This study revealed that ubiquitously expressed transcript (UXT)-V1 is recruited to tumor necrosis factor (TNF) receptor complex I by interacting with TNF receptor-associated factor 2. UXT-V1 is a short-half-life protein, the degradation of which facilitates the formation of the apoptotic receptor complex II in response to TNF treatment. This study uncovers UXT-V1 as a novel regulator of TNF-induced apoptosis.
Abstract
Proteins that directly regulate tumor necrosis factor (TNF) signaling have critical roles in determining cell death and survival. Previously we characterized ubiquitously expressed transcript (UXT)-V2 as a novel transcriptional cofactor to regulate nuclear factor-κB in the nucleus. Here we report that another splicing isoform of UXT, UXT-V1, localizes in cytoplasm and regulates TNF-induced apoptosis. UXT-V1 knockdown cells are hypersensitive to TNF-induced apoptosis. We demonstrated that UXT-V1 is a new component of TNF receptor signaling complex. We found that UXT-V1 binds to TNF receptor-associated factor 2 and prevents TNF receptor–associated death domain protein from recruiting Fas-associated protein with death domain. More importantly, UXT-V1 is a short-half-life protein, the degradation of which facilitates the formation of the apoptotic receptor complex II in response to TNF treatment. This study demonstrates that UXT-V1 is a novel regulator of TNF-induced apoptosis and sheds new light on the underlying molecular mechanism of this process.
INTRODUCTION
Tumor necrosis factor (TNF)-α is a potent cytokine that regulates critical cellular processes, including apoptosis, proliferation, and inflammation (Wajant et al., 2003). Aberrant TNF-α signaling has been correlated to the pathogenesis of cancer, sepsis, diabetes, and autoimmune diseases in humans (Aggarwal, 2003). After binding to its receptor TNFR1 (TNF receptor 1), TNF-α triggers the activation of a complex network of signal transduction pathways. These include mitogen-activated protein kinase (MAPK) pathway, (nuclear factor-κB (NF-κB) pathway, and the apoptosis pathway (Liu, 2005). The past few decades have witnessed intensive characterization of the individual signaling pathways and the reciprocal cross-talks among them. It remains to be elucidated, however, how cells regulate the divergence of the corresponding signaling pathways from the common receptor.
It is postulated that TNF-α induces the formation of TNFR1 trimers, which subsequently initiate the recruitment of the TNFR1 signaling complex. TNF receptor–associated death domain protein (TRADD), TRAF2 (TNF receptor–associated factor 2), and receptor interacting protein (RIP) have been found in this signaling complex. They are responsible for mediating “downstream” activation of the NF-κB and Jun N-terminal kinase pathways (Hsu et al., 1996; Schneider-Brachert et al., 2004). In certain conditions, the TRADD-RIP-TRAF2 complex (complex I) dissociates from TNFR1 and recruits another TNF effector protein, Fas-associated protein with death domain (FADD), to form an “apoptotic complex” (complex II), which activates caspase-8 and leads to apoptosis (Micheau and Tschopp, 2003; Wang et al., 2008).
To maintain the balance between survival and death in cells, the formation of complex II is strictly regulated. The activity of complex II is normally inhibited by cellular FLICE-inhibitory protein (c-FLIP), a protease-dead caspase-8 homologue that competes for caspase-8 binding to FADD (Irmler et al., 1997; Guiet et al., 2002; Kataoka, 2005). This inhibition is relieved through either activation of a c-FLIP degrading ubiquitin E3 ligase (Itch) or termination of the NF-κB transcriptional activity by protein synthesis inhibitors that shut down c-FLIP, cellular inhibitor of apoptosis (cIAP) 1, and cIAP2 synthesis (Wang et al., 1998; Micheau et al., 2001; Chang et al., 2006). Protein synthesis inhibitor, however, is needed for cells to undergo apoptosis efficiently, although they have a defective NF-κB pathway. The mechanism of the action of the protein synthesis inhibitor in TNF-induced apoptosis is still unclear.
Ubiquitously expressed transcript (UXT) is demonstrated to be widely present in human and mouse tissues. Its expression is markedly elevated in some human tumors (Schroer et al., 1999). The functional characterization of UXT is scarce, however. UXT is suggested to interact with the N terminus of the androgen receptor and regulate androgen-responsive genes (Taneja et al., 2004). It is also implicated as a component of the centrosome (Zhao et al., 2005). Our recent study demonstrates that UXT positively modulates NF-κB transcriptional enhanceosome in nucleus (Sun et al., 2007).
Notably, bioinformatics analysis reveals two splicing isoforms of UXT in cells. As of now, all studies have focused exclusively on the possible functions of the short form of UXT (157 aa), which we name UXT-V2 (UXT isoform 2). No report, however, has addressed the potential function of the long form of UXT (169 aa), which we name UXT-V1. In this study, we show that UXT-V1 binds to TRAF2 and prevents the TRAF2-RIP-TRADD complex from recruiting FADD and caspase-8, thus protecting cells against TNF-induced apoptosis. Importantly, UXT-V1 is a short-half-life protein, the degradation of which facilitates the formation of the apoptotic receptor complex II in response to TNF treatment.
RESULTS
Two isoforms of UXT localize in nucleus and cytoplasm, respectively
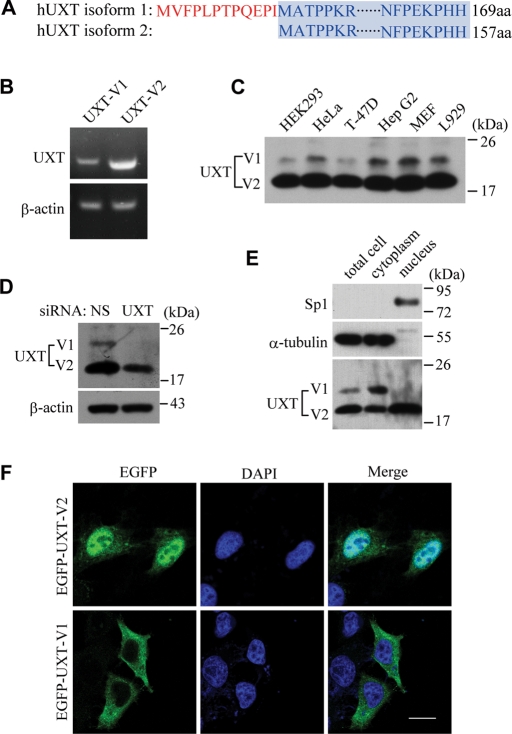
It came to our attention that there were two mRNA splicing isoforms of UXT when we explored its function in the context of NF-κB enhanceosomes. To avoid confusion, we refer to them hereafter as UXT-V1 and UXT-V2, respectively. As shown in Figure 1A, UXT-V1 harbors 12 more amino acids on its N terminus than does UXT-V2. The rest of the two proteins are identical.
FIGURE 1:
Two isoforms of UXT localize in nucleus and cytoplasm, respectively. (A) Sequence alignment of UXT-V1 against UXT-V2. (B) RT-PCR analysis of UXT mRNA in HeLa cells. (C) Immunoblot of UXT in several cell lines. (D) Immunoblot of UXT in UXT-RNAi HeLa cells or controls. (E) Subcellular fractionation analysis in HeLa cells. Control antibodies indicate accuracy of fractionation (Sp1, nucleus; α-tubulin, cytoplasm). (F) Confocal microscopy analysis of HeLa cells transfected with UXT-V1 or UXT-V2, with EGFP tagged at the N terminus. Scale bar: 10 μm.
We confirmed the presence of UXT-V1 by reverse transcription (RT)-PCR analysis and immunoblot analysis in HeLa cells (Figure 1, B and D), respectively. Using UXT siRNA effective for both UXT-V1 and UXT-V2, the observed 20-kDa band was further established as UXT-V1 (Figure 1D). Interestingly, the genomic structure of UXT was conserved among humans and mice. In addition, we used the National Center for Biotechnology Information’s (NCBI’s) Basic Local Alignment Search Tool (BLAST) to explore mouse EST (expressed sequence tag) sequences that are homologous to UXT. We identified GenBank AW988142.1 and GenBank CB203880.1 as mouse UXT-V1, which is highly conserved with human UXT-V1. We confirmed the ubiquitous expression of UXT-V1 in both human and mouse cell lines (e.g., HEK293, T-47D, HepG2, MEF, and L929) (Figure 1C). Apparently, UXT-V1 was expressed less abundantly than was UXT-V2. As expected, UXT-V2 was observed to reside predominantly in nucleus by subcellular fractionation analysis and confocal microscopy. To our surprise, UXT-V1 was reproducibly found to localize in cytoplasm (Figure 1, E and F), which suggested that UXT-V1 could perform functions different from those of UXT-V2 in cells.
UXT-V1 is a critical repressor in TNF-induced apoptosis
To explore the potential function of UXT in cellular processes, we took the RNAi approach by targeting both UXT-V1 and UXT-V2. Cell death was observed by phase-contrast microscopy when knocking down UXT in HeLa cells and in other cell lines (unpublished data). To investigate what type of cell death was induced, HeLa cells were analyzed with propidium iodide (PI) staining, followed by flow cytometry (Figure 2A). The cleavage of caspase-8 and poly (ADP-ribose) polymerase (PARP) was also measured (Figure 2B). It turned out that ∼20% of cells were dead when reducing endogenous expression of the two UXT isoforms simultaneously (Figure 2A). Consistently, both caspase-8 and PARP were activated under the same condition (Figure 2B), indicating that UXT is functionally linked to cell apoptosis.
FIGURE 2:
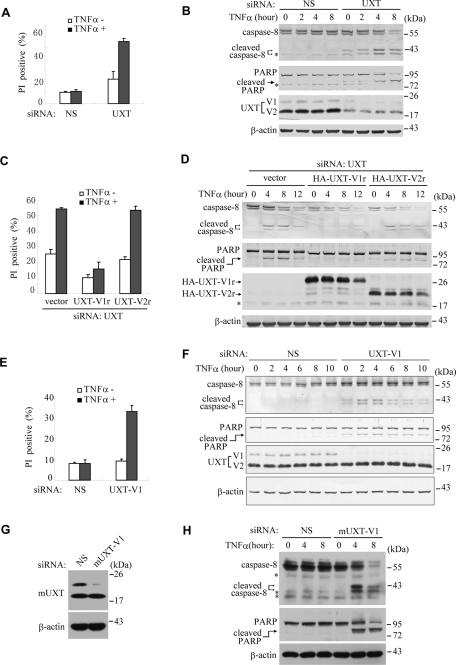
UXT-V1 is critical for protecting cells against TNF-induced apoptosis. (A) HeLa cells were transfected with nonspecific (NS) siRNA or UXT siRNA, and then treated with or without TNF-α (10 ng/ml) for 18 h. Cell death was quantified using PI staining on flow cytometry. (B) Both UXT-V1 and UXT-V2 in HeLa cells were knocked down with UXT-siRNA. The cells were treated with TNF-α for the indicated time. Cell lysates were collected for Western blot analysis of caspase-8, PARP, UXT, and β-actin levels. (C and D) HeLa cells were transfected with UXT siRNA and then rescued with siRNA-resistant UXT-V1r or UXT-V2r plasmids, followed by treatment with or without TNF-α. PI staining analysis and Western blot analysis were done as described. (E and F) HeLa cells were transfected with siRNA specifically knocking down UXT-V1, followed by PI staining analysis and Western blot analysis. (G) NIH3T3 cells were transfected with NS or mouse UXT-V1-specific siRNA, and then protein level of mouse UXT and β-actin was measured by Western blot. (H) Both control and mUXT-V1 knockdown NIH3T3 cells were treated with TNF-α as indicated. Cell lysates were collected for Western blot analysis of caspase-8, PARP, and β-actin. Data in A, C, and E are shown as mean + SD of three independent experiments. * indicates NS band.
It is well known that most cell types (including HeLa cells) would undergo apoptosis when treated with TNF-α plus cycloheximide (CHX). TNF-α alone, however, could not do so. Interestingly, TNF-α alone could significantly potentiate cell apoptosis up to more than 50% in UXT knockdown cells. Similarly, TNF-α synergized the activation of both caspase-8 and PARP when reducing endogenous UXT expression. In contrast, TNF-α alone had no effect on cell apoptosis in control cells (Figure 2, A and B).
To explore the potential functional difference between UXT-V1 and UXT-V2, two siRNA-resistant UXT plasmids (UXT-V1r and UXT-V2r) were generated, in which silent mutations were introduced into the corresponding sequence targeted by siRNA UXT, without changing the amino acid sequence of the proteins expressed. As shown in Figure 2, C and D, ectopic expression of UXT-V1r attenuated the cell apoptosis and the activation of caspase-8 and PARP, in cells having endogenous UXT knocked down. This rescue effect held true in the presence or absence of TNF-α, and was more dramatic upon TNF-α stimulation. In contrast, ectopic expression of UXT-V2r displayed barely such rescue effects, which indicated that UXT-V1 played a major role in protecting cells against TNF-induced apoptosis.
Alternatively, we screened out an siRNA (UXT-V1 siRNA) that could specifically knock down UXT-V1 but had no effects on UXT-V2. Consistent with the observations just mentioned, cell apoptosis apparently took place when knocking down endogenous UXT-V1 alone; and the HeLa cells became highly sensitive to TNF-induced apoptosis (Figure 2, E and F).
In addition, we screened out an siRNA that specifically knocked down mouse UXT-V1 (Figure 2G). Consistently, NIH3T3 mouse cells became sensitive to TNF-induced apoptosis when mUXT-V1 was knocked down (Figure 2H). Interestingly, knockdown of UXT induced low levels of stimulation-independent cell death in HeLa cells (Figure 2, A–C and F), whereas this effect was not observed in NIH3T3 cells (Figure 2H). Because the stimulation-independent cell death was weak, we speculate that the different sensitivities to apoptosis were due to the intrinsic difference between cell lines. The underlying mechanism, however, remains unknown. Collectively, these data demonstrate that UXT-V1 is a novel apoptosis repressor during TNF-induced apoptosis.
UXT-V1 interacts specifically with TRAF2
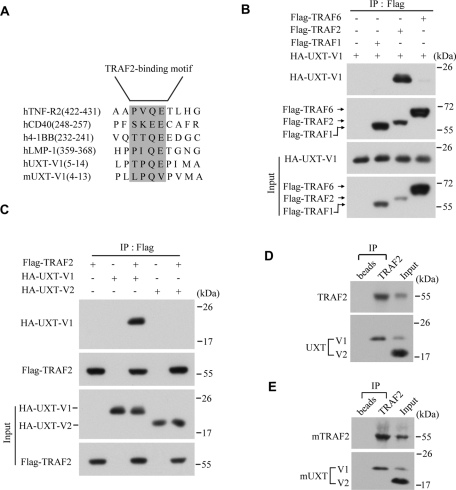
Given that UXT-V1 and UXT-V2 localized in different subcellular compartments, we went on to address how UXT-V1 regulated TNF signaling in cytoplasm. Because UXT-V1 has an additional 12-amino-acid peptide on its N terminus as compared with UXT-V2, the unique function of UXT-V1 is presumed to arise from this difference. Through the Bioinformatics method, a potential TRAF2 binding motif, –TPQE, was identified in this segment of UXT-V1, both in human and in mouse (Figure 3A). The consensus sequence, (P/S/A/T) × (Q/E) E, is highly conserved in proteins that were reported to interact specifically with TRAF2, such as TNFR2, CD40, 4–1BB, LMP-1, and so on (Park et al., 1999; Ye et al., 1999).
FIGURE 3:
UXT-V1 interacts specifically with TRAF2. (A) Sequence alignment of the TRAF2-binding motifs in hUXT-V1, mUXT-V1, TNFR2, CD40, 4–1BB, and LMP-1. (B) HA-UXT-V1 was cotransfected into HEK293T cells along with Flag-TRAF1, Flag-TRAF2, or Flag-TRAF6. Cell lysates were subjected to immunoprecipitation assays using anti-Flag antibody, followed by Western blot analysis using anti-HA antibody and anti-Flag antibody, respectively. (C) HEK293T cells were transfected with the indicated plasmids, and then the cell lysates were immunoprecipitated by anti-Flag antibody, followed by Western blot analysis using anti-HA and anti-Flag antibody, respectively. (D and E) HeLa cell lysates or NIH3T3 cell lysates were immunoprecipitated with anti-TRAF2 antibody or control protein A/G beads, followed by Western blot analysis with anti-TRAF2 antibody and anti-UXT antibody.
Therefore an immunoprecipitation assay was performed to explore the possible interactions between UXT-V1 and TRAF family proteins in HEK293T cells. Interestingly, UXT-V1 could bind strongly to TRAF2. In contrast, it interacted marginally with TRAF6, and not with TRAF1 at all (Figure 3B). Neither could UXT-V2 bind to TRAF2 (Figure 3C). The endogenous interaction between UXT-V1 and TRAF2 was also confirmed both in human cells (Figure 3D) and in mouse cells (Figure 3E). These results suggested that UXT-V1 regulated TNF signaling via targeting TRAF2.
TRAF2-binding motif of UXT-V1 is critical for its anti-apoptosis function
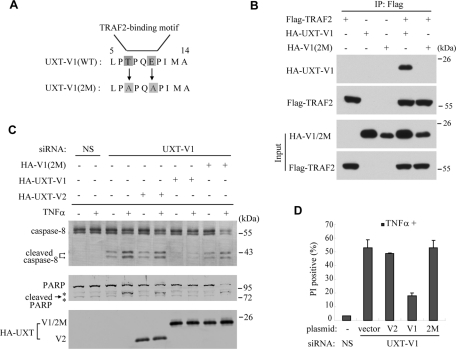
To elucidate the functional significance of this interaction, UXT-V1(2M) was generated, in which threonine (T) and glutamic acid (E) were mutated to glycine (A) at residue 7 and 10, respectively (Figure 4A). Consequently, UXT-V1(2M) was deprived of the ability to interact with TRAF2 (Figure 4B), confirming that the N-terminal 12–amino-acid motif of UXT-V1 was TRAF2-binding motif.
FIGURE 4:
TRAF2-binding motif is critical for UXT-V1’s anti-apoptosis function. (A) The mutation sites of UXT-V1(2M) in reference to UXT-V1(WT). (B) HEK293T cells were transfected with indicated combinations of plasmids. Cell lysates were immunoprecipitated with anti-Flag antibody, then probed by anti-HA and anti-Flag antibodies, respectively. (C and D) HeLa cells were transfected with UXT-V1 siRNA, then rescued with UXT-V1(WT), UXT-V1(2M), or UXT-V2 expression plasmids, respectively, followed by PI staining analysis and Western blot analysis. WT, wild type. * indicates nonspecific band. Data in (D) are shown as mean + SD of three independent experiments.
To determine whether this motif mediated UXT-V1’s anti-apoptotic function, wild-type UXT-V1, UXT-V1(2M), or UXT-V2 was individually introduced into the cells that had endogenous UXT-V1 already knocked down. Because the siRNA used in these cells was UXT-V1 siRNA, which specifically targeted the 5′ untranslated region of UXT-V1 mRNA, all these plasmids could express normally in UXT-V1 knockdown cells (Figure 4C). As expected, wild-type UXT-V1 could significantly inhibit TNF-induced apoptosis in these cells. In contrast, UXT-V1 (2M) and UXT-V2 could not attenuate TNF-induced apoptosis, leaving UXT-V1 knockdown cells highly sensitive to TNF-α (Figure 4, C and D). Thus the TRAF2-binding motif is essential for UXT-V1’s anti-apoptotic function, and UXT-V1 functions by modulating TRAF2.
UXT-V1 regulates the assemblage of complex II
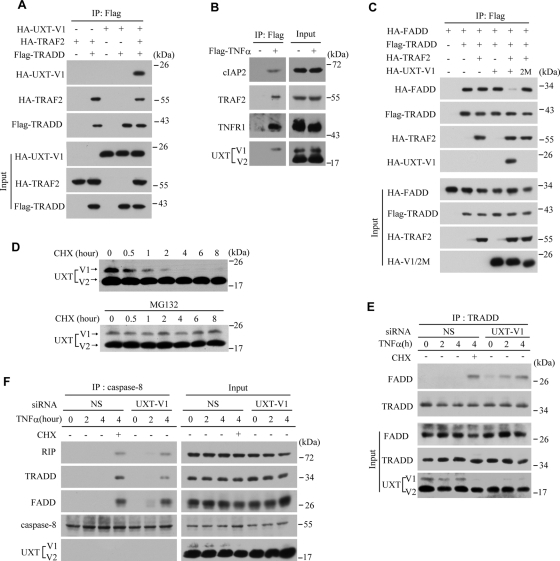
The “complex I” in apoptosis signaling is composed of TRADD, RIP, TRAF2, and other proteins. Because UXT-V1 could interact with TRAF2, we speculated that UXT-V1 was a new component in this complex. To test this hypothesis, TRADD, TRAF2, and UXT-V1 were ectopically expressed in HEK293T cells with indicated combinations, followed by immuneoprecipitating TRADD. It was observed that TRADD alone could not immuneoprecipitate UXT-V1. Notably, UXT-V1 was coimmunoprecipitated with TRADD in the presence of TRAF2 (Figure 5A), suggesting that TRAF2 served as a bridge linking UXT-V1 to TRADD complex. In addition, HeLa cells were stimulated by Flag-TNF-α, and then the receptor complex was immunoprecipitated by anti-Flag antibodies. Consistently, UXT-V1 was detected in the complex I (Figure 5B).
FIGURE 5:
UXT-V1 regulates the assemblage of the “complex II.” (A) HEK293T cells were transfected with the indicated combination of plasmids. Cell lysates were immunoprecipitated using anti-Flag antibody. The level of Flag-tagged TRADD was determined by Western blot analysis using anti-Flag antibody, and the levels of HA-tagged TRAF2 and UXT-V1 using anti-HA antibody. (B) HeLa cells were treated or not with Flag-TNF-α for 5 min. Cell lysates were immunoprecipitated by anti-Flag antibody, followed by Western blot assay with anti-TNFR1, -TRAF2, -cIAP2, and -UXT antibodies, respectively. (C) HEK293T cells were transfected with the indicated plasmids, and then immunoprecipitation assays were processed as described previously. (D) HeLa cells were treated with CHX (5 μg/ml) in the presence or absence of MG132 (10 μM) for the indicated times. Equal amounts of cell lysates were subjected to Western blot analysis of endogenous UXT-V1 and UXT-V2 levels. (E) HeLa cells transfected with nonspecific (NS) or UXT-V1 siRNAs were treated with TNF-α (10 ng/ml) for the indicated times, and CHX (5 μg/ml) was added as indicated. Cell lysates were immunoprecipitated by anti-TRADD antibody, followed by Western blot analysis with anti-TRADD, -FADD, or -UXT antibodies, respectively. (F) HeLa cells transfected with NS or UXT-V1 siRNAs were treated with TNF-α and/or CHX for the indicated times. Cell lysates were immunoprecipitated by anti-caspase-8 antibody, followed by Western blot analysis with anti-RIP, -caspase-8, -TRADD, -FADD, or -UXT antibodies, respectively.
The “apoptotic complex” (complex II) is formed when FADD is recruited to “complex I,” which in turn activates caspase-8 and initiates cell apoptosis. It has been reported that when TRADD and FADD were expressed together in HEK293T cells, TRADD could recruit FADD constitutively. Given that UXT-V1 had anti-apoptotic activity, it is conceivable that UXT-V1 may regulate the assemblage of the “apoptotic complex.” To explore this possibility, UXT-V1 and/or TRAF2 were expressed together with both TRADD and FADD in indicated combinations. It was observed that UXT-V1 alone could not attenuate the interaction between TRADD and FADD. TRAF2 alone did not influence this interaction, either. Interestingly, expression of TRAF2 and UXT-V1 together dramatically disrupted the interaction between TRADD and FADD (Figure 5C). In contrast, UXT-V1(2M) failed to display this effect. Taken together, UXT-V1 inhibits the assemblage of complex II via preventing FADD from being recruited to the TRAF2-RIP-TRADD complex.
CHX has been widely used in apoptosis study. The molecular mechanism of CHX action in potentiating apoptosis is largely unknown, however. We observed that UXT-V1 knockdown cells initiated the apoptosis program in response to TNF-α, but without the addition of CHX (Figures 2 and 4). We treated HeLa cells with CHX (5 μg/ml) as was commonly used. To our surprise, endogenous UXT-V1 was quickly degraded, in a time window overlapping with apoptosis progress. In contrast, CHX had no detectable effect on the stability of UXT-V2. When MG132 (an inhibitor of proteasome) was added, the degradation of UXT-V1 was rescued (Figure 5D). These data demonstrated that UXT-V1 was a short-half-life protein and that the degradation of UXT-V1 was possibly through the ubiquitin-proteasome pathway.
We went on to investigate the relationship between the degradation of UXT-V1 and TNF-induced apoptosis. TNF-α alone could not trigger FADD and caspase-8 to be recruited to complex I in HeLa cells. Addition of CHX could trigger the degradation of UXT-V1, which facilitated TRADD recruiting FADD; in UXT-V1 knockdown cells, complex I could recruit FADD upon the stimulation of TNF-α alone (Figure 5E). We also used anti-caspase-8 antibody to immunoprecipitate complex II upon the stimulation of TNF-α. Either knockdown of UXT-V1 or addition of CHX could initiate the assemblage of complex II (Figure 5F). These observations strongly suggest that TNF-induced apoptosis depends on the absence of UXT-V1 and that the degradation of UXT-V1 on CHX facilitates the formation of the apoptotic receptor complex II.
UXT-V1 attenuates cell apoptosis independent of NF-κB activation
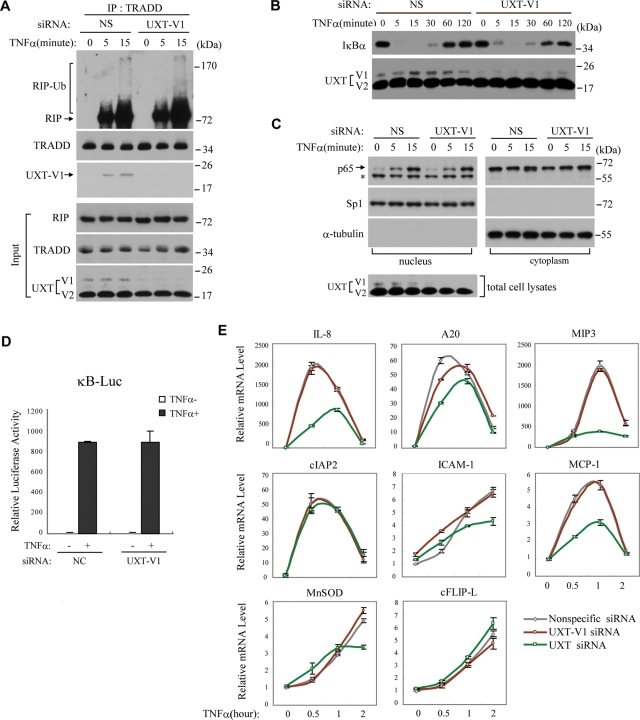
Formation of complex I would cause RIP poly-ubiquitination and recruits IκB kinase complex, leading to the activation of NF-κB (Ea et al., 2006; Wu et al., 2006). The transcription factor NF-κB regulates the induction of several critical anti-apoptotic genes, such as c-FLIP, cIAP2, and so on (Haas et al., 2009). To explore the relationship between UXT-V1 and NF-κB signaling, we used anti-TRADD antibody to immunoprecipitate complex I. There was no observable difference in the formation of complex I in the presence or absence of UXT-V1. The poly-ubiquitination of RIP was not modulated by UXT-V1 (Figure 6A). Consistently, UXT-V1 was also detected in this complex, confirming that UXT-V1 was indeed a new component of complex I (Figure 6A).
FIGURE 6:
UXT-V1 attenuates apoptosis independent of NF-κB signaling. (A) Control or UXT-V1 knockdown HeLa cells were treated with TNF-α (10 ng/ml) as indicated, and then cell lysates were immunoprecipitated with anti-TRADD antibody, followed by Western blot analysis of RIP, TRADD, and UXT. (B) HeLa cells with endogenous UXT-V1 knockdown or not were stimulated by TNF-α (10 ng/ml) for the indicated times. Equal amounts of cell lysates were subjected to Western blot analysis using anti-IκBα and anti-UXT antibodies. (C) Control or UXT-V1 knockdown HeLa cells were treated with TNF-α (10 ng/ml), and then cytoplasmic and nuclear fractions were extracted for immunoblot of p65, Sp1, and α-tubulin. The levels of Sp1 and α-tubulin were applied to indicate accuracy of fractionation. The total cell lysates were collected for immunoblot of UXT. *: nonspecific band. (D) NS or UXT-V1 siRNAs were transfected into HEK293T cells together with κB-Luc and pTK-Renilla reporters. 48 h after transfection, cells were stimulated with TNF-α for 8 h before luciferase assays were performed. pTK-Renilla reporter was used to normalize data. (E) 48 h after transfection with NS siRNA, UXT-V1 siRNA, or UXT siRNA, respectively, HeLa cells were stimulated with TNF-α for the indicated times. Induction of IL-8, A20, cIAP2, ICAM-1, MIP-3, MnSOD, MCP-1, and cFLIP-L mRNAs was measured by quantitative PCR. Data are shown as mean ± SD of three independent experiments.
Apparently, knockdown of UXT-V1 influenced neither the degradation nor resynthesis of NF-κB inhibitor α (IκBα) (Figure 6B). Furthermore, knockdown of UXT-V1 affects neither the p65 translocation to nucleus (Figure 6C) nor the activation of NF-κB reporter (Figure 6D). We also measured the induction of NF-κB–responsive genes (interleukin 8 [IL-8], A20, cIAP2, intercellular adhesion molecule 1 [ICAM-1], macrophage inflammatory protein 3 [MIP-3], manganese superoxide dismutase [MnSOD], monocyte chemoattractant protein-1 [MCP-1], and cellular FLICE-inhibitory protein long form [cFLIP-L]) by TNF-α, in cells that had UXT-V1 specifically knocked down or that had both isoforms of UXT knocked down. Consistently, knockdown of UXT-V1 did not influence expression of NF-κB–responsive genes. When knocking down both isoforms of UXT, however, a subset of NF-κB–responsive genes were obviously impaired (Figure 6E), which was due to the regulatory function of UXT-V2 (Sun et al., 2007). Importantly, several NF-κB–responsive genes, such as cIAP2, ICAM-1, and cFLIP-L, were not affected by UXT knockdown (Figure 6E), which indicated that the regulatory activity of UXT-V2 for NF-κB was selective.
Taken together, UXT-V1 did not modulate the formation of complex I and the activation of NF-κB. Instead, UXT-V1 regulated TNF-induced apoptosis via targeting the formation of complex II.
DISCUSSION
Although the molecular mechanism of TNF-induced apoptotic cell death has been well documented, it is still not fully understood what determines life or death in response to TNF for a given cell. In the current study, we report that one of the splicing isoforms of the UXT protein, UXT-V1, is a key regulator of TNF-induced apoptosis. We found that knocking down UXT-V1 renders cells hypersensitive to TNF-induced apoptosis. Our data suggest that UXT-V1 is a new component of the TNF receptor signaling complex. UXT-V1 binds to TRAF2 and prevents TRADD from recruiting FADD. More importantly, UXT-V1 is a short-half-life protein, the degradation of which facilitates the formation of the apoptotic receptor complex II in response to TNF treatment. This study demonstrates that UXT-V1 is a novel regulator of TNF-induced apoptosis, and sheds new light on the underlying molecular mechanism of this process.
In most cell types, the default TNF signaling induces the activation of signaling pathways such as NF-κB and MAPKs rather than apoptosis. Under normal conditions, most cell types are resistant to apoptosis. Under special circumstances, however, such as in the presence of protein synthesis inhibitor, CHX, the balance between life and death will be disrupted and allow cells to undergo apoptosis. Also, mice injected with d-galactosamine quickly die from massive liver necrosis after administration of TNF (Bradham et al., 1998). It has been suggested that NF-κB is responsible for such a protective effect against TNF-induced apoptosis through its target genes, such as XIAP, cIAP1 and 2, and cFLIP. In cells, in which NF-κB activation is compromised or the de novo protein synthesis is blocked, the levels of cIAPs and cFLIP will not be sufficient to inhibit the formation of TNF receptor complex II (Kreuz et al., 2001; Aota et al., 2002). It is still not clear why protein synthesis inhibitor is needed for cells, such as p65–/– cells, to undergo apoptosis efficiently although they have a defective NF-κB pathway (Van Antwerp et al., 1996; Wang et al., 1996). Our finding that UXT-V1 protects cells against TNF-induced apoptosis through regulating the formation of complex II provides a possible answer for this puzzle. The UXT gene is not a target of the NF-κB pathway. Therefore blocking of NF-κB has no effect on UXT expression. As we demonstrated, the presence of UXT-V1 in TNF receptor complex I prevents its conversion to the receptor complex II. But because it is a short-half-life protein, the removal of UXT-V1 will permit the recruitment of FADD into complex II and the subsequent activation of caspase cascade. Recent studies have reported that ABIN-1 protein protects cells against TNF-induced apoptosis through blocking caspase-8 being recruited to FADD (Oshima et al., 2009). Clearly, UXT-V1 functions differently from ABIN-1 and works apparently upstream of ABIN-1. This finding further suggests that the induction of apoptosis is finely regulated and that there are multiple checkpoints before the death pathway is engaged.
On TNF treatment, the TNF receptor signaling complex is formed by recruiting many effector proteins, such as TRAF2 and RIP (Liu, 2005; Wertz and Dixit, 2010). RIP is a death domain protein and is critical for TNF-induced activation of NF-κB pathway and three MAPK pathways as well as the induction of necrotic cell death (Kelliher et al., 1998; Holler et al., 2000). Ubiquitination and deubiquitination of RIP are key regulations of RIP-mediated TNF signaling (Bertrand et al., 2008; Wertz and Dixit, 2010). We found that the presence of UXT-V1 does not have any effect on the recruitment of RIP to the TNF receptor complex I and the ubiquitination of RIP. TRAF2 is a critical effector of TNF signaling and is also essential for transducing TNF signaling to the downstream targets. We found that UXT-V1 is recruited to TNF receptor complex I through its interaction with TRAF2. In contrast, UXT-V1 did not regulate Fas-induced apoptosis, in which TRAF2 is not involved (unpublished data, Huang and Wang).
Apparently, UXT-V1 plays a major role in the process of TNF-induced apoptosis, although UXT-V2 contributes indirectly and marginally to the modulation of apoptosis via regulating NF-κB activity. The only difference between UXT-V1 and UXT-V2 is the MVFPLPTPQEPI motif at the N terminus of these isoforms (Figure 1A). We have made a chimera of enhanced green fluorescent protein (EGFP) containing the indicated 12-aa peptide, and have explored the cellular localization of the fusion protein by confocal microscopy. Notably, this fragment is not sufficient to target GFP to cytoplasm (unpublished data, Huang and Wang), suggesting that the 12-aa peptide motif is not a signal for protein targeting. It is probable that the presence of the 12-aa portion in UXT-V1 blocks the nuclear localization signal/interaction that exposed in UXT-V2 isoform. Currently, it is not clear how the turnover of UXT-V1 is regulated, although our data indicate that UXT-V1 is degraded by the proteasome. Because several TNF effector proteins are regulated by ubiquitination, it is possible that the protein level of UXT-V1 is also controlled by the similar mechanism. This possibility needs further investigation.
It is known that UXT protein is highly expressed in several types of cancer. Based on our current findings, it is important to investigate whether UXT-V1 protein level is high in cancer. At least, as we showed in HeLa cells, knocking down of UXT-V1 will render cells to TNF-induced apoptosis. In addition, UXT-V1 has a short half-life, and it could be a valuable therapeutic target for treating certain types of cancer.
MATERIALS AND METHODS
Reagents
Recombinant human TNF-α was obtained from R&D Systems (Minneapolis, MN). CHX was purchased from Sigma (St. Louis, MO). MG132 was obtained from Boston Biochem (Cambridge, MA). The monoclonal anti-UXT antibody was provided by Wilhelm Krek (ETH-Hönggerberg, Zürich, Switzerland). The following antibodies were used for Western blot or immunoprecipitation: Sp1 (S9809; Sigma), α-tubulin (T9026; Sigma), β-actin (A5316; Sigma), PARP (sc-7150; Santa Cruz Biotechnology, Santa Cruz, CA), caspase-8 (9746; Cell Signaling Technology, Danvers, MA, and T341; Bioworld Technology, St. Louis Park, MN), hemagglutinin (HA) (sc-7392; Santa Cruz Biotechnology, and ab13834; Abcam, Cambridge, MA), Flag (F1804; Sigma, and SP7017; Sun Biomedical Technology, Beijing, China), TRAF2 (sc-876 and sc-7346; Santa Cruz Biotechnology), TRADD (sc-46653; Santa Cruz Biotechnology), FADD (sc-5559; Santa Cruz Biotechnology), IκBα (9242; Cell Signaling Technology), RIP (610458; BD Biosciences, Sparks, MD), TNFR1 (BS1478; bio-WORLD), and cIAP2 (ab23423; Abcam).
Cell culture
HeLa, HEK293, T47D, HepG2, L929, HEK293T, and NIH3T3 cell lines were obtained from the American Type Culture Collection and cultured according to the manufacturer’s instructions.
Plasmids, siRNA oligos, and cell transfection
Human or mouse UXT-V1 cDNA was amplified by RT-PCR from total RNA of HEK293T cells or NIH3T3 cells, respectively. The siRNA oligos against UXT (effective for both UXT-V1 and UXT-V2) or UXT-V1 were synthesized by GenePharma (UXT siRNA: 5′CCAAGGA CUCCAUGAAUAUTT3′; UXT-V1 siRNA: 5′GGCUGAACCUCCAGCUUGATT3′; mUXT-V1 siRNA: 5′GAAGUUUAAAUAGAGCGCUTT3′). The cells were transfected with siRNA oligos using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), and were incubated for 48 h before further analysis. In the rescue analysis, the plasmids were introduced into cells after these cells were transfected with siRNA for 24 h, and then were maintained for another 24 h before further analysis.
Nuclear extraction
Nuclear extractions of HeLa cells were prepared according to the procedure of Dignam et al. (1983).
Western blot and immunoprecipitation
The cell pellet was collected and resuspended in RIPA buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA; 0.5% NP-40; 0.25% sodium deoxycholate; 1 mM Na3VO4; 0.1 mM phenylmethylsulfonyl fluoride; Roche complete protease inhibitor set) for immunoprecipitation, or in RIPA buffer plus 0.1% SDS for Western blot. The resuspended cell pellet was vortexed for 20 s and then incubated on ice for 20 min and centrifuged at 20,000 × g for 20 min. The supernatants were collected for Western blot analysis or immunoprecipitation.
For immunoprecipitation, cell lysates were precleared with Protein A/G Plus-Agarose (Santa Cruz Biotechnology) at 4°C for 2 h, then antibody or control immunoglobulin G was added and incubated overnight. The next day, the cell lysates were incubated for another 2 h before the Protein A/G Plus-Agarose beads were added. The beads were washed with Tris-buffered saline buffer containing 0.5% NP-40; then the beads were boiled using 1× SDS loading buffer, and the supernatants were prepared for Western blot analysis.
Confocal microscopy
Twenty-four hours after being transfected with EGFP-tagged plasmids, cells cultured on coverslips were fixed with 4% paraformaldehyde (Sigma), and nuclei were stained with DAPI (Sigma). Slides were mounted by Aqua-Poly/Mount (Polysciences, Warrington, PA). Images were captured using a confocal microscope (TCS SP2 AOBS; Leica, Mannheim, Germany) with a 63× NA 1.4 oil objective.
PI staining and flow cytometry
After treatment or not, floating and adherent cells were all collected and stained with PI (BD Biosciences) according to the manufacturer’s instructions. Cell death was detected using FACSCalibur (BD Biosciences), and the data were analyzed with FlowJo software (Ashland, OR).
Quantitative PCR
Total cellular RNA was isolated with TRIzol (Invitrogen) according to the manufacturer’s instructions. Reverse transcription of purified RNA was performed using oligo(dT) primer. The quantification of gene transcripts was performed by real-time PCR using SYBR Green PCR mix (Applied Biosystems, Carlsbad, CA). All values were normalized to the level of β-actin mRNA. The primers used were as follows: β-actin, sense (AAAGACCTGTACGCCAACAC) and antisense (GTCAT ACTCCTGCTTGCTGAT); IL-8, sense (AGGTGCAGTTTTGCCAAGGA) and antisense (TTTCTGTGTTGGCGCAGTGT); A20, sense (GCGTTCAGGACACA GACTTG) and antisense (GCAAAGCCCCGTTTCAACAA); cIAP2, sense (TCCGT CAAGTTCAAGCCAGTT) and antisense (TCTCCTGGGCTGTCTGATGTG); MnSOD, sense (AACGTCACCGAGGAGAAGTACC) and antisense (CCTTGGA CACCAACAGATGC); ICAM-1, sense (GCAATGTGCAAGAAGATAGCCA) and antisense (CAGCGTAGGGTAAGGTTCTT); MIP-3, sense (TTGCTCCTGGCTGCT TTG) and antisense (GATAGCATTGATGTCACA); MCP-1, sense (GATCTCAGTG CAGAGGCTCG) and antisense (TGCTTGTCCAGGTGGTCCAT); cFLIP-L, sense (CTTGGCCAATTTGCCTGTAT) and antisense (GGCAGAAACTCTGCTGTTCC).
Acknowledgments
We thank Wilhelm Krek (ETH Zürich), Lin Li (Shanghai Institutes for Biological Sciences [SIBS]), and Zhengjun Chen (SIBS) for reagents. Zhenggang Liu and Chen Wang are members of the Center for Signal Transduction, Chinese Academy of Sciences. This work was supported by grants from Ministry of Science and Technology of Shanghai (09XD1404800); the National Natural Science Foundation of China (31030021); the Ministry of Science and Technology of China (2010CB529703, 2011CB910904, 2007CB914504); and the Chinese Academy of Sciences (KSCX1-YW-R-06).
Abbreviations used:
- BLAST
Basic Local Alignment Search Tool
- cFLIP
cellular FLICE-inhibitory protein
- cFLIP-L
cellular FLICE-inhibitory protein long form
- CHX
cycloheximide
- cIAP
cellular inhibitor of apoptosis
- EGFP
enhanced green fluorescent protein
- FADD
Fas-associated protein with death domain
- ICAM-1
intercellular adhesion molecule 1
- IκBα
NF-κB inhibitor α
- IL-8
interleukin 8
- MCP-1
monocyte chemoattractant protein-1
- MIP-3
macrophage inflammatory protein 3
- MnSOD
manganese superoxide dismutase
- NF-κB
nuclear factor-κB
- PARP
poly (ADP-ribose) polymerase
- PI
propidium iodide
- RIP
receptor interacting protein
- TNF
tumor necrosis factor
- TNFR1
TNF receptor 1
- TRADD
TNF receptor–associated death domain protein
- TRAF
TNF receptor-associated factor
- UXT
ubiquitously expressed transcript
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-10-0827) on February 9, 2011.
REFERENCES
- Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- Aota K, Azuma M, Tamatani T, Yamashita T, Ashida Y, Sato M. Stable inhibition of NF-kappa B in salivary gland cells does not enhance sensitivity to TNF-alpha-induced apoptosis due to upregulation of TRAF-1 expression. Exp Cell Res. 2002;276:111–119. doi: 10.1006/excr.2002.5515. [DOI] [PubMed] [Google Scholar]
- Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Bradham CA, Plumpe J, Manns MP, Brenner DA, Trautwein C. Mechanisms of hepatic toxicity. I. TNF-induced liver injury. Am J Physiol. 1998;275:G387–G392. doi: 10.1152/ajpgi.1998.275.3.G387. [DOI] [PubMed] [Google Scholar]
- Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K, Liu YC, Karin M. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11. 1983:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Guiet C, Silvestri E, De Smaele E, Franzoso G, Vito P. c-FLIP efficiently rescues TRAF-2-/- cells from TNF-induced apoptosis. Cell Death Differ. 2002;9:138–144. doi: 10.1038/sj.cdd.4400947. [DOI] [PubMed] [Google Scholar]
- Haas TL, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S, Bodmer JL, Schneider P, Seed B, Tschopp J. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol. 2000;1:489–495. doi: 10.1038/82732. [DOI] [PubMed] [Google Scholar]
- Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- Irmler M, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- Kataoka T. The caspase-8 modulator c-FLIP. Crit Rev Immunol. 2005;25:31–58. doi: 10.1615/critrevimmunol.v25.i1.30. [DOI] [PubMed] [Google Scholar]
- Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- Kreuz S, Siegmund D, Scheurich P, Wajant H. NF-kappaB inducers upregulate cFLIP, a cycloheximide-sensitive inhibitor of death receptor signaling. Mol Cell Biol. 2001;21:3964–3973. doi: 10.1128/MCB.21.12.3964-3973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZG. Molecular mechanism of TNF signaling and beyond. Cell Res. 2005;15:24–27. doi: 10.1038/sj.cr.7290259. [DOI] [PubMed] [Google Scholar]
- Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J. NF-kappaB signals induce the expression of c-FLIP. Mol Cell Biol. 2001;21:5299–5305. doi: 10.1128/MCB.21.16.5299-5305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Oshima S, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YC, Burkitt V, Villa AR, Tong L, Wu H. Structural basis for self-association and receptor recognition of human TRAF2. Nature. 1999;398:533–538. doi: 10.1038/19110. [DOI] [PubMed] [Google Scholar]
- Schneider-Brachert W, et al. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity. 2004;21:415–428. doi: 10.1016/j.immuni.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Schroer A, Schneider S, Ropers H, Nothwang H. Cloning and characterization of UXT, a novel gene in human Xp11, which is widely and abundantly expressed in tumor tissue. Genomics. 1999;56:340–343. doi: 10.1006/geno.1998.5712. [DOI] [PubMed] [Google Scholar]
- Sun S, Tang Y, Lou X, Zhu L, Yang K, Zhang B, Shi H, Wang C. UXT is a novel and essential cofactor in the NF-kappaB transcriptional enhanceosome. J Cell Biol. 2007;178:231–244. doi: 10.1083/jcb.200611081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja SS, Ha S, Swenson NK, Torra IP, Rome S, Walden PD, Huang HY, Shapiro E, Garabedian MJ, Logan SK. ART-27, an androgen receptor coactivator regulated in prostate development and cancer. J Biol Chem. 2004;279:13944–13952. doi: 10.1074/jbc.M306576200. [DOI] [PubMed] [Google Scholar]
- Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- Wajant H, Pfizenmaier K, Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- Wang CY, Mayo MW, Baldwin AS Jr. TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS Jr. NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Wertz IE, Dixit VM. Regulation of death receptor signaling by the ubiquitin system. Cell Death Differ. 2010;17:14–24. doi: 10.1038/cdd.2009.168. [DOI] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- Ye H, Park YC, Kreishman M, Kieff E, Wu H. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol Cell. 1999;4:321–330. doi: 10.1016/s1097-2765(00)80334-2. [DOI] [PubMed] [Google Scholar]
- Zhao H, Wang Q, Zhang H, Liu Q, Du X, Richter M, Greene MI. UXT is a novel centrosomal protein essential for cell viability. Mol Biol Cell. 2005;16:5857–5865. doi: 10.1091/mbc.E05-08-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]