Delta/Notch-dependent lateral inhibition becomes enhanced in neuroepithelial cells undergoing mitosis. We report that the RNA-binding protein Elavl1/HuR localizes within the cytoplasm of these cells, interacts with the 3′-untranslated region of Dll1, and stabilizes transcripts containing this sequence, thus favoring Notch signaling.
Abstract
In the vertebrate neuroepithelium, the decision to differentiate is made by neural precursors soon after mitosis, when they are apically located. This process is controlled by lateral inhibitory signals triggered by the Delta/Notch pathway. During mitosis, the capacity of neural precursors to express the neurogenic genes Dll1 and Notch1 is maximal due to mRNA stabilization, but the mechanism controlling this process remains unknown. Here we show that Elav-like (Elavl1)/HuR becomes enriched in the cytoplasm of neuroepithelial cells undergoing mitosis and that this ribonucleoprotein interacts with the 3′ untranslated region (UTR) of Dll1 mRNA. This interaction is functionally relevant because RNAi against Elavl1 reduces the stability of Dll1–3′UTR-containing transcripts in mitosis-arrested neuroepithelial cells, and Elavl1 null-mutant heterozygous mice show decreased Dll1 expression in the developing brain in vivo. We also show that RNAi against Elavl1 diminishes the capacity of brain precursors to trigger lateral inhibitory signals to their neighbors, an observation consistent with the increase in the rate of neurogenesis which can be detected in vivo in the developing retina of Elavl1 heterozygous mice. We conclude that Elavl1/HuR facilitates the exposure of vertebrate neuronal precursors to apically located Delta/Notch signals.
INTRODUCTION
Most vertebrate neurons arise from a pseudostratified neuroepithelium constituted by neural precursors the nuclei of which occupy a basal position during S-phase while they displace to the apical region during mitosis (M) (Sauer, 1935; Frade, 2002). The decision of coming out from the cell cycle and becoming a neuron is made by neural precursors during or soon after their last M, when they are apically located and the capacity to express determination proneural genes, known to initiate a cascade of events leading to neuronal differentiation (Bertrand et al., 2002), is maximal (Murciano et al., 2002; Cisneros et al., 2008). Whether or not a neural precursor comes out from the cell cycle and differentiates is tightly regulated by intercellular lateral inhibitory signals involving the Delta ligand and its Notch receptor (Louvi and Artavanis-Tsakonas, 2006). Notch activation by Delta can repress the expression of determination proneural genes, thereby preventing neuronal differentiation. Moreover, such activation prevents the expression of Delta itself. Therefore the Delta/Notch signaling pathway gives rise to a feedback loop that can amplify differences between adjacent cells, because the receipt of inhibition (i.e., activation of Notch) diminishes the ability to deliver inhibition (i.e., to produce Delta) (Collier et al., 1996).
We have previously demonstrated that the capacity of expression of both Notch1 and Delta-like 1 (Dll1) mRNA in the chick and mouse neuroepithelium becomes increased in precursor cells undergoing G2, M, and early G1 (Murciano et al., 2002; Cisneros et al., 2008) when they are apically located. This temporal expression pattern suggests that lateral inhibition is restricted to neural precursors that occupy the apical portion of the neuroepithelium. Accordingly, lateral inhibitory signaling was observed to be maximal in dissociated neural precursors synchronized in M (Cisneros et al., 2008). These results have been substantiated by independent studies demonstrating higher Notch activity at the apical portion of the neuroepithelium, where neural precursors undergo M (Del Bene et al., 2008; Ochiai et al., 2009), thus stressing the importance that the three-dimensional structure of the neuroepithelium has on the neurogenic process (Murciano et al., 2002; Xie et al., 2007; Del Bene et al., 2008; Agathocleous and Harris, 2009; Kageyama et al., 2009; Latasa et al., 2009; Schenk et al., 2009; Ge et al., 2010).
In our previous work (Cisneros et al., 2008), we showed that the capacity of neural precursors to express high levels of Notch1 and Dll1 mRNA during M was due to enhanced stability of these transcripts at this cell-cycle stage. In the leech Helobdella robusta, the 3′-untranslated region (3′UTR) of Notch mRNA has been shown to modulate transcript stability (Gonsalvez and Weisblat, 2007), likely due to the seven AU-rich elements (AREs) contained in its sequence. These elements, often defined by the sequence AUUUA, are known to promote mRNA deadenylation and decay (Xu et al., 1997; Barreau et al., 2005). In silico analysis demonstrated that Notch1 and Dll1 mRNAs contain conserved AREs within their 3′UTRs in all species studied (Cisneros et al., 2008), but the question as to whether Notch1–3′UTR and Dll1–3′UTR sequences participate in cell cycle–dependent stabilization of these transcripts remains unanswered.
Several RNA binding proteins (RBPs) can interact with ARE sequences and regulate the steady-state levels of their target mRNA (Barreau et al., 2005), being potential regulators of Notch1 and Dll1 stability in the neuroepithelium. We have focused our study on the RBP embryonic lethal abnormal vision (Elav)-like 1, also known as HuR. Elavl1/HuR can enhance the stability of many mRNAs, including the cell-cycle regulators cyclin A, B1, and D1 (Ccna, Ccnb1, and Ccnd1) (Wang et al., 2000; Lal et al., 2004), as well as the basic helix-loop-helix transcription factors MyoD and myogenin (Myod1 and Myog) (Figueroa et al., 2003), which are closely related to the proneural genes.
RESULTS
Generation of H2-b2T neuroepithelial cells constitutively expressing Egfp-Dll1–3′UTR or Egfp-Notch1–3′UTR
We have shown that the transcripts encoding Notch1 and Dll1 are differentially expressed along the cell cycle both in the mouse and chick neuroepithelia, resulting from the enhancement of the steady-state levels of these transcripts during M (Cisneros et al., 2008). One possibility is that the 3′UTRs of the Notch1 and Dll1 mRNAs, known to contain conserved AREs (Cisneros et al., 2008), are required for their stabilization during this stage of the cell cycle. To test this possibility, we created expression vectors containing the 3′UTRs of mouse Notch1 or Dll1 downstream of the coding sequence of enhanced green fluorescent protein (EGFP) (Figure 1). These constructs were transfected in H2-b2T neuroepithelial cells, an immortalized cell line established from the hindbrain of mouse transgenic embryos expressing a mutated version of the simian virus 40 (SV40) T antigen (Nardelli et al., 2003). In total, three polyclonal cell lines derived from H2-b2T neuroepithelial cells were created: a control cell line transfected with a vector containing the SV40 polyadenylation sequence downstream of the EGFP coding region (EC cells), and two cell lines transfected with the vectors containing the 3′UTRs of either Notch1 or Dll1 downstream of the EGFP coding sequence (EN and ED cells, respectively).
FIGURE 1:
Scheme of the constructs used for creating H2-b2T neuroepithelial cells constitutively expressing Egfp-specific transcripts. Transcripts with the SV40 polyadenylation sequence (EGFP-control), or with the 3′UTRs of mouse Dll1 (EGFP-Dll1–3′UTR) or Notch1 (EGFP-Notch1–3′UTR) mRNAs are shown. Note that the Dll1- and Notch1–3′UTR sequences have been subcloned at the NotI site of the EGFP-control vector. The SV40 polyadenylation signal is not used in these latter constructs as it lies downstream of the Dll1 or Notch1 polyadenylation signals.
H2-b2T neuroepithelial cells show characteristics of early neural precursors, constituting a good model system for the analysis of molecular pathways present in neuroepithelial cells (Nardelli et al., 2003). Cell-cycle kinetics in these cells was studied using a modification of a previously described method based on 5-bromo-2′-deoxyuridine (BrdU) incorporation during S-phase (Takahashi et al., 1993). We observed that approximately 30% of the H2-b2T neuroepithelial cells incorporated BrdU at the earliest time point analyzed (1 h), and then they continued incorporating BrdU in a linear manner during the first 17 h, a time point at which they reached a plateau (Supplemental Figure S1C). These observations allowed us to estimate the duration of the cell cycle (TC) and the S-phase (TS), concluding that in these cells TC lasts approximately 25 h, whereas TS lasts approximately 7.5 h (Supplemental Figure S1, B–D). Further analyses indicated that the first mitotic cells that are positively stained with BrdU were observed 2 h after addition of this nucleotide analog (Supplemental Figure S1A), thus indicating that this time is sufficient for these cells to undergo the G2 phase (Supplemental Figure S1D). Finally, 1.14% of the H2-b2T neuroepithelial cells were observed to undergo M (Supplemental Figure S1A), providing evidence that this cell-cycle stage lasts <20 min in these cells (Supplemental Figure S1D). Because TC occurs in ∼25 h and S+G2+M takes place in <10 h, the duration of G1 was calculated to be approximately 15.5 h (Supplemental Figure S1D). The duration of the cell cycle in H2-b2T neuroepithelial cells is in the same range as the half-life of EGFP, which has been estimated to last 24 h (Tamberg et al., 2007). G1 is therefore long enough to allow a substantial proportion of the EGFP produced during M to become degraded by the time the cells undergo S-phase. We conclude therefore that the levels of EGFP in H2-b2T cells can be used as an estimation of the cell-cycle–dependent stability of their chimeric mRNAs.
The 3′UTR sequence of Dll1 mRNA facilitates EGFP expression during G2/M/early G1
The levels of EGFP expression at different stages of the cell cycle were analyzed by flow cytometry in the H2-b2T neuroepithelial cell lines stably expressing the different vectors described earlier in the text. Cells were grown asynchronously, fixed, and labeled with propidium iodide (PI) to define the phases of the cell cycle (Figure 2A, bottom panels). PI labeling demonstrated that most H2-b2T neuroepithelial cells were octoploid (as evidenced by comparison with diploid mouse cells that were used as a reference, Supplemental Figure S2), with a small subpopulation of cells being tetraploid. This observation is consistent with the known polyploidy-inducing effect of the SV40 T antigen (Levine et al., 1991). Most H2-b2T neuroepithelial cells were in G1, in accordance with the long duration of this cell-cycle stage in these cells (Supplemental Figure S1D), whereas the proportion of cells in G2/M was extremely low as expected from the cell-cycle kinetics described earlier in the text (Figure 2A, bottom panels). Flow cytometry cannot distinguish between tetraploid cells in G2/M and octoploid cells in G1. Nevertheless, because the proportion of the former cells is quite reduced, their presence should not interfere with the main conclusions of this analysis. Flow cytometry demonstrated that the expression of EGFP in the EC cells was highest during late G1 and S-phase, in accordance with the general decrease of mRNA stability during M due to cdk1-dependent reduction in the activity of poly(A) polymerase (Colgan et al., 1996). In contrast, EGFP expression in ED cells was observed to be moderately, although nonsignificantly, increased during G2/M (3.96 ± 0.40% of ED cells showed high levels of EGFP, whereas 2.08 ± 0.91% of EC cells contained intense green labeling; n = 3) and to be highest during early G1 (7.17 ± 0.36% of ED cells contained intense green labeling, whereas 2.62 ± 0.45% of EC cells showed high levels of EGFP; n = 3, p < 0.005; Student’s t test) (Figure 2A). These results are consistent with a delay in EGFP expression after stabilization of its mRNA in ED cells undergoing M. The increase of EGFP expression during G1 was primarily associated with tetraploid ED cells (Figure 2B). To confirm the data obtained by flow cytometry, EC and ED cells were given a short pulse (1 h) of BrdU. This analysis demonstrated that both cell lines showed a similar proportion of BrdU-positive cells (i.e., cells in S-phase) containing high levels of EGFP. In contrast, the proportion of ED cells lacking BrdU (i.e., in phases of the mitotic cycle other than S) and expressing high levels of EGFP was significantly increased as compared with this value in the EC cells (Figure 2C; see Figure 2D for an example of an ED cell with high EGFP expression).
FIGURE 2:
Cell-cycle–dependent regulation of EGFP expression controlled by the 3′UTRs of mouse Notch1 and Dll1 mRNAs. (A) The levels of expression of EGFP (top panels) was analyzed by flow cytometry in parental H2-b2T neuroepithelial cells (Control cells), EC cells, ED cells, or EN cells treated with PI to reveal the amount of DNA. The proportion of cells with high EGFP expression (n = 3) increases during both G1 and G2/M in ED cells as compared with EC cells (rectangles). The horizontal line represents the threshold below which cells are negative for EGFP expression. PI analysis demonstrated that most H2-b2T neuroepithelial cells were octoploid (8C) (see Supplemental Figure S2), whereas a small proportion of these cells had a 4C DNA amount (bottom panels). *Note that G1 refers to the octoploid cells; all tetraploid cells are included in this interval, but those undergoing S and G2 represent just a minority in absolute terms (Supplemental Figure S1D). (B) Distribution of EGFP-positive EC and ED cells (i.e., those present above the baseline illustrated in A, top panels) in terms of PI labeling. An increase of EGFP-positive ED cells in G1 (arrows) or G2/M (arrowheads) was observed. (C) ED or EC cells were treated with BrdU for 1 h and then fixed and immunostained for BrdU. Quantification of ED cells (gray bars) or EC cells (white bars) expressing high levels of EGFP depending on their capacity to incorporate BrdU. BrdU+: cells in S-phase; BrdU–: cells in cell-cycle stages other than S-phase. The results represent the mean ± SEM (n = 3). *p < 0.05 (Student’s t test). (D) A representative example of ED cells treated with BrdU (red) for 1 h. BrdU-negative cells (arrow) express high levels of EGFP (green). Arrowhead: a BrdU-positive cell lacking EGFP expression. Bar: 3 μm. (E) Plasmids (1 μg/μl) expressing Egfp-control (open bars, H) or Egfp-Dll1–3′UTR (gray bars, I) were electroporated into the brain ventricle of HH12 chick embryos (Hamburger and Hamilton, 1951), and 4 h later the distribution of EGFP along the apicobasal axis of the neuroepithelium was analyzed. EGFP-positive cells were classified into three categories depending of the position of their cell soma within either the basal third, the middle of the neuroepithelium, or the apical third. The results represent the mean ± SEM (n = 3); p < 0.005 (two-way ANOVA). (F) Representative image of a section throughout the mesencephalon of an HH12 chick embryo electroporated with an Egfp-control expression vector. Cell somas are mostly located at the basal portion (arrowhead). Arrow: apical surface. v: ventricle. Inset: region inside the dotted square. For a quantification, see E. Bar: 50 μm. (G) Representative image of a section throughout the mesencephalon of a HH12 chick embryo electroporated with a Egfp-Dll1–3′UTR expression vector. Cell somas are equally located at both the basal portion (arrowhead) and the apical surface (arrow). v: ventricle. Inset: region inside the dotted square. For a quantification, see E. Bar: 50 μm. (H) qPCR on cDNA obtained from EC or ED cells previously synchronized in M with 1 μg/ml colchicine (Mitosis), in S-phase with 1 mM HU (S-phase) or left untreated (Asynchr.). Levels of Ccnb1 (open bars, Cyclin B1) or Egfp (gray bars), normalized to 18S rRNA, are shown. The results represent the mean ± SEM (n = 3); **p < 0.01; ***p < 0.005 (Student’s t test). (I) EC (blue diamonds), ED (green circles), or EN (red triangles) cells were synchronized in M for 24 h with 1 μg/ml colchicine, and then transcription was blocked in these cultures with 5 μg/ml actinomycin D for the indicated time points. Levels of Egfp mRNA, normalized to 18S rRNA, were then measured in triplicate by qPCR. The results represent the mean ± SEM (n = 3). **p < 0.01;***p < 0.005 (Student’s t test).
To study whether these observations are relevant in vivo, we performed electroporation experiments in stage HH12 chick embryos (Hamburger and Hamilton, 1951). In these experiments, Egfp-Dll1–3′UTR or Egfp-control expression vectors were electroporated into the brain neuroepithelium, and then chick embryos were killed 4 h after plasmid delivery. This short time point was chosen to avoid accumulation of EGFP, thus restricting the presence of this protein to neuroepithelial cells recently expressing the Egfp transcript. This analysis demonstrated that the presence of Egfp-Dll1–3′UTR resulted in a significant increase of apically located neuroepithelial cells (i.e., cells in G2/M/early G1) (Murciano et al., 2002) positive for EGFP (Figure 2, E and G), when compared with the expression pattern resulting from the Egfp-control plasmid (Figure 2, E and F).
In contrast to the ED cells, flow cytometric analysis demonstrated a general decrease of EGFP expression in the EN cells, which was cell-cycle–independent (Figure 2A). These results suggest that the 3′UTR of Notch1 confers instability to the Egfp chimeric mRNA, as previously observed for the 3′UTR of Notch in H. robusta (Gonsalves and Weisblat, 2007). In sum, the 3′UTR of Dll1, but not that of Notch1, seems to induce mRNA stabilization during G2/M/early G1 in neuroepithelium-derived cells.
The 3′UTR sequence of Dll1 confers mRNA stability during M
The observed increase of EGFP in ED cells undergoing G2/M/early G1 correlates with elevated Egfp mRNA expression. This observation was evidenced by real-time quantitative PCR (qPCR) analysis on cDNA obtained from ED cells synchronized in M, as compared with cDNAs from either unsynchronized or S-phase–synchronized ED cells (Figure 2H, right panel). We used colchicine for synchronizing these cells in M and hydroxyurea (HU) to synchronize them in S-phase, as previously described (Murciano et al., 2002; Cisneros et al., 2008). In contrast, M-synchronized EC cells showed Egfp mRNA levels similar to those of EC cells synchronized in S-phase or just left unsynchronized (Figure 2H, left panel). Ccnb1 expression was observed to be enriched in both EC and ED cells in M (Figure 2H), thus indicating that our cell synchronization protocol was effective in these experiments.
To demonstrate that the increase of Egfp during M derives from Dll1–3′UTR-dependent mRNA stabilization, mitotically synchronized EC or ED cells were treated with actinomycin D to block transcription. Subsequently, total RNA obtained from these cells at different time points was subjected to qPCR with Egfp-specific primers to follow the degradation kinetics of the Egfp transcript. This analysis revealed that Egfp mRNA diminished rapidly in the EC cells, whereas this transcript was highly stable in the ED cells (Figure 2I). As expected, Egfp was also rapidly degraded in the EN cells (Figure 2I). These data demonstrate that Dll1–3′UTR, but not Notch1–3′UTR, induces mRNA stabilization during G2/M/early G1 in neuroepithelium-derived cells.
Elavl1/HuR is expressed by mouse neuroepithelial cells in a cell-cycle–dependent manner
Elavl1/HuR is one of a few RBPs known to stabilize their target ARE-containing mRNAs (Barreau et al., 2005). As its expression pattern in the vertebrate neuroepithelium is currently unknown, we decided to perform immunostaining with an Elavl1/HuR-specific antibody in telencephalic cryosections obtained from E13.5 mouse embryos. This analysis revealed that Elavl1/HuR was highly expressed in the cytoplasm of fusiform cells located close to the ventricular surface as well as in apically located, round cells undergoing M (Figure 3A). Interestingly, such fusiform cells were not in S-phase, as indicated by the absence of BrdU incorporation in these cells after a short BrdU pulse (Figure 3A). Elavl1/HuR was also detected surrounding the nuclei of apically located precursors lacking the mitotic marker pH3 (Figure 3B), suggesting that this protein is expressed not only during M but also during late G2 and/or early G1. These data indicate that Elavl1/HuR becomes enriched during M in the cytoplasm of neuroepithelial cells in vivo. Elavl1/HuR protein was also observed to be expressed by H2-b2T neuroepithelial cells in a cell-cycle–dependent manner, because it was enriched in the cytoplasm of H2-b2T neuroepithelial cells undergoing M (i.e., labeled with pH3 specific antibodies) (Supplemental Figure S3A). Furthermore, high expression levels of EGFP were consistently observed in ED cells showing high Elavl1/HuR immunolabeling (Supplemental Figure S3B), suggesting that this RBP participates in the stabilization of the Egfp-Dll1–3′UTR transcript. To obtain an estimation of the percentage of H2-b2T neuroepithelial cells expressing high levels of Elavl1/HuR during M or S-phase, these cells were synchronized with colchicine or HU, as mentioned earlier in the text. This analysis demonstrated that approximately 97% of M-synchronized H2-b2T neuroepithelial cells expressed Elavl1/HuR at high levels (94 of 97 cells). In contrast, the percentage of S-phase–synchronized H2-b2T neuroepithelial cells expressing Elavl1/HuR was reduced (14 cells of 129 S-phase–synchronized cells expressed this RBP). The cell-cycle-dependent expression of Elavl1/HuR both in vivo and in the H2-b2T neuroepithelial cells, along with the presence of this protein in the cytoplasm, strongly suggests that it may prevent mRNA degradation in neuroepithelial cells at stages of the cell cycle different from S-phase, thus becoming a strong candidate for the stabilization of Dll1 mRNA during G2/M/early G1.
FIGURE 3:
Expression of Elavl1 in the embryonic murine cortex. (A) Cortical cryosections (15 μm) from E13.5 mouse previously treated with BrdU for 1 h immunostained for Elavl1/HuR (red) and BrdU (green), counterstained with bisbenzimide (Bisb.) to localize the nuclei (blue). Cells expressing Elavl1/HuR at high levels are located close to the ventricle (arrows), and they do not incorporate BrdU. (B) Cortical cryosections (15 μm) from E13.5 mouse immunostained for Elavl1/HuR (red) and pH3 (green), counterstained with bisbenzimide (Bisb.) to define the nuclei (blue). Most cells expressing Elavl1/HuR close to the ventricle are not in M. All cells in M show Elavl1/HuR immunoreactivity (arrows). V: ventricle; bar: 5 μm (A), 10 μm (B).
Elavl1/HuR is able to interact with mouse Dll1–3′UTR
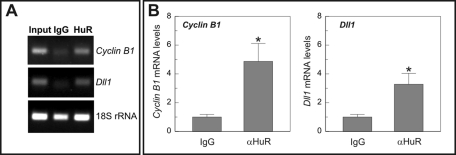
We thus tested whether Elavl1/HuR can interact with the 3′UTR of mouse Dll1 mRNA. Because the Egfp sequence does not contain AREs (accession number U55762; Figure 1), we reasoned that any interaction of the Egfp-Dll1–3′UTR mRNA with this RBP should depend on the Dll-3′UTR sequence. The endogenous association between Elavl1/HuR and Egfp-Dll1–3′UTR mRNA was analyzed in M-arrested ED cells by means of ribonucleoprotein complex IP (RNP-IP) assays using the anti-Elavl1/HuR monoclonal antibody (mAb) 3A2 followed by reverse transcriptase (RT)-PCR–based detection of the Egfp-Dll1–3′UTR mRNA. Although, theoretically, the 3A2 mAb can recognize all members of the Elavl/Hu family, in these experiments it only identifies Elavl1/HuR because the other neuronal Elavl/Hu members (Elavl2/HuB, Elavl3/HuC, and Elavl4/HuD) are not detected in the ED cells, even after performing 35 rounds of RT-PCR amplification (Supplemental Figure S4). Association between Elavl1/HuR and Egfp-Dll1–3′UTR mRNA was evidenced by a strong enrichment of Egfp-Dll1–3′UTR mRNA in Elavl1/HuR IP samples compared with IgG control IP (Figure 4A). As a positive control, we tested the association between Elavl1/HuR and endogenous Ccnb1, an mRNA the stability of which is known to be regulated by Elavl1/HuR (Wang et al., 2000; Lal et al., 2004). Ccnb1 mRNA was detected in anti-Elavl1/HuR antibody IP but not in control IgG IP (Figure 4A). A further control included detection of 18S rRNA, which is not a target of Elavl1/HuR and was found as a contaminating transcript in all IPs (Figure 4A). The assessment of 18S rRNA further ensured that equal amounts of cellular lysate were used in all IP reactions. Taken together, these results show that Elavl1/HuR interacts with Egfp-Dll1–3′UTR mRNA and thus strongly suggest that this RBP controls the steady-state levels of Dll1 mRNA.
FIGURE 4:
Binding of Elavl1/HuR to the Dll1–3′UTR sequence. Extracts from ED (A and B) or EC (B) cells synchronized in M were used for RNP-IP analysis. (A) The association of endogenous Elavl1/HuR (HuR) with Ccnb1 (Cyclin B1) and Egfp-Dll1–3′UTR (Egfp) was tested by RT-PCR after IP with an anti-Elavl1/HuR antibody. Mouse IgG was used as a control for IP. RNA extracted from cell lysates before IP (Input) was amplified in parallel. Amplification of contaminating traces of 18S rRNA was performed as an internal control. (B) Verification by qPCR of the association between endogenous Elavl1/HuR and Ccnb1 (Cyclin B1) or Egfp-Dll1–3′UTR (Egfp) after IP with an anti-Elavl1/HuR antibody (αHuR) in cell extracts from EC or ED cells. Mouse IgG was used as a control for IP. RNA extracted from cell lysates was converted to cDNA and amplified to show the levels of the analyzed RNAs (normalized to 18S rRNA). The results represent the mean ± SEM (n = 3). **p < 0.01; ***p < 0.005 (Student’s t test). (C) Digoxigenin-based pull-down assay using lysates prepared from H2-b2T cells. The binding of HuR to the DIG Dll1–3′UTR (Dll1 3′UTR) sequence was specific. In contrast, HuR did not bind either an irrelevant DIG sequence (Control) or the DIG Notch1–3′UTR sequence (Notch1 3′UTR).
The enrichment of Egfp-Dll1–3′UTR mRNA in the samples immunoprecipitated with the anti-Elavl1/HuR antibody, compared with the control IPs, was further confirmed by qPCR. This analysis demonstrated that, in extracts from M-arrested ED cells, Egfp mRNA levels (normalized to 18S rRNA) were fivefold higher in the Elavl1/HuR IP samples than in the control IPs (Figure 4B). This effect was specific for the 3′URT-Dll1 sequence because the levels of Egfp mRNA (normalized to 18S rRNA) were not significantly augmented in the Elavl1/HuR IP samples derived from EC cell extracts, compared with the control IPs (Figure 4B). As expected, Ccnb1 mRNA was observed to specifically interact with Elavl1/HuR in both ED and EC cells (Figure 4B).
To directly test the interaction between Dll1–3′UTR and Elavl1/HuR, we performed RNA pull-down experiments using synthetic digoxigenin-labeled (DIG) RNAs added to lysates from H2-b2T cells. Addition of DIG-Dll1–3′UTR to these extracts, followed by incubation with a digoxigenin-specific antibody allowed to pull down the Elavl1/HuR protein, which was detected by Western blot (Figure 4C). In contrast, neither DIG-Notch1–3′UTR nor a DIG-control sequence lacking ARE motifs were able to specifically pull down Elavl1/HuR from the H2-b2T cell lysates (Figure 4C), thus demonstrating the specificity of the interaction of this RBP with the Dll1–3′UTR sequence.
Finally, the interaction of endogenous Dll1 mRNA with Elavl1/HuR was demonstrated to occur in primary neuronal precursor cells. IP performed in lysates of E12 brain precursors synchronized in M, followed by qPCR (Figure 5, A and B), demonstrated that Dll1 mRNA (normalized to 18S rRNA) was threefold increased in the Elavl1/HuR IP samples compared with the control IPs. This pattern was similar to that of Ccnb1 mRNA (Figure 5, A and B).
FIGURE 5:
Binding of Elavl1/HuR to Dll1 in E12 brain precursor cells. Extracts from E12 brain precursor cells synchronized in M were used for RNP-IP analysis. (A) The association of endogenous Elavl1/HuR (HuR) with Ccnb1 (Cyclin B1) and Dll1 (Dll1) was tested by RT-PCR after IP with an anti-Elavl1/HuR antibody. Mouse IgG was used as a control for IP. RNA extracted from cell lysates before IP (Input) was amplified to show the expression levels of the analyzed RNAs. Amplification of contaminating traces of 18S rRNA was performed as an internal control. (B) Verification by qPCR of the association between endogenous Elavl1/HuR and Ccnb1 (Cyclin B1) or Dll1 (Dll1) after IP with an anti-Elavl1/HuR antibody (αHuR) in lysates from E12 brain precursor cells. Mouse IgG was used as a control for IP. RNA extracted from immunoprecipitates was converted to cDNA and amplified to show the levels of the analyzed RNAs (normalized to 18S rRNA). The results represent the mean ± SEM (n = 3). *p < 0.05 (Student’s t test).
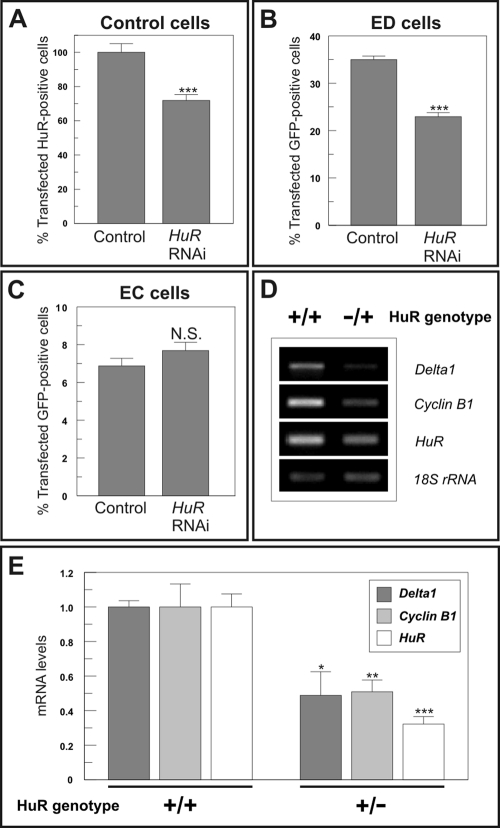
RNAi against Elavl1 reduces the steady-state levels of Egfp mRNA in ED cells synchronized in M
To study the effect of Elavl1/HuR on the steady-state levels of Dll1 mRNA in the H2-b2T neuroepithelial cells, two pairs of RNAi constructs against sequences specific for Elavl1 mRNAs were cloned in the pSilencer 1.0-U6 plasmid. The capacity of these constructs to interfere with Elavl1 was evaluated by transient transfection along with red fluorescent protein (RFP) in H2-b2T neuroepithelial cells synchronized in M, followed by immunostaining with Elavl1/HuR antibodies. Then the proportion of RFP-positive H2-b2T neuroepithelial cells expressing Elavl1/HuR was estimated in these cultures. This analysis demonstrated that Elavl1-specific RNAi constructs significantly reduced the expression of Elavl1/HuR in M-synchronized H2-b2T neuroepithelial cells when compared with the control pSilencer 1.0-U6 plasmid (Figure 6A and Supplemental Figure S5A). The capacity of the Elavl1-specific RNAi constructs to reduce the expression of Elavl1/HuR in H2-b2T neuroepithelial cells was confirmed by Western blot analysis in cell extracts from H2-b2T cells lipofected with RFP plus either the Elavl1-specific RNAi construct or the control vector, and then mitotically synchronized with colchicine. To avoid the presence in the cell extracts of Elavl1/HuR expressed in nonlipofected cells, RFP-positive cells were isolated by fluorescence-activated cell sorting (FACS) and then used for protein extraction. This analysis demonstrated that the levels of Elavl1/HuR become reduced in cells expressing the Elavl1-specific RNAi construct (Supplemental Figure S5B). Importantly, decreased Elavl1/HuR expression is correlated with a decreased number of M-synchronized ED cells expressing EGFP (Figure 6B), as compared with the ED cells transfected with the control pSilencer 1.0-U6 plasmid. These results are in sharp contrast with those obtained in M-synchronized EC cells where Elavl1-specific RNAi constructs did not alter the proportion of cells expressing EGFP (Figure 6C).
FIGURE 6:
Elavl1 regulates Dll1 expression both in vitro and in vivo. (A) Parental H2-b2T neuroepithelial cells (Control cells) were cotransfected with RFP and a pSilencer-Elavl1 RNAi construct (HuR RNAi) or a control pSilencer 1.0-U6 plasmid (Control), and then synchronized in M with 1 μg/ml colchicine. Shown are the percentages of RFP-positive cells expressing high levels of Elavl1/HuR in both conditions. Normalized data are presented. ***p < 0.005 (n = 3; Student’s t test). (B) ED cells were transfected with RFP and a pSilencer-Elavl1 construct (HuR RNAi) or a control pSilencer 1.0-U6 plasmid (Control) and then synchronized in M with 1 μg/ml colchicine. The percentage of cells expressing high levels of EGFP in both situations is shown. ***p < 0.005 (n = 3; Student’s t test). (C) EC cells were transfected with RFP and a pSilencer-Elavl1 construct (HuR RNAi) or a control pSilencer 1.0-U6 plasmid (Control), and then synchronized in M with 1 μg/ml colchicine. The percentage of cells expressing high levels of EGFP in both situations is shown (n = 3). N.S.: non significant. (D) cDNA obtained from E10.5 brains of heterozygous Elavl1/HuR null-mutant mice (+/−) or WT mice (+/+) was used for PCR amplification using primers specific for Dll1 (Delta1), Ccnb1 (Cyclin B1), Elavl1 (HuR), or 18S rRNA (18S rRNA). (E) cDNA obtained from E10.5 brains of heterozygous Elavl1/HuR null-mutant mice (+/−) or WT mice (+/+) was used for qPCR using primers specific for Dll1 (Delta1), Ccnb1 (Cyclin B1), or Elavl1 (HuR). Values were normalized to 18S rRNA. The results represent the mean ± SEM (n = 4); *p < 0.05; **p < 0.001; ***p < 0.005 (Student’s t test).
In some instances, Elavl1/HuR has been shown to enhance translation (Mazan-Mamczarz et al., 2003). To verify that the observed reduction of EGFP levels in response to the Elavl1-specific RNAi construct was due, at least partially, to a reduction of the steady-state levels of the Egfp-Dll1–3′UTR mRNA, ED cells were cotransfected with RFP along with either Elavl1 RNAi or the control RNAi construct. Then these cells were synchronized in M for 24 h and either left untreated or subjected to a 4-h treatment with actinomycin D to block transcription. Those cells expressing high levels of RFP (i.e., with high levels of Elavl1-specific RNAi) were subsequently isolated by FACS, and the levels of Egfp mRNA (normalized to 18S rRNA) were estimated by qPCR. This analysis demonstrated that the interference against Elavl1 resulted in a significant reduction of Egfp mRNA levels after 4-h treatment with actinomycin D, as compared with the control situation (Supplemental Figure S6). Taken together, these results stress the importance of Elavl1/HuR for maintaining high levels of Dll1–3′UTR–containing transcripts in the H2-b2T neuroepithelial cells.
Reduced Dll1 mRNA levels in the developing brain of Elavl1/HuR knockout mice
We next studied the capacity of Elavl1/HuR to enhance the steady-state levels of Dll1 mRNA in vivo. To this end, we compared the levels of Dll1 mRNA expression in brains from E10.5 wild-type (WT) or Elavl1 heterozygous knockout (Elavl1+/−) embryos. Elavl1 knockout mice lack exon 2 from the Elavl1 murine gene, which contains the ATG for the initiation of translation. This mutation results in the absence of Elavl1/HuR protein expression and embryonic lethality from E10.5 to E12.5 due to placental failure (Katsanou et al., 2009). At E10.5, Elavl1−/– mice display a developmental lag of ∼1.0–1.5 d postcoitum (Katsanou et al., 2009). Because this delay could compromise the analysis of developmental-dependent gene expression in Elavl1−/– mice due to differences in brain maturation, we decided to focus the study on WT versus Elavl1+/− mice littermates, which display no morphological abnormalities when compared with each other (Katsanou et al., 2009). Both RT-PCR (Figure 6D) and qPCR (Figure 6E) analysis revealed that Dll1 mRNA expression was reduced in the brain of Elavl1+/– mice, in parallel with a decrease in Ccnb1 mRNA expression levels. Altogether, these results indicate that reducing Elavl1/HuR expression (either by knockdown or knockout) affects the level of Dll1 mRNA expression.
Elavl1 RNAi reduces the capacity of brain precursors to trigger Dll1-dependent lateral inhibitory signals
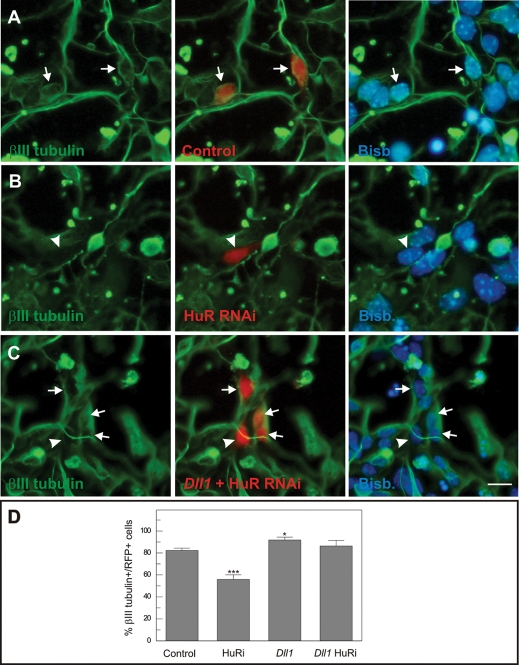
Neurogenesis is controlled by lateral inhibitory signals triggered by the Delta/Notch signaling pathway. This mechanism amplifies and stabilizes any stochastic difference in Dll1 expression between neighbor cells, thus resulting in neuronal differentiation of precursors with higher amount of Dll1 (Louvi and Artavanis-Tsakonas, 2006). It is therefore expected that any manipulation aimed to reduce Dll1 expression in a neural precursor surrounded by normal precursor cells should result in its inability to differentiate due to an increase of lateral inhibitory signals delivered by adjacent cells. To analyze the capacity of Elavl1/HuR to regulate this process, Elavl1-specific RNAi constructs were coelectroporated along with RFP in brain precursors from E12 mouse embryos, which subsequently were cocultured for 18 h with nontransfected E12 brain precursors at high density to allow lateral inhibition to take place. Cells were then fixed and immunostained with anti-βIII tubulin–specific antibodies to reveal neuronal commitment. Cells were classified as positive for this marker when βIII tubulin–positive immunostaining was observed clearly surrounding the nucleus, which was labeled with bisbenzimide. The percentage of βIII tubulin–positive neurons was significantly diminished in brain precursors expressing Elavl1-specific RNAi constructs, as compared with precursors transfected with the control plasmid (Figure 7, B and D). The reduced neurogenic capacity of Elavl1 RNAi-transfected cells is likely to derive from their diminished capacity to deliver lateral inhibitory signals to their neighbors due to decreased expression of Dll1, and not from side effects on alternative signaling pathways involved in the process of neurogenesis. To test this hypothesis, brain precursors were cotransfected with an expression vector containing the coding sequence of Dll1 together with the Elavl1-specific RNAi constructs. Expression of Dll1 significantly increased the proportion of brain precursors showing βIII tubulin–specific immunoreactivity in the presence of Elavl1-specific RNAi (Figure 7, C and D). Altogether these results indicate that Elav1/HuR is necessary for Dll1-dependent lateral inhibition to occur, likely through regulation of Dll1 expression.
FIGURE 7:
RNAi against Elavl1 results in Dll1-dependent decreased neurogenesis. Neuronal precursors from E12 mouse brain were coelectroporated with different plasmid combinations, including a Dll1 expression vector and an Elavl1/HuR-specific RNAi vector plus RFP (red), and cultured at high density (200,000 cells/cm2) to allow cell–cell contacts and lateral inhibition to occur. After 18 h, the cells were fixed and immunostained with anti–βIII tubulin antibodies (green), known to specifically label neurons. Nuclei were counterstained with bisbenzimide (blue). (A) An example of cells transfected with control plasmids (arrows) showing βIII tubulin–specific immunoreactivity. (B) An example of a cell transfected with the Elavl1/HuR-specific RNAi vector (arrowhead) lacking βIII tubulin–specific immunoreactivity. (C) An example of cells transfected with the Dll1 expression vector and the Elavl1/HuR-specific RNAi vector. Most cells showed βIII tubulin–specific immunoreactivity (arrows). A transfected cell lacking βIII tubulin–specific immunoreactivity is indicated (arrowhead). (D) In the presence of the Elavl1/HuR-specific RNAi vector, the proportion of βIII tubulin–positive neurons was significantly reduced (see arrowhead in B). The expression of Dll1 significantly increased the proportion of brain precursors showing βIII tubulin–specific immunoreactivity, an effect that could not be reduced by the Elavl1/HuR-specific RNAi vector. *p < 0.05; ***p < 0.005 (n = 3; Student’s t test). Bar: 10 μm (A and B), 14 μm (C).
Elavl1+/− embryos show increased neuronal production in the developing retina
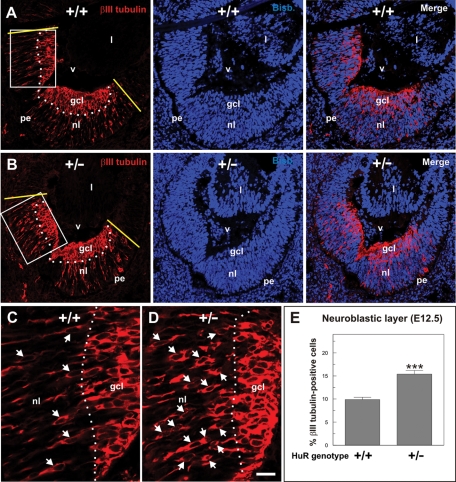
To confirm in vivo that the absence of Elavl1/HuR reduces the capacity of neuronal precursors to trigger lateral inhibitory signals, thus favoring neuronal differentiation, we focused on the E12.5 mouse retina. At this early stage, the retina contains two major layers: the ganglion cell layer (GCL), which contains differentiated retinal ganglion cells, and the neuroblastic layer, constituted by proliferating neuroepithelial cells and recently born neuroblasts migrating to the GCL (Figure 8, A–D). In addition, a differentiation gradient exists in this tissue that restricts neurogenesis to its most central portion (illustrated in Figure 8, A and B, as the neurogenic region located between the two yellow lines). The rate of neurogenesis in this tissue was estimated as the proportion of differentiating neurons (i.e., βIII tubulin–positive cells) with respect to the total number of cells (i.e., bisbenzimide-labeled nuclei) that are present in the portion of the neuroblastic layer located inside the neurogenic region. This analysis indicated that a small, although significant, increase in the proportion of βIII tubulin–positive cells can be observed in the Elavl1 heterozygous embryos, as compared with their control littermates (Figure 8, C–E).
FIGURE 8:
Neuronal production is enhanced in the differentiating retina of Elavl1+/– mice. Retinal cryosections (15 μm) from E12.5 mouse embryos (A and C: WT mice; B and D: Elavl1 heterozygous mice) immunostained for βIII tubulin (red in A–D), and counterstained with bisbenzimide (blue in A and B) to reveal cell nuclei. (A) A cryosection of a WT retina (+/+) is shown. The dotted line represents the boundary between the ganglion cell layer (gcl) and the neuroblastic layer (nl). Yellow lines represent the boundaries between the neurogenic region in the central retina, where βIII tubulin–positive cells can be observed, and the nonneurogenic region in the peripheral retina, a region lacking βIII tubulin expression. (B) A cryosection of a Elavl1+/− retina (+/−) is shown. For specific details, see A. (C) Inset shown in A. (D) Inset shown in B. (E) Percentage of differentiating βIII tubulin–positive neuroblasts present in the neuroblastic layer located within the neurogenic region (surrounded by the yellow lines in A and B). pe: pigment epithelium; v: vitreous body; l: lens. Arrows in C and D: βIII tubulin–positive cells. ***p < 0.005 (n = 4; Student’s t test). Bar: 50 μm (A and B), 16 μm (C and D).
DISCUSSION
The RBP Elavl1/HuR is believed to have ubiquitous expression patterns in most tissues (Ma et al., 1996; Lu and Schneider, 2004). We have further analyzed the expression of this RBP in the developing neuroepithelium. This analysis demonstrated that Elavl1/HuR displays a distinct expression pattern, with strong cytoplasmic localization in apically located cells. It was also detected in apically located, fusiform neuroepithelial cells that do not incorporate BrdU, resembling the expression pattern of Dll1-positive cells in the mouse developing brain (Cisneros et al., 2008). Elavl1/HuR has been shown to be primarily localized to the nucleus of NIH-3T3 cells, but in response to different stimuli it can shuttle to the cytoplasm (Atasoy et al., 1998), thus protecting its target mRNAs from degradation (Fan and Steitz, 1998; Doller et al., 2008; Kim and Gorospe, 2008). The expression pattern of Elavl1/HuR is consistent with its ability to enhance the steady-state levels of specific transcripts in neural precursors undergoing M, including those from Dll1. Interestingly, Elavl1/HuR function is linked to the cell cycle. As such, this RBP has the capacity to modulate the expression of key cell-cycle regulators (Barreau et al., 2005). Furthermore, Elavl1/HuR can undergo cell-cycle–dependent regulation because it can be phosphorylated by the G2/M-specific kinase Cdk1, thus leading to retention of this RBP in the nucleus of HeLa cells (Kim et al., 2008). In contrast with this finding, we have observed in vivo and in vitro that neuroepithelial cells are characterized by cytoplasmic translocation of Elavl1/HuR during M, indicating a complex regulation of nuclear export of this RBP.
We have shown that Elavl1/HuR can interact with Egfp-Dll1–3′UTR mRNA in H2-b2T neuroepithelial cells synchronized in M. This interaction likely takes place through the Dll1–3′UTR sequence because Egfp does not contain AREs in its sequence and a synthetic Dll1–3′UTR RNA can specifically pull down Elavl1/HuR from H2-b2T neuroepithelial cell lysates. Furthermore, we have presented evidence that Elavl1/HuR can interact with endogenous Dll1 mRNA in E12 brain precursors, indicating that the observed interaction between Elavl1/HuR and Dll1–3′UTR is functionally significant. From the RNP-IP experiments performed in lysates of E12 neuronal precursors, we cannot exclude that Dll1 mRNA could interact with Elavl2/HuB, Elavl3/HuC, and Elavl4/HuD, because the anti-Elavl1/HuR antibody used for these assays can recognize these other members of the Elavl/Hu family. Nevertheless, Elavl2/HuB, Elavl3/HuC, and Elavl4/HuD are mainly expressed in the differentiated layers of the embryonic brain (see Gene Expression Nervous System Atlas at www.ncbi.nlm.nih.gov/projects/gensat/), whereas Dll1 is restricted to the neuroepithelium at these embryonic stages (Lindsell et al., 1996; Cisneros et al., 2008). The expression patterns of Elavl2/HuB, Elavl3/HuC, and Elavl4/HuD suggest that Elavl1/HuR is the major Elavl/Hu member interacting with Dll1 in the undifferentiated neuroepithelium.
Our data demonstrate an enhancement of the steady-state levels of both EGFP and Egfp mRNA in ED cells undergoing M when compared with control H2-b2T cells. This effect is likely due to mRNA stabilization because Elavl1-specific RNAi reduces the levels of Egfp-Dll1–3′UTR transcript in these cells. These observations are consistent with the enhanced capacity of apically located precursors to express Dll1 in vivo (Murciano et al., 2002; Cisneros et al., 2008), concomitant with the expression of Elavl1/HuR in these cells. This role seems to be conserved through evolution, because we provide evidence that the murine Dll1–3′UTR sequence is also functional in chick neuroepithelial cells in vivo. In contrast, the presence of the 3′UTR of Notch1 mRNA correlated with reduced expression levels of EGFP in EN cells, supporting the conclusion that the Dll1–3′UTR sequence specifically confers mRNA stability during M.
Notch1–3′UTR–induced mRNA destabilization occurs at all stages of the cell cycle and seems to be independent of Elavl1/HuR because this RBP does not interact with the Notch1–3′UTR sequence in pull-down assays. Nevertheless, we cannot rule out that the apparent lack of interaction between Elavl1/HuR and the 3′UTR of Notch1 in vitro may be due to competition of the Notch1–3′UTR sequence with other RBPs or with specific micro RNAs. The dramatic inhibition of EGFP expression triggered by the 3′UTR of Notch1 in EN cells is consistent with the observation that the Notch-3′UTR sequence is sufficient for degradation of zygotic Notch in the leech Helobdella robusta (Gonsalves and Weisblat, 2007). Our data are also consistent with the expression pattern of Notch1 mRNA in the vertebrate neuroepithelium in vivo, which is basically absent from the basal neuroepithelial cells (Murciano et al., 2002; Cisneros et al., 2008). Its absence from the basal neuroepithelium suggests that Notch1 mRNA is rapidly degraded in the absence of stabilizing RBPs in cells undergoing S-phase.
Our study provides genetic evidence for the involvement of Elavl1/HuR in the stabilization of Dll1 mRNA. As such, the expression of this gene was observed to be down-regulated in the brain of E10.5 mouse embryos containing one null allele for murine Elavl1 (Katsanou et al., 2009). At this stage, most of the brain-specific Dll1 transcripts are restricted to the neuroepithelium (Lindsell et al., 1996; Cisneros et al., 2008). Therefore genetic evidence supports the hypothesis that Elavl1/HuR is required for stabilization of Dll1 in neural precursors undergoing M, thus facilitating Delta/Notch signaling in these cells. Elavl1 deficiency also caused a reduction of Ccnb1 mRNA expression. Although this observation may suggest that part of the effect observed on Dll1 expression is due to reduced proliferative capacity of the neuroepithelial cells, it is important to stress that no gross morphological alterations are observed in the heterozygous mice (Katsanou et al., 2009). Interference against Elavl1 mRNA in M-synchronized ED cells also resulted in the decrease of the levels of EGFP-Delta1–3′UTR mRNA, further indicating that Elavl1/HuR has an intrinsic capacity to stabilize Dll1 during M.
Interference against Elavl1 mRNA correlated with reduced capacity of brain precursors to deliver lateral inhibitory signals. This effect, observed in RNAi-transfected precursors cocultured at high density with nontransfected precursor cells, is likely to depend on the destabilization of Dll1 mRNA resulting from reduced Elavl1/HuR expression because Dll1 overexpression prevented the effect of Elavl1 interference on lateral inhibition. These results were confirmed by using a genetic approach, as we have shown that Elavl1 heterozygous mice exhibit increased neuronal production in the developing retina, in accordance with the reduction of Dll1 expression observed in the developing nervous system of these mice. Such a reduction of Dll1 expression in Elavl1 heterozygous mice is likely to enhance proneural gene expression in precursor cells, thus favoring neuronal differentiation. Because no obvious phenotype has been detected in the Elavl1 heterozygous mice (Katsanou et al., 2009), neuronal overproduction in these mice could be counterbalanced by programmed cell death at later stages of development.
In sum, our results stress the importance of Elavl1/HuR in vertebrate neurogenesis, a concept that was previously unsuspected despite the importance of the Elav locus for the development and maintenance of the nervous system in Drosophila (Robinow et al., 1988). Unlike Elav, which is expressed by postmitotic neurons, Elavl1/HuR has an important role in mitotic neural precursors as it regulates Delta/Notch signaling in apical neuroepithelial cells. Dll1 mRNA stabilization may contribute to the maintenance of the Delta ligand in differentiating neuroblasts as they initiate their migration to the differentiated brain regions.
MATERIALS AND METHODS
Mice
C57BL6/J and Elavl1 mice (Katsanou et al., 2009) were used in this study. Elavl1 genotypes were determined by genomic PCR as previously described (Katsanou et al., 2009). Experimental procedures were approved by the CSIC and CNRS animal ethics committees.
Primary antibodies
The anti-Elavl1/HuR mouse mAb 19F12 (CLONEGENE, Hartford, CT) was diluted 500-fold (immunocytochemistry and immunohistochemistry). The anti-Elavl1/HuR mAb 3A2 (from I. Gallouzi, McGill University) was used at 1:500 (vol:vol) (Western blot). The rabbit anti-GFP polyclonal antiserum (Invitrogen, Carlsbad, CA) was used at 1:1000 (vol:vol) (immunocytochemistry). The anti-BrdU mouse mAb G3G4 (Developmental Studies Hybridoma Bank [DSHB], University of Iowa, Iowa City, IA) was diluted 4000-fold. The anti-BrdU rat mAb BU1/75 (ICR1; AbD Serotec, Raleigh, NC) was used at 1:200 (vol:vol). The mouse mAb against neuron-specific βIII tubulin (clone 5G8; Millipore, Billerica, MA) was diluted 1000-fold (immunohistochemistry) or 10,000-fold (Western blot). The anti-phospho-Histone H3 (pH3) rabbit polyclonal antibody (Upstate Biotechnology, Lake Placid, NY) is a well-characterized marker of M, and it was diluted 1:400 for immunostaining. The anti-digoxigenin mAb (Roche, Basel, Switzerland) was used at 1:500 (vol:vol) dilution for pull-down experiments.
Plasmids
The pEGFP-N1 expression vector (Clontech, Mountain View, CA) was used to generate the pEGFP-Notch1–3′UTR and pEGFP-Dll1–3′UTR vectors. The Notch1–3′UTR sequence was amplified with Pfu DNA polymerase (BioTools, Jupiter, FL) from a plasmid containing full-length Notch1 (Hansson et al., 2006) using the Notch1 up and Notch1 down oligonucleotides (Supplemental Table S1). The Dll1–3′UTR sequence was amplified with Pfu DNA polymerase (BioTools) from E13.5 mouse brain cDNA using the Dll1 up1 and Dll1 down1 oligonucleotides (Supplemental Table S1). The amplified fragments were sequenced, cloned into the pGEM-Teasy vector (Promega, Madison, WI), and subcloned into the NotI site of pEGFP-N1, thus giving rise to the pEGFP-Notch1–3′UTR and pEGFP-Dll1–3′UTR vectors, respectively. The pSilencer 1.0-U6 siRNA vector (Ambion, Austin, TX) was used to construct vectors to interfere with mouse Elavl1 (accession number NM_010485), using as target sequences: base pairs 265–283 (pSilencer-Elavl1/1) and base pairs 1130–1148 (pSilencer-Elavl1/2). Two oligonucleotide pairs were used to generate the target sequences (Elavl1/1 sense and Elavl1/1 antisense; Elavl1/2 sense and Elavl1/2 antisense) (Supplemental Table S1), following the instructions of the manufacturer. The RFP-expressing vector pRFPRNAiC was provided by Stuart Wilson (Das et al., 2006). A pCEP4 expression vector containing the full-length sequence of mouse Dll1 (pCEP4-Dll1) was provided by Kohzo Nakayama (Shinshu University, Nagano, Japan).
In vivo BrdU treatment
Pregnant female C57BL6/J mice were intraperitoneally injected with BrdU in phosphate-buffered saline (PBS; 50 μg/g body weight) and killed 1 h later.
RT-PCR
mRNA was extracted using the QuickPrep Micro mRNA purification kit (GE Healthcare, San Diego, CA), from which cDNA was prepared using the First-Strand cDNA Synthesis Kit (GE Healthcare). Total RNA was extracted with TRIzol Reagent (Invitrogen), and reverse transcribed with Superscript II RNase H- Reverse Transcriptase (Invitrogen). Amplification of cDNAs was performed using standard procedures, and data were obtained when amplification was linear. Amplifications performed in the absence of reverse transcriptase lacked any specific band. The PCR primers used (Supplemental Table S1) were Dll1 up2 and Dll1 down2 for Dll1, Elavl1 up and Elavl1 down for Elavl1/HuR, Elavl2 up and Elavl2 down for Elavl2/HuB, Elavl3 up and Elavl3 down for Elavl3/HuC, Elavl4 up and Elavl4 down for Elavl4/HuD, Ccnb1 up and Ccnb1 down for Ccnb1, Egfp up and Egfp down for Egfp, and 18S rRNA up and 18S rRNA down for 18S rRNA.
qPCR
Total RNA from dissociated neuroepithelial cell cultures, from E10.5 brain, or from immunoprecipitated material was extracted using TRIzol Reagent (Invitrogen). Using 100 ng of this total RNA sample as a template, cDNAs were prepared with SuperScript II Reverse Transcriptase (Invitrogen) in 20-μl reactions. The RT-PCR reactions were run in triplicate on 96-well reaction plates with SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA), using an ABI PRISM 7700 Sequence Detection System (Applied Biosystems). The qPCR reaction was performed with 1 μl of cDNA for Egfp, Ccnb1, Dll1, and Elavl1, or with 1 μl of a 10-fold dilution for 18S rRNA to normalize the mRNA levels (Schmittgen and Zakrajsek, 2000). The primers (0.2 μM) used for qPCR (Supplemental Table S1) were Egfp forward and Egfp reverse for Egfp, Ccnb1 forward and Ccnb1 reverse for Ccnb1, Dll1 forward and Dll1 reverse for Dll1, 18S forward and 18S reverse for 18S rRNA, and Elavl1 forward and Elavl1 reverse for Elavl1. More than 40 cycles of amplification were performed, including a denaturation (95°C; 15 s) and annealing/extension (60°C; 1 min) step. Data acquisition and the analysis of the qPCR assays were performed using the 7000 System SDS Software (Version 1.2.3; Applied Biosystems). SYBR Green/dsDNA complex signal was normalized to the passive reference dye (ROX) during data analysis to correct for nonPCR-related well-to-well fluorescent fluctuations.
RNP-IP
IP of endogenous Elavl1-mRNA complexes was performed as previously described (Lal et al., 2004) with the following modifications. Briefly, PBS-washed pellets from 20 × 106 Dll1-H2-b2T neuroepithelial cells or 2 × 107 E12 brain cells, synchronized in M, were resuspended in 600 μl of RSB (50 mM Tris-HCl, pH 7.4, 50 mM KCl, 10 mM magnesium acetate) plus 200 μl of glass beads (Sigma), vortexed twice for 30 s, and centrifuged for 10 min at 3000 rpm in a minifuge. Protease (Roche) and RNAse inhibitors (rRNAsin; Promega) were added. The supernatant was precleared with Protein A sepharose beads (Sigma) for 1 h at 4°C. Half of the supernatant was incubated with antibody-coated, prewashed Protein A sepharose beads (20 μg of anti-Elavl1/HuR mAb 3A2 or control IgG) and left overnight at 4°C. The beads were washed five times in RSB complemented with protease and RNAse inhibitors. Total RNA in the immunoprecipitated material or input cytoplasmic extracts was extracted with TRIzol Reagent, and RT-PCR or qPCR amplified. The presence of contaminating traces of RNAs that do not interact with the studied RBPs is commonly used as a negative control for the RNP-IP assays (Lal et al., 2004; López de Silanes et al., 2004). In this study, 18S rRNA was used as a negative control.
In vitro RNA pull-down assay
DIG RNA corresponding to the 3′ UTR sequences of mouse Notch1 or mouse Dll1, or an irrelevant sequence lacking ARE motifs (base pairs 347–804, accession number AF032966), was synthesized by using the DIG RNA-labeling kit (Roche). DIG RNA pull-down experiments were performed following a modification of a previously described procedure (Hsu et al., 2009). Briefly, cell extracts (2.5 × 107 H2-b2T neuroepithelial cells/ml) were prepared in extraction buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Triton X-100, 15 mg/ml tRNA, 1× protease inhibitor [Roche], 1000 U/ml Protector RNase inhibitor [Roche]), and then precleared with preequilibrated Protein G Sepharose 4 Fast Flow beads (GE Healthcare). DIG RNA (10 μm) was mixed with 400 μl of extract and incubated on a rotator for 1 h at 4°C. Anti-digoxigenin was added (1:500 [vol:vol] dilution) to each binding reaction, and the mixture was further incubated on a rotator overnight at 4°C. Preequilibrated Protein G Sepharose 4 Fast Flow beads (20 μl) were then added to each binding reaction, and the mixture was incubated for 90 min at 4°C. Beads were then isolated by centrifugation, washed five times with extraction buffer, and boiled in SDS sample buffer. In parallel, aliquots of the cell extract (INPUT) were ten times concentrated using Ultracel-3K centrifugal filter units (Millipore), and boiled in SDS sample buffer. Immunoprecipitates and inputs were resolved on a 12% SDS-protein gel and subjected to immunoblot analysis using an anti-Elavl1/HuR–specific antibody.
Electroporation
In ovo electroporation was performed following standard procedures. Plasmids (1 μg/μl) were injected into the brain ventricle of stage HH12 chick embryos (Hamburger and Hamilton, 1951). Electroporation was then performed with a TSS20 OVODYNE electroporator (Intracel, Royston, UK), programmed to deliver five 50-ms, 12-V pulses at a 300-ms frequency. The eggs were sealed and allowed to develop for another 4 h. At this point, embryos were removed and fixed for 3 h at room temperature with 4% paraformaldehyde (PFA).
Explant electroporation was performed as previously described (Morillo et al., 2010). E12 mouse brains were dissected in the presence of 2.4 U/ml dispase (Invitrogen) to facilitate the removal of the mesenchyme. The tissue was fragmented into small pieces of approximately 10 mm2, which were laid onto glass coverslips (Menzel-Gläser, Braunschweig, Germany) and immersed in 4 μl of PBS containing (i) 250 ng/μl pSilencer 1.0-U6 plus 250 ng/μl pRFPRNAiC, (ii) 250 ng/μl pSilencer-Elavl1/1 plus 250 ng/μl pRFPRNAiC, (iii) 250 ng/μl pSilencer 1.0-U6 plus 125 ng/μl pRFPRNAiC and 125 ng/μl pCEP4-Dll1, or (iv) 250 ng/μl pSilencer-Elavl1/1 plus 125 ng/μl pRFPRNAiC and 125 ng/μl pCEP4-Dll1. Electroporation was performed with a TSS20 OVODYNE electroporator using four 50-ms pulses of 25 V, at a 500-ms frequency. After electroporation, the explants were grown in suspension for 4 h in DMEM/F12 medium (Sigma) containing N2 supplement (DMEM/F12/N2; Invitrogen). Explants were then dissociated as previously described (Frade and Rodríguez-Tébar, 2000) and cultured for 18 h as described later in the text.
Cell culture
H2-b2T immortalized neuroepithelial cells (Nardelli et al., 2003) were maintained at 37ºC in RPMI 1640 medium containing GlutaMAX I, 25 mM HEPES (Invitrogen), 10% fetal calf serum (Invitrogen), and streptomycin/penicillin (Invitrogen). Cultures were synchronized in S-phase for 24 h with 10 mM HU (Sigma), or in M for 24 h with 1 μg/ml colchicine (Sigma). In some cases, BrdU (0.5 μg/ml) was added to label cells in S-phase, whereas in other cases, transcription was blocked by adding 5 μg/ml actinomycin D (Sigma). Transfection of H2-b2T neuroepithelial cells (40,000–45,000 cells/cm2) was performed with Lipofectin 2000 (Invitrogen). Polyclonal cell lines constitutively expressing EGFP were generated after transfection of pEGFP-N1, pEGFP-Notch1–3′UTR, or pEGFP-Dll1–3′UTR, followed by selection with 1.5 mg/ml Geneticin-418 (G-418; Invitrogen) during the first 5 d, and then they were maintained with 3 mg/ml G-418. Parental cells were observed to die by 3 d after 1.5 mg/ml G-418 treatment. Dissociated precursor cells from E12 mouse brain were cultured at high density (200,000 cells/cm2) as previously described (Cisneros et al., 2008). In some instances, the brain precursors were cultured in the presence of 1 μg/ml colchicine for 20 h and then used for RNP-IP.
RNAi
H2-b2T neuroepithelial cells, seeded at 80–90% confluence on coverslips previously coated with 20 μg/ml poly-l-lysine (Sigma), were lipofected with pRFPRNAiC (expressing RFP) plus pSilencer interference or pSilencer control plasmids, and synchronized with 1 μg/ml colchicine (Sigma) for 24 h. Similar results were obtained with both pairs of RNAi constructs.
Immunocytochemistry
H2-b2T cells or E12 brain precursors cultured as described earlier in the text were fixed with 4% PFA for 15 min, and then permeabilized for 30 min at room temperature with 0.5% Triton X-100 in PBS (PBT). Immunolabeling was performed as previously described (Cisneros et al., 2008). For BrdU immunolabeling, cultures were previously subjected to DNA denaturation by incubation for 30 min with 2N HCl/0.33× PBS at room temperature, followed by a neutralization step consisting of three 15-min washes with 0.1 M sodium borate, pH 8.9, and two washes of 5 min each with PBT. Nuclei were counterstained with 1 μg/ml bisbenzimide. Images were recorded using a DXM 1200 digital camera (Nikon, Melville, NY), and they were processed only minimally using Adobe Photoshop (version 11.0). Color balance was applied to all parts of the image, as well as to the controls, equally.
Immunohistochemistry
Cryosections (15 μm) were washed with PBS and permeabilized for 30 min at room temperature in the presence of PBT. Immunostaining was performed as previously described (Murciano et al., 2002). Nuclei were counterstained with 1 μg/ml bisbenzimide. Images were recorded using confocal microscopy (Leica TCS-SP5; Leica, Wetzlar, Germany), and they were processed as described earlier in the text.
Western blot
Cell extracts were separated by SDS–PAGE on 11% acrylamide gels under reducing conditions and transferred to Immun-Blot polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). Membranes were blocked for 2 h with 2% ECL Advance blocking agent (ECL Advance Western Blotting Detection Kit; GE Healthcare) in PBS with 0.1% Tween 20 (PBTW), and then incubated successively with the appropriate antibodies. After washing with PBTW several times, membranes were incubated with peroxidase-coupled secondary antibodies and washed several times again, then finally specific signal was revealed with ECL Advance reagent.
Flow cytometric analysis
H2-b2T neuroepithelial cells were trypsinized and fixed with 2% PFA at 4ºC for 40 min. The cells were then resuspended in PBS, treated with 25 μg/ml ribonuclease A (Sigma) for 1 h at room temperature, and stained with 25 μg/ml PI (Sigma). Diploid mouse cells obtained from adult retina were used as a reference. Argon-ion laser excitation at 488 nm was then used to measure PI fluorescence through a band-pass 620-nm filter using an Epics XL flow cytometer (Beckman Coulter, Fullerton, CA). Debris and duplets were always excluded from the analysis. In each experiment 10,000 cells were counted. Statistical analyses were performed using EXPO32 software (Beckman Coulter).
FACS
For cell sorting, a FACSAria cytometer (BD Biosciences, San Diego, CA) equipped with a double argon (488 nm) and helium–neon laser (633 nm) was used. Data were collected by using a linear digital signal process. The emission filters used were BP 530/30 for EGFP (FL1) and BP 585/42 for RFP (FL2). Appropriate values of electronic compensation were adjusted between each fluorescence spectrum. Debris and duplets were always excluded from the analysis. Cell populations were isolated in autoclaved 12 × 75 mm, 5 ml polystyrene tubes, using 20 psi pressure and a 100 μm nozzle aperture. The identity of the isolated cells was confirmed by epifluorescence microscopy. Data were analyzed with FACSDiva data analysis software (BD Biosciences) and displayed using biexponential scaling. As an average, 5000–20,000 RFP-positive cells were isolated.
Analysis of cell-cycle kinetics in vitro
The duration of the different stages of the cell cycle was estimated using a modification of a previously described method (Takahashi et al., 1993). Cumulative labeling with 0.5 μg/ml BrdU was given for progressive time lapses, and then cells were fixed and BrdU immunolabeled. For each time point, an average labeling index (LI) (representing BrdU-labeled cells as a fraction of total cells) was calculated. The different LI values (y-axis) were plotted against the different time points at which LI values were calculated (x-axis), thus giving rise to the “LI profile.” LI increases linearly until a particular value in which it remains constant (growth fraction [GF]). GF represents the fraction of proliferating cells that are present in the culture. In this plot, the duration of the cell cycle (TC) and the duration of the S-phase (TS) can be calculated from the y-intercept, the value of which is equal to (TS/TC) × GF, and from the time required to label the complete GF, which is TC – TS (i.e., at TC – TS, LI = GF). The slope of the LI linear increase was estimated by means of least-squares fit. The duration of M (TM) was estimated from the mitotic index (MI), which represents the percentage of cells showing mitotic figures (TM = Tc × MI/100). The duration of G2 (TG2) was calculated from the shorter time lapse at which mitotic figures start to contain BrdU-specific labeling. Finally, the duration of G1 (TG1) was estimated as the difference between TC and TS + TG2 + TM.
Cell counting
Cell counting was performed with a Nikon E80i microscope (Nikon, Melville, NY) with phase contrast and epifluorescence illumination. On average, 100 positive cells were analyzed per experimental point. To quantify cells expressing high levels of EGFP, randomly taken pictures were analyzed with ImageJ software using 54 as a threshold value for all experimental points. The analysis of βIII tubulin expression in the retina from E12.5 mouse embryos was performed in at least three retinal cryosections from each embryo. The average percentage of βIII tubulin cells was obtained for each embryo, and the mean ± SEM from these average values was obtained from four different embryos per genotype.
Statistical analysis
Quantitative data are shown as the mean ± SEM from at least three independent experiments. Statistical differences were analyzed using analysis of variance (ANOVA) or Student’s t test.
Supplementary Material
Acknowledgments
We thank F. Pituello, J.R. Martínez-Morales, and N. López-Sánchez for comments on the manuscript; J. Nardelli for the H2-b2T neuroepithelial cells; I. Gallouzi for the 3A2 mAb; U. Lendahl for the pCAG-FLN1-IRES-Puro expression vector; S. Wilson for the pRFPRNAiC vector; K. Nakayama for the Dll1 expression vector; and P. Lastres and E. Abanto for technical assistance. The mAb G3G4, developed by S Kaufman, was obtained from the DSHB (University of Iowa). This study was supported by grants from the MICINN (J.M.F.), La Caixa Foundation (J.M.F.), FUNDALUCE (J.M.F.), and ARC (D.M.).
Abbreviations used:
- 3′UTR
3′-untranslated region
- ARE
AU-rich element
- BrdU
5-bromo-2′-deoxyuridine
- DIG
digoxigenin-labeled
- Dll1
Delta-like 1
- EGFP
enhanced green fluorescent protein
- Elav
embryonic lethal abnormal vision
- Elavl
Elav-like
- FACS
fluorescence-activated cell sorting
- GCL
ganglion cell layer
- GF
growth fraction
- HU
hydroxyurea
- IP
immunoprecipitation
- LI
labeling index
- M
mitosis
- mAb
monoclonal antibody
- MI
mitotic index
- PBS
phosphate-buffered saline
- PBT
0.5% Triton X-100 in PBS
- PBTW
PBS with 0.1% Tween 20
- PFA
paraformaldehyde
- pH3
phospho-Histone H3
- PI
propidium iodide
- qPCR
real time quantitative PCR
- RBP
RNA binding protein
- RFP
red fluorescent protein
- RNP-IP
ribonucleoprotein complex IP
- RT-PCR
reverse transcriptase-PCR
- SV40
simian virus 40
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-10-0808) on February 23, 2011.
REFERENCES
- Agathocleous M, Harris WA. From progenitors to differentiated cells in the vertebrate retina. Annu Rev Cell Dev Biol. 2009;25:45–69. doi: 10.1146/annurev.cellbio.042308.113259. [DOI] [PubMed] [Google Scholar]
- Atasoy U, Watson J, Patel D, Keene JD. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Cisneros E, Latasa MJ, García-Flores M, Frade JM. Instability of Notch1 and Dll1 mRNAs and reduced Notch activity in vertebrate neuroepithelial cells undergoing S-phase. Mol Cell Neurosci. 2008;37:820–831. doi: 10.1016/j.mcn.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Murthy KG, Prives C, Manley JL. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- Collier JR, Monk NA, Maini PK, Lewis JH. Pattern formation by lateral inhibition with feedback: a mathematical model of delta-notch intercellular signalling. J Theor Biol. 1996;183:429–446. doi: 10.1006/jtbi.1996.0233. [DOI] [PubMed] [Google Scholar]
- Das RM, et al. A robust system for RNA interference in the chicken using a modified microRNA operon. Dev Biol. 2006;294:554–563. doi: 10.1016/j.ydbio.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doller A, Akool ES, Huwiler A, Müller R, Radeke HH, Pfeilschifter J, Eberhardt W. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol Cell Biol. 2008;28:2608–2625. doi: 10.1128/MCB.01530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XC, Steitz JA. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Muñoz-Canoves P, Gorospe M, Muñoz A. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frade JM. Interkinetic nuclear movement in the vertebrate neuroepithelium: encounters with an old acquaintance. Prog Brain Res. 2002;136:67–71. doi: 10.1016/s0079-6123(02)36007-2. [DOI] [PubMed] [Google Scholar]
- Frade JM, Rodríguez-Tébar A. Neuroepithelial differentiation induced by ECM molecules. Methods Mol Biol. 2000;139:257–264. doi: 10.1385/1-59259-063-2:257. [DOI] [PubMed] [Google Scholar]
- Ge X, Frank CL, Calderon de Anda F, Tsai LH. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron. 2010;65:191–203. doi: 10.1016/j.neuron.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalves FC, Weisblat DA. MAPK regulation of maternal and zygotic Notch transcript stability in early development. Proc Natl Acad Sci USA. 2007;104:531–536. doi: 10.1073/pnas.0609851104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Hansson EM, Teixeira AI, Gustafsson MV, Dohda T, Chapman G, Meletis K, Muhr J, Lendahl U. Recording Notch signaling in real time. Dev Neurosci. 2006;28:118–127. doi: 10.1159/000090758. [DOI] [PubMed] [Google Scholar]
- Hsu RJ, Yang HJ, Tsai HJ. Labeled microRNA pull-down assay system: an experimental approach for high-throughput identification of microRNA-target mRNAs. Nucleic Acids Res. 2009;37:e77. doi: 10.1093/nar/gkp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I. Dynamic regulation of Notch signaling in neural progenitor cells. Curr Opin Cell Biol. 2009;21:733–740. doi: 10.1016/j.ceb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Katsanou V, Milatos S, Yiakouvaki A, Sgantzis N, Kotsoni A, Alexiou M, Harokopos V, Aidinis V, Hemberger M, Kontoyiannis DL. The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol Cell Biol. 2009;29:2762–2776. doi: 10.1128/MCB.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Gorospe M. Phosphorylated HuR shuttles in cycles. Cell Cycle. 2008;7:3124–3126. doi: 10.4161/cc.7.20.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, et al. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 2008;22:1804–1815. doi: 10.1101/gad.1645808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latasa MJ, Cisneros E, Frade JM. Cell cycle control of Notch signaling and the functional regionalization of the neuroepithelium during vertebrate neurogenesis. Int J Dev Biol. 2009;53:895–908. doi: 10.1387/ijdb.082721ml. [DOI] [PubMed] [Google Scholar]
- Levine DS, Sánchez CA, Rabinovitch PS, Reid BJ. Formation of the tetraploid intermediate is associated with the development of cells with more than four centrioles in the elastase-SV40 tumor antigen transgenic mouse model of pancreatic cancer. Proc Natl Acad Sci USA. 1991;88:6427–6431. doi: 10.1073/pnas.88.15.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lu JY, Schneider RJ. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J Biol Chem. 2004;279:12974–12979. doi: 10.1074/jbc.M310433200. [DOI] [PubMed] [Google Scholar]
- Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Galbán S, López de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo SM, Escoll P, de la Hera A, Frade JM. Somatic tetraploidy in specific chick retinal ganglion cells induced by nerve growth factor. Proc Natl Acad Sci USA. 2010;107:109–114. doi: 10.1073/pnas.0906121107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murciano A, Zamora J, López-Sanchez J, Frade JM. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol Cell Neurosci. 2002;21:285–300. doi: 10.1006/mcne.2002.1174. [DOI] [PubMed] [Google Scholar]
- Nardelli J, Catala M, Charnay P. Establishment of embryonic neuroepithelial cell lines exhibiting an epiplastic expression pattern of region specific markers. J Neurosci Res. 2003;73:737–752. doi: 10.1002/jnr.10716. [DOI] [PubMed] [Google Scholar]
- Ochiai W, et al. Periventricular Notch activation and asymmetric Ngn2 and Tbr2 expression in pair-generated neocortical daughter cells. Mol Cell Neurosci. 2009;40:225–233. doi: 10.1016/j.mcn.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Robinow S, Campos AR, Yao KM, White K. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science. 1988;242:1570–1572. doi: 10.1126/science.3144044. [DOI] [PubMed] [Google Scholar]
- Sauer FC. Mitosis in the neural tube. J Comp Neurol. 1935;62:377–405. [Google Scholar]
- Schenk J, Wilsch-Bräuninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc Natl Acad Sci USA. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS, Jr. Cell cycle parameters and patterns of nuclear movement in the neocortical proliferative zone of the fetal mouse. J Neurosci. 1993;13:820–833. doi: 10.1523/JNEUROSCI.13-02-00820.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamberg N, Lulla V, Fragkoudis R, Lulla A, Fazakerley JK, Merits A. Insertion of EGFP into the replicase gene of Semliki Forest virus results in a novel, genetically stable marker virus. J Gen Virol. 2007;88:1225–1230. doi: 10.1099/vir.0.82436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Moy LY, Sanada K, Zhou Y, Buchman JJ, Tsai LH. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Chen CY, Shyu AB. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.