Summary
Background
The leading actin network in motile cells is composed of two compartments, the lamellipod and the lamellum. Construction of the lamellipod requires a set of conserved proteins that form a biochemical cycle. The timing of this cycle and the roles of its components in determining actin network architecture in vivo, however, are not well understood.
Results
We performed fluorescent speckle microscopy on spreading Drosophila S2 cells by using labeled derivatives of actin, the Arp2/3 complex, capping protein, and tropomyosin. We find that capping protein and the Arp2/3 complex both incorporate at the cell edge but that capping protein dissociates after covering less than half the width of the lamellipod, whereas the Arp2/3 complex dissociates after crossing two thirds of the lamellipod. The lamellipodial actin network itself persists long after the loss of the Arp2/3 complex. Depletion of capping protein by RNAi results in the displacement of the Arp2/3 complex and disappearance of the lamellipod. In contrast, depletion of cofilin, sling-shot, twinfilin, and tropomyosin, all factors that control the stability of actin filaments, dramatically expanded the lamellipod at the expense of the lamellum.
Conclusions
The Arp2/3 complex is incorporated into the lamellipodial network at the cell edge but de-branches well before the lamellipodial network itself is disassembled. Capping protein is required for the formation of a lamellipodial network but dissociates from the network precisely when filament disassembly is first detected. Cofilin, twinfilin, and tropomyosin appear to play no role in lamellipodial network assembly but function to limit its size.
Introduction
Amoeboid cell motility requires construction of dynamic networks of actin filaments at the cell’s leading edge. These networks form characteristic compartments, defined by (i) proximity to the membrane, (ii) rates of filament assembly and disassembly, and (iii) velocity of filament movement relative to the cell edge [1–3]. Salmon et al. identified four distinct compartments in the actin cytoskeleton of migrating newt lung epithelial cells [3]. At the very front of the cell, in a compartment called the lamellipod, actin filaments are nucleated and cross-linked and treadmill rapidly toward the cell body as they elongate from their membrane-proximal barbed ends. Filaments in this compartment are relatively short lived and depolymerize not far from the cell edge. Behind the lamellipod is a more slowly moving actin network called the lamellum. A third population of filaments, located in the cell body, undergoes anterograde flow toward the cell edge. At the boundary between the lamellum and the cell body, termed the convergence zone, myosin contraction causes actin filaments from the lamellum and cell body to meet.
We previously proposed, based on dendritic nucleation by the Arp2/3 complex, a biochemical cycle for assembly and disassembly of actin filaments in motile networks [4]. In our model, the Arp2/3 complex is activated at the membrane of the leading edge by interaction with nucleation-promoting factors and preexisting actin filaments. There, it nucleates new daughter filaments that form an interconnected, branching network [5]. The new filaments elongate from their free barbed ends, and the free energy of this polymerization is converted into work, pushing the cell membrane forward. Each filament elongates until capping protein binds to its barbed end and terminates its growth [6].Actin disassembly is catalyzed in part by cofilin, which together with profilin promotes recycling of actin monomers [7]. This mechanism is thought to drive actin dynamics primarily in the lamellipod, in part because tropomyosin prevents binding of both cofilin and the Arp2/3 complex to actin filaments in the lamellum [8, 9]. This view is consistent with the morphologies seen by electron microscopy of actin networks of motile cells. Svitkina et al. found the lamellipodial network to be composed primarily of very short (100 nm), densely branched actin filaments that contain significant amounts of the Arp2/3 complex, whereas they found lamellar regions of the cell to contain long, unbranched filaments and much less Arp2/3 complex [10].
In this study, we used fluorescent speckle microscopy of actin, Arp2/3, capping protein, and tropomyosin to correlate the spatial and temporal dynamics of the actin network at the leading edge of Drosophila S2 cells with those of the proteins that construct it. These proteins are visible only when bound to filamentous actin, and so imaging their association with the network provides a dynamic map of when and where they exert their influence on cytoskeletal architecture. We coupled this dynamic mapping approach with dsRNA-induced knockdown of protein expression to determine the specific effects of filament capping and disassembly on the structure of leading-edge networks. We found that the lamellipodial compartment is more complex than previously appreciated and contains at least three subcompartments: (i) a distal zone, near the cell edge, in which the Arp2/3 complex and capping protein associate with the network, (ii) a middle zone lacking capping protein but containing the Arp2/3 complex, and (iii) a region that is near the boundary with the lamellum and lacks both proteins. We also found that assembly of the lamellipod depends critically on the presence of both the Arp2/3 complex and capping protein. Loss of cofilin, the cofilin phosphatase slingshot, or tropomyosin leads to expansion of the lamellipod at the expense of the lamellum. Together, our data reveal the precise timing of molecular events at the leading edge and indicate that cofilin and capping protein play profoundly different roles in determining the architecture of motile actin networks in vivo.
Results
Speckle Dynamics Reveal Multiple Actin Networks in Drosophila S2 Cells
Rogers et al. previously used Drosophila S2 cells to study the function of actin regulatory proteins [11]. Here we used fluorescent speckle microscopy to study S2 cytoskeletal dynamics in live cells. When plated on concanavilin A, S2 cells spread into a circular morphology with a 4- to 5-µm wide, actin-rich cortex surrounding a central cell body. The cortex is less than 200 nm thick, making it well suited for speckle microscopy. We expressed Drosophila actin fused to EGFP under control of a copper-inducible promoter. In the absence of added copper, promoter leakiness produces low, stable levels of GFP-actin and produces fluorescent speckles.
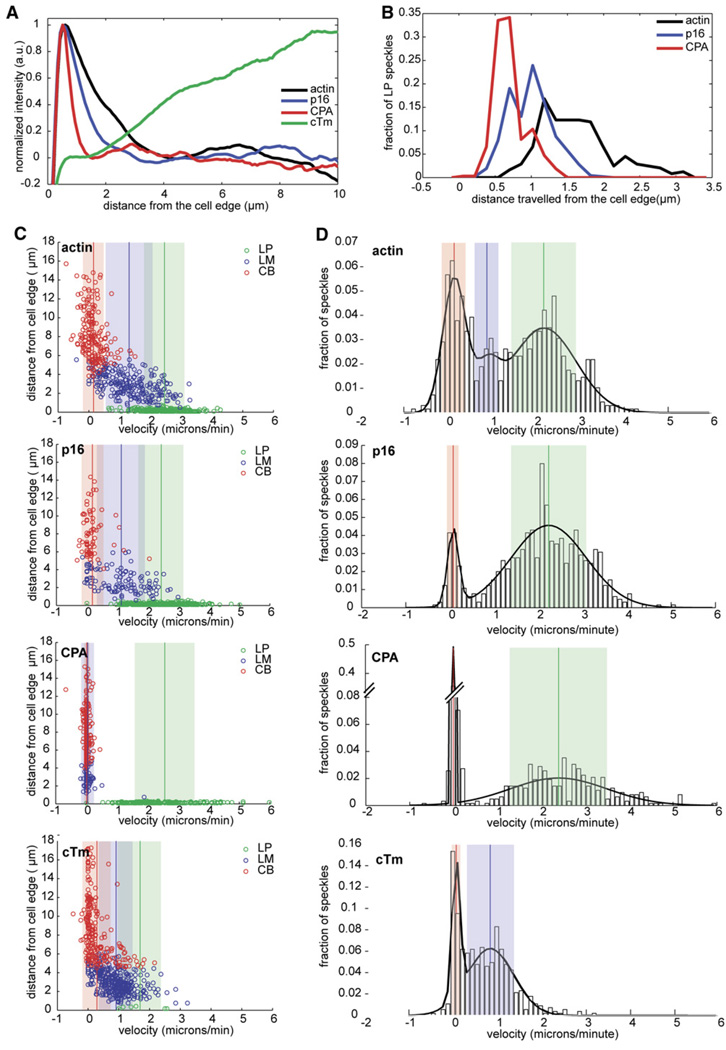
Consistent with phalloidin staining of S2 cells, GFP-actin speckles appeared primarily in the peripheral cortex (Movie S1 in the Supplemental Data available online). The highest speckle density occurs within one micron of the cell edge, and the lowest density occurs in the cell body (Figures 1A and 1B). We used kymographs to analyze speckle trajectories (11 cells, 42 kymographs, 600 speckles; Figure 1B, top panel) and plotted the initial distance of each speckle from the cell edge versus its average velocity (Figure 2C). This analysis revealed three classes of actin speckles (Figure 2B; also Table S1): (i) rapidly moving (2.46 ± 0.04 µm/min) speckles near the cell edge (<1 µm) that travel an average distance of 1.40 ± 0.03 µm; (ii) more slowly moving (1.31 ± 0.04 µm/min) speckles between the cell edge and the cell body (1–6 µm from the edge) that travel an average distance of 0.77 ± 0.03 µm, and (iii) immobile (0.15 ± 0.02 µm/min) speckles in the cell body (>6 µm from the edge). Individual actin speckles did not undergo significant changes in velocity. Three distinct populations also appeared in a histogram of actin-speckle velocities, even when location was not taken into account (Figure 2D), and their mean values matched the velocities calculated from bins based on distance from the cell edge (Figures 2C and 2D). Thus, based on location and velocity, we could distinguish three compartments in the S2 cytoskeleton, similar to those previously described by Ponti et al. [12]. These authors referred to the membrane-proximal actin network as the lamellipod and the slower network immediately behind it as the lamellum. We follow the same convention here.
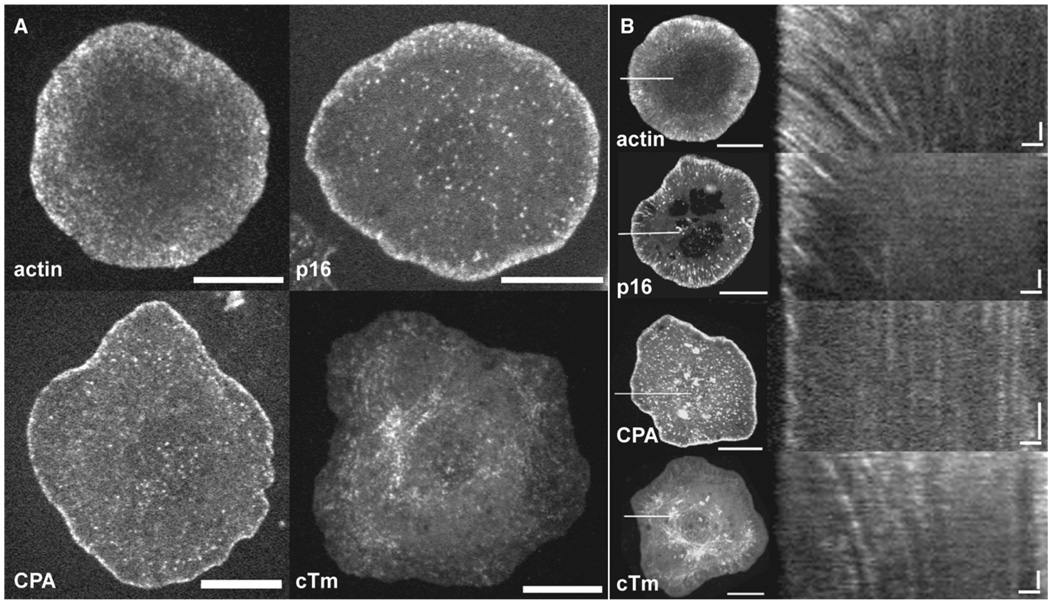
Figure 1. GFP-Tagged Actin and Actin-Regulatory Proteins Occupy Unique Compartments at the Leading Edge.
(A) S2 cells expressing GFP-actin, GFP-p16 (Arp2/3 subunit), GFP-capping protein α (CPA), and GFP-cytoskeletal tropomyosin (cTm) were imaged by confocal microscopy. Images are a single frame from time-lapse movies of live S2 cells plated on concanavilin A for approximately 1 hr. The scale bar represents 10 µm.
(B) Kymographs of GFP speckles reveal dynamic compartmentalization of actin and actin-binding proteins. Left, maximum-intensity projections of time-lapse movies of S2 cells expressing GFP-actin (top), GFP-p16, GFP-CPA, and GFP-cTm. The scale bar represents 10µm. The white line at the 9 o’clock position indicates the cell area from which kymographs (right) were constructed. Right panel: The x axis scale bar represents 1 µm; the y axis scale bar represents 30 s.
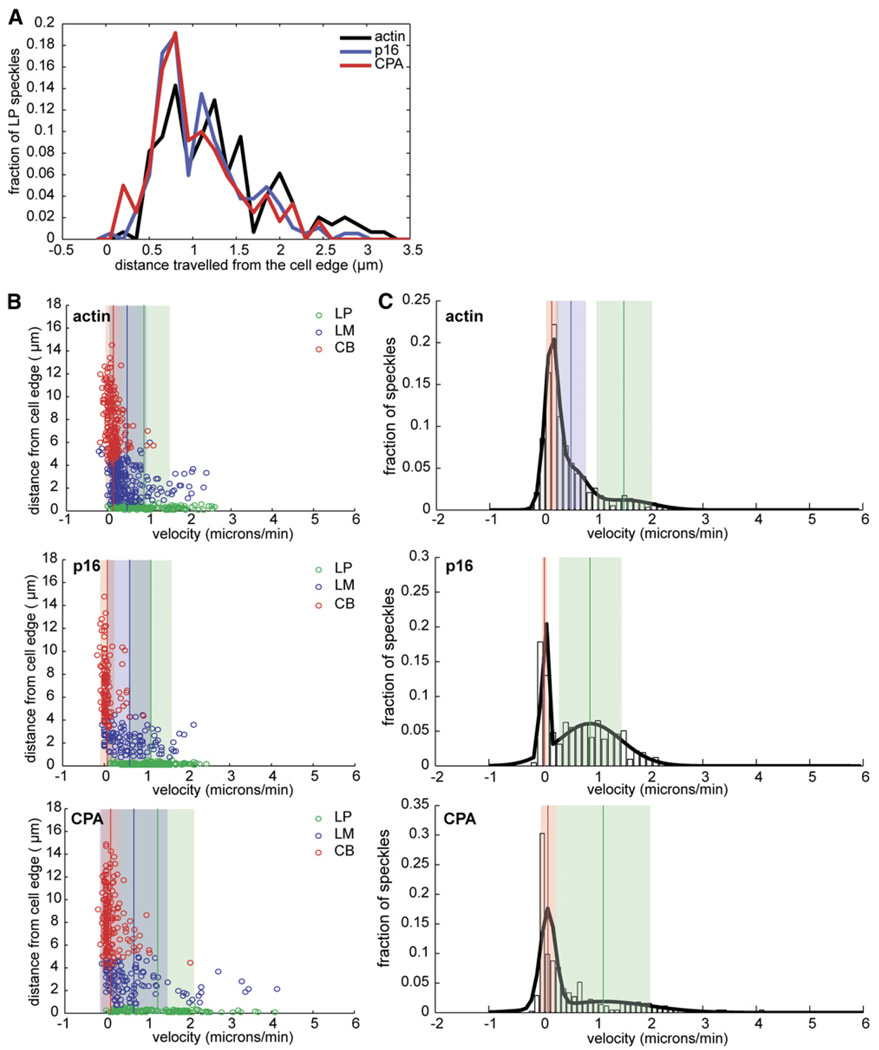
Figure 2. Quantitative Analysis of GFP-actin, GFP-p16, GFP-CPA, and GFP cTm Speckle Localization and Dynamics.
(A) Normalized average-fluorescence-intensity line scan of GFP fluorescence taken from a representative movie of an S2 cell expressing GFP-actin, GFP-p16, GFP-CPA, or GFP-cTm and displayed as a function of distance from the cell edge, as described in the Experimental Procedures. (B–D) Individual speckle trajectories from at least ten cells were analyzed via kymograph analyses. (B) Line histogram showing the distance traveled by GFP-actin, GFP-p16, and GFP-CPA speckles originating in the LP. Trajectories used in this histogram are shown as green circles in each of the scatter plots in (C). (C) Scatter plots of the distance from the cell edge versus velocity. From top to bottom: actin, cTm, p16, and CPA. Speckle trajectories were labeled as originating in the lamellipod (LP, green circles), lamellum (LM, blue circles), or cell body (CB, red circles) based on their starting distance from the cell edge. Vertical lines indicate the mean velocity for each population, and shaded areas indicate the standard deviation. (D) Probability density function of the velocities of all GFP speckles of (top to bottom) actin, cTm, p16, and CPA. The probability density function is overlayed with the best-fit line (black line) derived from cumulative density-function-curve fitting (data not shown). Vertical lines indicate means, and shaded areas indicate the standard deviation for each population.
Actin-Regulatory Proteins Define Additional Subcompartments of the Peripheral Cytoskeleton
We next used speckle microscopy to study the dynamics of key actin-regulatory factors. We expressed low levels of EGFP fused to subunits of the Arp2/3 complex, capping protein (CPA), or cytoskeletal tropomyosin (cTm) in S2 cells. Previous studies suggest that Arp2/3 subunits and capping protein can be fluorescently tagged without disrupting in vivo function [13–15]. We find that GFP-cTm is incorporated into the actin network normally and can partially rescue the phenotype of cTm-depleted cells (Figure S3).
The localization and dynamics of GFP-Arp3 and GFP-p16 speckles were identical, but GFP-p16 typically generated lower background fluorescence and higher contrast, and so we used it for most experiments presented here. The distribution of fluorescently tagged Arp2/3 complex in S2 cells was similar to that of previous immunolocalization studies [11]. Based on speckle location and velocity, we can conclude that the complex associates almost exclusively with the lamellipodial actin network within 1 µm of the cell edge (Figure 1; also Movie S2). When we plotted distance from the cell edge versus velocity (ten cells, 81 kymographs, more than 650 speckles) we observed three populations of Arp2/3 speckles that corresponded to those observed with labeled actin. Arp2/3 speckles, however, were born predominantly in the lamellipod and underwent retrograde flow at the same velocity as lamellipodial actin speckles (2.44 ± 0.03 µm/min versus 2.46 ± 0.04 µm/min). Arp2/3 speckles, however, had a significantly shorter average lifetime than actin speckles (21.6 ± 0.6 s versus 35.4 ± 0.6 s) and did not travel as far from the cell edge (0.83 ± 0.01 µm versus 1.40 ± 0.03 µm; see also Table S1). We confirmed these differences by simultaneously expressing GFP-actin and RFP-p16 in the same cells (our unpublished observations).
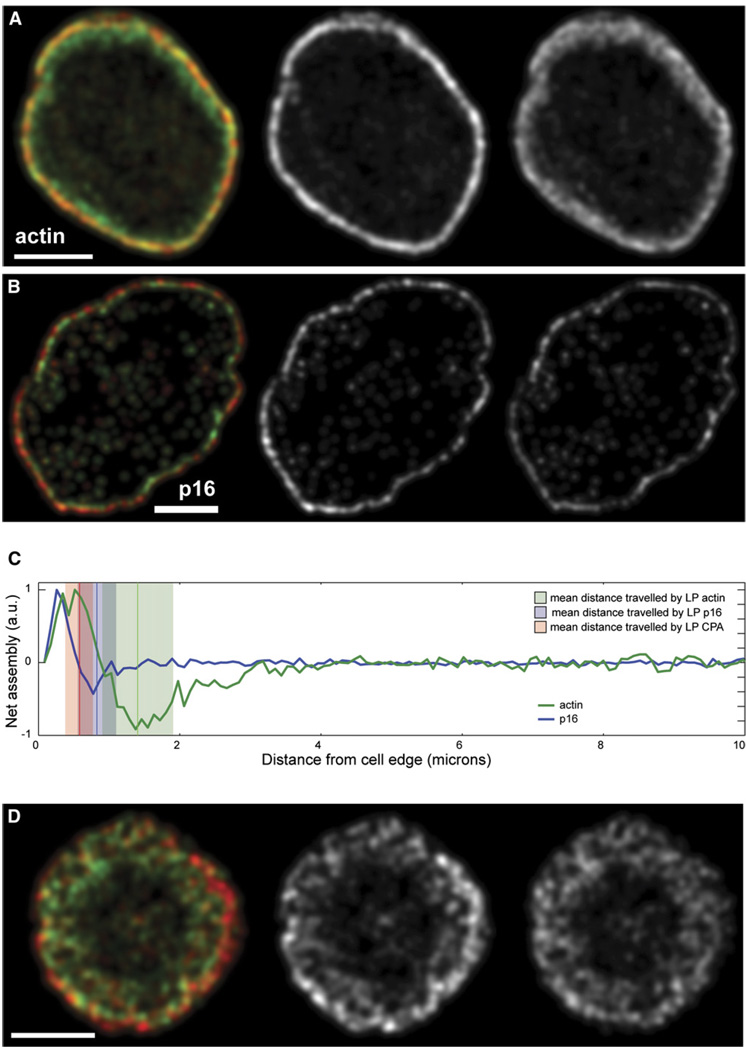
Ponti et al. [12] used quantitative image analysis to map the kinetics of the appearance and disappearance of actin speckles and found that the lamellipod contains a narrow (1 µm) band of rapid speckle appearance near the cell edge and then a band of rapid speckle disappearance. We used the same speckle-tracking software to create maps of both actin and Arp2/3 speckle dynamics (Figures 3A and 3B). Both actin and Arp2/3 speckle maps revealed sharp, polarized bands of assembly and disassembly near the cell edge but, although the appearance of Arp2/3 speckles coincided with the peak of actin filament assembly, Arp2/3 speckle disappearance preceded actin disassembly by 0.6 µm (Figure 3C). Interaction of the Arp2/3 complex with the actin network, therefore, defines a distinct subcompartment comprising the membrane-proximal two-thirds of the lamellipod.
Figure 3. Visualization of Network Assembly in S2 cells.
(A and B) Time-averaged turnover map of F-actin (A) and p16 (B) calculated from speckle-tracking analysis of an S2 cell expressing GFP-actin or GFP-p16. Speckles were tracked and analyzed with fsmCenter, created by the Danuser group (Scripps Research Institute), with algorithms previously described [14]. In the left panels, red marks the locations of rapid actin (A) and Arp2/3 (B) speckle appearance. Green marks rapid actin (A) and Arp2/3 (B) speckle disappearance. In the middle and right panels, the rates of actin (A) and Arp2/3 (B) speckle birth (middle) and death (right) are plotted as grayscale values. The scale bar represents 10 µm.
(C) Net actin and Arp2/3 speckle turnover rates are plotted versus distance from the cell edge. Data are from single S2 cells expressing either GFP-actin (A) or GFP-p16 (B). The thick green line indicates net actin filament assembly/disassembly calculated from the cell in (A). The thick blue line indicates net Arp2/3 association/dissociation to the actin network; this was calculated from the cell in (B). Mean (vertical line) and standard deviation (shaded rectangle) of the distance traveled by capping protein (red), Arp2/3 (blue) and actin (green) were calculated from kymograph analyses. The figure was created with cytoProbe software from the Danuser group (Scripps).
(D) A turnover map of F-actin was calculated from speckle-tracking analysis of an S2 cell depleted of cTm and expressing GFP-actin. Actin assembly is shown in red (center), and actin disassembly is shown in green (right). The scale bar represents 10 µm.
Capping protein (CPA) speckles were restricted to a smaller portion of the lamellipod than Arp2/3 speckles and generally appeared as a thin ring at the very cell edge (Figure 1A; see also Movie S3). CPA speckles were too short lived for automated analysis, but kymograph analysis (12 cells, 127 kymographs, >500 speckles) revealed that they were restricted to within 0.58 ± 0.01 µm of the cell edge and had an average lifespan of 15.0 ± 0.6 s. Their average velocity was 2.57 ± 0.05 µm/min, similar to that of lamellipodial actin and Arp2/3 complex speckles (Figures 2C and 2D). The distribution of CPA speckle velocities revealed two populations with means matching the average velocities of actin in the lamellipod and cell body (Figure 2D). Capping protein association thus defines an additional lamellipodial subcompartment that is smaller than, and nested within, that defined by the Arp2/3 complex.
Finally, we analyzed the localization and dynamics of cytoplasmic tropomyosin (cTm) speckles (ten cells, 103 kymographs, >550 speckles). GFP-cTm speckles were all born at least 1 µm from the cell edge (Figure 2C; also Movie S4) and had velocities matching those of GFP-actin speckles in the lamellum and cell body (0.91 ± 0.03 µm/min, 0.29 ± 0.03 µm/min; Figure 2D). The zones in which tropomyosin and Arp2/3 complex speckles appeared were almost completely nonover-lapping.
Depletion of Tropomyosin, Slingshot, or Twinfilin Causes Expansion of the Lamellipod at the Expense of the Lamellum
To understand the molecular mechanisms underlying construction of the peripheral actin network, we incubated cells with double-stranded (ds) RNAs designed to deplete various actin-regulatory proteins. We observed the effects on cell morphology and on dynamics of actin, capping protein, tropomyosin, and the Arp2/3 complex by using speckle microscopy.
In samples treated with cTm dsRNA, 50% of cells appeared to be wild-type, 35% had ragged leading edges, and 15% were stellate or were otherwise not well spread. To determine whether the range of phenotypes observed represents cell-to-cell variation in efficacy of cTm RNAi, we treated an S2 cell line stably expressing GFP-cTm with cTm dsRNA for 7 days. Cells retaining high GFP-cTm fluorescence appeared to be wild-type, whereas cells expressing little or no GFP-cTm displayed irregular leading edges (Figure S3B). We focused on cells exhibiting the ragged-edged phenotype produced by the partial knockdown of cTm (Figure S1A).
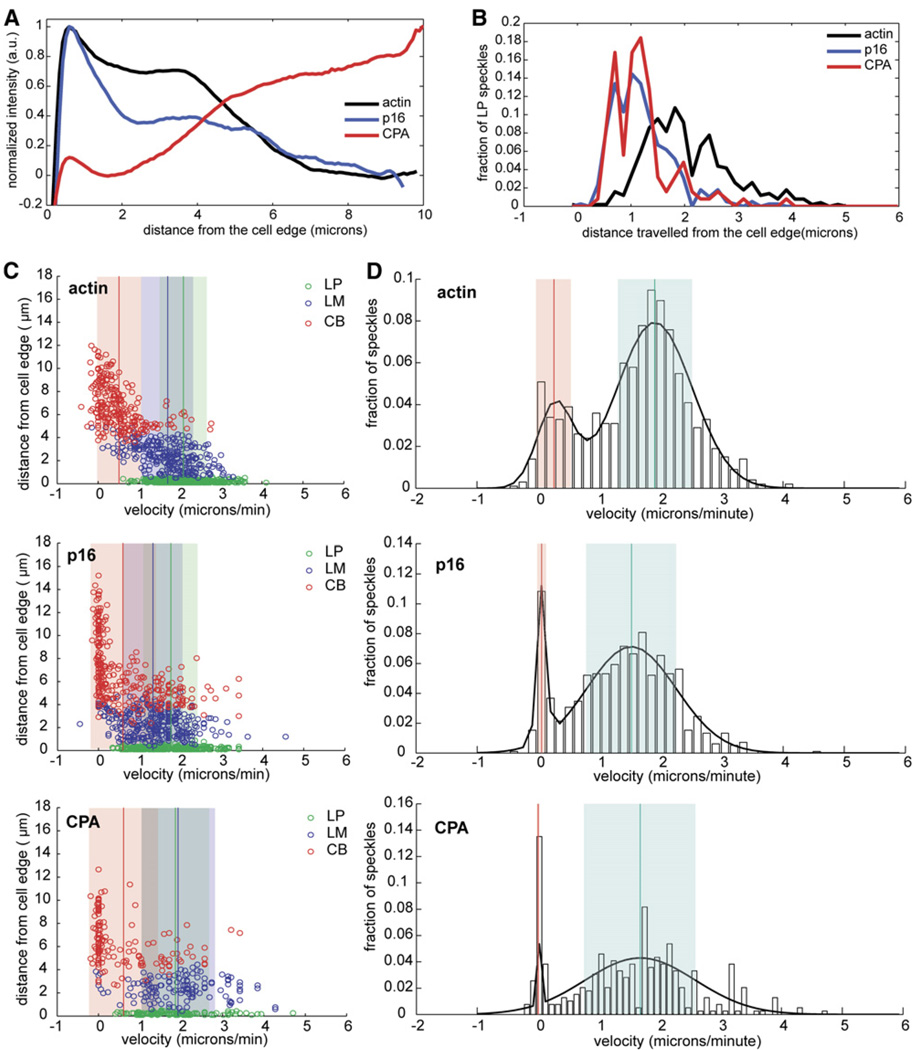
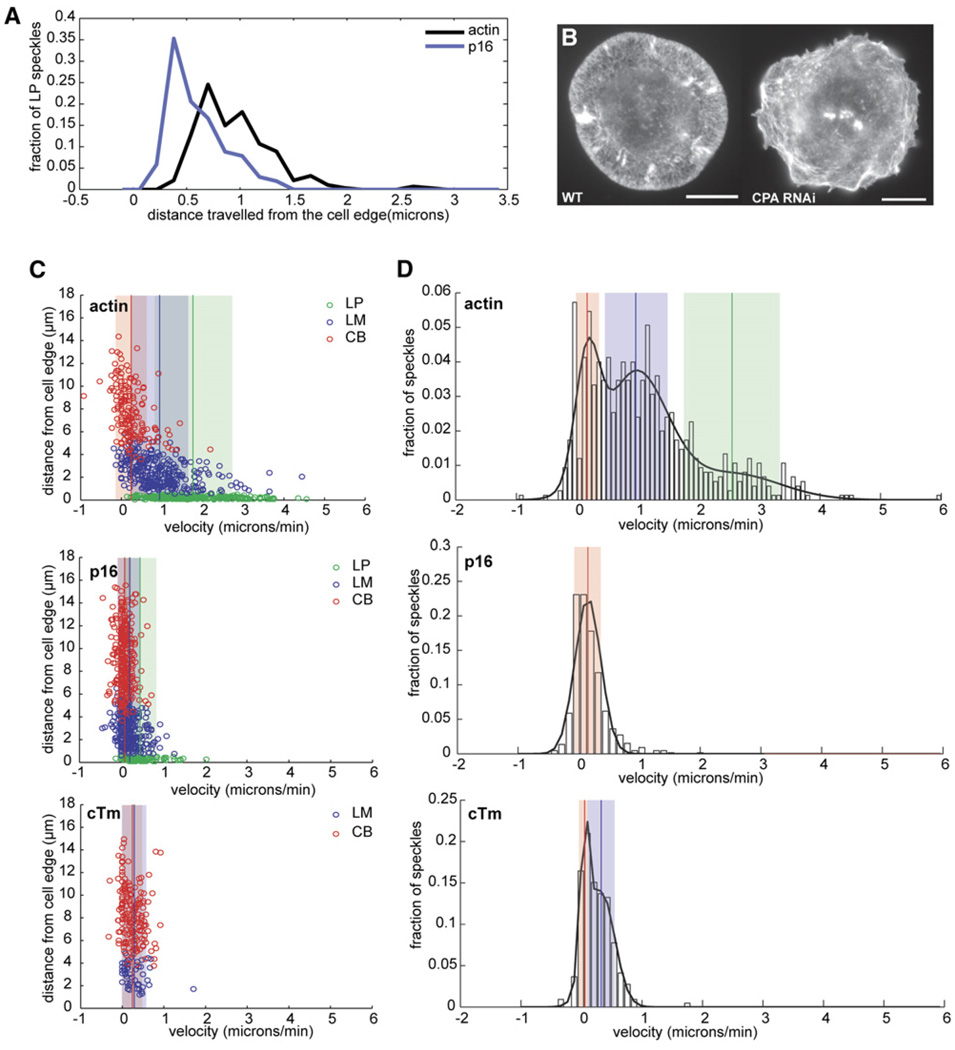
The leading edges of cTm-depleted cells were more dynamic than untreated cells and underwent rapid protrusion and retraction (Figure S1B). We observed only two classes of actin speckles in these cells: (i) rapidly moving speckles distributed uniformly around the cell periphery and (ii) immobile speckles in the cell body. From kymograph analysis, we determined that the velocity of rapidly moving actin speckles (2.08 ± 0.03 µm/min) was intermediate between the lamellipodial and lamellar velocities of untreated cells and had a distribution that is best described by two Gaussians rather than three (Figure 4D). In addition, the lifetime of the rapidly moving actin speckles was 64% longer than that of lamellipodial actin speckles in untreated cells (Table S1) and appeared to comprise a single broad network (Figures 4C–4E).
Figure 4. Tropomyosin RNAi Causes Expansion of the Lamellipod at the Expense of the Lamellum.
(A) Normalized average-fluorescence-intensity line scan of GFP fluorescence taken from a representative movie of a cTm-depleted S2 cell expressing GFP-actin, GFP-p16, or GFP-CPA and displayed as a function of distance from the cell edge.
(B) Line histogram showing distance traveled by GFP-actin, GFP-p16, and GFP-CPA speckles originating in the LP. Speckles used in this histogram are shown as green circles in each of the scatter plots in (C).
(C) Scatter plots were created as described in Figure 2C. Tropomyosin-depleted cells expressing (from top to bottom) GFP-actin, GFP-p16, and GFP-CPA were analyzed.
(D) Probability density functions were created as described in Figure 2D. Tropomyosin-depleted cells expressing (from top to bottom) actin, p16, or CPA were analyzed.
To determine whether this network represents a rapidly moving lamellum or a greatly expanded lamellipod, we examined the rates of actin-filament turnover and the dynamics of lamellipod-specific proteins. By automated image analysis, we observed broad, polarized bands of actin assembly and disassembly spanning the cortex (Figure 3D). In addition, the spatial distributions of Arp2/3 and capping protein speckles were no longer restricted to subcompartments and extended throughout the cortex (Figures 4C–4E), and some p16 and CPA speckles now reached the boundary of the cell body. The peripheral actin network of cTm-treated cells, therefore, appears to be an expanded lamellipodial network.
Interestingly, depletion of the cofilin phosphatase slingshot or the actin-sequestering and -severing protein twinfilin produced the same phenotype as depletion of tropomyosin (Table S1 and Figure S4 and Figure S5). Neither slingshot nor twinfilin knockdown affected the spreading or gross morphology of S2 cells. Speckle microscopy of labeled actin, capping protein, and Arp2/3, however, revealed that the peripheral actin cytoskeleton in slingshot- or twinfilin-depleted cells was a single, homogeneous, lamellipodial network with retrograde flow velocities slightly slower than the lamellipodial velocities of untreated cells (Table S1). Thus, loss of tropomyosin, slingshot, and twinfilin all produce similar expansions of the lamellipod.
Cofilin Depletion Decreases Retrograde Velocities of Peripheral Actin Networks and Causes Lamellipod Expansion
We depleted cofilin in S2 cells by using dsRNA against Drosophila cofilin/ADF (twinstar). Consistent with a previous report [11], most of the treated cells (76%) failed to spread on concanavilin A. Given sufficient time (1.5–2 hr), 20%–30% of cells eventually spread, and the gross morphology of these cells appeared to be normal. By staining with phalloidin, we observed elevated levels of filamentous actin (data not shown). Confocal microscopy of live cells expressing GFP actin showed that the leading-edge networks of the well-spread cofilin-depleted cells moved significantly more slowly than controls (Figure 5). GFP-actin speckles averaged 0.88 ± 0.05 µm/min in the lamellipodial region (and 2.46 ± 0.04 µm/min in untreated cells) and 0.48 ± 0.03 µm/min in the lamellar region (and 1.31 ± 0.04 µm/min in untreated cells). Similarly, GFP-p16 and GFP-CPA also traversed the lamellipod at slower rates; velocities were 1.11 ± 0.04 µm/min and 1.25 ± 0.08 µm/min, respectively (Table S1). Tropomyosin decoration of actin filaments was greater in cofilin-depleted cells but was still most pronounced in the cell body and relatively sparse near the cell edge (data not shown).
Figure 5. Cofilin RNAi Causes Decreased Velocities and Lamellipodial Expansion.
(A) Line histogram showing the distance traveled by GFP-actin, GFP-p16, and GFP-CPA speckles originating in the LP in cofilin-depleted cells. Speckles used in this histogram are shown as green circles in each of the scatter plots in (B).
(B) Scatter plots were created as described in Figure 2C. Cofilin-depleted cells expressing (from top to bottom) GFP-actin,GFP-p16, or GFP-CPA were analyzed.
(C) Probability density functions were created as described in Figure 2D. Cofilin-depleted cells expressing (from top to bottom) actin, p16, or CPA were analyzed.
Cofilin-depleted cells, like cells depleted of tropomyosin, slingshot, and twinfilin, exhibited expanded zones of capping-protein and Arp2/3-complex speckle movement as well as increased speckle lifespans (Table S1; also Figures 5A–5C). Because of the speckles’ slow speed relative to the length of the time-lapse sequences, many trajectories were truncated in the kymographs (Figure 5A), and this resulted in the underestimation of lifespan and distance traveled. Nonetheless, the coincident localization of lamellipodial actin, Arp2/3 complex, and capping protein are indicative of an expanded lamellipod.
Depletion of Capping Protein Abolishes the Lamellipod but has Little Effect on the Lamellum
We next observed the effect of depleting the α subunit of capping protein on peripheral actin networks. Depletion of the β subunit or both subunits of capping protein produced identical phenotypes (data not shown). After 7 day RNAi treatment, 95% of cells showed intense membrane ruffling but spread well on concanavilin A. In most CPA-depleted cells, actin filaments were long and bundled, curved around the margins of the cell, and often formed clumps or tangles (Figure 6B).
Figure 6. Depletion of Capping Protein Abolishes the Lamellipod but has Little Effect on Lamellar Dynamics.
(A) Line histogram showing the distance traveled by GFP-actin and GFP-p16 speckles originating in the LP in CPA-depleted cells. Speckles used in this histogram are shown as green circles in each of the scatter plots in (C).
(B) S2 cells were fixed and F-actin and visualized with Alexa488 phalloidin. Left: untreated cell. Right: CPA-depleted cell.
(C) Scatter plots were created as described in Figure 2C. Capping-protein-depleted cells expressing (from top to bottom) GFP-actin, GFP-p16, and GFP-cTm were analyzed. (D) Probability density functions (PDF) were created as described in Figure 2D. Capping protein depleted cells expressing (from top to bottom) GFP-actin, GFP-p16 or GFP-cTm were analyzed.
Despite the pronounced ruffling at the edges of CPA-depleted cells, GFP-actin speckles at the cell edge moved 25% more slowly and for a significantly shorter distance (0.70 ± 0.02 µm) than lamellipodial actin speckles in untreated cells. Farther from the cell edge, actin speckles moved at velocities characteristic of lamellar actin in untreated cells (0.94 ± 0.04 µm/min). The distribution of actin-speckle velocities fit well to a three-population model, and means were similar to those in untreated cells. However, the population moving at lamellar-like speeds was much larger than in untreated cells and were often found close to the cell edge (Figures 6C and 6D).
The effect of CPA depletion on Arp2/3 speckles was dramatic. Almost all observed speckles were immobile and aggregated in bright puncta, often in areas distal from the cell edge. Kymograph analysis showed that the velocities of GFP-p16 speckles averaged less than 0.5 µm/min (Figure 6C). The velocities of GFP-cTm speckles in the lamellum and cell body averaged 0.2–0.3 µm/min (Table S1; also Figure 6C), but GFP-cTm speckles were still excluded from the very edge of the cell.
Discussion
Association of Capping Protein and the Arp2/3 Complex with the Lamellipodial Actin Network
We detected two distinct actin networks at the leading edge of Drosophila S2 cells; these networks were similar to those identified by Ponti et al. [12] in other cell types. Actin filaments in the most membrane-proximal network (the lamellipod) assemble and disassemble in a spatially polarized fashion and undergo rapid retrograde translocation toward the cell body at rates comparable to those observed in fish epidermal keratocytes [16]. The majority of these filaments are nucleated at the cell edge and disassembled 35 s later, approximately 1.5 µm from where they were born ([12] and this study).
We observed that most Arp2/3 complex speckles are confined to the lamellipod, typically appear at the very edge of the cell, undergo retrograde flow at the same rate as lamellipodial actin, and have an average lifetime of 22 s before they disappear in a narrow zone approximately 0.8 µm from the cell edge. This zone is distinct from the region where lamellipodial actin speckles begin disappearing. Arp2/3 speckles always disappeared at the same place, regardless of speckle density, indicating that the observed spatial differences are not an artifact of relative differences in labeling. This result has several important implications. (1) The lamellipod network is not held together solely by the Arp2/3 complex. Further studies will be required to determine which crosslinkers maintain the integrity of the lamellipod in the absence of the Arp2/3 complex. One excellent candidate is ABP-280, which supports leading-edge protrusion and motility in several cell types [17, 18]. (2) In vivo, filament debranching and filament depolymerization are separate processes. This finding differs from those of in vitro reconstitution studies in which the Arp2/3 complex dissociates from motile actin networks only when the filaments to which it is attached are disassembled [19]. The mechanism of Arp2/3 dissociation in vivo requires further study.
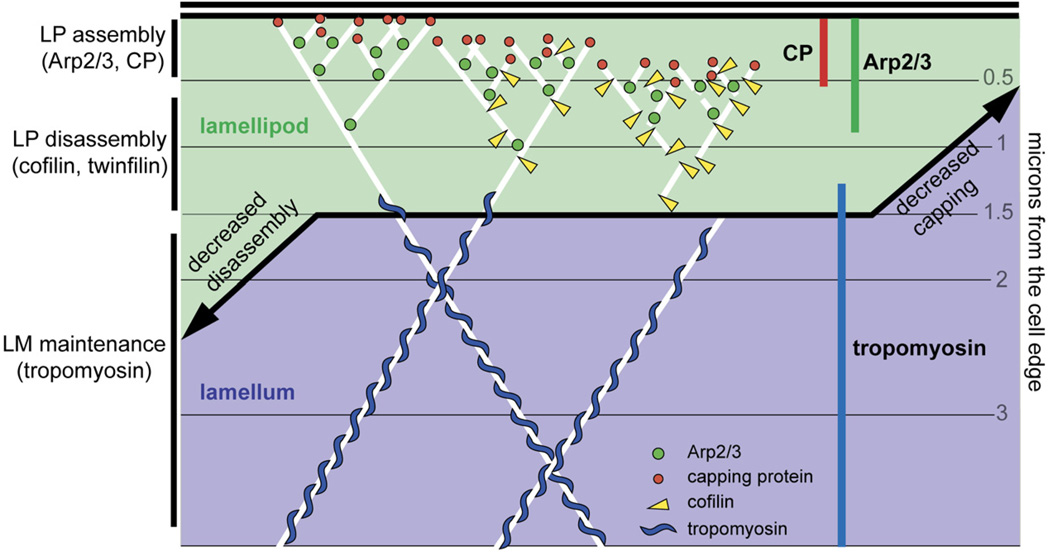
Capping protein also begins life at the cell edge but disappears even more quickly than Arp2/3 speckles. If the Arp2/3 complex nucleates actin filaments at the cell edge and capping protein binds their barbed ends, why do the two proteins dissociate from the network with different kinetics? Formally, there are three possibilities: (1) Filaments are specifically uncapped by a factor within the lamellipod [20]; (2) filaments bound to capping protein are preferentially targeted for early disassembly; and (3) the mechanism of filament disassembly in the lamellipod releases capping protein from the network before the Arp2/3 complex. Because knockdown of the actin-disassembly factor cofilin results in expansion and complete overlap of both the Arp2/3 and capping-protein zones (Figure 5), we favor the third idea. When overlaid on the map of actin speckle dynamics, we find that the disappearance of capping protein corresponds to the earliest point at which actin disassembly is observed. Because the pointed ends of filaments in the lamellipod are anchored to the network by the Arp2/3 complex, cofilin-mediated filament severing will increase the mobility of the barbed end more than that of the pointed end (Figure 7).
Figure 7. Schematic Model of Actin and Actin-Regulatory Proteins at the Leading Edge.
Arp2/3 (green circles), capping protein (red circles), cofilin (yellow triangles), and actin (white lines) build lamellipodial actin networks (green area), whereas tropomyosin (blue “S” shapes) is associated with lamellar actin networks (blue area). A capping decrease due to depletion of capping protein leads to the collapse of the lamellipod and expansion of the lamellum. A disassembly decrease due to cofilin, slingshot, twinfilin, or tropomyosin depletion leads to the expansion of the lamellipod at the expense of the lamellum.
Depletion of capping protein results in loss of Arp2/3-complex activity, as judged from association with the actin network, and almost complete loss of the lamellipod. Actin speckles still undergo slow retrograde movement from the leading edge, indicating that the underlying lamellar actin network is intact. Mejillano et al. found that depletion of capping protein in mouse melanoma cells caused explosive filopodium formation and mislocalization of the Arp2/3 complex [21] and speculated that, in the absence of capping protein, filopodium-forming factors such as Ena/VASP-family proteins might displace the Arp2/3 complex. The likely explanation for the difference in phenotypes is that S2 cells express lower amounts of the filopodial machinery than those used by Mejillano et al. Our results therefore argue that filopodium-forming factors cannot account for the effect of capping protein on Arp2/3-complex activity. One possible explanation is that removal of capping protein perturbs the global balance between filamentous and monomeric actin. In the absence of capping protein, individual filaments polymerize longer and deplete soluble, monomeric actin, which in turn causes inhibition of Arp2/3-complex activity. Regardless of the mechanism, our results indicate that limiting filament growth at the cell edge plays a specific role in determining lamellipodial architecture and promoting efficient Arp2/3-complex activity.
Both the Arp2/3 complex and capping protein are required to reconstitute actin-dependent motility similar to that of the intracellular pathogen Listeria monocytogenes [22]. In the absence of capping protein, the Arp2/3 complex generates disorganized clouds of filamentous actin around Listeria [23] or polystyrene micro-spheres coated with the Arp2/3-activating domains of ActA or N-WASP [24]. The addition of capping protein converts these clouds into organized, polarized, and motile networks ([25] and our unpublished observations). These results reveal an important kinetic synergy between the Arp2/3 complex and capping protein and suggest that self-sustaining lamellipod formation requires a precise balance between the rates of filament nucleation and capping. This is also consistent with theoretical and experimental studies indicating that efficiency of protrusion driven by growth of dendritically branched networks is highly sensitive to the average filament length [26, 27].
Cofilin-Mediated Actin Disassembly Limits the Size of the Lamellipod
In most cell types, cofilin localizes to a zone near the lamellipod/lamellum junction [10], but its precise role in determining dynamic network architecture has not been established. In vitro, cofilin severs and depolymerizes actin filaments that have hydrolyzed their bound ATP and released the inorganic phosphate [28–30]. Consistent with this activity, several groups have reported that loss of cofilin function in vivo causes accumulation of filamentous actin [11, 31, 32]. Other studies report that filament severing by cofilin at the cell edge promotes actin assembly and is required for rapid membrane protrusion [33, 34].
Based on speckle microscopy of actin, Arp2/3 complex, and capping protein, we found that cofilin depletion specifically expands the lamellipodial actin network at the expense of the lamellar network. We observed similar but even more obvious phenotypes when we depleted the cofilin-activating phosphatase, slingshot or twinfilin. Twinfilin was initially described as an actin-monomer-sequestering protein [35], but budding-yeast twinfilin has recently been shown to sever actin filaments in vitro [36]. Our results suggest that twinfilin and cofilin work synergistically to disassemble lamellipodial actin networks and that loss of either enables the expansion of the lamellipod and loss of a two-network system.
Tropomyosin, an actin side-binding protein, inhibits both Arp2/3 binding and actin severing by cofilin [8, 37]. Several studies have confirmed that tropomyosin and the Arp2/3 complex localize to distinct compartments in vivo [8, 38], and we found that knockdown of tropomyosin led to expansion of the lamellipod. The simplest explanation for this result is that, in the absence of tropomyosin, the Arp2/3 complex is no longer limited to the leading edge but is free to associate with filaments throughout the cell. The fact that knockdown of tropomyosin produces the same phenotype as knockdown of slingshot or twinfilin suggests another possibility— namely, that the effect of tropomyosin knockdown may be mediated by actin-depolymerizing factors. In wild-type cells, cofilin activity may be directed toward the lamellipod by the activity of tropomyosin in the lamellum. In the absence of tropomyosin, all ADP actin filaments in the cell would become potential cofilin substrates, and this might act to disperse cofilin throughout the cell and reduce its concentration in the lamellipod.
Conclusions
Using fluorescent speckle microscopy, we reconstructed the timing of critical events in the construction of actin-filament networks at the leading edge of motile cells. Capping protein and the Arp2/3 complex occupy unique, nested subcompartments within the lamellipod and are both absolutely required for lamellipod formation. Cofilin, twinfilin, slingshot, and tropomyosin appear to play no role in the constructing the lamellipodial network but function to limit its size.
Experimental Procedures
Cell Culture and RNAi Treatment
Drosophila Schneider S2 cells were cultured and treated with dsRNA as previously described [12]. CPA RNAi was done with dsRNA specific to the α subunit. Cells were depleted of cTm by treatment with dsRNA specific to type II tropomyosin isoforms. Cofilin RNAi was performed with dsRNA against the entire coding sequence.
Plasmid Construction and Transfection
Gateway cloning technology (Invitrogen) was used for creating vectors for GFP expression in S2 cells. Drosophila genes for p16 (ArpC5), Arp3, cTm, capping protein α and β, and cofilin (twinstar) were cloned from an S2 cDNA library, sequenced, and inserted into p-Entr-D-TOPO plasmids. Genes were then cloned into destination vectors that included GFP under the control of a copper promoter. Actin, Arp3, p16, cofilin, and CPA were tagged with an N-terminal fluorescent tag, whereas cTm was labeled at its C terminus. S2 cells were transfected with Cellfectin (Invitrogen), 1–2 µg of destination plasmid, and 0.5 µg of pCoHygro plasmid (Invitrogen) (for construction of stable cell lines only). After 2 days, we observed fluorescent protein in transient cell lines or added hygromycin to select for stably transfected cells.
Speckle Microscopy
S2 cells transiently or stably expressing low quantities of fluorescent protein were plated on concanavilin-A-coated glass-bottomed dishes (MatTek) and observed between 30 min and 1.5 hr after plating. Images were taken with an Orca ER II camera (Hamamatsu) mounted to an Axiovert microscope (Zeiss) and Metamorph software (Molecular Devices) at 1 or 3 s intervals, and integration times were between 300 and 800 ms; usually, 2x binning was used.
Immunofluorescence and fixation
Cells were fixed as previously described [12]. Actin filaments were visualized with 200 nM Alexa-488 or Alexa-568 phalloidin (Molecular Probes/Invitrogen). Images were acquired with an Orca II cooled CCD camera (Hamamatsu) equipped on a Nikon TE300 inverted microscope and with Simple PCI software (Compix).
Kymograph Analysis
Kymograph analyses were done with ImageJ (http://rsb.info.nih.gov/ij) with a plugin written by J. Rietdorf and A. Seitz (http://www.embl.de/eamnet/html/body_kymograph.html). For sparsely speckled cells, 1-µm-thick lines were used for kymograph creation. For densely speckled cells, such as GFP-actin, kymographs were constructed with a 4-µm-thick line. Individual speckle trajectories and leading-edge outlines were drawn by hand in ImageJ and were analyzed with Matlab (Mathworks). Background cytoplasmic fluorescence, which is significant in the green channel, was utilized for manually determining the cell edge in maximal-intensity projections. Between 4 and 12 kymographs were created per cell, and between 6 and 12 cells were used for every RNAi and control condition.
Actin Speckle-Tracking and -Assembly Maps
We used FsmCenter, a software package designed by the Danuser group (Scripps, La Jolla, CA), to track GFP-actin and p16 speckles in S2 cells and to create actin-assembly maps. For assembly profiles, we reduced spatial averaging to a 1 pixel width in order to accurately visualize the narrow range in which GFP-p16 speckles appear. We used CytoProbe, written by Matthias Machacek, to create profiles showing activity or fluorescence intensity from the cell edge. To compare the fluorescence-intensity profiles of different proteins as shown in Figure 2A, we normalized the profiles such that the maximum value was set to 1 and the minimum value was set to 0. This allowed for a direct comparison of the localization of lamellipodial and lamellar proteins despite differences in cellular abundance.
Supplementary Material
Acknowledgments
We thank members of the Mullins laboratory, and especially Orkun Akin and Mark Dayel, for their insights, support, and helpful discussions. We are grateful to Ron Vale and his laboratory members for technical assistance and for the use of microscopes and reagents. Special thanks are due to Gaudenz Danuser and Matthias Machacek for training and the use of fsmCenter and cytoProbe, as well as for helpful discussions. Finally, we thank Tapio Heino, Katherine Miller, and Tadashi Uemura for generously providing antibodies. This work was supported by grants to R.D.M. from the National Institutes of Health (RO1 grant GM61010) and the Sandler Family Supporting Foundation and by funding from the University of California, San Francisco/University of California, Berkeley Nanomedicine Development Center and a National Science Foundation Graduate Research Fellowship to J.H.I.
Footnotes
Supplemental Data
Five figures, one table, and four movies are available online at http://www.current-biology.com/cgi/content/full/17/5/395/DC1/.
References
- 1.Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- 2.Vallotton P, Gupton SL, Waterman-Storer CM, Danuser G. Simultaneous mapping of filamentous actin flow and turnover in migrating cells by quantitative fluorescent speckle microscopy. Proc. Natl. Acad. Sci. USA. 2004;101:9660–9665. doi: 10.1073/pnas.0300552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salmon WC, Adams MC, Waterman-Storer CM. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J. Cell Biol. 2002;158:31–37. doi: 10.1083/jcb.200203022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 5.Welch MD, Mullins RD. Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 2002;18:247–288. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
- 6.Carlier MF, Pantaloni D. Control of actin dynamics in cell motility. J. Mol. Biol. 1997;269:459–467. doi: 10.1006/jmbi.1997.1062. [DOI] [PubMed] [Google Scholar]
- 7.Bamburg JR. Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 8.Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr. Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- 9.DesMarais V, Ichetovkin I, Condeelis J, Hitchcock-DeGregori SE. Spatial regulation of actin dynamics: A tropomyosin-free, actin-rich compartment at the leading edge. J. Cell Sci. 2002;115:4649–4660. doi: 10.1242/jcs.00147. [DOI] [PubMed] [Google Scholar]
- 10.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- 13.Aizawa H, Fukui Y, Yahara I. Live dynamics of Dictyostelium cofilin suggests a role in remodeling actin latticework into bundles. J. Cell Sci. 1997;110:2333–2344. doi: 10.1242/jcs.110.19.2333. [DOI] [PubMed] [Google Scholar]
- 14.Schafer DA, Welch MD, Machesky LM, Bridgman PC, Meyer SM, Cooper JA. Visualization and molecular analysis of actin assembly in living cells. J. Cell Biol. 1998;143:1919–1930. doi: 10.1083/jcb.143.7.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helfman DM, Berthier C, Grossman J, Leu M, Ehler E, Perriard E, Perriard JC. Nonmuscle tropomyosin-4 requires coexpression with other low molecular weight isoforms for binding to thin filaments in cardiomyocytes. J. Cell Sci. 1999;112:371–380. doi: 10.1242/jcs.112.3.371. [DOI] [PubMed] [Google Scholar]
- 16.Jurado C, Haserick JR, Lee J. Slipping or gripping? Fluorescent speckle microscopy in fish keratocytes reveals two different mechanisms for generating a retrograde flow of actin. Mol. Biol. Cell. 2005;16:507–518. doi: 10.1091/mbc.E04-10-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flanagan LA, Chou J, Falet H, Neujahr R, Hartwig JH, Stossel TP. Filamin A, the Arp2/3 complex, and the morphology and function of cortical actin filaments in human melanoma cells. J. Cell Biol. 2001;155:511–517. doi: 10.1083/jcb.200105148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sokol NS, Cooley L. Drosophila filamin is required for follicle cell motility during oogenesis. Dev. Biol. 2003;260:260–272. doi: 10.1016/s0012-1606(03)00248-3. [DOI] [PubMed] [Google Scholar]
- 19.Samarin S, Romero S, Kocks C, Didry D, Pantaloni D, Carlier MF. How VASP enhances actin-based motility. J. Cell Biol. 2003;163:131–142. doi: 10.1083/jcb.200303191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen PG. Actin filament uncapping localizes to ruffling lamellae and rocketing vesicles. Nat. Cell Biol. 2003;5:972–979. doi: 10.1038/ncb1059. [DOI] [PubMed] [Google Scholar]
- 21.Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 22.Carlier MF, Wiesner S, Le Clainche C, Pantaloni D. Actin-based motility as a self-organized system: mechanism and reconstitution in vitro C. R. Biol. 2003;326:161–170. doi: 10.1016/s1631-0691(03)00067-2. [DOI] [PubMed] [Google Scholar]
- 23.Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- 24.Noireaux V, Golsteyn RM, Friederich E, Prost J, Antony C, Louvard D, Sykes C. Growing an actin gel on spherical surfaces. Biophys. J. 2000;78:1643–1654. doi: 10.1016/S0006-3495(00)76716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der, Gucht J, Paluch E, Plastino J, Sykes C. Stress release drives symmetry breaking for actin-based movement. Proc. Natl. Acad. Sci. USA. 2005;102:7847–7852. doi: 10.1073/pnas.0502121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys. J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 28.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J. Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGough A, Chiu W. ADF/cofilin weakens lateral contacts in the actin filament. J. Mol. Biol. 1999;291:513–519. doi: 10.1006/jmbi.1999.2968. [DOI] [PubMed] [Google Scholar]
- 30.Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 32.Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan AY, Bailly M, Zebda N, Segall JE, Condeelis JS. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 2000;148:531–542. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 35.Palmgren S, Vartiainen M, Lappalainen P. Twinfilin, a molecular mailman for actin monomers. J. Cell Sci. 2002;115:881–886. doi: 10.1242/jcs.115.5.881. [DOI] [PubMed] [Google Scholar]
- 36.Moseley JB, Okada K, Balcer HI, Kovar DR, Pollard TD, Goode BL. Twinfilin is an actin-filament-severing protein and promotes rapid turnover of actin structures in vivo. J. Cell Sci. 2006;119:1547–1557. doi: 10.1242/jcs.02860. [DOI] [PubMed] [Google Scholar]
- 37.Nishida E, Maekawa S, Sakai H. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry. 1984;23:5307–5313. doi: 10.1021/bi00317a032. [DOI] [PubMed] [Google Scholar]
- 38.Gupton SL, Anderson KL, Kole TP, Fischer RS, Ponti A, Hitchcock-DeGregori SE, Danuser G, Fowler VM, Wirtz D, Hanein D, et al. Cell migration without a lamellipodium: Translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 2005;168:619–631. doi: 10.1083/jcb.200406063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.