Abstract
Using a system that accelerates the serendipitous discovery of new reactions by evaluating hundreds of DNA-encoded substrate combinations in a single experiment, we explored a broad range of reaction conditions for new bond-forming reactions. We discovered reactivity that led to a biomolecule-compatible, Ru(II)-catalyzed, visible light-induced azide reduction reaction. In contrast with current azide reduction methods, this reaction is highly chemoselective and is compatible with alcohols, phenols, acids, alkenes, alkynes, aldehydes, alkyl halides, alkyl mesylates, and disulfides. The remarkable functional group compatibility and mild conditions of this reaction enabled azide reduction to be performed on nucleic acid and oligosaccharide substrates without the detectable occurrence of side reactions. The reaction was also performed in the presence of a protein enzyme without loss of enzymatic activity, in contrast with two commonly used azide reduction methods. The visible light dependence of this reaction provides a means of photouncaging functional groups such as amines and carboxylates on biological macromolecules without using UV irradiation.
Reaction discovery provides new tools for chemical synthesis, illuminates new modes of reactivity, and can accelerate the discovery of molecules with desired properties in many fields of science. Traditional approaches to reaction discovery focus on one desired transformation at a time and typically search different reaction conditions for their ability to effect the desired transformation starting with one set of substrates. Recently, several research groups have taken a complementary approach to reaction discovery that searches many substrate combinations for new reactivity under various reaction conditions without focusing on any particular type of transformation.1-3 Our group previously developed a DNA-templated reaction discovery system using DNA hybridization to organize substrate pairs in a single solution such that bond formation transfers a biotin group to the DNA template that encodes the identities of the reactive substrates.1 In vitro selection, PCR amplification, and DNA microarray analysis subsequently revealed the identities of reactive substrate pairs. A second-generation DNA-encoded reaction discovery system expanded the set of compatible reaction conditions to include those that do not support DNA hybridization including organic solvents and elevated temperatures.3 Here we report results from the application of this second-generation system to explore regions of transition metal and reaction condition space, and describe the development of a resulting Ru(II)-catalyzed, visible light-induced azide reduction that is compatible with a wide variety of functional groups, including those present on biological macromolecules.
The reduction of azides to amines is an important transformation in organic synthesis. Readily prepared in regio- and stereoselective processes, azides are widely used as amine precursors.4 Current methods for the reduction of azides to amines include metal hydride reduction,4 catalytic hydrogenation,5 treatment with thiols,6 and the phosphine-based Staudinger reduction,7 as well as additional methods that require harsher conditions.4 While widely used, these methods have significant limitations. In general, azide reduction reactions involving metal hydrides or catalytic hydrogenation cannot be used on substrates containing acidic groups such as alcohols and acids, or unsaturated groups such as alkenes, alkynes, and carbonyls.5 Similarly, the Staudinger reduction is not compatible with disulfides and alkyl halides,8,9 and can require elevated temperature and lengthy reaction times.7 The limitations of existing azide reduction methods may explain the near absence of examples of azide reduction in biological macromolecules10 despite the common use of both azides11-14 and amines15-17 as chemical handles in proteins, nucleic acids, and oligosaccharides. In addition to being highly tolerant of a variety of acidic and basic functional groups, a reaction compatible with biological macromolecules must also operate efficiently in neutral aqueous environments.18 The azide reduction discovered and developed in this work exhibits these key features of a biological macromolecule-compatible reaction.
Results and Discussion
Choice of Substrates and Reaction Conditions
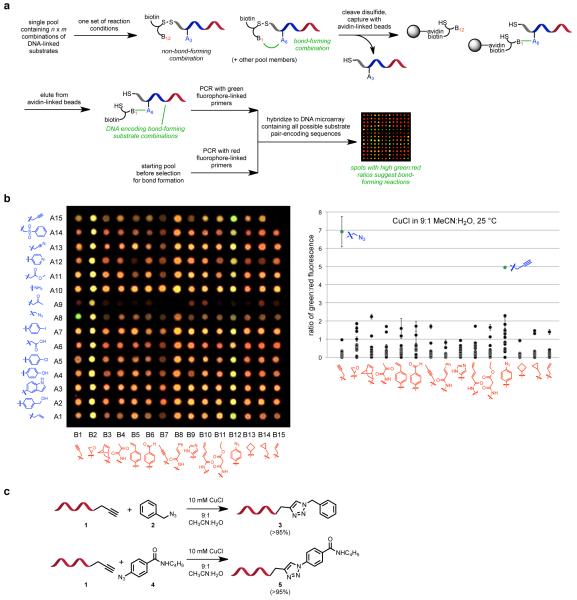
The second-generation DNA-encoded reaction discovery system3 requires a pool of n × m DNA-linked substrate pairs that is assembled from two separate sub-pools of n and m substrates (Fig. 1a). We selected 30 substrates (two groups of 15) comprising 28 distinct functional groups based on two criteria: (i) substrates with high latent energy that also exhibit kinetic stability in unactivated form, and (ii) substrates anticipated to be compatible with DNA and pool assembly conditions (Fig. 1b). Collectively, these substrates represent 15 × 15 = 225 potentially reactive substrate pairs. We assembled the system using a modified version of our previously reported protocol starting with 5 nmol of DNA-linked substrates.3 Following enzymatic ligation and primer extension steps, we obtained ~1 nmol of the fully assembled pool of 15 × 15 pairwise substrate combinations. Because each reaction discovery experiment requires only 0.5 pmol of the substrate pool, this quantity of material is sufficient to assess the reactivity of these substrate combinations under ~2,000 different reaction conditions, collectively representing the evaluation of ~450,000 potential reactions. See the Supplementary Information for detailed procedures used to assemble and characterize the substrate pool.
Figure 1.
DNA-encoded reaction discovery system and validation experiments. (a) Selection and analysis method for the detection of bond-forming reactions between DNA-linked small-molecule substrates. (b) Detection of the known Cu(I)-catalyzed cycloaddition19 between alkynes and azides. Conditions: 10 mM CuCl in CH3CN, 25 °C, 10 min. The green:red ratio of each spot on the DNA microarray (left) is plotted on the right. Error bars represent the standard deviation of signals from three microarray replicates. (c) The reaction products of DNA-linked alkynes and non-DNA-linked azides are consistent with the anticipated triazole products. Conversion efficiencies estimated by LC/MS analysis are shown in parentheses.
We performed control experiments to validate the system using the known Cu(I)-catalyzed cycloaddition between alkynes and azides.19 We confirmed the ability of the system to detect alkyne-azide coupling under these conditions in the DNA-encoded library format and verified the formation of expected triazole products using DNA-linked substrates (Figs. 1b-c).
Next we determined regions of reaction condition space that are compatible with the reaction discovery system. Since the second-generation system does not rely on DNA hybridization to organize substrates into pairwise combinations, compatible reaction conditions include any that do not rapidly degrade nucleic acids. Reaction conditions that lead to the chemical modification or partial degradation of DNA are not necessarily excluded given the high sensitivity of PCR amplification and the tolerance of PCR to some types of chemical modifications; indeed, conditions observed to degrade DNA were shown to be compatible with our earlier reaction discovery system.1,3
Conditions that preserved the ability of the remaining DNA to be readily detected following 25 cycles of PCR were considered potentially compatible reaction conditions. These conditions included the presence of transition metals (Fe(II), Fe (III), Co(II), Ni(0), Ni(II), Cu(I), Cu(II), Ru(0), Ru(II), Ru(III), Rh(I), Rh(II), Pd(0), Pd(II), La(III), and Au(III)), common metal ligands (anions, phosphines, dienes, carbon monoxide, and nitrogen-based ligands), organic catalysts (amines and thiazolium salts), common oxidants (benzoquinones and peroxides), and single electron transfer reagents (hypervalent iodine, SmI2, Mn(OAc)3, and cerium ammonium nitrate (CAN)). A detailed list of potentially compatible reaction conditions is provided in Supplementary Fig. S2.1.
In Vitro Selection for Bond Formation and DNA Microarray Analysis
We treated 0.5 pmol of the substrate pool with a variety of compatible reaction conditions in 200 μL of aqueous or organic solvent (Figs. 1b and 2; details are provided in the Methods section).3 After exposure to reaction conditions, the disulfide bonds in the substrate pool members were cleaved with tris(2-carboxyethyl)phosphine (TCEP), such that only DNA templates linked to (and encoding) substrates that have undergone bond formation remain covalently attached to biotin. These biotinylated DNA templates were captured with streptavidin-linked magnetic particles, washed, then eluted by heating in deionized water at 70 °C.20 The eluted DNA was amplified by PCR using fluorophore-linked primers (Cy3 for the samples subjected to reaction conditions and in vitro selections, and Cy5 for starting substrate pools), then hybridized in triplicate to DNA microarrays containing all 225 possible DNA sequences complementary to each pool member (Fig. 1a).
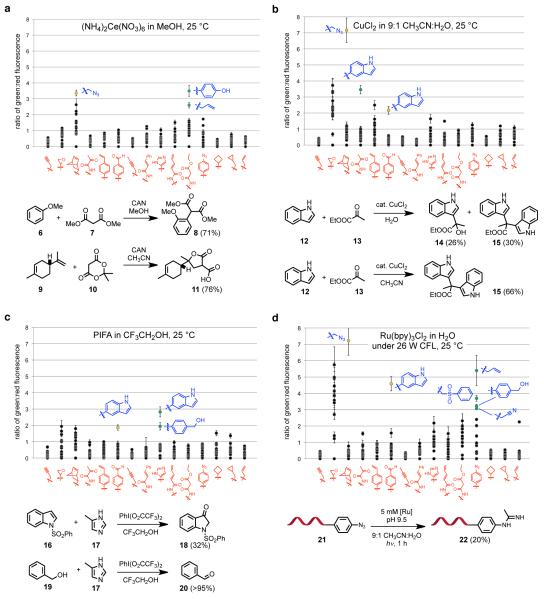
Figure 2.
Selection results from four reaction conditions. The scatter plots show green:red fluorescence ratios of substrate combinations in the DNA-encoded library. The schemes show the outcomes of reactions in a flask. (a) In the presence of CAN coupling was observed between malonic esters and electron-rich π systems. 21,22 Conditions: 10 mM CAN in MeOH, 25 °C, 2 h. (b) CuCl2 induced coupling of indoles and pyruvates, consistent with Friedel-Crafts-type coupling. Conditions: 1 mM CuCl2 in H2O, 25 °C, 16 h. (c) Phenyliodine bis(trifluoroacetate) (PIFA) generated positives consistent with imidazole-indole and imidazole-benzyl alcohol coupling, but in a non-DNA-linked format only substrate oxidation was observed. Conditions: 10 mM PIFA in CF3CH2OH, 25 °C, 2 h. (d) Ru(bpy)3Cl2 under visible light irradiation resulted in several positives including azide-nitrile coupling. Conditions: 10 mM Ru(bpy)3Cl2 in 100 mM aqueous sodium carbonate, pH 9.5, 20 cm from a 26 W CFL bulb, 25 °C, 1 h. For (a), (b), and (c), isolated yields are shown in parentheses; for (d), the conversion efficiency by LC/MS analysis is shown in parentheses.
We observed a number of spots on the DNA microarrays with reproducibly high Cy3:Cy5 (green:red) ratios suggestive of potential bond-forming reactions between two substrates (Fig. 2). We grouped these initial positives into several categories. Some positives correspond to substrate combinations known to undergo bond formation under the reaction conditions tested (green circles in Fig. 2a). While these positives did not represent new reactions, they provided additional validation that the second-generation reaction discovery system can detect bond-forming reactivity. For example, we observed phenol A4 + malonic ester B11 and alkene A1 + malonic ester B11 when the system was treated with CAN in methanol (Fig. 2a), consistent with previous reports that CAN mediates oxidative coupling reactions between malonic esters and electron-rich π systems (see Fig. 1b for the structures of substrates A1-A15 and B1-B15).21,22
A second group of positives represented reactions that are potentially new but consistent with known reaction mechanisms. For example, copper (II) chloride resulted in a positive signal corresponding to coupling between indole A3 and pyruvamide B4 (green circle in Fig. 2b). In the flask, treatment of indole 12 and ethyl pyruvate 13 in the presence of CuCl2 or Cu(OTf)2 led to the formation of a Friedel-Crafts-type coupling product (14) together with the bis-indole adduct (15) in aqueous solvents, while in organic solvents exclusive bis-indole formation was observed (Fig. 2b).23 We also observed substrate combinations that could be facilitated by a variety of Lewis acids or transition metals (orange circles in Figs. 2a-d). For instance, coupling between indole A3 and aldehyde B6, consistent with a Friedel-Crafts-type reaction (orange circles in Figs. 2b-d), and coupling between azide A8 and norbornene B3, consistent with a cycloaddition (orange circles in Figs. 2a, 2b and 2d), were observed under several tested reaction conditions involving transition metals including Fe(III), Co(II), Ni (II), Cu (II) and Ru(II).24,25
A third category of positives corresponded to potential coupling reactions that were reproducible in the DNA-encoded system but when tested in an intermolecular format did not result in the formation of a coupling product. For example, hypervalent iodine26 resulted in robust positive signals corresponding to the coupling of indole A3 with imidazole B9, as well as the coupling of benzyl alcohol A4 and imidazole B9 (green circles in Fig. 2c). When the corresponding reactions were attempted in flasks using small-molecule substrates, however, only the formation of oxidation byproducts 3-oxoindole 18 and benzaldehyde 20 were observed (Fig. 2c). We speculate that for this category of positives, bond-forming reactivity in the DNA-linked reaction discovery system is dependent in part on the highly dilute, intramolecular format of the system, or on interactions with groups in DNA that are absent when these reactions are explored in the flask.
A fourth category of positives correspond to bond-forming reactions that have not been previously reported, that are not easily predicted based on known reactivity, and that we observed to take place in an intermolecular format with either DNA-linked or non-DNA-linked substrates. For example, when the system was treated with Ru(bpy)3Cl2 in the presence of a visible light source (a 26 W compact fluorescent lamp (CFL) placed ~20 cm from the tube containing the substrate library), we observed positives that corresponded to coupling of aryl azide B12 and several functional groups (alkene A1, sulfone A14, phenol A4, and nitrile A13) (green circles in Fig. 2d). Although Ru(bpy)3Cl2 as a visible-light photoredox catalyst is well-studied in inorganic chemistry,27,28 its exploration in synthetic organic chemistry is more recent.29-32 Given the emerging importance of visible light-mediated organic transformations and the lack of previous reports of azide reactivity under these conditions, we chose to explore these reactions in greater detail.
Visible Light-Induced Azide Coupling and Reduction Reactions
We first studied the reaction between DNA-linked aryl azides and nitriles. Under conditions similar to those used in the DNA-encoded system (5 mM Ru(bpy)3Cl2 in 200 μL of 1:9 water:acetonitrile with 30 mM Na2CO3 20 cm from a 26 W CFL bulb), DNA-linked aryl azide 21 reacted with acetonitrile to generate a product consistent by LC/MS analysis with imidate adduct 22 (Fig. 2d). No significant product formation was observed in the absence of ruthenium complex or in the absence of a CFL light source.
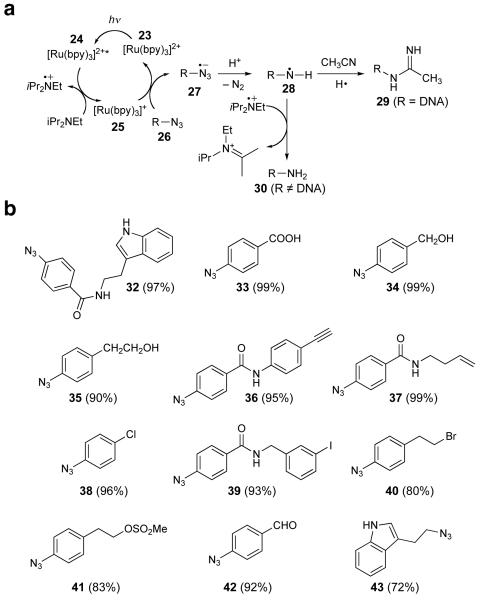
Since bond formation between aryl azides and nitriles induced by visible light catalysis was unprecendented, we developed a mechanistic hypothesis to explain the above observations (Fig. 3a). Ground-state [Ru(bpy)3]2+ (23) is known to absorb visible light (λmax = 452 nm) to generate the excited triplet [Ru(bpy)3]2+* (24).27,28 When an electron donor is present, 24 can be reductively quenched to [Ru(bpy)3]+ (25).30,31,33 We speculate that 25 may reduce azide 26 through a one-electron transfer to generate azide radical anion 27. Following nitrogen gas extrusion and protonation, the resulting aminyl radical 28 may undergo a radical addition to the nitrile.34,35
Figure 3.
Development of the Ru(bpy)3Cl2-mediated, visible light-induced azide reduction reaction. (a) Proposed mechanism for the azide-nitrile coupling reaction and the azide reduction reaction. (b) Substrate scope of the azide reduction reaction under the conditions shown in Table 1, entry 8 (except for substrate 43, which was reduced with iPr2NEt and Hantzsch ester in 24 h using conditions described in the Methods section). Isolated yields are shown in parentheses.
With this mechanistic hypothesis in mind, we further studied these reactions in a non-DNA-linked format on a 0.1 mmol scale (Table 1). While the conditions that generated the coupling product of the DNA-linked azide and nitrile for unknown reasons did not lead to significant coupling product formation with azide 4 in the flask, the addition of the commonly used sacrificial electron donor diisopropylethylamine30,31 to these reaction conditions resulted in moderate yields (~60%) of the azide reduction product (31) after 96 h (Table 1, entry 1). The amine can form through formal hydrogen abstraction by the proposed aminyl radical intermediate (Fig. 3a),35,36 although future studies are needed to probe these and other mechanistic possibilities. We speculated that this reaction may offer significant advantages over existing azide reduction methods and sought to further develop the transformation.
Table 1.
Optimization of the azide reduction reaction. Reported yields above 5% are isolated yields.
Optimization of a Visible Light-Induced Azide Reduction Reaction
While the initial azide reduction reaction proceeded slowly and in moderate yields, the addition of formic acid or the Hantzsch ester (diethyl 1,4-dihydro-2,6-dimethyl-3,5-pyridinedicarboxylate) as hydrogen donors, supplementing or replacing tertiary amines as hydrogen donors,31 decreased the reaction time to 12 h and increased yields to ~90% (Table 1, entries 2 and 3). Without the ruthenium complex or the CFL light source, no azide reduction was observed under the above conditions (Table 1, entries 4 and 5). Further optimization revealed that the tertiary ammonium salt provided a neutral reaction media and when combined with the Hantzsch ester improved the reaction efficiency to 92-96% yields in 2 h in a variety of solvents (DMF, acetonitrile, or dichloromethane) (Table 1, entries 6-8). The use of acetate rather than formate as the ammonium counterion generated similar excellent product yields in 2 h (Table 1, entry 9). As little as 1 mol% Ru(bpy)3Cl2 was sufficient to afford the reduction product in 95% yield in 2 h (Table 1, entry 10).
Next we evaluated the functional group compatibility of the optimized azide reduction under the conditions shown in Table 1, entry 8 (5 mol% Ru(bpy)3Cl2, 10 equiv. iPr2NEt/HCOOH, 1.5 equiv. Hantzsch ester in CH2Cl2). We observed that a variety of substrates containing protic functional groups including free indoles, acids, and alcohols are compatible with the reaction (Fig. 3b, substrates 32-35). In addition, functional groups sensitive to hydrogenation including alkenes, alkynes, and aryl halides are also not affected by these reaction conditions (Fig. 3b, substrates 36-39). Likewise, functional groups sensitive to nucleophiles including alkyl halides, alkyl mesylates, and aldehydes also emerged intact under the reaction conditions (Fig. 3b, substrates 40-42). An alkyl azide was also reduced in 72% yield in 24 h under modified conditions (Fig. 3b, substrate 43).
Encouraged by the remarkable chemoselectivity of this reaction, we sought to develop an aqueous version to explore the possibility that this azide reduction might be applicable to biological molecules. Under the above optimized conditions (Table 1 entry 8) but in 1:1 CH3CN:H2O, the efficiency of the reaction was drastically reduced (Table 1, entry 11). Efficient product formation was restored when formic acid was removed, presumably increasing the availability of an unprotonated tertiary amine as a competent electron donor (Table 1, entry 12). The water-soluble reducing agent NADH could replace the Hantzsch ester with only a modest reduction in reaction efficiency (Table 1, entry 13). While effective, we speculated that the above basic conditions may compromise the reaction’s compatibility with biological macromolecules. We discovered however that sodium ascorbate can act as both an electron donor and as a hydrogen donor37 without significantly lowering yields or lengthening reaction times (Table 1, entry 14). In the presence of sodium ascorbate the reduction proceeds efficiently in buffers of varying acidity (pH 4.0, pH 7.4, and pH 9.5; see Supplementary Fig. S3.1).
Compatibility of the Azide Reduction Reaction with DNA, Oligosaccharide, and Protein
Previous reports have demonstrated the use of Ru(bpy)3Cl2 to mediate reactions on protein substrates in crude cell extracts,38 and related complexes have been used as cell-permeable agents to image DNA in living eukaryotic and prokaryotic cells.39 These observations coupled with the excellent functional group tolerance and mild reaction conditions of the azide reduction raised the possibility that this reaction could be performed on functional group-rich biomolecules without inducing any side reactions. We tested this possibility by performing the reduction on, or in the presence of, nucleic acids, a protein, and an oligosaccharide and characterizing these macromolecules after each reaction.
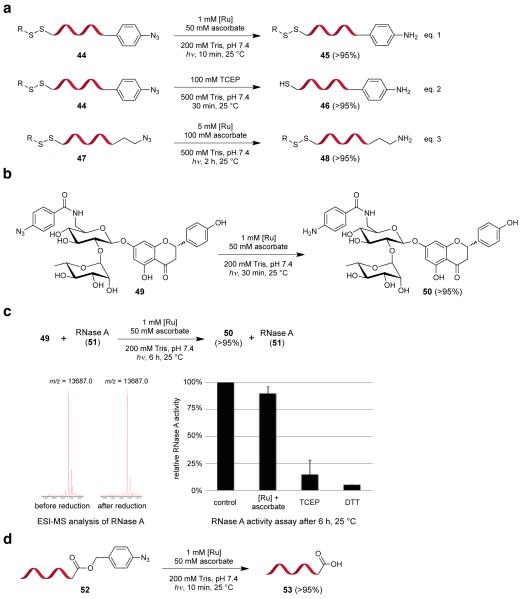
We prepared DNA oligonucleotide 44 containing both an aryl azide and a disulfide and treated 0.5 μM of this substrate with 1 mM Ru(bpy)3Cl2 and 50 mM of sodium ascorbate in aqueous 200 mM Tris, pH 7.4 positioned 20 cm from a 26 W CFL bulb (standard aqueous reduction conditions). Azide reduction proceeded cleanly in 10 min without disulfide bond reduction (eq. 1 in Fig. 4a). In the absence of light, no azide reduction was observed after 15 h, and exposure to light after 15 h of darkness resulted in complete azide reduction in 10 min. In comparison, tris(2-carboxyethyl)phosphine (TCEP), a common Staudinger reaction agent, reduced both the azide and disulfide non-selectively (eq. 2 in Fig. 4a). Likewise, DNA oligonucleotide 47 containing an alkyl azide and a disulfide underwent selective azide reduction in 2 h when treated with 5 mM Ru(bpy)3Cl2 and 100 mM ascorbate (eq. 3 in Fig. 4a).
Figure 4.
Compatibility of the azide reduction reaction with biological molecules. Conversion efficiencies are shown in parentheses. (a) Azide- and disulfide-linked DNA oligonucleotides (0.5 μM each) were reduced to the corresponding amines under the aqueous reaction conditions shown without inducing side reactions such as disulfide reduction. In contrast, TCEP reduced both the azide and disulfide groups. Reaction conversions were calculated by LC/MS analysis. (b) A functional group-rich azide analog of the disaccharide naringin (100 μM) was reduced cleanly to the corresponding amine. Conversion was calculated by HPLC analysis. (c) Ru(bpy)3Cl2-mediated, visible light-induced reduction of the naringin azide analog (100 μM) in the presence of the protein enzyme RNase A (10 μM in the reaction, 0.83 μM when assayed) did not alter the enzyme’s covalent structure or activity. In contrast, thiol- or phosphine-induced azide reduction both resulted in loss of RNase A activity. Conditions are as follows. Control: 200 mM Tris, pH 7.4, 20 cm from 26 W CFL, 6 h, 25 °C. [Ru]+ascorbate: 1 mM [Ru], 50 mM ascorbate, 200 mM Tris, pH 7.4, 20 cm from 26 W CFL, 6 h, 25 °C. TCEP: 100 mM TCEP in 1 M phosphate, pH 8.0, 6 h, 25 °C. DTT: 500 mM DTT in 1 M phosphate, pH 8.0, 6 h, 25 °C. (d) Visible light-triggered photouncaging of carboxylic acid functional group in oligonucleotide substrates. Conversion was calculated by LC/MS analysis.
To test the compatibility of the azide reduction reaction with oligosaccharide substrates we prepared an azide-containing variant of naringin, a flavanone disaccharide (49) (Fig. 4b). When 100 μM of 49 was treated with standard aqueous reduction conditions, we observed complete reduction in 30 min with no other transformations detected by HPLC, 1H-NMR, or mass spectrometry (Fig. 4b).
Finally, we performed the azide reduction in the presence of a protein enzyme. We treated a solution containing 100 μM of naringin azide (49) and 10 μM of RNase A with standard aqueous reduction conditions. After 6 h, azide 49 was completely reduced by HPLC (Fig. 4c). We isolated the RNase A from the completed reaction by size-exclusion chromatography. No new species consistent with covalent modification of RNase A were observed by ESI mass spectrometry of the macromolecular fraction. Likewise, enzymatic assay of RNase A activity, which requires intact disulfide bonds,39 revealed ≤ 10% loss of catalytic activity from the azide reduction reaction developed in this work, while thiol-based reduction and the Staudinger azide reduction both resulted in the loss of 85-100% of enzyme activity (Fig. 4c).
Visible Light-Triggered Photouncaging of Carboxylic Acids
We hypothesized that the reaction described above may also enable azide-protected functional groups other than amines on functional group-rich biomolecules to be uncaged efficiently with visible light under mild conditions. Current photouncaging40,41 reactions typically require UV light that can induce undesired side reactions including protein degradation, photobleaching, inhibition of photosynthesis, and nucleic acid damage.42
We prepared a DNA oligonucleotide-linked carboxylic acid caged as a 4-azidobenzyl ester10,43 (52) and treated it with standard aqueous reduction conditions. Photouncaging of the free carboxylic acid through azide reduction and 1,6-elimination was complete within 10 min without any detected byproducts by LC/MS (Fig. 4d). As a deprotection strategy, this reaction proceeds rapidly under very mild conditions and is orthogonal to most currently used carboxyl deprotection strategies such as acid or base hydrolysis.44
Conclusion
Using a second-generation DNA-encoded reaction discovery system, we explored a broad range of transition metals and reaction conditions to identify combinations of substrates and reaction conditions that result in bond formation between substrates. From the reactivity revealed by resulting hits we developed a Ru(II)-catalyzed, visible light-induced azide reduction reaction that is efficient and can be conducted in organic or aqueous solvents, open to the air, at room temperature, and under neutral conditions. This reaction exhibits remarkable chemoselectivity in contrast with existing azide reduction methods and is compatible with alcohols, phenols, acids, alkenes, alkynes, aldehydes, alkyl halides, mesylates, and disulfides. The unusual functional group compatibility and mild required conditions of the reaction enabled azide reduction to be performed on oligonucleotide and oligosaccharide substrates, and in the presence of a protein enzyme, without compromising the structure or the enzymatic activity of the biomolecules. This reaction can also be used to photouncage groups such as amines and carboxylates on biomolecules without requiring UV light.
Methods
See the Supplementary Information for additional experimental details.
General Reaction Discovery Procedure
Each reaction discovery experiment was performed in 200 μL total volume containing 0.5 pmol of the total substrate pool corresponding to 2.2 fmol of each unique substrate combination. Following exposure to the reaction conditions shown in Fig. 2, each solution was precipitated with ethanol to recover DNA-linked species. The pellet was dissolved in 30 μL of water and 150 μL of 0.1 M tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in 1.0 M aqueous sodium phosphate, pH 8.0 was added to effect disulfide cleavage. After 30 min at 25 °C, this solution was combined with streptavidin-linked magnetic particles (14 μL, corresponding to 20 pmol biotin-binding capacity, Roche Biosciences) suspended in 300 μL of 10 mM Tris-Cl, 0.1 M NaCl, 1 mM EDTA, pH 7.5. After incubation 15 min at 25 °C, the supernatant was removed and the particles were captured by a magnetic separator, then rinsed once with 200 μL 10 mM Tris-Cl, 1 M NaCl, 1 mM EDTA, pH 7.5, and once with 200 μL H2O. The particles were suspended in 40 μL deionized water and incubated at 70 °C for 5 min to elute captured DNA.20 The supernatant was collected and the elution step was repeated once. The combined supernatants were used directly in PCR reactions for microarray analysis as previously described3 and detailed in Supplementary Information.
Representative Small-Molecule Azide Reductions
For aryl azides in organic solvent: to a solution of Ru(bpy)3Cl2 (7.5 mg, 0.01 mmol, 0.05 equiv.), Hantzsch ester (76 mg, 0.3 mmol, 1.5 equiv.), iPr2NEt (350 μL, 2.0 mmol, 10 equiv.) and HCOOH (75 μL, 2.0 mmol, 10 equiv.) in 1 mL CH2Cl2 was added azide 4 (43.6 mg, 0.2 mmol, 1.0 equiv.). The dark orange solution was stirred at 25 °C at a distance of 20 cm from a 26 W CFL bulb until TLC indicated the complete consumption of azide 4 (usually ≤ 2 h). The reaction mixture was directly subjected to silica gel chromatography. Isolated yields for other aryl azides are shown in Fig. 3b.
For aryl azides in mixed aqueous/organic solvent: to a solution of Ru(bpy)3Cl2 (3.8 mg, 0.005 mmol, 0.05 equiv.) and azide 4 (21.8 mg, 0.1 mmol, 1.0 equiv.) in 1 mL DMF/H2O (1:1 ratio) was added sodium ascorbate (198 mg, 1.0 mmol, 10 equiv.) in two equal portions (the second portion was added after 24 h). The orange solution was stirred at 25 °C at a distance of 20 cm from a 26 W CFL bulb until TLC indicated the complete consumption of azide 4 (~48 h). The resulting reaction mixture was diluted with ethyl acetate, washed with water and brine, dried over Na2SO4, concentrated in vacuo and subjected to silica gel chromatography.
For alkyl azides: To a solution of Ru(bpy)3Cl2 (15 mg, 0.02 mmol), Hantzsch ester (102 mg, 0.4 mmol) and iPr2NEt (105 μL, 0.6 mmol) in 2 mL DMF/MeOH (3:1 ratio) was added alkyl azide 43 (37.2 mg, 0.2 mmol). The dark orange suspension was stirred at 25 °C at a distance of 20 cm from a 26 W CFL bulb until TLC indicated the complete consumption of azide 43 (~24 h). To the reaction mixture was added di-tert-butyl dicarbonate (87.3 mg, 0.4 mmol) and triethylamine (56 μL, 0.4 mmol) and stirred for 12 h. The resulting solution was diluted with ethyl acetate, washed with water and brine, dried over Na2SO4, concentrated in vacuo and subjected to silica gel chromatography.
Representative DNA-Linked Azide Reduction
To a solution of 1 mM Ru(bpy)3Cl2 and 50 mM sodium ascorbate in 200 μL of aqueous 200 mM Tris-Cl, pH 7.4 was added DNA-linked azide to a final concentration of 0.5 μM. After incubation at 25 °C at a distance of 20 cm from a 26 W CFL bulb for 10 min, the reaction was quenched by precipitation with ethanol. The pellet containing DNA-linked small molecules was dissolved in 20 μL of 10 mM phosphate buffer, pH 8 and subjected to LC/MS analysis.
Naringin Azide Reduction in the Presence of RNase A
To a solution of 1 mM Ru(bpy)3Cl2 and 50 mM sodium ascorbate in 200 μL of aqueous 200 mM Tris-Cl, pH 7.4 was added naringin azide and RNase A to final concentrations of 100 μM and 10 μM, respectively. After incubation at 25 °C at a distance of 20 cm from a 26 W CFL bulb for 6 h, RNase A was collected through size-exclusion chromatography (Centri-Sep column, Princeton Separations) and characterized by mass spectrometry. After sample dilution 12-fold, RNase A enzyme activity was assayed by following cytidine 2′,3′-cyclic phosphate hydrolysis spectrophotometrically as previously described.45 The small-molecule fraction from polyethersulfone membrane ultrafiltration (Corning Life Sciences) was directly analyzed by reverse-phase HPLC to assess naringin azide reduction.
Supplementary Material
Acknowledgements
This work was supported by NIH grant R01GM065865 and the Howard Hughes Medical Institute. We thank Drs. Yinghua Shen and Christoph Dumelin for mass spectrometry assistance, and Dr. David Gorin for helpful discussions.
Footnotes
Competing Financial Interests The authors declare no competing financial interests.
References Cited
- 1.Kanan MW, Rozenman MM, Sakurai K, Snyder TM, Liu DR. Reaction discovery enabled by DNA-templated synthesis and in vitro selection. Nature. 2004;431:545–549. doi: 10.1038/nature02920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beeler AB, Su S, Singleton CA, Porco JA. Discovery of chemical reactions through multidimensional screening. J. Am. Chem. Soc. 2007;129:1413–1419. doi: 10.1021/ja0674744. [DOI] [PubMed] [Google Scholar]
- 3.Rozenman MM, Kanan MW, Liu DR. Development and initial application of a hybridization-independent, DNA-encoded reaction discovery system compatible with organic solvents. J. Am. Chem. Soc. 2007;129:14933–14938. doi: 10.1021/ja074155j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scriven EFV, Turnbull K. Azides - their preparation and synthetic uses. Chem. Rev. 1988;88:297–368. [Google Scholar]
- 5.Johnstone RAW, Wilby AH, Entwistle ID. Heterogeneous catalytic transfer hydrogenation and its relation to other methods for reduction of organic-compounds. Chem. Rev. 1985;85:129–170. [Google Scholar]
- 6.Bayley H, Standring DN, Knowles JR. Propane-1,3-dithiol - selective reagent for efficient reduction of alkyl and aryl azides to amines. Tetrahedron Lett. 1978;19:3633–3634. [Google Scholar]
- 7.Gololobov YG, Kasukhin LF. Recent advances in the Staudinger reaction. Tetrahedron. 1992;48:1353–1406. [Google Scholar]
- 8.Burns JA, Butler JC, Moran J, Whitesides GM. Selective reduction of disulfides by tris(2-carboxyethyl)phosphine. J. Org. Chem. 1991;56:2648–2650. [Google Scholar]
- 9.Maryanoff BE, Reitz AB. The Wittig Olefination Reaction and Modifications Involving Phosphoryl-Stabilized Carbanions - Stereochemistry, Mechanism, and Selected Synthetic Aspects. Chem. Rev. 1989;89:863–927. [Google Scholar]
- 10.Sakurai K, Snyder TM, Liu DR. DNA-templated functional group transformations enable sequence-programmed synthesis using small-molecule reagents. J. Am. Chem. Soc. 2005;127:1660–1661. doi: 10.1021/ja0432315. [DOI] [PubMed] [Google Scholar]
- 11.Agard NJ, Baskin JM, Prescher JA, Lo A, Bertozzi CR. A comparative study of bioorthogonal reactions with azides. ACS Chem. Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 12.Brase S, Gil C, Knepper K, Zimmermann V. Organic azides: An exploding diversity of a unique class of compounds. Angew. Chem. Int. Ed. 2005;44:5188–5240. doi: 10.1002/anie.200400657. [DOI] [PubMed] [Google Scholar]
- 13.Kohn M, Breinbauer R. The Staudinger ligation - a gift to chemical biology. Angew. Chem. Int. Ed. 2004;43:3106–3116. doi: 10.1002/anie.200401744. [DOI] [PubMed] [Google Scholar]
- 14.Kolb HC, Sharpless KB. The growing impact of click chemistry on drug discovery. Drug Discov. Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 15.Valeur E, Bradley M. Amide bond formation: beyond the myth of coupling reagents. Chem. Soc. Rev. 2009;38:606–631. doi: 10.1039/b701677h. [DOI] [PubMed] [Google Scholar]
- 16.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermanson GT. Bioconjugate Techniques. 2nd edn Academic Press; 2008. pp. 169–181. [Google Scholar]
- 18.Basle E, Joubert N, Pucheault M. Protein chemical modification on endogenous amino acids. Chem. Biol. 2010;17:213–227. doi: 10.1016/j.chembiol.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Meldal M, Tornoe CW. Cu-catalyzed azide-alkyne cycloaddition. Chem. Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 20.Holmberg A, et al. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis. 2005;26:501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- 21.Solabannavar SB, Helavi VB, Desai UV, Mane RB. A novel short synthesis of norbisabolide. Tetrahedron Lett. 2002;43:4535–4536. [Google Scholar]
- 22.Baciocchi E, Dellaira D, Ruzziconi R. Dimethyl arylmalonates from cerium(IV) ammonium-nitrate promoted reactions of dimethyl malonate with aromatic-compounds in methanol. Tetrahedron Lett. 1986;27:2763–2766. [Google Scholar]
- 23.Poulsen TB, Jorgensen KA. Catalytic asymmetric Friedel-Crafts alkylation reactions-copper showed the way. Chem. Rev. 2008;108:2903–2915. doi: 10.1021/cr078372e. [DOI] [PubMed] [Google Scholar]
- 24.Bandini M, Melloni A, Umani-Ronchi A. New catalytic approaches in the stereoselective Friedel-Crafts alkylation reaction. Angew. Chem. Int. Ed. 2004;43:550–556. doi: 10.1002/anie.200301679. [DOI] [PubMed] [Google Scholar]
- 25.Scheiner P, Schomake JH, Deming S, Libbey WJ, Nowack GP. Addition of aryl azides to norbornene. A kinetic investigation. J. Am. Chem. Soc. 1965;87:306–311. [Google Scholar]
- 26.Zhdankin VV, Stang PJ. Chemistry of polyvalent iodine. Chem. Rev. 2008;108:5299–5358. doi: 10.1021/cr800332c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juris A, et al. Ru(II) polypyridine complexes: photophysics, photochemistry, electrochemistry, and chemiluminescence. Coord. Chem. Rev. 1988;84:85–277. [Google Scholar]
- 28.Kalyanasundaram K. Photophysics, photochemistry and solar-energy conversion with tris(bipyridyl)ruthenium(II) and its analogs. Coord. Chem. Rev. 1982;46:159–244. [Google Scholar]
- 29.Nicewicz DA, MacMillan DWC. Merging photoredox catalysis with organocatalysis: The direct asymmetric alkylation of aldehydes. Science. 2008;322:77–80. doi: 10.1126/science.1161976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ischay MA, Anzovino ME, Du J, Yoon TP. Efficient visible light photocatalysis of [2+2] enone cycloadditions. J. Am. Chem. Soc. 2008;130:12886–12887. doi: 10.1021/ja805387f. [DOI] [PubMed] [Google Scholar]
- 31.Narayanam JMR, Tucker JW, Stephenson CRJ. Electron-transfer photoredox catalysis: development of a tin-free reductive dehalogenatioin reaction. J. Am. Chem. Soc. 2009;131:8756–8757. doi: 10.1021/ja9033582. [DOI] [PubMed] [Google Scholar]
- 32.Yoon TP, Ischay MA, Du JN. Visible light photocatalysis as a greener approach to photochemical synthesis. Nature Chem. 2010;2:527–532. doi: 10.1038/nchem.687. [DOI] [PubMed] [Google Scholar]
- 33.Delaive PJ, Sullivan BP, Meyer TJ, Whitten DG. Applications of light-induced electron-transfer reactions - coupling of hydrogen generation with photo-reduction of ruthenium(II) complexes by triethylamine. J. Am. Chem. Soc. 1979;101:4007–4008. [Google Scholar]
- 34.Su W, Li YS, Zhang YM. Samarium diiodide induced reductive coupling of nitriles with azides. J. Chem. Res. -S. 2001:32–33. [Google Scholar]
- 35.Benati L, et al. Radical reduction of aromatic azides to amines with triethylsilane. J. Org. Chem. 2006;71:5822–5825. doi: 10.1021/jo060824k. [DOI] [PubMed] [Google Scholar]
- 36.Hays DS, Fu GC. Development of Bu3SnH-catalyzed processes: efficient reduction of azides to amines. J. Org. Chem. 1998;63:2796–2797. [Google Scholar]
- 37.Borak JB, Falvey DE. A new photolabile protecting group for release of carboxylic acids by visible-light-induced direct and mediated electron transfer. J. Org. Chem. 2009;74:3894–3899. doi: 10.1021/jo900182x. [DOI] [PubMed] [Google Scholar]
- 38.Fancy DA, Kodadek T. Chemistry for the analysis of protein-protein interactions: rapid and efficient cross-linking triggered by long wavelength light. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6020–6024. doi: 10.1073/pnas.96.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gill MR, et al. A ruthenium(II) polypyridyl complex for direct imaging of DNA structure in living cells. Nature Chem. 2009;1:662–667. doi: 10.1038/nchem.406. [DOI] [PubMed] [Google Scholar]
- 40.Lee HM, Larson DR, Lawrence DS. Illuminating the chemistry of life: design, synthesis, and applications of “caged” and related photoresponsive compounds. ACS Chem. Biol. 2009;4:409–427. doi: 10.1021/cb900036s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayer G, Heckel A. Biologically active molecules with a “light switch”. Angew. Chem. Int. Ed. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 42.Sinha RP, Hader DP. UV-induced DNA damage and repair: a review. Photochem. Photobiol. Sci. 2002;1:225–236. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- 43.Griffin RJ, et al. The 4-azidoberazyloxycarbonyl function; application as a novel protecting group and potential prodrug modification for amines. J. Chem. Soc., Perkin Trans. 1996;1:1205–1211. [Google Scholar]
- 44.Wuts PGM, Greene TW. Greene’s Protective Groups in Organic Synthesis. 4th edn Wiley-Interscience; 2007. pp. 533–646. [Google Scholar]
- 45.Crook EM, Mathias AP, Rabin BR. Spectrophotometric assay of bovine pancreatic ribonuclease by the use of cytidine 2′-3′-phosphate. Biochem. J. 1960;74:234–238. doi: 10.1042/bj0740234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.