing conditions as described above. Differentiated TH cells were used in ChIP assay after 6 h PMA+I stimulation. The ChIP assay was performed as described previously45. The primers sets used for real-time PCR to quantitate the ChIP enriched DNA are listed in the Supplementary Table 2.The antibodies used for ChIP were polyclonal rabbit anti-T-bet (clone 9856) and anti-Stat3 (C-20; Santa Cruz) antibodies.
Luciferase assays
The mouse Rorc(T) promoterwas cloned into pGL3-basic plasmid upstreamof the firefly luciferase gene (Promega). To study the effects of T-bet and Runx1 on the Rorc promoter activity, HEK293 cells were transfected with Rorc-luc plasmid, increasing concentrations of Runx1,T-bet or pCDNA3.1- (empty vector control) and pRL-TK (Promega) using Fugene (Roche). To determine the dose-dependent effect of T-bet on Runx1 activity, HEK293 cells were transfected with Rorc-luc plasmid, Runx1, increasing concentration of T-bet and pRL-TK. Fireflyand Renilla luciferase activities were measured 48 h aftertransfection with the Dual Luciferase System (Promega).
Coimmunoprecipitation
HEK293 cells were transiently transfected with WT T-bet, or T-bet point mutants, Myc-Runx1 or empty vector using Fugene (Roche). 48 h after transfection, cells were lysed in TNT lysis buffer (1% triton X-100, 50mM Tris pH 7.5, 200mM NaCl, 1mM DTT, protease and phosphatase inhibitors). For endogenous Runx1 and T-bet coimmunoprecipitations, 3~4×108 of differentiated TH0, TH1 and TH17 cells were stimulated for 6 h with PMA+I after which cells were lysed in TNT lysis buffer. Whole cell lysates were immunoprecipitated withthe anti-Runx1 antibody (ab23980; Abcam) or IgG and protein A-sepharose coupled beads at 4°C. Immune complexes were immunoblotted using polyclonal rabbit anti-T-bet antibody. In the reverse co-immunoprecipitation assay, T-bet was immunoprecipitated using polyclonal rabbit anti-T-bet antibody (H-210; Santa Cruz) and separated immune complexes were immunoblotted with polyclonal rabbit anti-Runx1 antibody (ab23980; Abcam). To minimize interference of Runx1 or T-bet detection by the heavy chains of the immunoprecipitating T-bet, Trueblot ULTRA kit (eBioscience) was used for interaction analysis of Runx1 with T-bet in the reverse co-immunoprecipitation assay.
DNA binding assay
DNA binding assay was performed as described previously46. HEK293 cells were transfected with Myc-Runx1 expression plasmid in the absence or presence of increasing concentrations of T-bet expression vector. 150 μg of nuclear protein was incubated with 50 nt biotinylated probe containing wild-type, mutant T-bet binding site or mutant Runx1 binding site [− 2kb of exon 1 encoding Rorc(T)] plus streptavidin-agarose (Invitrogen) for 1 hour at 4°C in Binding buffer (100 mM NaCl, 10 mM Tris-HCl pH 7.6, 0.1 mM EDTA, 1 mM DTT, 5% glycerol, 1 mg/ml BSA, 20 μg/ml poly dI/dC plus protease inhibitors). Streptavidin-beads were washed in Binding buffer, and bound proteins were immunoblotted for over-expressed Myc-Runx1 and T-bet.
Supplementary Material
Acknowledgments
We thank Deneen Kozoriz for help in cell sorting, Dr. Warren Strober (NIAID, NIH) for providing us with Runx1 and Runx1-DN retroviral plasmids, Drs. Steven Reiner (University of Pennsylvania) and Christopher Wilson (The Bill and Melinda Gates Foundation) for providing us with Tbx21F/F and CD4cre mice, respectively. This work has been supported by NIH grant P01 NS038037 (L.H.G), NCRC program R15-2006-020 (E.S.H.) and an Irvington Institute/Bristol-Myers Squibb Fellowship from the Cancer Research Institute (V.L.). The authors thank Drs. Ann-Hwee Lee, Tracy Staton-Winslow, Matthew Greenblatt and Marc Wein for critical review of the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS:
V.L. designed and executed experiments and prepared the manuscript. X.C. performed ChIP assays and J.H.S. performed DNA pull-down and co-immunoprecipitation assays. E.S.H. created T-bet mutant retroviral constructs, E.J. performed doxycycline transgenic T cell experiment and M.O. generated IL-23R.GFP mice. V.K.K. contributed to discussions and manuscript preparation. A.N.B. provided technical assistance. L.H.G. supervised the research, designed experiments and participated in the manuscript preparation.
COMPETING INTERESTS STATEMENT
L.H.G. is on the Board of Directors and holds equity in Bristol Myers Squibb Pharmaceutical Company.
References
- 1.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 2.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 3.Harrington LE, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 4.Langrish CL, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mangan PR, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 6.Park H, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 8.Liang SC, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Zhou L, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 11.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 12.Korn T, et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448:484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGeachy MJ, et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djuretic IM, et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 15.Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- 16.Burrell BE, Csencsits K, Lu G, Grabauskiene S, Bishop DK. CD8+ Th17 mediate costimulation blockade-resistant allograft rejection in T-bet-deficient mice. J Immunol. 2008;181:3906–3914. doi: 10.4049/jimmunol.181.6.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rangachari M, et al. T-bet negatively regulates autoimmune myocarditis by suppressing local production of interleukin 17. J Exp Med. 2006;203:2009–2019. doi: 10.1084/jem.20052222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan X, et al. A novel role of CD4 Th17 cells in mediating cardiac allograft rejection and vasculopathy. J Exp Med. 2008;205:3133–3144. doi: 10.1084/jem.20081937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo S, Cobb D, Smeltz RB. T-bet inhibits the in vivo differentiation of parasite-specific CD4+ Th17 cells in a T cell-intrinsic manner. J Immunol. 2009;182:6179–6186. doi: 10.4049/jimmunol.0803821. [DOI] [PubMed] [Google Scholar]
- 20.Hegazy AN, et al. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity. 2010;32:116–128. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Lee YK, et al. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szabo SJ, et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 23.Bettelli E, et al. Loss of T-bet, but not STAT1, prevents the development of experimental autoimmune encephalomyelitis. J Exp Med. 2004;200:79–87. doi: 10.1084/jem.20031819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGeachy MJ, et al. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, et al. Crucial role of interleukin-7 in T helper type 17 survival and expansion in autoimmune disease. Nat Med. 2010;16:191–197. doi: 10.1038/nm.2077. [DOI] [PubMed] [Google Scholar]
- 26.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang ES, Hong JH, Glimcher LH. IL-2 production in developing Th1 cells is regulated by heterodimerization of RelA and T-bet and requires T-bet serine residue 508. J Exp Med. 2005;202:1289–1300. doi: 10.1084/jem.20051044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abromson-Leeman S, Bronson RT, Dorf ME. Encephalitogenic T cells that stably express both T-bet and ROR gamma t consistently produce IFNgamma but have a spectrum of IL-17 profiles. J Neuroimmunol. 2009;215:10–24. doi: 10.1016/j.jneuroim.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta DS, Wurster AL, Weinmann AS, Grusby MJ. NFATc2 and T-bet contribute to T-helper-cell-subset-specific regulation of IL-21 expression. Proc Natl Acad Sci U S A. 2005;102:2016–2021. doi: 10.1073/pnas.0409512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brustle A, et al. The development of inflammatory T(H)-17 cells requires interferon- regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 32.Schraml BU, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durant L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenner RG, et al. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc Natl Acad Sci U S A. 2009;106:17876–17881. doi: 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gocke AR, et al. T-bet regulates the fate of Th1 and Th17 lymphocytes in autoimmunity. J Immunol. 2007;178:1341–1348. doi: 10.4049/jimmunol.178.3.1341. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, et al. T-bet is essential for encephalitogenicity of both Th1 and Th17 cells. J Exp Med. 2009;206:1549–1564. doi: 10.1084/jem.20082584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghoreschi K, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 39.Veldhoen M, Hirota K, Christensen J, O'Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med. 2009;206:43–49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris TJ, et al. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol. 2007;179:4313–4317. doi: 10.4049/jimmunol.179.7.4313. [DOI] [PubMed] [Google Scholar]
- 41.Liu X, Lee YS, Yu CR, Egwuagu CE. Loss of STAT3 in CD4+ T cells prevents development of experimental autoimmune diseases. J Immunol. 2008;180:6070–6076. doi: 10.4049/jimmunol.180.9.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathur AN, et al. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol. 2007;178:4901–4907. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- 43.Mukasa R, et al. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32:616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 45.Cawley S, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 46.Jones DC, et al. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312:1223–1227. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.