Abstract
Gram-negative bacteria, such as Escherichia coli, expel toxic chemicals via tripartite efflux pumps spanning both the inner and outer membranes. The three parts are: 1) a membrane fusion protein connecting 2) a substrate-binding inner membrane transporter to 3) an outer membrane-anchored channel in the periplasmic space. A crystallographic model of this tripartite efflux complex has been unavailable simply because co-crystallization of different components of the system has proven to be extremely difficult. We previously described the crystal structures of both the inner membrane transporter CusA1 and membrane fusion protein CusB2 of the CusCBA efflux system3,4 from E. coli. We here report the co-crystal structure of the CusBA efflux complex, revealing the trimeric CusA efflux pump interacts with six CusB protomers at the upper half of the periplasmic domain. These six CusB molecules form a channel extending contiguously from the top of the pump. The affinity of the CusA and CusB interaction was found to be in the micromolar range. Finally, we predicted a three-dimensional structure of the trimeric CusC outer membrane channel, and develop a model of the tripartite efflux assemblage. This CusC3-CusB6-CusA3 model presents a 750 kDa efflux complex spanning the entire bacterial cell envelope to export Cu(I)/Ag(I) ions.
In Gram-negative bacteria, efflux systems of the resistance-nodulation-division (RND) family play major roles in the intrinsic and acquired tolerance of antibiotics and toxic compounds.5,6 They represent key components for Gram-negative pathogens to use in overcoming toxic environments unfavorable for their survival. An RND efflux pump7-14 works in conjunction with a periplasmic membrane fusion protein,15-18 and an outer membrane channel to form a functional protein complex.19,20 In Escherichia coli, one such tripartite efflux system CusCBA is responsible for extruding biocidal Cu(I) and Ag(I) ions.3,4 CusA is a large proton-motive-force-dependent inner membrane RND efflux pump comprising 1,047 amino acids.3,4 CusC is a 457 amino acid protein that forms an outer membrane channel.3,4 The membrane fusion protein CusB, containing 379 amino acids, bridges between CusA and CusC to form a tripartite efflux complex CusCBA.3,4 This three-component system spans the entire cell envelope of E. coli to export Ag(I)/Cu(I).
Between the cusC and cusB genes, there is a small chromosomal gene that encodes a periplasmic protein CusF.4 CusF functions as a chaperone carrying Cu(I)/Ag(I) to the CusCBA efflux pump.21,22
We previously reported the crystal structure of the full-length CusB membrane fusion protein at a resolution of 3.40 Å, revealing four linearly-arranged domains (Domains 1-4) comprising approximately 80% of the protein.2 Overall, CusB is folded into an elongated structure, ~120 Å long and ~40 Å wide. The first three domains (Domains 1-3) of the protein are mostly β-strands. However, the fourth domain (Domain 4) is all α-helices and is folded into a three-helix bundle structure.
We also determined the crystal structure of the full-length CusA RND pump (at the 3.52-Å resolution) that includes approximately 98% of the amino acids.1 The structure suggests that CusA exists as a homotrimer. Each subunit of CusA consists of 12 transmembrane helices (TM1-TM12) and a large periplasmic domain formed by two periplasmic loops between TM1 and TM2, and TM7 and TM8, respectively. The periplasmic domain of CusA can be divided into a pore domain (comprising sub-domains PN1, PN2, PC1, PC2) and a CusC docking domain (containing sub-domains DN and DC). Through the use of lysine-lysine cross-linking and mass spectrometry, it was determined that Domain 1 of CusB directly contacts the upper portion of sub-domain PN1 of the CusA efflux pump.2
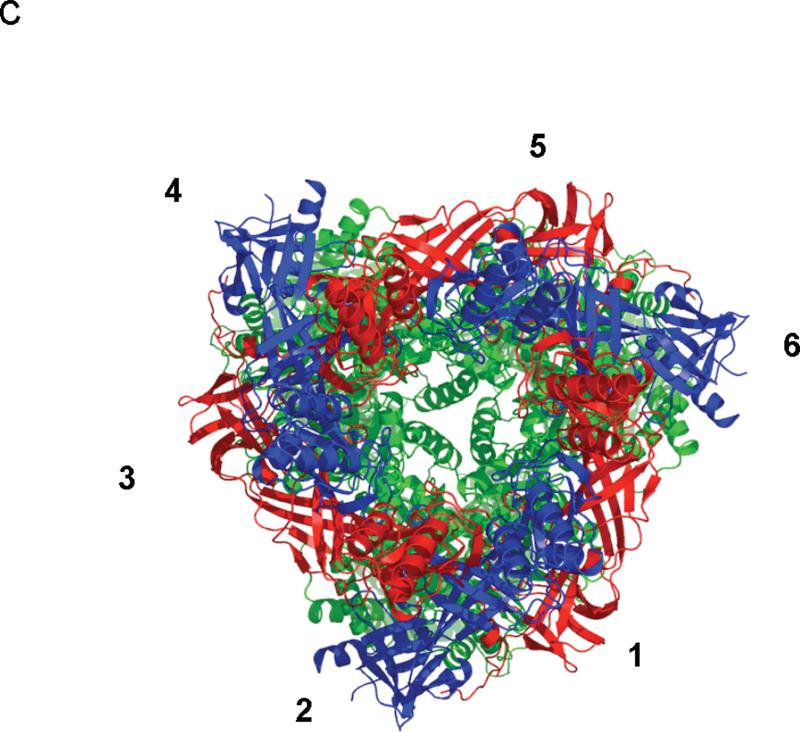
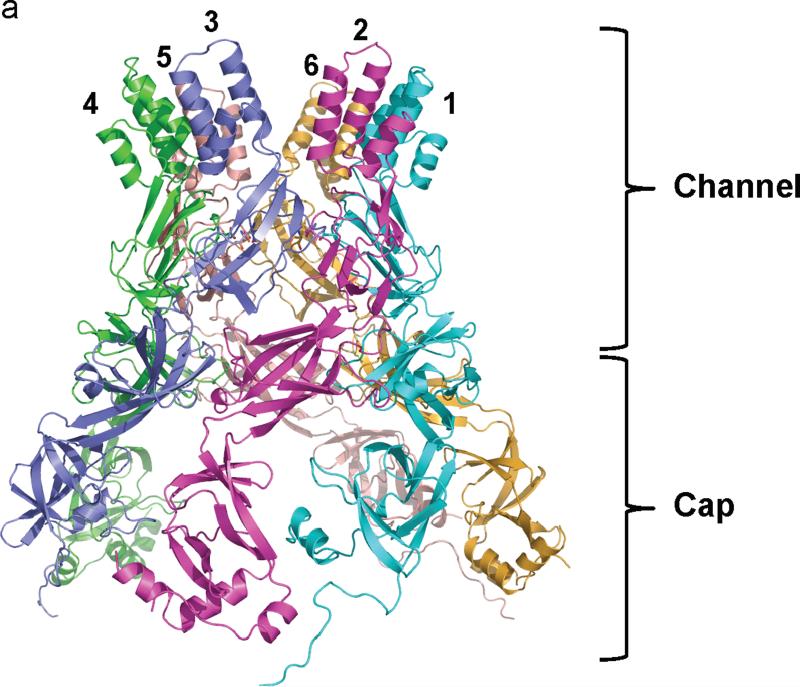
Here, we describe the co-crystal structure of the CusBA heavy-metal efflux complex. We used molecular replacement with single-wavelength anomalous dispersion (MRSAD) to determine the structure (Table S1 and Fig. S1), revealing each protomer of CusA interacts specifically with two elongated molecules of CusB (molecules 1 and 2) at the upper half portion of the periplasmic domain (Fig. 1). The two CusB adaptors are tilted at an angle of ~50° with respect to the membrane surface, and establish a close fit with the transporter at the concave surface formed by Domains 1 and 2 of the adaptor. Molecule 1 of CusB contacts mainly the upper regions of PN2 and PC1, and the DN sub-domain of CusA. Molecule 2 of CusB, however, predominantly bridges to the upper regions of PC1 and PC2, and also the sub-domain DC of the pump. These two adaptor molecules are also seen to specifically contact one another, primarily through Domains 1, 2 and 3 of these two elongated molecules. The trimeric CusA pump therefore directly contacts six CusB adaptor molecules, which form a channel at the top of the CusA trimer (Figs. 1 and 2).
Fig. 1.
Structure of the CusBA efflux complex. (a) Ribbon diagram of the structures of one CusA protomer (green) and two CusB protomers (red and blue) in the asymmetric unit of the crystal lattice. (b) Side view of the CusBA efflux complex. Each subunit of CusA is colored green. Molecules 1, 3 and 5 of CusB are colored red. Molecule 2, 4 and 6 of CusB are in blue. (c) Top view of the CusBA efflux complex. Each subunit of CusA is in green. Molecules 1, 3 and 5 of CusB are in red. Molecules 2, 4 and 6 of CusB are in blue.
Fig. 2.
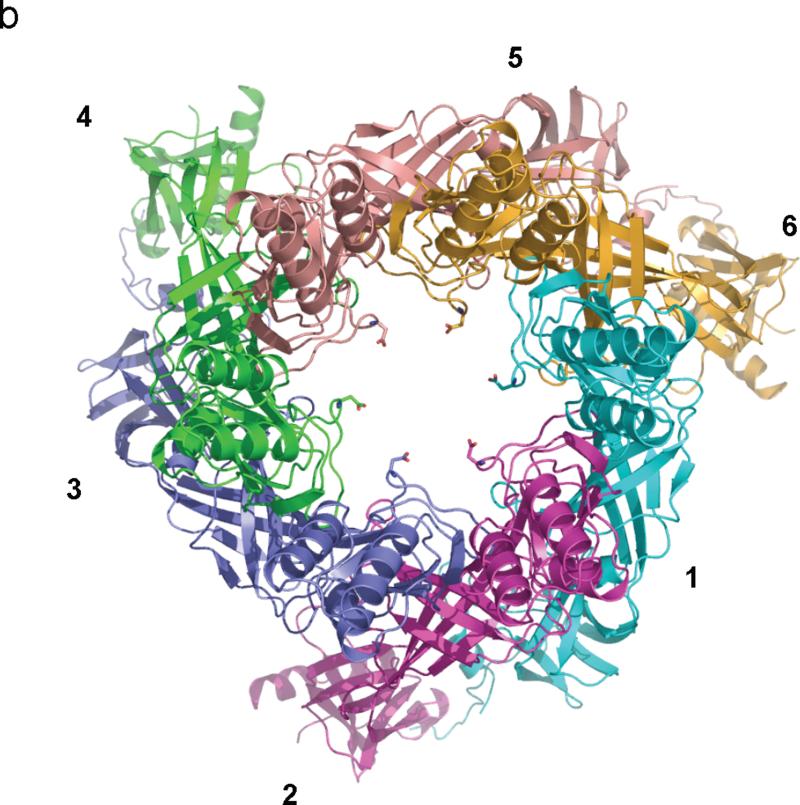
Structure of the hexameric CusB channel. (a) Side view of the hexameric CusB channel. The six molecules CusB are shown in ribbons (cyan, molecule 1; magenta, molecule 2; slate, molecule 3; green, molecule 4; pink, molecule 5; orange, molecule 6). (b) Top view of the hexameric CusB channel. The six molecules CusB are shown in ribbons (cyan, molecule 1; magenta, molecule 2; slate, molecule 3; green, molecule 4; pink, molecule 5; orange, molecule 6). Residue D232, which forms the narrowest region of the central channel, from each subunit of CusB is in sticks. The internal diameter of this narrowest region is ~18 Å.
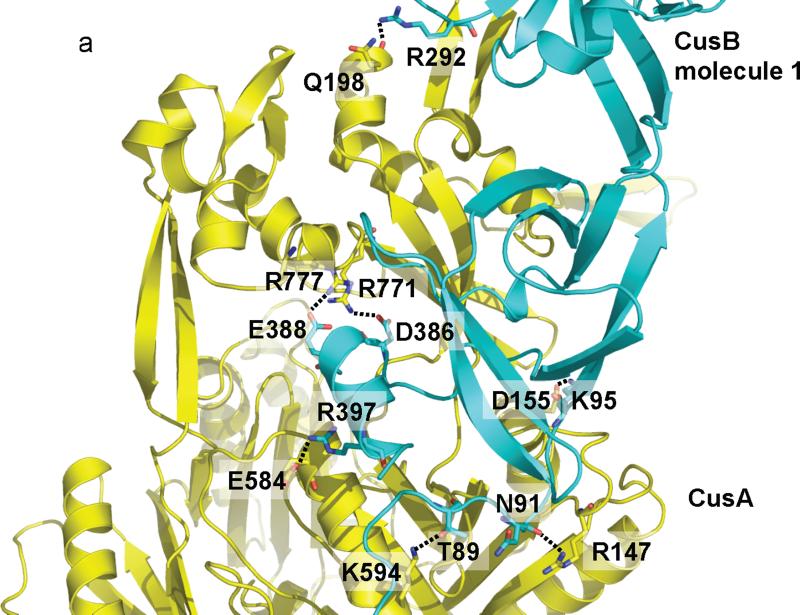
Intriguingly, molecule 1 of CusB interacts predominantly with CusA through charge-charge interactions. Residues K95, D386, E388 and R397 of this CusB molecule form four salt bridges with D155, R771, R777 and E584 of CusA, respectively. In addition, T89, the backbone oxygen of N91, and R292 of molecule 1 of CusB form hydrogen bonds with K594, R147, and the backbone oxygen of Q198 of CusA to secure the interaction (Fig. 3a). However, the interaction between molecule 2 of CusB and CusA appears to be governed principally by charge-dipole and dipole-dipole interactions. Specifically, Q108, S109, S253 and N312 of CusB (molecule 2) form hydrogen bonds with Q785, Q194, D800 and Q198 of CusA, respectively. The backbone oxygens of L92 and T335 of this CusB molecule also contribute two additional hydrogen bonds with the side chains of K591 and T808 of the CusA pump to anchor the proteins (Fig. 3b).
Fig. 3.
CusA-CusB interactions. (a) The interactions between molecule 1 of CusB and CusA. Residues K95, D386, E388 and R397 of this CusB molecule form four salt bridges with D155, R771, R777 and E584 of CusA, respectively. In addition, T89, the backbone oxygen of N91, and R292 of molecule 1 of CusB form hydrogen bonds with K594, R147, and the backbone oxygen of Q198 of CusA. (b) The interactions between molecule 2 of CusB and CusA. Residues Q108, S109, S253 and N312 of molecule 2 of CusB form hydrogen bonds with Q785, Q194, D800 and Q198 of CusA, respectively. The backbone oxygens of L92 and T335 of this CusB molecule also contribute two additional hydrogen bonds with the side chains of K591 and T808 of the CusA pump to anchor the proteins.
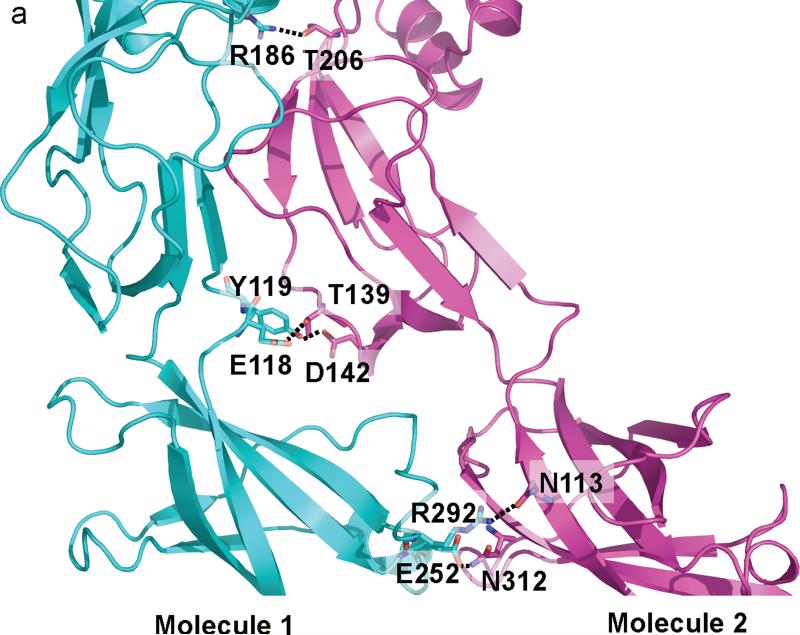
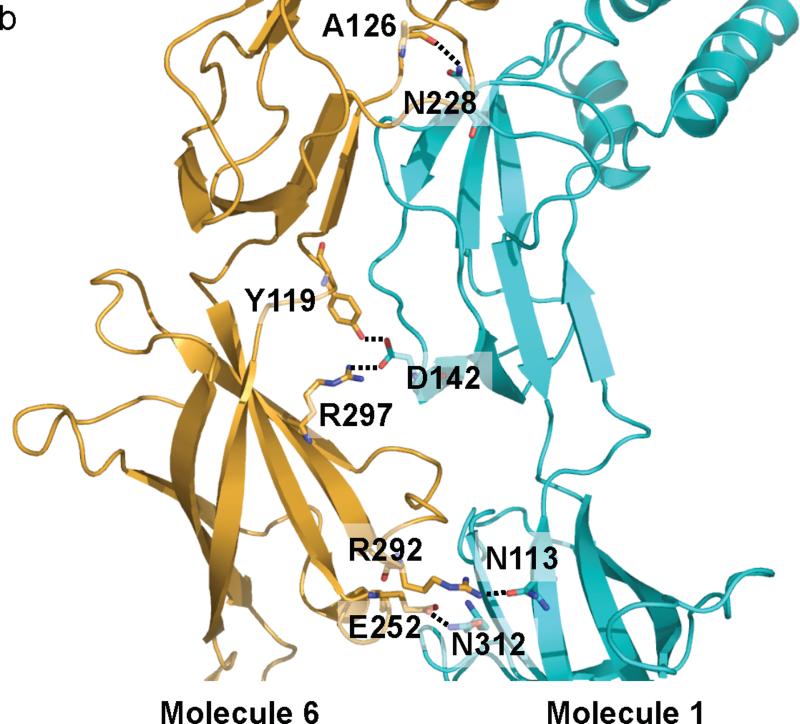
For CusB-CusB interactions, molecule 1 of CusB makes a close contact with molecule 2 of CusB. Domains 1-3 of these two molecules are involved in the binding. E118, Y119, R186, E252 and E292 of molecule 1 of CusB participate to form hydrogen bonds with T139, D142, T206, N312 and N113 of molecule 2 of CusB, respectively (Fig. 4a). Further, molecule 1 of CusB also contributes to contact molecule 6 of CusB, which is anchored to the next subunit of CusA. The majority of the interactions come from Domains 2 and 3 of these two molecules. Particularly, N113, N228, and N312 of molecule 1 of CusB pair up with R292, the backbone oxygen of A126, and E252 of molecule 6 of CusB to form three hydrogen bonds. In addition, D142 of molecule 1 of CusB participates to form two hydrogen bonds with Y119 and R297 of molecule 6 of CusB to secure the binding (Fig. 4b).
Fig. 4.
CusB-CusB interactions. (a) The interactions between molecules 1 and 2 of CusB. Residues E118, Y119, R186, E252 and E292 of molecule 1 of CusB participate to form hydrogen bonds with residues T139, D142, T206, N312 and N113 of molecule 2 of CusB, respectively. These hydrogen-bonded distances are 2.7, 2.7, 3.0, 3.0 and 3.0 Å, respectively. (b) The interactions between molecules 1 and 6 of CusB. Residues N113, N228, and N312 of molecule 1 of CusB pair up with R292, the backbone oxygen of A126, and E252 of molecule 6 of CusB to form three hydrogen bonds. D142 of molecule 1 of CusB also participates to form two hydrogen bonds with Y119 and R297 of molecule 6 of CusB. These hydrogen-bonded distances are 2.7, 2.8, 3.1, 2.7 and 2.8 Å, respectively.
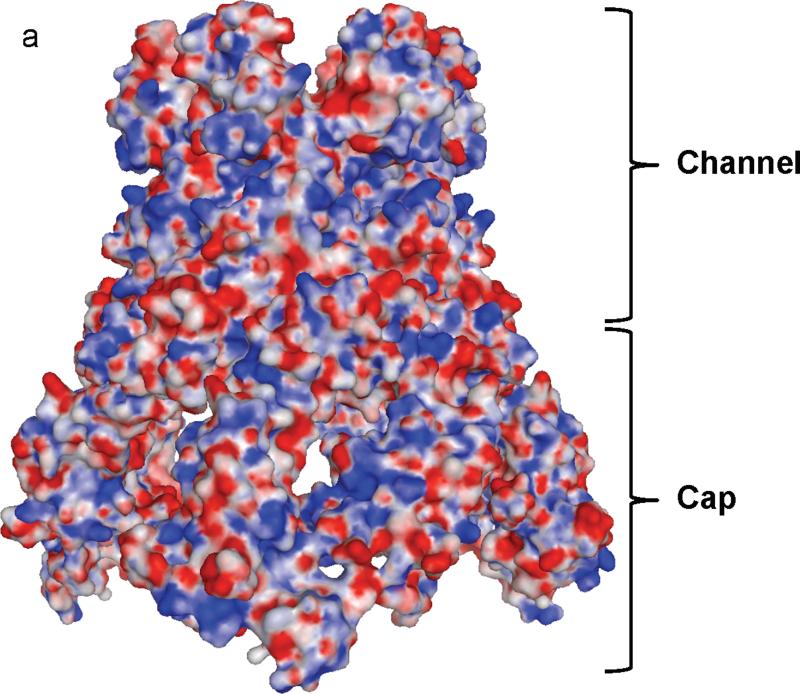
The hexameric arrangement of CusB allows this protein to create a funnel-like structure with the central channel formed along the crystallographic threefold symmetry axis. These protomers utilize a side-by-side packing arrangement to form this funnel (Fig. 2). Domain 1 and the lower half of Domain 2 of CusB primarily create a cap-like structure whereas the upper half of Domain 2, Domain 3 and Domain 4 of the adaptor contribute to the central channel of the funnel. The inner surface of the cap fits closely with the outer surface of the upper portion of the periplasmic domain of the CusA trimer. The channel formed above the cap of the adaptor is ~62 Å long with an average internal diameter of ~37 Å. Thus, the interior of the channel gives rise to a large elongated cavity with a volume of ~65,000 Å3. The lower half of the channel is primarily created by β-barrels whereas the upper half is an all α-helical tunnel. The diameter of the channel is gradually constricted and then dilated as it approaches the outer membrane. Thus, the α-helices of Domain 4 create an inverted conical structure. The narrowest section of the central channel is located at residue D232, which is close to the hinge region between Domains 3 and 4 of the membrane fusion protein. The widest section of the channel appears to form at the top edge with an inner diameter of ~56 Å. The inner surface of the channel is predominantly negatively charged, as indicated by the electrostatic surface diagram (Fig. 5), suggesting that the interior surface of the channel may have the capacity to bind positively charged metal ions.
Fig. 5.
Electrostatic surface potential of CusB. (a) Side view of the electrostatic surface potential of the hexameric CusB channel. This view shows the cap and channel regions formed by the CusB hexamer. Blue (+15 kBT) and red (-15 kBT) indicate the positively and negatively charged areas, respectively, of the protein. (b) Top view of the electrostatic surface potential of the hexameric CusB channel. The widest section of the hexameric channel appears to form at the top edge with its internal diameter of ~56 Å. Blue (+15 kBT) and red (-15 kBT) indicate the positively and negatively charged areas, respectively, of the protein.
We used isothermal titration calorimetry (ITC) to determine the binding affinity of CusB to the CusA pump. The ITC data indicates an equilibrium dissociation constant of 5.1 ± 0.3 μM (Fig. S2).
Previously, it was found that the N and C-terminal ends (residue 29-88 and 386-407) of the CusB adaptor are intrinsically disordered and cannot be identified in the electron density maps of the CusB crystals.2 Here, the co-crystal structure suggests that this region forms several short α-helices. In molecule 1 of CusB, residues 392-399 at the C-terminal end form a short α-helix. However, residues 79-95 of the N-terminus feature a long random coil and these amino acids are located immediately outside the cleft formed between sub-domains PC1 and PC2 of the CusA pump. For molecule 2 of CusB, the N-terminal residues 79-85 and 86-92 participate to form a random coil and a short α-helix, respectively. However, the C-terminal residues 382-392 and 394-400 appear to create two short α-helices. Like molecule 1 of CusB, the N-terminus of molecule 2 of CusB is near the periplasmic cleft of the pump (Fig. 1).
It has been proposed that the N-terminal residues M49, M64 and M66 of CusB form a three-methionine metal-binding site.23 Although these three methionine residues cannot be identified in the electron density maps of our co-crystal, the co-crystal structure does show that the N-terminal tails of both molecule 1 and 2 of CusB are located outside the cleft formed between PC1 and PC2 of the CusA pump. Thus, it is possible that CusB might help to transfer the metal ions via the N-terminal three-methionine binding site into the periplasmic cleft of CusA. Indeed, a similar suggestion has been made with the adaptor protein AcrA whereas this protein might assist in transporting a drug from the periplasm into the pump.24
The co-crystal structure presented here highlights the structural importance of the periplasmic membrane fusion protein. Given the fact that six CusB molecules assemble to form a channel at the CusA funnel top, this suggests that the adaptor is likely to be involved in the active extrusion of metal ions.
We then constructed a CusCBA model based on the CusBA crystal complex structure and the predicted CusC model. The final CusC3-CusB6-CusA3 structural model represents a 750 kDa tripartite efflux complex spanning both the inner and outer membranes of E. coli to extrude Cu(I)/Ag(I) (Fig. S3).
We believe that CusA can take up metal ions from both the periplasm and cytoplasm utilizing the methionine-residue ion relay network.1 Metal ions could enter the three-methionine binding-site, formed by M573, M623 and M672, inside the cleft between sub-domains PN2 and PC1 on the periplasmic portion of CusA or via the methionine pairs within the transmembrane domain of the pump. It has been demonstrated that the chaperone CusF can directly transfer its bound Cu(I) to the CusB membrane fusion protein.25 Thus, it is likely that CusF is responsible for delivering metal ions to the CusCBA tripartite efflux system in the periplasm. There is a chance that the initial step of metal transport through the periplasmic cleft of the CusA pump may involve a direct transfer from CusF to the previously proposed three-methionine metal binding site23 (M49, M64 and M66) at the long N-terminal tail of CusB, situating near the PN2/PC1 cleft of the CusA pump. Thus, the second step could well be the delivery of the bound metal ion from CusB to the three-methionine cluster (M573, M623 and M672) inside the periplasmic cleft of CusA.1 The bound metal ion could then be released to the nearest methionine pair (M271-M755) located directly above the three-methionine metal binding site,1 from which the ion could be released into the central funnel of CusA and eventually the CusC channel for final extrusion. It is not yet known whether the interior of the hexameric CusB channel forms part of the extrusion pathway for Cu(I) and Ag(I). Exactly what this tripartite efflux system's mechanism is must await confirmation by elucidation of additional crystal structures of the CusCBA tripartite complex.
Methods Summary
Crystallization of CusA
The procedures for cloning, expression and purification of the CusA and CusB proteins have been described previously.1,2 Co-crystals of the CusBA complex were obtained using sitting-drop vapor diffusion. A 2 μl protein solution containing 0.1 mM CusA and 0.1 mM CusB in buffer solution containing 20 mM Na-HEPES (pH 7.5) and 0.05% (w/v) CYMAL-6 was mixed with 2 μl of reservoir solution containing 10% PEG 6000, 0.1 M Na-HEPES (pH 7.5), 0.1 M ammonium acetate and 20% glycerol. The resultant mixture was equilibrated against 500 μl of the reservoir solution. The co-crystallization conditions for CusA (native)-CusB (SeMet) were the same as those for the native CusBA complex. Co-crystals of CusBA grew to full size in the drops within two months. Cryoprotection was achieved by raising the glycerol concentration stepwise to 30% in 5% increments.
Structural determination and refinement
Data of the CusA (native)-CusB (SeMet) co-crystal were collected (Table S1). SAD phasing using the program PHASER26 was employed to obtain experimental phases in addition to the phases from the structural model of apo-CusA. Phases were subjected to density modification and phase extension to 2.90 Å-resolution using the program RESOLVE.27 The full-length CusB protein contains nine methionine residues, and six selenium sites per CusB molecule (12 selenium sites per asymmetric unit) were identified. After tracing the initial model manually using the program Coot,28 the model was refined against the native data at 2.90 Å-resolution using TLS refinement adopting a single TLS body as implemented in PHENIX29 leaving 5% of the reflections in the Free-R set. Iterations of refinement using PHENIX29 and CNS30 and model building in Coot28 lead to the current model, which consists of 1,686 residues (residues 4-1043 of CusA; residues 79-400 of molecule 1 of CusB; and residues 79-402 of molecule 2 of CusB).
Supplementary Material
Acknowledgements
This work is supported by NIH Grants R01GM074027 (E.W.Y.), R01GM086431 (E.W.Y.), R01GM081680 (R.L.J.) and R01GM072014 (R.L.J.). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Methods
Isothermal Titration Calorimetry
We used isothermal titration calorimetry to examine the binding of the purified CusB adaptor to the purified CusA pump. Measurements were performed on a VP-Microcalorimeter (MicroCal, Northampton, MA) at 25 °C. Before titration, the CusA and CusB proteins were thoroughly dialyzed against buffer containing 20 mM Na-HEPES pH 7.5 and 0.05% CYMAL-6, respectively. The protein concentrations were determined using the Bradford assay. The CusA protein sample was then adjusted to a final monomeric concentration of 14 μM. The CusB protein solution consisting of 350 μM monomeric CusB in 20 mM Na-HEPES pH 7.5 and 0.05% CYMAL-6 was prepared as the titrant. The samples were degassed before they were loaded into the cell and syringe. Binding experiments were carried out with the CusA protein solution (1.5 ml) in the cell and the CusB protein solution as the injectant. Ten microliter injections of the CusB solution were used for data collection.
Injections occurred at intervals of 300 s, and the duration time of each injection was 10 s. Heat transfer (μcal/s) was measured as a function of elapsed time (s). The mean enthalpies measured from injection of the ligand in the buffer were subtracted from raw titration data before data analysis with ORIGIN software (MicroCal). Titration curves were fitted by a nonlinear least squares method to a function for the binding of a ligand to a macromolecule. Nonlinear regression fitting to the binding isotherm provided us with the equilibrium binding constant (KA = 1/KD) and enthalpy of binding (ΔH). Based on the values of KA, the change in free energy (ΔG) and entropy (ΔS) were calculated with the equation: ΔG = - RT lnKA = ΔH - TΔS, where T is 273 K and R is 1.9872 cal/K per mol. Calorimetry trials were also carried out in the absence of CusA in the same experimental conditions. No change in heat was observed in the injections throughout the experiment.
Structural prediction and docking of CusC onto CusBA
A homology model of CusC was generated using the crystal structure of OprM (PDB ID: 1wp1)20 as a template through the I-TASSER server.31 Alignment of protein sequences suggests that these two channel proteins share 42% identity. A steered molecular dynamics simulation was then used to rigidly dock the trimeric CusC channel to the crystal form of the hexameric CusB channel bound to the trimeric CusA pump. During the rigid-body docking process, the molecules were aligned with the z-axis. CusB was then held rigidly, allowing CusC to pull along the z-axis toward CusB until no further change in the center of mass was measured.
We next investigated the possible interaction of CusC with Domain 2 of CusB, which also brings it into close proximity to CusA. First, the trimeric CusA pump and Domains 1 and 2 of the hexameric CusB channel were held rigidly, while Domains 3 and 4 of each CusB monomer were pulled outward using customized harmonic forces to open the annulus of the hexameric channel. Next, we manually inserted CusC into the opened CusB ring. Finally, simulated annealing followed by energy minimization was performed to relax CusB and CusC from these positions and allow them to assume a lower energy conformation. All simulations were performed using NAMD32 and the CHARMM27+CMAP force field.33
Footnotes
Author Information
Atomic coordinates and structure factors have been deposited with the Protein Data Bank under the code 3NE5.
References
- 1.Long F, Su C-C, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, Yu EW. Crystal structures of the CusA heavy-metal efflux pump suggest methionine-mediated metal transport mechanism. Nature. 2010;467:484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su C-C, Yang F, Long F, Reyon D, Routh MD, Kuo DW, Mokhtari AK, Van Ornam JD, Rabe KL, Hoy JA, Lee YJ, Rajashankar KR, Yu EW. Crystal structure of the membrane fusion protein CusB from Escherichia coli. J. Mol. Biol. 2009;393:342–355. doi: 10.1016/j.jmb.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franke S, Grass G, Nies DH. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiol. 2001;147:965–972. doi: 10.1099/00221287-147-4-965. [DOI] [PubMed] [Google Scholar]
- 4.Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development protein. J. Mol. Microbiol. Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 6.Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 7.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 8.Yu EW, McDermott G, Zgruskaya HI, Nikaido H, Koshland DE., Jr. Structural basis of multiple drug binding capacity of the AcrB multidrug efflux pump. Science. 2003;300:976–980. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
- 9.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 10.Seeger MA, Schiefner A, Eicher T, Verrey F, Dietrichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 11.Sennhauser G, Amstutz P, Briand C, Storchengegger O, Grütter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu EW, Aires JR, McDermott G, Nikaido H. A periplasmic-drug binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J. Bacteriol. 2005;187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sennhauser G, Bukowska MA, Briand C, Grütter MG. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 2009;389:134–145. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Su C-C, Li M, Gu R, Takatsuka Y, McDermott G, Nikaido H, Yu EW. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J. Bacteriol. 2006;188:7290–7296. doi: 10.1128/JB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins MK, Bokma E, Koronakis E, Hughes C, Koronakis V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. USA. 2004;101:9994–9999. doi: 10.1073/pnas.0400375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akama H, Matsuura T, Kashiwag S, Yoneyama H, Narita S, Tsukihara T, Nakagawa A, Nakae T. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:25939–25942. doi: 10.1074/jbc.C400164200. [DOI] [PubMed] [Google Scholar]
- 17.Mikolosko J, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14:577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Symmons M, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. USA. 2009;106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 20.Akama H, Kanemaki M, Yoshimura M, Tsukihara T, Kashiwag T, Yoneyama H, Narita S, Nakagawa A, Nakae T. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:52816–52819. doi: 10.1074/jbc.C400445200. [DOI] [PubMed] [Google Scholar]
- 21.Xue Y, Davis AV, Balakrishnan G, Stasser JP, Staehlin BM, Focia P, Spiro TG, Penner-Hahn JE, O'Halloran TV. Cu(I) recognition via cation-π and methionine interactions in CusF. Nat. Chem. Biol. 2008;4:107–109. doi: 10.1038/nchembio.2007.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loftin IR, Franke S, Blackburn NJ, McEvoy MM. Unusual Cu(I)/Ag(I) coordination of Escherichia coli CusF as revealed by atomic resolution crystallography and X-ray absorption spectroscopy. Prot. Sci. 2007;16:2287–2293. doi: 10.1110/ps.073021307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bagai I, Liu W, Rensing C, Blackburn N, McEvoy MM. Substrate-linked conformational change in the periplasmic component of Cu(I)/Ag(I) efflux system. J. Biol. Chem. 2007;282:35695–35702. doi: 10.1074/jbc.M703937200. [DOI] [PubMed] [Google Scholar]
- 24.Gerken H, Misra R. Genetic evidence for functional interactions between TolC and AcrA proteins of a major antibiotic efflux pump of Escherichia coli. Mol. Microbiol. 2004;54:620–631. doi: 10.1111/j.1365-2958.2004.04301.x. [DOI] [PubMed] [Google Scholar]
- 25.Bagai I, Rensing C, Blackburn NJ, McEvoy MM. Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry. 2008;47:11408–11414. doi: 10.1021/bi801638m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger TC. Maximum-likelihood density modification using pattern recognition of structural motifs. Acta Cryst. 2001;D57:1755–1762. doi: 10.1107/S0907444901013737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCroy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 30.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feller SE, MacKerell AD., Jr. An improved empirical potential energy for molecular simulations of phospholipids. J. Phys. Chem. B. 2000;104:7510–7515. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.