As Arf6 is key to coordinating plasma membrane trafficking and regulates cellular invasion by several microorganisms, the authors studied Arf6 function during early HIV-1 infection. The data suggest that HIV-1 requires Arf6-driven plasma membrane dynamics and depends on GTP/GDP activity to efficiently fuse, enter, and infect CD4+ T lymphocytes.
Abstract
As the initial barrier to viral entry, the plasma membrane along with the membrane trafficking machinery and cytoskeleton are of fundamental importance in the viral cycle. However, little is known about the contribution of plasma membrane dynamics during early human immunodeficiency virus type 1 (HIV-1) infection. Considering that ADP ribosylation factor 6 (Arf6) regulates cellular invasion via several microorganisms by coordinating membrane trafficking, our aim was to study the function of Arf6-mediated membrane dynamics on HIV-1 entry and infection of T lymphocytes. We observed that an alteration of the Arf6–guanosine 5′-diphosphate/guanosine 5′-triphosphate (GTP/GDP) cycle, by GDP-bound or GTP-bound inactive mutants or by specific Arf6 silencing, inhibited HIV-1 envelope–induced membrane fusion, entry, and infection of T lymphocytes and permissive cells, regardless of viral tropism. Furthermore, cell-to-cell HIV-1 transmission of primary human CD4+ T lymphocytes was inhibited by Arf6 knockdown. Total internal reflection fluorescence microscopy showed that Arf6 mutants provoked the accumulation of phosphatidylinositol-(4,5)-biphosphate–associated structures on the plasma membrane of permissive cells, without affecting CD4-viral attachment but impeding CD4-dependent HIV-1 entry. Arf6 silencing or its mutants did not affect fusion, entry, and infection of vesicular stomatitis virus G–pseudotyped viruses or ligand-induced CXCR4 or CCR5 endocytosis, both clathrin-dependent processes. Therefore we propose that efficient early HIV-1 infection of CD4+ T lymphocytes requires Arf6-coordinated plasma membrane dynamics that promote viral fusion and entry.
INTRODUCTION
There is increasing evidence suggesting that membrane dynamics, like the traffic of vesicles and their spatial reorganization, is required for several biological functions such as cytokinesis (Albertson et al., 2005), cellular migration (Sabe, 2003; Schmoranzer et al., 2003; Letinic et al., 2009), regulation of plasma membrane morphology and polarization (Mellman and Warren, 2000; Folsch et al., 2009), and phagocytosis (Faurschou and Borregaard, 2003; Nordenfelt et al., 2009). Furthermore, some bacteria and viruses regulate membrane traffic in target cells to generate compartments to accomplish their replication process, which in many cases is regulated by Rab and ADP ribosylation factor (Arf) guanosine 5′-triphosphatases (GTPases) (Belov and Ehrenfeld, 2007; Pierini et al., 2009).
As the initial barrier to viral entry, the plasma membrane along with membrane-trafficking machinery is also of fundamental importance in the first stages of the viral cycle (Marsh and Helenius, 2006; Mudhakir and Harashima, 2009). It is thought that certain enveloped viruses such as herpes simplex virus 1 (HSV-1), Sendai virus, human immunodeficiency virus type 1 (HIV-1), and many retroviruses have pH-independent viral fusion proteins that allow the virus to penetrate into cells by fusing directly with the plasma membrane (Stein et al., 1987; Earp et al., 2005; Kielian and Rey, 2006; Marsh and Helenius, 2006). HIV-1 interacts with target cells through cell-surface CD4 and CXCR4 or CCR5 coreceptors, a process that is cooperative and requires cell signaling such as actin polymerization, reorganization (Iyengar et al., 1998; Jimenez-Baranda et al., 2007; Yoder et al., 2008; Barrero-Villar et al., 2009; Liu et al., 2009), and stabilization of microtubules (Valenzuela-Fernandez et al., 2005; Malinowsky et al., 2008) to achieve pore fusion formation. Although cytoskeleton reorganization and dynamics have well-documented roles in HIV-1 fusion and entry events, the contribution of plasma membrane dynamics is less clear during these early viral infection steps. It has been reported that HIV-1 fusion and entry could occur in micropinosomes and endosomes (Pauza and Price, 1988; Marechal et al., 2001), a process described as being clathrin dependent (Daecke et al., 2005), pH independent, and dynamin dependent (Miyauchi et al., 2009). On the other hand, HIV-1 internalization and infection in polarized trophoblasts appear to be pH-dependent processes (Vidricaire and Tremblay, 2005) that are driven by a clathrin-, caveolae-, and dynamin-independent endocytic pathway and require free membrane cholesterol (Vidricaire and Tremblay, 2007). Therefore HIV-1 entry and infection are orchestrated by viral and cellular membrane interaction, which appears to occur through complexes and associated membrane traffic events.
We have recently reported that the fluidity of plasma membrane, regulated by phosphatidylinositol-4-phosphate 5-kinase Iα (PI4P5-K Iα)–mediated phosphatidylinositol-(4,5)-biphosphate (PIP2) production, is crucial for HIV-1 entry and the early steps of infection in permissive lymphocytes (Barrero-Villar et al., 2008). Interestingly, plasma membrane morphology and dynamics are also regulated by the traffic of PIP2-associated membranes from the cell surface to a nonclathrin intracellular compartment, which in turn relies on the membrane transport activity of ADP ribosylation factor 6 (Arf6) (Radhakrishna and Donaldson, 1997; Franco et al., 1999; Donaldson, 2003; Naslavsky et al., 2003; Aikawa and Martin, 2005; Donaldson and Honda, 2005). In fact, Arf6 is the only member of the Ras-related Arf family of small GTPases that affects cell-surface dynamics, thereby regulating plasma membrane/endosome trafficking and cortical actin reorganization (D’Souza-Schorey et al., 1995; Radhakrishna and Donaldson, 1997; Franco et al., 1999; Al-Awar et al., 2000; Donaldson, 2003; Naslavsky et al., 2003; Donaldson and Honda, 2005). Arf6 appears to be associated with a tubular endosomal compartment in its inactive GDP-bound form and with the plasma membrane in its active GTP-bound form, thereby regulating membrane movement between these two compartments through its GTPase cycle (D’Souza-Schorey et al., 1995, 1998; Radhakrishna and Donaldson, 1997). On the other hand, it seems that the GDP/GTP cycle of Arf6 occurs mainly at the plasma membrane (Pasqualato et al., 2001), suggesting that Arf6 coordinates membrane dynamics on the cell surface (Macia et al., 2004). Therefore Arf6-dependent membrane movement is a complex process that has still not been elucidated.
Considering that Arf6 is key to coordinating plasma membrane dynamics and has functional implications for cellular invasion by several microorganisms (Criss et al., 2001; Nishi and Saigo, 2007; Laakkonen et al., 2009; Marchant et al., 2009), we decided to study the role of Arf6-mediated membrane dynamics during early HIV-1 entry and infection of T lymphocytes. In the present work, we observed that perturbation of Arf6-driven PIP2-associated membrane movement inhibits HIV-1 fusion, entry, and infection of permissive cells, regardless of the viral tropism. Therefore it appears that efficient early HIV-1 infection of CD4+ T lymphocytes requires Arf6-mediated plasma membrane dynamics.
RESULTS
Alteration of Arf6-dependent membrane trafficking impairs HIV-1 infection in permissive lymphocytes
We used the Arf6 wild-type construct (WT Arf6), which could cycle between its GDP-bound inactive and GTP-bound active forms, and the Arf6 mutants Q67L and T44N (Arf6-Q67L and Arf6-T44N), which are respectively locked in their GTP- and GDP-bound conformations (D’Souza-Schorey et al., 1995; Donaldson, 2003; Macia et al., 2004), in order to explore the functional role of Arf6-mediated membrane dynamics in HIV-1 entry and infection of permissive lymphocytes. These Arf6 mutants, defective in GTP-bound (T44N) and GTP hydrolysis (Q67L), have been used to identify Arf6’s cellular locations and to define its cellular functions (Radhakrishna et al., 1996; Al-Awar et al., 2000; Donaldson, 2003).
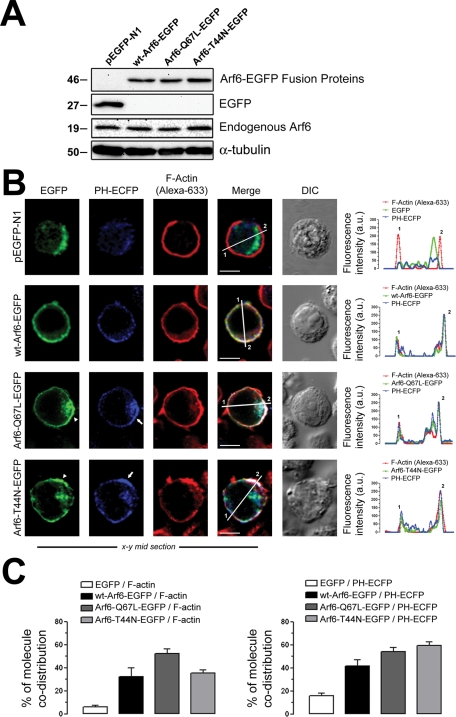
We used C-terminal hemagglutinin (HA)– and enhanced green fluorescent protein (EGFP)–tagged WT Arf6 or mutant constructs to explore the functional involvement of Arf6 in HIV-1 entry and infection. First, we analyzed the endogenous level of Arf6 expression and the different C-terminal EGFP-tagged Arf6 constructs by Western blot (Figure 1A), transiently expressed in the permissive CEM-CCR5 cell line. Furthermore, we studied the cellular localization of these EGFP-labeled Arf6 constructs in these permissive cells (Figure 1B). We observed that WT Arf6–EGFP localized mainly at the plasma membrane. The Arf6-Q67L–EGFP and Arf6-T44N–EGFP mutants were localized at the plasma membrane and cytoplasm of permissive T-cells (Figure 1B, EGFP images). These Arf6 constructs colocalized with PIP2-associated structures (Figure 1B, PH-ECFP and merged images), as monitored by the pleckstrin homology–enhanced cyan fluorescent protein (PH-ECFP) probe, previously described in Barrero-Villar et al. (2008). As a control, we transfected a construct codifying the EGFP protein, which presented a clear-cut cytoplasmic distribution pattern, excluded from plasma membrane as monitored by cortical F-actin localization (Figure 1B, pEGFP-N1 images).
FIGURE 1:
Pattern of expression of endogenous Arf6 and Arf6-EGFP constructs on permissive lymphocytes. (A) Western blot analysis of endogenous Arf6, WT Arf6–, Arf6-Q67L–, and Arf6-T44N–EGFP expression in CEM-CCR5 cells. α-Tubulin and pEGFP-N1 are the controls for total protein and EGFP expression, respectively. (B) Left, a series of confocal images, x–y midsections, show the expression pattern for WT Arf6–, Arf6-Q67L–, and Arf6-T44N–EGFP molecules in CEM-CCR5 cells. PIP2 (PH-ECFP probe), F-actin (Alexa 633–labeled phalloidin), free EGFP distribution, and merged and differential interference contrast (DIC) images are shown. White arrowheads and arrows indicate Arf6 mutants and PH-ECFP plasma-membrane localization, respectively. Bar, 5 μm. Right, Arf6-EGFP constructs, F-actin, and PH-ECFP distribution were quantified along lines drawn through the diameter of the cells (1 and 2 indicate measurement points in merged pictures). (C) Codistribution quantification for each Arf6-EGFP construct or EGFP with F-actin (left) or with the PH-ECFP probe (right) in whole cells. Data are mean ± standard error of the mean (SEM) (n = 15 different cells).
It is thought that Arf6 localizes at the plasma membrane and in endosome-associated membranes in many cells (Cavenagh et al., 1996; D’Souza-Schorey et al., 1998; Donaldson, 2003), as it is involved in membrane/vesicle trafficking from the plasma membrane (D’Souza-Schorey et al., 1998; Donaldson, 2003; D’Souza-Schorey and Chavrier, 2006). The Arf6-Q67L mutant has been reported to impede recycling of the internalized vesicles, provoking the accumulation of PIP2- and F-actin–coated membrane structures (Donaldson, 2003; Naslavsky et al., 2003). Furthermore, the inactive GDP-bound Arf6-T44N mutant distributed with F-actin in many cell-surface structures, such as large membrane folds and small membrane extensions, where it codistributed with PIP2, as monitored by the PH domain of the phospholipase C δ1 (PLCδ1) (Macia et al., 2004). Similarly, we observed that the Arf6-Q67L– and Arf6-T44N–EGFP mutants provoked accumulation of PIP2-associated structures (Figure 1B, PH-ECFP and merged images in Arf6-mutant-EGFP and line scans at right), which did not occur with control or WT Arf6–transfected cells (Figure 1B, pEGFP-N1 or WT Arf6–EGFP images and line scans at right). Moreover we also observed a plasma membrane ruffling effect in permissive lymphocytes overexpressing the Arf6-EGFP mutants, as is shown by the cortical F-actin protrusive distribution (Figure 1B, F-actin images in Arf6-Q67L– and Arf6-T44N–EGFP conditions), when compared with control-, WT Arf6–EGFP–, and EGFP-transfected lymphocytes (Figure 1B, F-actin images in pEGFP-N1 and WT Arf6–EGFP conditions). Qualitative and quantitative analyses revealed the specific codistribution of WT and mutant Arf6 molecules with F-actin and the PH-ECFP probe. Codistribution was absent for free EGFP and appeared to be slightly higher for the Arf6 mutants (Figure 1, B, line scans at right, and C). These data indicate that Arf6 mutants negatively affect PIP2-associated membrane trafficking in permissive lymphocytes.
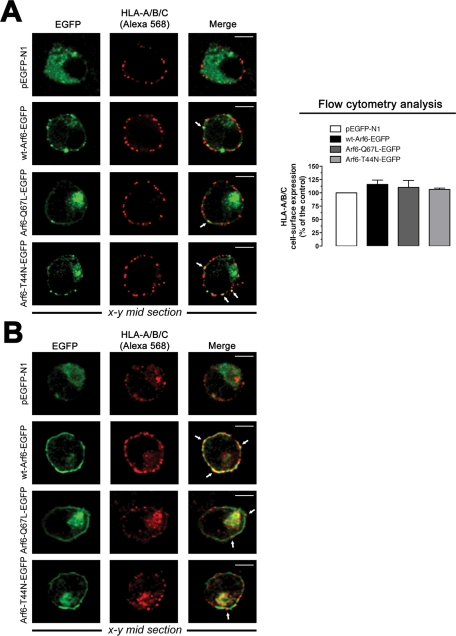
To further characterize these Arf6 constructs, we analyzed their distribution with class I molecules of the major histocompatibility complex (MHC-I), which is thought to traffic and recycle to plasma membrane from an Arf6-tubulovesicular network (Blagoveshchenskaya et al., 2002; Caplan et al., 2002; Larsen et al., 2004; Massol et al., 2005; Barral et al., 2008; Yi et al., 2010). We first analyzed cell-surface expression of human leukocyte antigen (HLA)-A/B/C in nonpermeabilized CEM-CCR5 cells expressing Arf6-EGFP constructs. We observed a dotted, cell-surface pattern of expression for HLA-A/B/C molecules, with some degree of codistribution with Arf6-EGFP constructs (Figure 2A, left). Arf6-Q67L– and Arf6-T44N–associated structures accumulated in cytoplasm, whereas WT Arf6–EGFP localized mainly at the cell surface, as we observed earlier. Flow cytometry analysis indicated that Arf6-EGFP constructs did not alter cell-surface expression of HLA-A/B/C molecules, compared with free EGFP-transfected cells (Figure 2A, right). In permeabilized CEM-CCR5 cells, we observed cell-surface and cytoplasmic codistribution of endogenous HLA-A/B/C molecules with overexpressed Arf6-EGFP constructs (Figure 2B). These results agree with reported data indicating that Arf6 constructs did not induce per se down-regulation of cell-surface MHC-I molecules (Blagoveshchenskaya et al., 2002; Larsen et al., 2004; Yi et al., 2010) and that MHC-I recycles to plasma membrane from Arf6 intracellular compartments (Blagoveshchenskaya et al., 2002; Caplan et al., 2002; Naslavsky et al., 2003; Larsen et al., 2004; Massol et al., 2005; Barral et al., 2008; Yi et al., 2010). Although the role of Arf6 activity on Nef-mediated MHC-I down-regulation and recycling has been extensively studied in Nef-expressing cells (Blagoveshchenskaya et al., 2002; Larsen et al., 2004; Massol et al., 2005; Barral et al., 2008; Yi et al., 2010), the present work focuses on the role played by Arf6-mediated membrane dynamics during early HIV-1 infection and therefore in Nef-negative uninfected cells.
FIGURE 2:
Pattern of expression of Arf6-EGFP constructs and endogenous MHC-I molecules on permissive lymphocytes. (A) Left, a series of confocal images, x–y midsections, show the expression pattern for endogenous HLA-A/B/C molecules and overexpressed WT Arf6–, Arf6-Q67L–, and Arf6-T44N–EGFP constructs in nonpermeabilized CEM-CCR5 cells. Free EGFP distribution and merged images are shown. White arrows indicate Arf6-EGFP constructs and HLA-A/B/C codistribution at cell surface. Bar, 5 μm. Right, flow cytometry analysis of HLA-A/B/C cell-surface expression in Arf6-EGFP–transfected cells (control, 100% HLA-A/B/C expression in pEGFP-N1–transfected cells). Data are mean ± SEM, n = 9. (B) A series of confocal images, x–y midsections, show the expression pattern for endogenous HLA-A/B/C and overexpressed WT Arf6–, Arf6-Q67L–, and Arf6-T44N–EGFP constructs in permeabilized CEM-CCR5 cells. Free EGFP distribution and merged images are shown. Bar, 5 μm.
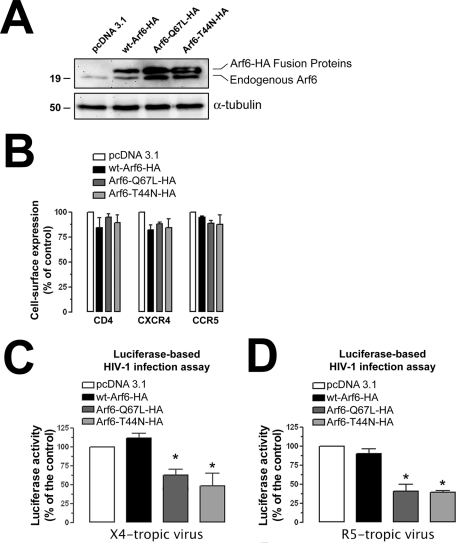
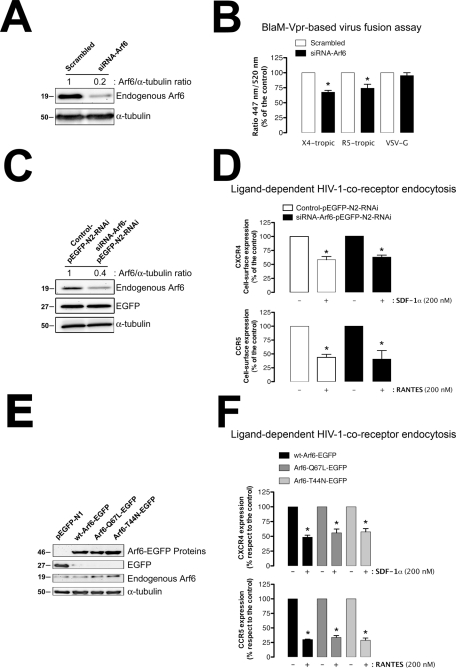
We conducted HIV-1 infection experiments, under these experimental conditions, with single-cycle viruses bearing the Luc-reporter gene, which allows the monitoring and quantifying of HIV-1 entry and infection (Barrero-Villar et al., 2008, 2009; Barroso-Gonzalez et al., 2009a). We first observed that overexpression of C-terminal HA-tagged WT Arf6, Q67L, or T44N constructs (Figure 3A) did not affect the cell-surface level of expression of CD4, CXCR4, and CCR5 molecules, the receptors for HIV-1 infection, in permissive lymphocytes (Figure 3B). Nonreplicative X4- or R5-tropic HIV-1 viral particles were then incubated with CEM-CCR5 permissive cells overexpressing either WT Arf6–, Arf6-Q67L–, or Arf6-T44N–HA mutant. HIV-1 entry and infection were impaired in cells overexpressing the Arf6-Q67L or Arf6-T44N mutant (Figure 3, C and D). Data obtained for these events were similar when using either X4- or R5-tropic HIV-1 viral strains (Figure 3, C and D; 40% or 52% of inhibition by Arf6-Q67L or Arf6-T44N, and 59% or 61% of inhibition by Arf6-Q67L or Arf6-T44N, respectively), indicating that alteration of Arf6-mediated membrane dynamics affects HIV-1 entry and infection regardless of viral tropism.
FIGURE 3:
Effect of the Arf6 constructs on HIV-1 entry and infection in permissive lymphocytes. (A) Western blot analysis of endogenous Arf6, WT Arf6–, Arf6-Q67L–, and Arf6-T44N–HA expression in CEM-CCR5 cells. α-Tubulin and pcDNA 3.1 are the controls for total protein and transfected cells, respectively. A representative experiment of the three is shown. (B) Flow cytometry analysis of CD4, CXCR4, and CCR5 cell-surface expression in Arf6-HA–transfected cells. Data are mean ± SEM, n = 9. (C and D) Luciferase-based assay of viral entry and infection by nonreplicative X4- and R5-tropic HIV-1 viral particles, respectively, in Arf6-HA–transfected CEM-CCR5 cells (control, 100% viral entry and infection in pcDNA3.1-transfected cells). Data are mean ± SEM of three independent experiments carried out in triplicate. Asterisk indicates p < 0.05, t test.
We also assayed the effect of Arf6-N48I/Q67L– and Arf6-T27N–HA constructs, two mutants for the Arf6-GTP/GDP cycling activity, on HIV-1 entry and infection (Figure 4, A and B). There are two possible explanations of why cytoplasmic Arf6-T27N is defective in GTP loading acting as a cytoplasmic dominant-negative protein: 1) It is thought to be locked in the GDP-bound state, where it interferes with the traffic through the intracellular vesiculotubular Arf6 network (Radhakrishna and Donaldson, 1997), or 2) it does not mimic a GDP-bound form, thereby affecting the binding of both GTP and GDP nucleotides (Macia et al., 2004). The Arf6-N48I/Q67L double mutant persists in the active GTP-bound state (Q67L mutation) and fails to activate the Arf6-downstream phospholipase D (PLD) (N48I mutation) (Vitale et al., 2002). Overexpression of these two mutants (Figure 4A) also inhibited X4- and R5-tropic HIV-1 entry and infection (Figure 4B). The results obtained with Arf6-Q67L, Arf6-T44N, and Arf6-T27N mutants together with the fact that the Arf6-N48I/Q67L construct bears the Q67L mutation, acting as Arf6-Q67L, reinforce the role of the Arf6 GTP/GDP cycle in early HIV-1 infection and suggest that the Arf6-mediated PLD signal is not involved during the first steps of viral infection. It is noteworthy that Arf6-induced PLD activity acts in exocytic events as reported with Arf6-N48I/Q67L and Arf6-N48I mutants (Vitale et al., 2002; Begle et al., 2009). More work needs to be performed to determine whether this downstream Arf6-effector is involved in early HIV-1 infection.
FIGURE 4:
Effect of different Arf6 constructs and the EFA6 factor on HIV-1 entry and infection in permissive lymphocytes. (A) Western blot analysis of endogenous Arf6, Arf6-N48I/Q67L–, and Arf6-T27N–HA expression in CEM-CCR5 cells. α-Tubulin and pcDNA 3.1 are the controls for total protein and transfected cells, respectively. A representative experiment of the three is shown. (B) Luciferase-based assay of viral entry and infection by nonreplicative X4- and R5-tropic HIV-1 viral particles, respectively, in Arf6-N48I/Q67L– and Arf6-T27N–HA–transfected CEM-CCR5 cells (control, 100% viral entry and infection in pcDNA3.1-transfected cells). Data are mean ± SEM of three independent experiments carried out in triplicate. Asterisk indicates p < 0.05, t test. (C) Western blot analysis of endogenous Arf6, WT Arf6–HA, and vsv-g-EFA6 expression in CEM-CCR5 cells. α-Tubulin and pcDNA 3.1 are the controls for total protein and transfected cells, respectively. A representative experiment of the three is shown. (D) Luciferase-based assay of viral entry and infection by nonreplicative X4- and R5-tropic HIV-1 viral particles, respectively, in WT Arf6–HA, vsv-g-EFA6, and double WT Arf6–HA/vsv-g-EFA6–transfected CEM-CCR5 cells (control, 100% viral entry and infection in pcDNA3.1-transfected cells). Data are mean ± SEM of three independent experiments carried out in triplicate.
Overexpression of WT Arf6 did not enhance HIV-1 entry and infection (Figure 3, C, 12% enhancement by WT Arf6 with respect to the control, and D), even in the presence of the cotransfected guanine nucleotide exchange factor EFA6 (Figure 4, C and D), described as being specific for Arf6 (Franco et al., 1999). Overexpression of EFA6 alone (Figure 4C) did not significantly enhance viral entry and infection, regardless of viral tropism (Figure 4D). In general, Arf6 functions have been determined by using their related Arf6 mutants, owing to the lack of functional effect of WT Arf6 overexpression (Blagoveshchenskaya et al., 2002; Donaldson, 2003; Larsen et al., 2004).
Although Arf6 activates PLD and PI4P5-K effectors (Brown et al., 2001; Vitale et al., 2002; D’Souza-Schorey and Chavrier, 2006; Gillingham and Munro, 2007), and overexpression of PI4P5-K Iα enhances HIV-1 viral fusion and infection (Barrero-Villar et al., 2008), the results we obtained with Arf6/EFA6 (Figure 4, C and D), single Arf6-GTP/GDP cycle mutants, and the Arf6-N48I/Q67L mutant (Figures 3 and 4, A and B, Figures 8 and 10 later in this paper, and Supplemental Figure 3), which fails to activate PLD, suggest that WT Arf6/EFA6 overexpression does not activate PI4P5-K Iα and that PLD activity is not directly involved in the regulation of viral fusion and entry. Therefore we propose that HIV-1 takes advantage of Arf6-coordinated plasma membrane movements, a dynamic process inhibited by Arf6-GTPase knockdown or Arf6 mutants, to efficiently fuse, enter, and infect CD4+ lymphocytes.
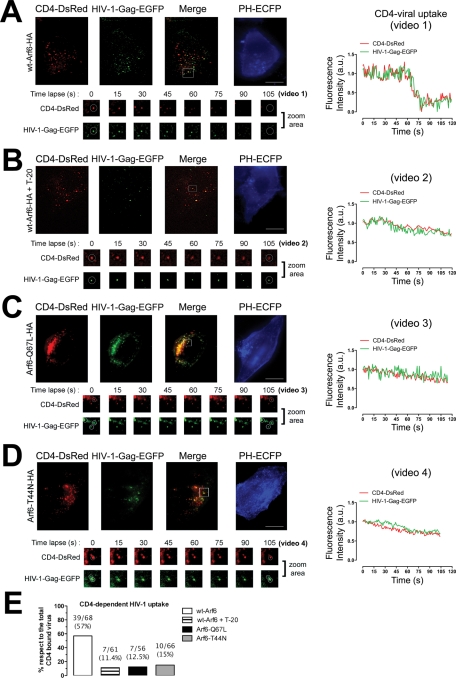
FIGURE 8:
Effects of the different Arf6-HA constructs on PIP2–plasma membrane distribution and HIV-1 entry analyzed by TIRFM. (A–D) A series of TIRFM images representing the expression pattern of cell-surface CD4-DsRed, HIV-1–Gag-EGFP virions, CD4-attached HIV-1–Gag-EGFP virions (merge), and the PH-ECFP probe (readout for PIP2) at the EF of TZMbl cells, under any experimental condition. T-20 treatment represents a control for the blockade of CD4-dependent fusogenic viral entry. White squares in merged images show a representative area (zoom area) where CD4-dependent HIV-1 uptake or blockade was observed, corresponding to Supplemental Videos 1–4, and a time lapse series of images (105 s, zoom area), under any experimental condition. White open circles, in a time lapse series of images, show representative events for CD4-dependent viral uptake or inhibition analyzed by time for their fluorescence intensities (right curves). Bar, 5 μm. (E) Histograms show the percentage of bound HIV-1–Gag-EGFP particles to CD4-DsRed that entered seven cells (ratio of CD4-dependent viral uptake events to the total number of CD4/HIV-1 interactions analyzed appears above the histograms), under any experimental condition.
FIGURE 10:
Arf6 regulates HIV-1 viral fusion with CD4+ lymphocytes, without affecting CCR5 and CXCR4 internalization. (A) Western blot analysis of endogenous Arf6 knockdown 24 h after siRNA nucleofection of CEM-CCR5 cells, quantified as the band intensity ratios to α-tubulin. Scrambled oligonucleotides represent the negative control for RNAi. A representative experiment of three is shown. (B) Specific silencing of endogenous Arf6 specifically affects the early steps of viral infection. Control (scrambled)– or siRNA-Arf6–treated CEM-CCR5 cells were incubated for 3 h with equivalent viral inputs (determined by standard p24-ELISA) of X4-tropic or R5-tropic pNL4-3.Luc.R-E- virions containing the BlaM-Vpr fusion protein. After adsorption for 3 h, cells were treated with CCF2-AM and analyzed by fluorescence spectrophotometry after 16 h. VSV-G virions containing the BlaM-Vpr fusion protein were used to control the specificity of Arf6-mediated effects on HIV-1 viral fusion. The percentages of HIV-1–fused cells were determined by measuring the ratio of blue (447 nm; cleaved CCF2) to green (520 nm; intact CCF2) fluorescence signals in target cells. Each assay was done in triplicate, and results are representative of three independent experiments. (C) Western blot analysis of endogenous Arf6 knockdown, 24 h after control- or siRNA-Arf6–pEGFP-N2-RNAi nucleofection of CEM-CCR5 cells, quantified as the band intensity ratios to α-tubulin. Control and siRNA oligonucleotides were transfected using the pEGFP-N2-RNAi plasmid; therefore treated cells expressed the EGFP protein, which serves as a control of cell treatment. A representative experiment of three is shown. (D) Effect of Arf6 knockdown on ligand-induced CXCR4 (SDF-1α) and CCR5 (RANTES) endocytosis in CEM-CCR5 cells. Control or siRNA-Arf6–pEGFP-N2-RNAi–treated cells were exposed to SDF-1α (200 nM) or RANTES (200 nM) for 1 h at 37°C. Then CXCR4 or CCR5 expression was analyzed by flow cytometry in control/EGFP+ and siRNA-Arf6/EGFP+ cells using PE-conjugated specific mAbs against cell-surface CXCR4 or CCR5. Data are mean ± SEM of three independent experiments carried out in triplicate and refer to CXCR4 or CCR5 expression in the absence of SDF-1α or RANTES, respectively, taken as 100%. Asterisk indicates p < 0.05, t test. (E) Western blot analysis of endogenous Arf6, WT Arf6–, Arf6-Q67L–, and Arf6-T44N–EGFP expression in CEM-CCR5 cells. α-Tubulin and pEGFP-N1 are the controls for total protein and intact Arf6-EGFP expression, respectively. (F). Effect of different Arf6-EGFP constructs on ligand-induced CXCR4 (SDF-1α) and CCR5 (RANTES) endocytosis in CEM-CCR5 cells. The experiments were carried out as indicated in (D) but in cells overexpressing each Arf6-EGFP construct. Data are mean ± SEM of three independent experiments carried out in triplicate and refer to cell-surface CXCR4 or CCR5 expression in WT Arf6–EGFP–transfected cells in the absence of SDF-1α or RANTES, respectively, taken as 100%. Asterisk indicates p < 0.05, t test.
Specific RNA interference (RNAi) of endogenous Arf6 inhibits HIV-1 infection in permissive T-cells
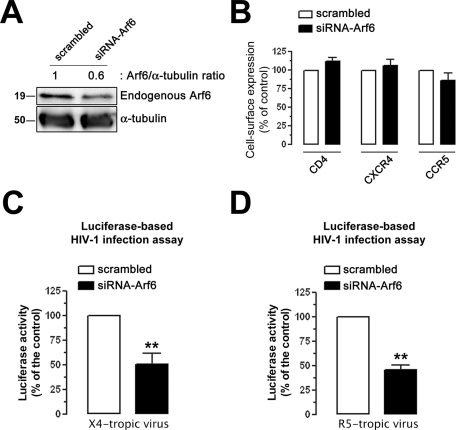
To further confirm the functional involvement of Arf6 during early HIV-1 infection, we infected permissive lymphocytes in which endogenous Arf6 protein expression was previously silenced by specific small interfering RNA (siRNA) (Figure 5A, siRNA-Arf6 band), without affecting cell-surface expression of HIV-1 receptors (Figure 5B). We observed that endogenous Arf6 knockdown negatively affected luciferase–HIV-1 entry and infection, as opposed to control scrambled-transfected cells. The extent of this blockade was similar to the level of Arf6 silencing achieved (Figure 5, C and D; 50% and 56% of inhibition, respectively). The inhibition of HIV-1 entry and infection again appeared to be independent of the viral tropism (Figure 5, C and D).
FIGURE 5:
Effect of knockdown of the endogenous Arf6 protein on HIV-1 entry and infection in permissive lymphocytes. (A) Western blot analysis of endogenous Arf6 knockdown in siRNA-Arf6–treated CEM-CCR5 cells, quantified as the band intensity ratios to α-tubulin. A representative experiment of three is shown. (B) Flow cytometry analysis of CD4, CXCR4, and CCR5 cell-surface expression in scrambled- or siRNA-Arf6–treated CEM-CCR5 cells. Data are the mean ± SEM of three independent experiments carried out in triplicate. (C and D) Luciferase-based assay of viral entry and infection by nonreplicative X4- and R5-tropic HIV-1 viral strains, respectively, in siRNA-Arf6–silenced CEM-CCR5 cells (control, 100% viral entry in scrambled-treated cells). Data are mean ± SEM of three independent experiments carried out in triplicate. Asterisks indicate p < 0.01, t test.
Therefore all these data suggest that efficient HIV-1 entry and infection require functional Arf6.
Specific silencing of endogenous Arf6 inhibits HIV-1 spreading and the infection of primary CD4+ T-cells
We also addressed the role of Arf6 in HIV-1 infection of primary cells. Preliminary work using stimulated primary lymphocytes showed long-term toxicity with Arf6 mutants or Arf6 silencing that hampered not only the maintenance of the siRNA silencing but also the analysis of cell-free HIV-1 infections over 4 d (unpublished data). Therefore to avoid long-term cultures of previously Arf6-transfected or Arf6-silenced HIV-1–infected and stimulated CD4+ T-cells, we analyzed the effect of endogenous Arf6 silencing on target nonactivated primary CD4+ T-cells during cell-mediated HIV-1 transmission, which is a fast and highly efficient mechanism of viral spread (Blanco et al., 2004; Jolly et al., 2004; Chen et al., 2007; Puigdomenech et al., 2008) that accounts for more than 90% of new CD4+ T-cell infections in vivo (Dixit and Perelson, 2004).
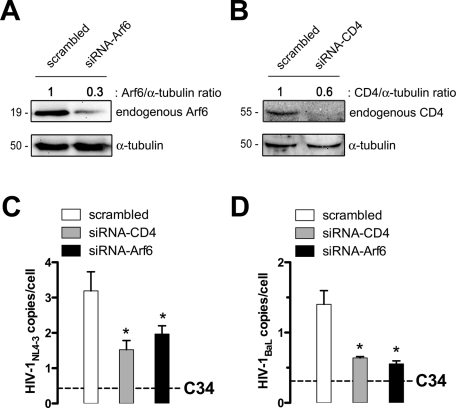
To do this, MOLT/CCR5 cells chronically infected with X4 or R5 viral isolates, MOLTNL4–3 or MOLTBaL, respectively, were used as effector cells and cocultured with unstimulated primary CD4+ T-cells, which were previously nucleofected with scrambled or specific siRNA for Arf6 silencing (Figure 6A). In addition, in this set of experiments, an siRNA directed against CD4 was used as a control of inhibition of HIV-1 transmission. After silencing, a reduction in the levels of Arf6 and CD4 expression was observed by Western blot as compared with the scrambled control (Figure 6, A and B). Specific Arf6 silencing did not affect CD4, CXCR4, and CCR5 cell-surface expression (unpublished data). Proviral HIV-1 DNA was measured after 24 h of coculture by quantitative PCR using primers to amplify the viral long terminal repeats (LTRs). It is worth mentioning that both Arf6 and CD4 knockdown partially but significantly inhibited cell-mediated HIV-1 transmission, irrespective of the viral tropism (Figure 6, C and D). Complete inhibition of viral transmission, which yielded background HIV DNA levels in MOLT/CCR5 cells, was achieved in C34 (anti-fusogenic peptide)–treated cultures (Figure 6, C and D, dashed line).
FIGURE 6:
Effect of Arf6 silencing on cell-to-cell HIV-1 transmission and infection of primary CD4+ T-cells. (A and B) Western blot analysis of Arf6 and CD4 knockdown in specific siRNA- or scrambled-nucleofected human primary CD4+ T-cells, 24 h after nucleofection, quantified as the band intensity ratios to α-tubulin. A representative experiment of three is shown. (C and D) After 24 h, nucleofected unstimulated primary CD4+ T-cells were cocultured with (C) MOLTNL4-3 or (D) MOLTBaL cells. Cell-to-cell HIV-1 transmission was analyzed at 24 h postcoculture by real-time PCR using a standard curve of a known number of HIV and CCR5 copies. Data represented HIV-1 DNA copies per cell, as values were normalized to the copy number of CCR5. Dashed lines represent the background levels of HIV-1 DNA in MOLT cells as determined in control cocultures in the presence of the fusion inhibitor peptide C34. Data are mean ± SD of three independent experiments. Asterisks indicate p < 0.05, t test.
Taken together these data suggest that Arf6-mediated membrane dynamics are involved in efficient cell-to-cell HIV-1 transmission to primary CD4+ T lymphocytes.
Arf6-mediated membrane dynamics regulate HIV-1 fusion, entry, and infection in permissive cells
Arf6-mediated membrane trafficking and its cellular characterization have been extensively described in HeLa cells (Radhakrishna and Donaldson, 1997; Donaldson, 2003). It appears that the Arf6-mediated membrane recycling system is different from the transferrin receptor recycling pathway and from its actin reorganization activity (Radhakrishna and Donaldson, 1997; Al-Awar et al., 2000; Donaldson, 2003). Permissive CXCR4+/CCR5+/CD4+-HeLa cells, such as HeLa-P5 and TZMbl cells, have been used to study and image HIV-1 infection, as envelope-mediated membrane fusion, entry, and viral egress events (Pleskoff et al., 1997; Valenzuela-Fernandez et al., 2005; Jouvenet et al., 2008; Barrero-Villar et al., 2009; Barroso-Gonzalez et al., 2009a; Ivanchenko et al., 2009; Miyauchi et al., 2009). Therefore we studied Arf6 cellular location and the functional consequences of Arf6 mutants and Arf6 silencing on Arf6-mediated membrane dynamics during early HIV-1 infection in permissive HeLa cells.
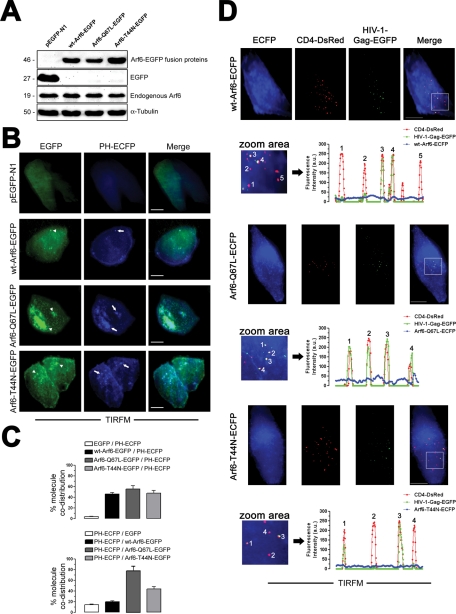
We further validated the data obtained in CD4+ T lymphocytes by first analyzing the expression pattern of C-terminal EGFP- or ECFP-tagged WT Arf6, Arf6 Q67L, and Arf6 T44N (mutants for the GTP/GDP cycle able to localize at the cell surface) constructs, transiently transfected in permissive TZMbl cells (Figure 7 and Supplemental Figure 1), and compared them with the distribution of cellular PIP2-associated structures and free or HIV-1–bound CD4 viral receptor. Fluorescence confocal microscopy showed that WT Arf6–EGFP was homogeneously localized on the plasma membrane as well as on cytoplasm in permissive TZMbl cells without promoting the accumulation of large PIP2-associated structures (Supplemental Figure 1, WT Arf6–EGFP and related PH-ECFP and merged images). We observed accumulation of PIP2-associated structures in cells overexpressing Arf6-Q67L– and Arf6-T44N–EGFP (Supplemental Figure 1, see arrowheads in Arf6 mutants and arrows in related PH-ECFP and merged images). We also observed colocalization of Arf6 constructs with F-actin (Supplemental Figure 1, see F-actin in related Arf6 images). The quantification of the codistribution pattern of each EGFP-labeled Arf6 construct or the free EGFP protein with the PH-ECFP probe or F-actin is shown in line scans (Supplemental Figure 1).
FIGURE 7:
TIRFM analysis for plasma membrane expression pattern of Arf6-EGFP or Arf6-ECFP constructs, and free or HIV-1–bound CD4-DsRed molecules on permissive HeLa cells. (A) Western blot analysis of endogenous Arf6, WT Arf6–, Arf6-Q67L–, and Arf6-T44N–EGFP expression in permissive TZMbl cells. α-Tubulin is the control for total protein. Free EGFP protein expression in pEGFP-N1–transfected cells. A representative experiment of the three is shown. (B) TIRFM analysis for the plasma membrane expression pattern of each Arf6-EGFP and the PH-ECFP probe. Merged images are shown. A representative experiment of the three is shown. Bar, 5 μm. White arrowheads or arrows indicate the distribution of Arf6 mutant or accumulation of PIP2-associated structures, respectively. (C) Quantification of the codistribution of each Arf6-EGFP or free EGFP molecule with PIP2 (PH-ECFP)–associated plasma membrane structures (top) or PIP2 (PH-ECFP) with each Arf6-EGFP or free EGFP molecule (bottom) from TIRFM images, as shown in (B). A representative experiment of three is shown. (D) A series of TIRFM images representing the expression pattern of cell-surface CD4-DsRed, HIV-1–Gag-EGFP virions, CD4-attached HIV-1–Gag-EGFP virions (merge), and WT Arf6–, Arf6-Q67L–, or Arf6-T44N–ECFP constructs, respectively, at the EF of TZMbl cells. The quantification of the pattern of distribution of free or HIV-1–Gag-EGFP–bound CD4-DsRed or Arf6-ECFP constructs is shown by line scan quantification, after background remove, through regions 1–4 or 5 indicated in zoom areas. Bar, 5 μm.
Furthermore, we analyzed the distribution of the different Arf6-EGFP constructs together with the PH-ECFP probe at the plasma membrane of these permissive cells using TIRFM. We observed that TZMbl cells expressing Arf6-Q67L– and Arf6-T44N–EGFP (Figure 7, A and B) presented accumulation of PIP2-associated structures on plasma membranes, as observed in the evanescent field (EF) (Figure 7B, see arrows in PH-ECFP images), where these mutants colocalized (Figure 7B, see arrowheads in Arf6-Q67L– and Arf6-T44N–EGFP images). In general, we observed that the Arf6-Q67L mutant induced a more extensive accumulation of PIP2-associated structures either on cytoplasm or on plasma membrane, compared with the effect exerted by the Arf6-T44N mutant (Figure 7, B and C, and Supplemental Figures 1 and 5; see related PH-ECFP or merged images). In this regard, we even observed the accumulation of PIP2-associated vacuole-like structures near the plasma membrane in some cells overexpressing the Arf6-Q67L–HA mutant (Supplemental Figure 2). This fact has also been previously reported in cells transfected with the GTP-bound Arf6-Q67L mutant (Naslavsky et al., 2003; Aikawa and Martin, 2005; Cohen et al., 2007). The quantification of the codistribution of each EGFP-labeled Arf6 construct or the free EGFP protein with the PH-ECFP probe on plasma membrane observed by TIRFM indicated that all Arf6 molecules similarly codistributed with PIP2-associated plasma membrane domains (Figure 7C, top). The quantification of the codistribution of the PH-ECFP probe with each EGFP-labeled Arf6 construct or the free EGFP protein on plasma membrane clearly indicated that Arf6 mutants provoked the accumulation of PIP2-associated membrane domains on cell-surface regions where the mutants are localized (Figure 7C, bottom). Therefore inhibition of the Arf6-GTP/GDP cycle seems to perturb the movement of PIP2-associated structures, provoking its accumulation on plasma membrane of Arf6-Q67L– or Arf6-T44N–treated cells.
TIRFM studies indicate that WT Arf6–, Arf6-Q67L–, and Arf6-T44N–ECFP constructs did not colocalize with cell-surface CD4-DsRed (Figure 7D). HIV-1 binding to CD4 did not promote codistribution of virus-bound or free CD4 with Arf6 constructs, and Arf6 constructs did not affect the first CD4/HIV-1 interaction (Figure 7D). We observed that free or HIV-1–bound CD4-DsRed molecules did not distribute with Arf6 structures (Figure 7D, line scans). This fact suggests that CD4 molecules do not internalize to or recycle from Arf6 compartments. Therefore the results obtained with Arf6-EGFP and Arf6-ECFP constructs point to the coordination of PIP2-associated membrane dynamics exerted by Arf6 on plasma membrane, which is perturbed by the Arf6-Q67L and Arf6-T44N mutants, without affecting CD4 cell-surface expression and the first HIV-1/CD4 interactions.
TIRFM studies indicate that functional Arf6 is required for efficient HIV-1 entry
We studied the ability of the CD4-bound HIV-1 virus to enter target cells with a perturbed Arf6-GTP/GDP cycle and, therefore, with accumulated PIP2-associated structures at the cell surface. TIRFM can dynamically study, at the plasma membrane, the fate of internalization or export of different cargos or cell-surface molecules (Barroso-Gonzalez et al., 2009a, 2009b). Furthermore, it has also been applied to the study of HIV-1 fusion and entry, viral assembly, and release (Jouvenet et al., 2006; Ivanchenko et al., 2009). In this regard, we performed TIRFM studies of the CD4-dependent HIV-1 uptake process by using nonreplicative, fluorescent HIV-1–Gag-EGFP viral particles in nonlymphoid permissive TZMbl (CD4+/CXCR4+/CCR5+) cells (see Materials and Methods) transiently expressing the fluorescent CD4-DsRed molecule together with one of the different Arf6-HA constructs and the PH-ECFP probe. This probe allowed us to monitor accumulated PIP2-associated structures at the EF of permissive cells and provided a clear-cut readout for effective Arf6 mutant–mediated inhibition of endogenous Arf6-coordinated PIP2-membrane traffic (Figure 8, PH-ECFP images). The study of CD4-dependent HIV-1 entry in cells presenting accumulation of PIP2-associated structures, provoked by Arf6 mutants that also codistributed with Arf6 mutants (Figures 1, 7, and 8 and Supplemental Figures 1, 2, and 5), gives a better guaranty of selecting cells where Arf6-dependent membrane dynamics have been altered, thereby avoiding a possible incorrect selection of cells for analysis of viral uptake based on Arf6 detection, which could lead to the selection of cells expressing critical levels of Arf6-fluorescent constructs without perturbing Arf6/PIP2-coordinated membrane dynamics.
We first assayed luciferase-based HIV-1 entry and infection experiments on HeLa-P5 cells, transfected with the different Arf6-HA constructs or siRNA-Arf6 oligonucleotides (Supplemental Figures 3A and 4A, respectively), in order to confirm that Arf6-HA mutants and siRNA-Arf6 treatment affect early HIV-1 infection in HeLa permissive cells. CD4, CXCR4, and CCR5 cell-surface expression was not affected by the expression of any Arf6-HA construct used (Supplemental Figure 3B). Furthermore, X4- and R5-tropic HIV-1 entry and infection appeared to be impaired by Arf6-GTP/GDP cycling mutants (Supplemental Figure 3, C and D, 40% and 42% inhibition by Arf6-Q67L and Arf6-T44N and 50% and 43% inhibition by Arf6-Q67L and Arf6-T44N, respectively). Similar inhibitory effects on viral infection were observed by the anti-fusogenic T-20 peptide (Supplemental Figure 3, C and D, T-20 bars in cells transfected with pcDNA3.1). However, Arf6 mutants did not affect cell infection by HIV-1 vectors pseudotyped with the vesicular stomatitis virus G (VSV-G) protein (Supplemental Figure 3E), an envelope protein that drives the entry and infection process in a clathrin-dependent endocytic manner (Matlin et al., 1982; Sun et al., 2005). Similarly, nonfunctional Arf6 mutants impaired HIV-1 envelope–mediated cell-to-cell fusion, independently of envelope-viral tropism (Supplemental Figure 3, F and G, 40% and 48% inhibition by Arf6-Q67L and Arf6-T44N and 39% and 42% inhibition by Arf6-Q67L and Arf6-T44N, respectively), when quantified by a β-galactosidase-based cellular model for membrane fusion as previously described (Pleskoff et al., 1997; Valenzuela-Fernandez et al., 2005; Barrero-Villar et al., 2009; Barroso-Gonzalez et al., 2009a). Furthermore, endogenous Arf6 knockdown (Supplemental Figure 4A), which did not affect cell-surface expression of CD4, CXCR4, or CCR5 viral receptors (Supplemental Figure 4B), negatively affected X4- and R5-tropic luciferase-based HIV-1 infection (Supplemental Figure 4, C and D, ∼50% inhibition). However, Arf6 knockdown did not affect cell infection by HIV-1 vectors pseudotyped with the VSV-G protein (Supplemental Figure 4E). Altogether these data lead us to suggest that Arf6-HA constructs and specific Arf6 silencing affect viral entry and infection in HeLa permissive cells, as observed in CD4+ lymphocytes, thus confirming the inhibitory effect of Arf6-HA mutants on HIV-1 entry and infection used in the following TIRFM studies.
Our results suggest that the Arf6-mediated effect on HIV-1 entry is independent of the viral tropism. Thus we present CD4-dependent viral uptake experiments performed by using X4-tropic HIV-1–Gag-EGFP virions in this section. Cell-surface CD4-DsRed molecules can monitor CD4-dependent HIV-1 uptake, whereas the PH-ECFP probe serves as a readout for the accumulation of PIP2-associated structures at the plasma membrane of cells expressing the GTP/GDP cycling Arf6-HA mutants (Figure 8, A–D). Of note, overexpression of each Arf6-HA construct did not affect the initial CD4/HIV-1 interaction (Figure 8, A–D, cell-surface CD4-DsRed/HIV-1–Gag-EGFP codistribution in merged images). Interestingly, PIP2 accumulation was clearly observed in the EF of cells transfected with either Arf6-Q67L–HA or Arf6-T44N–HA (Figure 8, C and D, PH-ECFP images), thereby representing an alteration of Arf6-coordinated plasma membrane dynamics. On the contrary, overexpression of the WT Arf6–HA construct did not provoke the accumulation of large PIP2-associated structures (Figure 8, A and B, PH-ECFP images), which could be indicative of normal membrane dynamics.
HIV-1 uptake was studied under each experimental condition as described in Materials and Methods. CD4-dependent viral uptake was clearly observed in control and WT Arf6–nucleofected cells (Figure 8A and quantified in E as 57% of CD4-dependent viral uptake), as monitored by the coordinated dimming of the intensity of fluorescence for the DsRed and EGFP fluorophores, respectively, associated with CD4 and viral particles (Figure 8A, fluorescence intensity curves and time lapse images from zoom area for CD4-DsRed and bound HIV-1–Gag-EGFP, and Supplemental Video 1). We performed CD4-dependent HIV-1–Gag-EGFP uptake experiments in cells overexpressing WT Arf6–HA in the presence of the anti-fusogenic peptide T-20 to analyze CD4-dependent productive viral entry. We observed an important inhibition of CD4-dependent viral uptake (Figure 8, B and quantified in E as only 11.4% of CD4-dependent viral uptake). Therefore the CD4-dependent viral uptake observed in the presence of T-20 could correspond to nonproductive viral entry, considering that CD4-dependent fusogenic entry is blocked by T-20 (Figure 8B, fluorescence intensity curves and time lapse images from zoom area for CD4-DsRed and bound HIV-1–Gag-EGFP, and Supplemental Video 2).
Our results obtained in permissive cells overexpressing Arf6-Q67L–HA or Arf6-T44N–HA mutants indicated that alteration of Arf6-mediated PIP2-membrane dynamics prevented CD4-dependent HIV-1 uptake (Figure 8, C or D and quantified in E as 12.5% and 15% of CD4-dependent viral uptake, respectively). Given all the data presented concerning the inhibitory effect of each Arf6 mutant on HIV-1 viral entry and infection, and the blockade of CD4-dependent viral uptake by T-20, it is conceivable that Arf6 mutants inhibited CD4-dependent productive viral entry as observed by TIRFM (Figure 8, C and D, fluorescence intensity curves and time lapse images from zoom area for CD4-DsRed and bound HIV-1–Gag-EGFP, and Supplemental Videos 3 and 4).
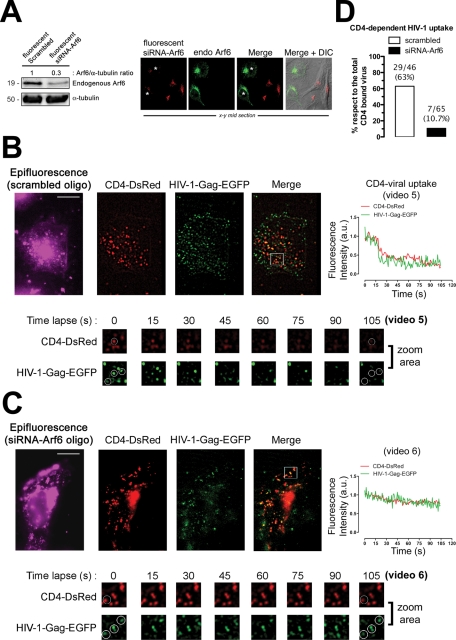
We performed similar experiments on TZMbl cells, transiently expressing CD4-DsRed, where endogenous Arf6 protein expression was previously silenced to further confirm the role of Arf6 function during HIV-1 entry (Figure 9). We used fluorescent siRNA-Arf6 oligonucleotides to select cells to be studied by TIRFM for this purpose. We first confirmed that fluorescent siRNA-Arf6 oligonucleotides silenced the expression of the endogenous Arf6 protein (Figure 9A), as did the nonfluorescent oligonucleotides (Figures 5A, 6A, and 10A and Supplemental Figure 4A). The presence of the fluorescent scrambled or siRNA-Arf6 oligonucleotides inside the cells was monitored by epifluorescence (Figure 9, B and C, epifluorescence images) during TIRFM-based CD4-dependent HIV-1 uptake experiments. Our results indicated that endogenous Arf6 knockdown did not affect the first CD4-DsRed/HIV-1–Gag-EGFP interaction (Figure 9, B and C, merged images) but prevented CD4-dependent viral uptake, compared with control and scrambled-treated cells (Figure 9D, 63% and 10.7% of CD4-dependent viral uptake for scrambled and siRNA-Arf6, respectively). Given the observed inhibitory effect of Arf6 knockdown on HIV-1 viral entry and infection (Figures 5 and 6 and Supplemental Figure 4), it is conceivable that siRNA-Arf6 inhibited CD4-dependent productive viral entry, as was observed by TIRFM (Figure 9, B and C, fluorescence intensity curves and time lapse images from zoom area for CD4-DsRed and bound HIV-1–Gag-EGFP, and Supplemental Videos 5 and 6).
FIGURE 9:
Effect of endogenous Arf6 knockdown on HIV-1 entry in TZMbl cells analyzed by TIRFM. (A) Left, Western blot analysis of endogenous Arf6 knockdown, 24 h after fluorescent-siRNA nucleofection of TZMbl cells, quantified as the band intensity ratios to α-tubulin. Fluorescent scrambled oligonucleotides represent the negative control for RNAi. A representative experiment of three is shown. Right, a series of x–y midsection images showing the specific silencing of endogenous Arf6 (green) by fluorescent siRNA-Arf6 oligonucleotides (red). Asterisks indicate cells where Arf6 was not silenced. Merge and merge/DIC images are shown from a representative experiment. (B and C) A series of TIRFM images indicating cell-surface CD4-DsRed, HIV-1–Gag-EGFP virions, and CD4-attached HIV-1–Gag-EGFP virions (merge) at the EF of TZMbl, both in scrambled- or siRNA-Arf6–transfected cells. Fluorescent scrambled or siRNA-Arf6 oligonucleotides are monitored by epifluorescence. White squares in the merged images show a representative area (zoom area) where CD4-dependent HIV-1 uptake or blockade was observed, corresponding to Supplemental Videos 5 and 6, and a time lapse series of images (105 s, zoom area). Bar 5 μm. White open circles, in a time lapse series of images, show representative events for CD4-dependent viral uptake or inhibition, analyzed by time for their fluorescence intensities (right curves). (D) Histograms show the percentage of bound HIV-1–Gag-EGFP particles to CD4-DsRed that entered seven cells (ratio of CD4-dependent viral uptake events to the total number of CD4/HIV-1 interactions analyzed appears above the histograms), under any experimental condition.
Arf6 regulates HIV-1 viral fusion and entry steps of the viral cycle
We performed viral fusion and entry experiments by using R5- and X4-tropic HIV-1 viral particles containing the BlaM-Vpr chimera in Arf6-silenced CD4+ T lymphocytes (Figure 10A) to ascertain the mechanism involved in Arf6-dependent efficient early HIV-1 infection. These chimera virions were designed specifically to study the first steps of viral infection (Cavrois et al., 2002) because β-lactamase activity directly correlates with viral fusion and entry (Cavrois et al., 2002; Barrero-Villar et al., 2008, 2009). Control (scrambled)– or siRNA-Arf6–treated CEM-CCR5 cells were incubated (3 h) with equivalent viral inputs of X4-tropic or R5-tropic virions containing the BlaM-Vpr fusion protein. Concurring with all the results presented earlier in this article and as observed by TIRFM (Figure 9), specific Arf6 silencing significantly inhibited viral fusion and entry, regardless of viral tropism, compared with the more susceptible scrambled-treated cells to viral fusion and entry (Figure 10B). Therefore it seems that Arf6 knockdown interfered with Arf6-mediated plasma membrane dynamics during the virus–cell fusion process.
It has been recently reported that HIV-1 enters cells in mainly a clathrin-dependent, endocytic pathway (Miyauchi et al., 2009). We used BlaM-Vpr virions pseudotyped with the VSV-G envelope protein as a control of the specificity of Arf6 knockdown regulation of HIV-1–induced membrane fusion to explore the Arf6 role on clathrin-dependent viral fusion and entry process. These pseudotyped viruses enter target cells using the clathrin-endocytic lower pH pathway due to the VSV-G envelope (Matlin et al., 1982; Sun et al., 2005). As observed above, VSV-G–mediated viral fusion and entry were independent of Arf6 activity (Figure 10B), thereby suggesting that Arf6 activity did not affect clathrin-mediated vesicle trafficking from the cell surface.
We next analyzed the possibility that Arf6 silencing or Arf6-GTP/GDP cycle mutants may affect the function of CXCR4 and CCR5, the main HIV-1 coreceptors, which may result in viral fusion, entry, and infection inhibition. For this purpose, we first assayed ligand-induced CXCR4 and CCR5 internalization in CD4+ lymphocytes, transiently transfected with control or siRNA-Arf6–pEGFP-N2-RNAi plasmids. Cells transfected with these vectors express both control and siRNA-Arf6 oligonucleotides, for Arf6 knockdown, and the EGFP protein (Figure 10C), which allows us to identify control and Arf6-silenced cells by EGFP-associated fluorescence. Ligand-induced endocytosis of viral coreceptors was then performed and analyzed by flow cytometry in EGFP-positive cells under any experimental condition. Arf6 knockdown by this plasmid did not affect CXCR4 or CCR5 cell–surface expression (Figure 10D), as presented for Arf6-silenced cells (Figure 5B and Supplemental Figure 4B). It is noteworthy that Arf6 silencing did not affect SDF-1α (CXCL12)– or RANTES (regulated on activation, normal T expressed and secreted) (CCL5)–mediated CXCR4 or CCR5 internalization, respectively, compared with control cells (Figure 10D). Similarly, overexpression of Arf6-Q67L– or Arf6-T44N–EGFP mutants (Figure 10E) neither affected cell–surface expression of CXCR4 and CCR5 nor prevented its ligand-induced internalization, compared with WT Arf6–EGFP–transfected cells (Figure 10F). These data were obtained by flow cytometry analysis of EGFP-positive cells expressing Arf6-EGFP constructs. Therefore ligand-induced CXCR4 and CCR5 internalization, a clathrin-dependent endocytic process (Borroni et al., 2010), does not seem to be affected by Arf6 activity (Figure 10, C–F). CXCR4 and CCR5 viral coreceptors appear to be functional under our experimental conditions.
Taken together these results prompted us to suggest that Arf6-coordianted PIP2-associated plasma membrane dynamics are required for efficient HIV-1 fusion and entry events, regardless of the viral tropism and without affecting CD4 expression and trafficking, HIV-1/CD4 interaction, and CXCR4 or CCR5 function. Arf6 activity does not seem to be involved in VSV-G–mediated fusion, entry, and infection or ligand-mediated viral coreceptor endocytosis; both are clathrin-dependent processes.
Arf6-dependent and clathrin/transferrin-dependent endocytic pathways have been reported to coexist as two separately trafficking routes from plasma membrane in HeLa cells and other cell types (Radhakrishna and Donaldson, 1997; Nichols and Lippincott-Schwartz, 2001; Donaldson, 2003; Naslavsky et al., 2003; Donaldson et al., 2009). We observed, using TIRFM, the distribution of clathrin-coated structures (CCS; i.e., clathrin-coated pits [CCP] and vesicles [CCV]) at the plasma membrane, under any experimental condition and as described in Barroso-Gonzalez et al. (2009b). It appears that neither CCP nor CCV formation at the plasma membrane was affected by an alteration of Arf6 activity, after overexpressing each Arf6 mutant, when compared with control conditions (Supplemental Figure 5, CCS-mLCa-DsRed images). In addition, we observed that each Arf6 mutant colocalized with and provoked the accumulation of PIP2-associated structures at the plasma membrane in nonclathrin plasma membrane regions (Supplemental Figure 5, EGFP, PH-ECFP, and merged images). Therefore it seems that Arf6 did not alter CCS at the plasma membrane; VSV-G-mediated virus fusion, entry, and infection; and ligand-induced endocytosis of HIV-1 main coreceptors.
Taking all the presented results together, we propose that efficient HIV-1 fusion, entry, and infection require Arf6-coordinated PIP2-associated membrane dynamics. This process depends on Arf6-GTP/GDP activity to promote viral fusion (i.e., pore fusion formation) at the cell surface of target cells and appears to be clathrin independent.
DISCUSSION
We have shown here that early HIV-1 infection of permissive CD4+ T lymphocytes was impaired by the modification of Arf6-dependent membrane dynamics, either by specific RNAi or by overexpressing GTP-bound or GDP-bound inactive mutants of Arf6. Arf6-Q67L and Arf6-T44N mutants induced the accumulation of PIP2-associated structures at the plasma membrane of permissive cells, as monitored by the PH-ECFP probe. This fact is indicative of a perturbed PIP2-associated membrane trafficking from the cell surface. Arf6-Q67L, Arf6-T44N, Arf6-T27N, and Arf6-N48I/Q67L mutants inhibited early viral infection, indicating that the Arf6-GTP/GDP cycle regulates efficient HIV-1 entry and infection.
Alteration of the GTPase cycle of Arf6 had no effect on the cell-surface level of expression and function of CD4, CCR5, and CXCR4 receptors for HIV-1. TIRFM studies for CD4-dependent HIV-1 uptake, in nonlymphoid permissive HeLa-derived cells expressing CD4, CCR5, and CXCR4 viral receptors, showed that the first virus/CD4 interactions are not affected by the inhibition of Arf6-coordinated plasma membrane dynamics. Arf6 mutants or specific Arf6 silencing inhibits viral entry and infection independently of viral tropism, suggesting that viral receptors are not affected or negatively involved in this process. Free or virus-bound CD4 does not distribute or localize with Arf6 constructs, suggesting that CD4 neither traffics from nor recycles to plasma membrane Arf6 dependently. Arf6 mutants or silencing do not appear to affect ligand-induced CXCR4 or CCR5 endocytosis, a clathrin-dependent process (Borroni et al., 2010).
Regulation of early HIV-1 infection by Arf6 activity seems to be related to fusion and entry steps of the viral cycle. We observed that either Arf6 mutants or specific Arf6 silencing inhibited HIV-1 Env–mediated membrane fusion, viral entry, and infection regardless of viral tropism. We observed, by TIRFM, that CD4-DsRed–preattached HIV-1–Gag-EGFP viral input internalized together with the associated cell-surface CD4 receptor in cells transfected by WT Arf6–HA. In fact, their respective intensity of fluorescence dimmed in the EF, representing CD4-dependent viral uptake. On the contrary, PIP2-associated plasma membrane structures accumulated in cells overexpressing each Arf6-HA mutant, rendering them refractory to CD4-dependent HIV-1 uptake. Comparable inhibition was observed in cells transfected by WT Arf6–HA pretreated by T-20, which prevents CD4-dependent HIV-1 fusion and productive entry. Transiently expressed CD4-DsRed attached HIV-1–Gag-EGFP virions under any experimental condition. This fact indicates that Arf6-mediated plasma membrane trafficking is required for efficient CD4-dependent HIV-1 entry, without affecting CD4/virus interaction. Blockade of Arf6-GTP/GDP activity by siRNA silencing inhibits efficient X4- or R5-tropic HIV-1 fusion and entry (i.e., pore fusion formation) of viruses containing the BlaM-Vpr chimera in CD4+ lymphocytes, without affecting ligand-induced CXCR4 or CCR5 endocytosis, and VSV-G-mediated fusion, entry, and infection, both clathrin-dependent processes. Therefore Arf6 knockdown appears to interfere with Arf6-mediated plasma membrane dynamics.
In general, viruses, including HIV, can disseminate within an infected host by cell-free viruses and via direct cell-to-cell transmission (Phillips, 1994; Johnson and Huber, 2002; McDonald et al., 2003; Gousset et al., 2008). The relative contribution of these modes of HIV-1 dissemination in vivo is not well established, but cell-to-cell spread probably occurs mainly in tissues densely populated with target cells, such as CD4+ T-cells in lymph nodes (Haase, 1999; McDonald et al., 2003). Our results indicate that efficient cell–HIV-1 transmission and infection of human primary CD4+ T-cells requires Arf6 activity in target lymphocytes because it is inhibited by Arf6-GTPase silencing independently of viral tropism. These data agree with Arf6 silencing–mediated inhibition of HIV-1 fusion, entry, and infection with luciferase- and BlaM-Vpr–bearing virions. BlaM-Vpr–based experiments are sensitive and specific to the detection of viral entry due to the quantitative measurement of the incorporated BlaM-Vpr chimeric virions present at cytoplasm after viral fusion (Cavrois et al., 2002), avoiding interference with viruses captured in vesicular compartments, which may account for 60–90% of total viral uptake (Marechal et al., 1998, 2001). Cellular signaling in target CD4+ T-cells, such as cell-surface receptors and actin cytoskeleton reorganization, occurred equally during early HIV-1 infection, both in cell-to-cell transmission (i.e., the virological synapse) and free HIV-1 particle models (Jolly et al., 2004; Jolly and Sattentau, 2004; Jimenez-Baranda et al., 2007; Yoder et al., 2008; Barrero-Villar et al., 2009; Liu et al., 2009). This finding is consistent with the fact that, at the virological synapse, HIV first buds from the infected donor cell and then binds to and fuses with the recipient cell, as in a free virus system (Blanco et al., 2004; Puigdomenech et al., 2009). Our results, obtained by using these two working models, indicate that Arf6-mediated membrane dynamics are required for efficient cell-viral transmission, HIV-1 fusion, entry, and infection of CD4+ T lymphocytes.
There are divergent data regarding the functional implication of Arf6 activity in different aspects of HIV-1 cell biology. Thus HIV-1 infection in polarized trophoblasts, which are thought to play a determinant role in HIV-1 transmission in utero, seems to rely on Rab5 and Rab7 without the contribution of Arf6 or Rab11. This process is an unusual clathrin/caveolae-independent endocytic pathway, independent of the HIV-1 envelope complex (Vidricaire and Tremblay, 2005, 2007). HIV-1 eludes its specific immune recognition by down-regulating the expression of MHC-I at the cell surface of infected cells (Schwartz et al., 1996). Arf6-activity, using the Arf6-Q67L mutant, is involved in this Nef-induced MHC-I internalization process (Blagoveshchenskaya et al., 2002). However, other authors indicate that Arf6 activity is not involved in Nef-mediated down-modulation of MHC-I (Larsen et al., 2004). Overexpression of WT Arf6 did not enhance or substitute the effects of Nef on MHC-I internalization (Blagoveshchenskaya et al., 2002; Larsen et al., 2004). We have not observed any functional effect for WT Arf6, EFA6, or WT Arf6/EFA6 overexpression on HIV-1 fusion, entry, and infection in permissive cells, indicating that Arf6-dependent signaling is not involved in this process. This lack of activity for full-length Arf6 has been described in several cellular processes, in which Arf6 implication was determined by using its different mutants (Blagoveshchenskaya et al., 2002; Donaldson, 2003; Larsen et al., 2004). Although Arf6 activates PLD and PI4P5-K effectors (Brown et al., 2001; Vitale et al., 2002; D’Souza-Schorey and Chavrier, 2006; Gillingham and Munro, 2007), and considering that overexpression of PI4P5-K Iα enhances HIV-1 viral fusion and infection (Barrero-Villar et al., 2008), our results with Arf6/EFA6, single Arf6-GTP/GDP cycle mutants, and the Arf6-N48I/Q67L mutant, which bears the Q67L mutation and fails to activate PLD, suggest that WT Arf6/EFA6 overexpression does not activate PI4P5-K Iα and that PLD activity is not directly involved in the regulation of viral fusion and entry.
It has been reported that HIV-1 fuses with and enters cells via endocytosis in a dynamin- and clathrin-dependent manner in HeLa permissive cells (Miyauchi et al., 2009). Arf6- and clathrin/transferrin-dependent endocytic pathways have been reported to coexist as two separately trafficking routes from plasma membrane in HeLa and other cell types (Radhakrishna and Donaldson, 1997; Nichols and Lippincott-Schwartz, 2001; Donaldson, 2003, Naslavsky et al., 2003; Donaldson et al., 2009). Our TIRFM studies indicate that Arf6 inhibition did not affect CCS formation at plasma membrane. Moreover, Arf6 mutants or specific Arf6 silencing did not inhibit cell infection by HIV-1 vectors pseudotyped with the VSV-G protein. Considering all the exposed data and discussion, we propose that Arf6-GTP/GDP activity has synergy with the key first HIV-1/receptors interactions by maintaining PIP2-associated membrane dynamics to promote efficient viral fusion and entry in a clathrin-independent manner.
Therefore efficient early HIV-1 infection of permissive CD4+ T lymphocytes requires Arf6-coordinated plasma membrane dynamics.
MATERIALS AND METHODS
Antibodies and reagents
The monoclonal antibody (mAb) RPA-T4 (clone) is directed against CD4, and the CD184 (clone 12G5) and CD195 (clone 2D7/CCR5), used as phycoerythrin (PE) conjugates (BD Bioscience/BD PharMingen, San Jose, CA), are directed against the second extracellular loop of CXCR4 or CCR5, respectively. The mAb L3T4 is a neutralizing antibody against CD4 (eBioscience, San Diego, CA). The anti-CD4 mAb Leu3a was from Becton Dickinson (Franklin Lakes, NJ). C34 is a fusion inhibitor covering the 628–661 amino-acid sequence of gp41 viral protein, similar to T-1249 (residues 628–663) and to T-20 (residues 638–673). Mouse mAb (8A6-4) against Arf6 (Londono et al., 1999) was kindly provided by Sylvain Bourgoin (Center de Recherche du CHUQ, pavillon CHUL, Rheumatologie et Immunologie, Ste-Foy, QC, Canada). The mAb recognizing Arf6 (ARF6 [3A-1]: sc-7971), anti-EGFP rabbit polyclonal antibody (pAb) (sc-8334), anti-HA mAb (sc-7392), anti–HLA-A/B/C mAb (3F10; sc-65288), and PE-conjugated goat anti-mouse immunoglobulin (Ig) G (sc-3738) came from Santa Cruz Biotechnology (Santa Cruz, CA). Anti–VSV-G tag (ab3861) goat pAb to detect vsv-g-EFA6 was from Abcam (Cambridge, UK). Anti–α-tubulin mAb was from Sigma-Aldrich (St. Louis, MO). Secondary horseradish peroxidase–conjugated anti-mAb was from ImmunoTools (Friesoythe, Germany), and secondary horseradish peroxidase–conjugated anti–rabbit and anti–goat antibody were from Dako (Glostrup, Denmark). Alexa Fluor 633 phalloidin and Alexa Fluor 568–goat anti-mouse were from Molecular Probes (Eugene, OR). Stromal cell–derived factor (SDF) 1α (CXCL12) was kindly synthesized and provided by Françoise Baleux (Institut Pasteur, Paris, France) (Valenzuela-Fernandez et al., 2001, 2002). RANTES (CCL5) was from R&D Systems (Minneapolis, MN).
Cells
The human CEM.NKR-CCR5 (CEM-CCR5) permissive cell line (catalogue no. 4376, NIH AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH]) was grown at 37°C in a humidified atmosphere with 5% CO2 in RPMI 1640 medium (Lonza, Verviers, Belgium) supplemented with 10% fetal calf serum (FCS) (Lonza), 1% l-glutamine, and 1% penicillin-streptomycin antibiotics. Cells were regularly passaged every 3 d. The 293T cell line was similarly cultured in supplemented DMEM (Lonza) and was regularly passaged every 2–3 d. Cells were harvested and resuspended at a density of 50–70% in fresh supplemented DMEM 24 h before cell transfection with viral DNA constructs. HeLa-P5 cells, stably transfected with human CD4 and C-terminal EGFP-tagged CCR5 cDNAs and with an HIV-LTR–driven β-galactosidase (β-Gal) reporter gene, were provided by M. Alizon (Hôpital Cochin, Paris, France) (Valenzuela-Fernandez et al., 2005; Barrero-Villar et al., 2009; Barroso-Gonzalez et al., 2009a). TZMbl (HeLa) cells are similar to HeLa-P5 cells but express nonfluorescent human CCR5 and contain integrated HIV-LTR–driven β-Gal and firefly luciferase reporter genes (TZMbl, catalogue no. 8129, NIH AIDS Research and Reference Reagent Program). HeLa 243 and HeLa ADA cells coexpressing the Tat and HIV-1 Env proteins were also provided by M. Alizon (Pleskoff et al., 1997; Valenzuela-Fernandez et al., 2005; Barrero-Villar et al., 2009; Barroso-Gonzalez et al., 2009a). Peripheral blood mononuclear cells were obtained by Ficoll density gradient of blood cells provided by the local blood bank and immediately used to purify CD4 T-cells (>95%) by immunomagnetic negative selection (Miltenyi Biotec, Bergisch Gladbach, Germany). MOLT-4/CCR5 (MOLT/CCR5) cells, which are highly permissive for R5-tropic HIV-1 infection either uninfected or chronically infected with the HIV-1NL4–3 and HIV-1BaL isolates (>90% producing HIV-1 particles) have been previously described (Blanco et al., 2004; Puigdomenech et al., 2008). Cells were maintained in RPMI 1640 (Life Technologies-BRL, Grand Island, NY) supplemented with 10% FCS (Life Technologies) and used without stimulation.
Human DNA constructs
Human cDNAs of the C-terminal HA-tagged WT Arf6–, Arf6-Q67L–, and Arf6-T27N–HA were kindly provided by Julie Donaldson (Laboratory of Cell Biology, National Heart, Lung, and Blood Institute, Bethesda, MD). Arf6-T44N–HA mutant and vsv-g–tagged EFA6 factor were gifts from Michel Franco (Institut de Pharmacologie Moléculaire et Cellulaire, Centre National de la Recherche Scientifique [CNRS] Unité Mixte de Recherche 6097, Valbonne Sophia-Antipolis, France). Arf6-N48I/Q67L–HA, control- and siRNA-Arf6–pEGFP-N2-RNAi plasmids were gifts from Nicolas Vitale (Institut des Neurosciences Cellulaires et Intégratives, CNRS, Strasbourg, France) (Vitale et al., 2002; Begle et al., 2009). Arf6-EGFP or Arf6-ECFP constructs were generated by PCR using each Arf6-HA construct as template and CGCTCGAGGCCACCATGGGGAAGGTGCTATC (sense) and CGGAATTCCAGATTTGTAGTTAGAGG (antisense) as 5′ and 3′ primers, respectively. The amplified products were respectively subcloned into pEGFP-N1 or pECFP-N1 (Clontech, Palo Alto, CA) after restriction with XhoI and EcoRI. For CD4-DsRed expression, total RNA from CEM.NKR-CCR5 T-cells was extracted, and first-strand cDNA was made using reverse transcriptase and random hexamers as primers. Thus the CD4 construct lacking the stop codon was generated by Expand High Fidelity PCR using cDNA as template and CTTCGAATTCGCCACCATGAACCGGGGAGTCCCTTTTAGGC (sense) and GCGGGGTACCACAATGGGGCTACATGTCTTCTGAAACC (antisense) as 5′ and 3′ primers, respectively. The amplified product was subcloned into pDsRed-NI (Clontech) after restriction with EcoRI and KpnI restriction enzymes.
The C-terminal ECFP-tagged PH domain of the PLCδ1 (PH-ECFP) was provided by Senena Corbalán-García (Universidad de Murcia, Spain) and was used as a PIP2 biosensor in the plasma membrane as described (Marin-Vicente et al., 2005; Barrero-Villar et al., 2008; Barroso-Gonzalez et al., 2009b). mLCa-DsRed was provided by Wolfhard Almers (Vollum Institute, Oregon Health and Science University, Portland, OR) and was used as described (Barroso-Gonzalez et al., 2009b). All constructs were verified by digestion with restriction enzymes.
Messenger RNA silencing
We have designed the following specific short interference oligonucleotides against the following mRNA (siRNA) sequence of Arf6: siRNA-Arf6 1 (position 699–622; sense: 5′-gacaacaauccuguacaag-3′-dTdT; antisense: 5′-guuguacaggauuguuguc-3′dTdT) and siRNA-Arf6 2 (position 930–622; sense: 5′-gcaccgcauuaucaaugaccg-3′-dTdT; antisense: 5′-cggugauugauaaugcggugc-3′-dTdT). We used a mix of these two oligonucleotides to induce specific siRNA-mediated silencing of the endogenous expression of the Arf6 protein, which is called siRNA-Arf6 in the present work. We have also designed the following siRNA oligonucleotides against human CD4 mRNA: siRNA-CD4 (position 769–787: sense: 5′-gaacaaggaagugucugua-3′-dTdT; antisense: 5′-uacagacacuuccuuguuc-3′-dTdT). Alexa 546–conjugated or nonfluorescence siRNA oligonucleotides, irrelevant scrambled siRNA (control), or siRNA-Arf6 oligonucleotides were from Sigma-Aldrich. cDNA fragments, kindly provided by Nicolas Vitale (Strasbourg, France), encoding 19-nucleotide siRNA sequence (GCTGCACCGCATTATCAAT) derived from the target transcript and separated from its reverse 19-nucleotide complement by a short spacer, were annealed and cloned in the BglII and HindIII sites in front of the H1 promoter of the pEGFP-N2-RNAi plasmid, as previously described (Begle et al., 2009) for siRNA targeting of human Arf6 by plasmids. Cells treated with control or siRNA-Arf6–pEGFP-N2-RNAi plasmid express free EGFP. The siRNAs for Arf6 and CD4 sustained specific interference of protein expression for at least 72 h, as monitored by Western blot (unpublished data).
Cellular transfection
All human permissive HeLa cell lines were grown at 37°C in a humidified atmosphere with 5% CO2 in DMEM (Lonza) supplemented with 10% FCS (Lonza), 1% l-glutamine, and 1% penicillin-streptomycin antibiotics. Cells were harvested and resuspended at a density of 50–70% in fresh supplemented DMEM 24 h before cell transfection with siRNA and/or DNA constructs. Specific Amaxa kits (Amaxa, Köln, Germany) were used for delivery of DNA constructs and/or siRNA into HeLa cells. Similarly, permissive T lymphocytes and human primary CD4+ T-cells were nucleofected according to the manufacturer’s instructions (Amaxa), as previously described (Barrero-Villar et al., 2008, 2009; Barroso-Gonzalez et al., 2009a), with 1 μM siRNA and/or 1 μg of each DNA construct used and assayed no more than 24 or 48 h later.
Western blotting
The extent of protein expression or gene silencing was assessed by Western blot of cell lysates. Cells nucleofected with scrambled or specific siRNA oligonucleotides against Arf6 or CD4, or with the different DNA constructs, were lysed 24 h later for 30 min at 4°C in 1% Triton X-100, 50 mM Tris HCl, and 150 mM NaCl with a protease and phosphatase inhibitor mixture (Roche Diagnostics, Mannheim, Germany) and centrifuged at 4ºC, 13,000 rpm, for 15 min. Equivalent amounts of proteins, measured using the bicinchoninic acid method (BCA protein assay kit from Pierce, Rockford, IL), were separated by SDS–PAGE using 12% gradient gels and electroblotted onto 0.45 μm polyvinylidene difluoride membranes (Millipore Corporation, Billerica, MA). Cell lysates were immunoblotted with specific antibodies, protein bands were detected by luminescence using an ECL System (Pierce), and protein bands were analyzed using a VersaDoc device and Quantity One 4.6.7 software (Bio-Rad, Hercules, CA).
Viral DNA constructs
The pNL4-3.Luc.R-E- provirus (catalogue no. 6070013), HXB2-env (catalogue no. 5040154), and pCAGGSSF162-gp160-env (catalogue no. 3041817) glycoprotein vectors for X4- and R5-tropic viral envelopes, and the pGag-EGFP (catalogue no. 11468; from Marilyn Resh) vector, allowing imaging of intracellular Gag trafficking and localization in live cells, which directs Rev-independent expression of an HIV-1–Gag-EGFP fusion protein (Schwartz et al., 1992), were obtained through the NIH AIDS Research and Reference Reagent Program. The pCMV-BlaM-Vpr vector was kindly provided by Warner C. Greene (University of California, San Francisco).
Production of viral particles
Replication-deficient luciferase–HIV-1 viral particles were obtained as previously described (Barrero-Villar et al., 2008, 2009; Barroso-Gonzalez et al., 2009a). Briefly, replication-deficient viral particles were derived by the luciferase-expressing reporter virus HIV/Δnef/Δenv/luc+ (which bears the luciferase gene inserted into the nef ORF and does not express envelope glycoprotein) with an X4-tropic (Lai) or R5-tropic (SF162) envelope glycoprotein. X4- or R5-tropic HIV-1 viral particles were produced by cotransfecting 293T cells (70% of confluence) in 10-cm2 dishes with pNL4-3.Luc.R-E- (10 μg) and X4-tropic (HXB2-env) or R5-tropic (pCAGGS SF162 gp160) envelope glycoprotein (10 μg) vector, as previously described (Barrero-Villar et al., 2008, 2009; Barroso-Gonzalez et al., 2009a). Viral plasmids were transduced in 293T cells by using linear polyethylenimine, with an average molecular mass of 25 kDa (PEI25k) (Polyscience, Warrington, PA). Viral plasmids were first dissolved in 1/20th of the final tissue culture volume of DMEM in 150 mM NaCl. The PEI25k was prepared as a 1 mg/ml solution in water and adjusted to neutral pH. After the addition of PEI25k, previously dissolved in 150 mM NaCl, to the viral plasmids (at a plasmids:PEI25k ratio of 1:3 [wt/wt]), the solution was mixed by vortexing, incubated for 20–30 min at room temperature, and then added to 293T cells in culture. After 6 h the medium was changed to RPMI 1640 and supplemented with 10% FCS and antibiotics, and the cells were cultivated for 48 h to allow viral production. Viruses were harvested 48 h posttransfection. The supernatant was clarified by centrifugation at 3000 × g for 30 min, filtered by 0.45 μm, and concentered by Amicon Ultra-4 Centrifugal filter devices (Millipore). Virions were then stored at −80°C. Viral stocks were normalized by p24-Gag content measured with an enzyme-linked immunosorbent assay test (Innogenetics, Gent, Belgium). HIV-1–Gag-EGFP virions were similarly obtained after cotransduction of the pGag-EGFP vector (5 μg) with Env vectors (10 μg). Therefore we used this pGag-EGFP vector to produce fluorescent viruslike particles with an efficiency equivalent to that of Gag (Schwartz et al., 1992), which allowed us to image HIV-1 entry by TIRFM. Nonreplicative viral particles, containing the BlaM-Vpr chimera, were similarly produced after cotransduction of the pCMV-BlaM-Vpr vector (5 μg) with pNL4-3.Luc.R-E- (10 μg) and Env vectors (10 μg), as described (Barrero-Villar et al., 2008, 2009).
Luciferase viral entry and infection assay
Untreated or nucleofected CEM-CCR5 cells (100,000 cells in 96-well plates) and untreated or nucleofected HeLa-P5 cells (20,000 cells in 96-well plates) were infected with a synchronous dose of luciferase-based X4- or R5-tropic HIV-1 viral inputs (500 ng of p24), in 200 μl RPMI 1640 medium for 2 h, as described (Barrero-Villar et al., 2008, 2009; Barroso-Gonzalez et al., 2009a). Virus was removed by washing infected cells. After 48 h of infection, luciferase activity was determined by using a luciferase assay kit (Biotium, Hayward, CA) with a microplate reader (VictorTM X5, PerkinElmer, Waltham, MA). When indicated T-20 (5 μM) was used as a control for the blockage of viral–cell fusion, preincubated in cells for 30 min at 37°C before infection. Data were analyzed using GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA).
Virion-based fusion assay
The 0.5 × 106 CEM-CCR5 permissive cells (scrambled or siRNA-Arf6 treated) were incubated for 3 h with equivalent viral inputs of BlaM-Vpr–containing virions (500 ng p24) in 500 μl RPMI-1640 medium. Cells were then extensively washed to remove free virions and incubated (1 h, room temperature) with CCF2-AM loading mix, as recommended by the manufacturer (GeneBLAzer detection kit; Invitrogen, Carlsbad, CA), as previously described (Barrero-Villar et al., 2008, 2009). Next excess dye was washed off and cells were incubated for 16 h at room temperature before fixation with 1.2% paraformaldehyde. The percentages of CCF2-loaded target-infected cells, fused with BlaM-Vpr–containing virions, were determined by measuring the fluorescence intensities of the intact and cleaved CCF2 probe in a fluorescence spectrophotometer (Cary Eclipse, Varian, Melbourne, Australia), as described (Barrero-Villar et al., 2009). Thus the percentage of 100% of infection was determined by measuring the fluorescence intensities of the intact and cleaved CCF2 probe in control infected cells (scrambled treated cells) and subtracting the background blue and green fluorescence ratio determined in noninfected cells (without β-lactamase activity), as proposed by the manufacturer (GeneBLAzer detection kit, Invitrogen). In siRNA-Arf6–treated cells, a decrease in the ratio of blue (447 nm; cleaved CCF2) to green (520 nm; intact CCF2) fluorescence signals compared with control (scrambled) cells indicates fewer virions fused to target cells. The background blue and green fluorescence was determined in noninfected CCF2-loaded cells (without β-lactamase activity), under any experimental condition.
HIV-1 Env–mediated cell-to-cell fusion assay
A β-Gal cell fusion assay was performed as previously described (Barrero-Villar et al., 2008, 2009; Barroso-Gonzalez et al., 2009a). Briefly, HeLa-243 or HeLa-ADA cells were mixed with HeLa-P5 cells or control, or previously transfected with Arf6 constructs, in 96-well plates in a 1:1 ratio (20,000 total cells). These cocultures were kept at fusion for 16 h at 37°C. The fused cells were washed with Hanks’ balanced salt solution and lysed, and the enzymatic activity was evaluated by chemiluminescence (β-Gal reporter gene assay, Roche Diagnostics). Anti-CD4 neutralizing mAb L3T4 was used as a control for the blockage of cell fusion (5 μg/ml was preincubated in HeLa-P5 cells for 30 min at 37°C before coculture with Env+-HeLa cells).
Cocultures of HIV-1–infected MOLT/CCR5 cells with primary CD4+ T-cells
Chronically infected MOLT/CCR5 cells (500,000 cells/well) were preincubated in 96-well plates, either alone (control) or with the fusion inhibitor peptide C34 (5 μg/ml) for 1 h at 37ºC, before adding 500,000 primary nonactivated, scrambled-, or siRNA-Arf6–treated CD4+ T-cells. C34 inhibitor concentrations were selected on the basis of complete inhibition of cell-to-cell HIV-1 transmission and infection. DNA was extracted after 24 h using the QIAmp DNA Blood Mini Kit (Qiagen, Hilden, Germany) and amplified in duplicate using a TaqMan universal PCR Master Mix, primers and probes for HIV-1 LTR, and human CCR5 genes (Applied Biosystems, Foster City, CA). Reactions (40 cycles of 15 s at 95ºC for melting and 1 min at 60ºC for annealing/extending) were conducted in an ABI 7000 Sequence Detection System (Applied Biosystems). HIV-1 infection was quantified by qPCR in which the relative proviral DNA synthesis was calculated using a standard curve generated with a plasmid that harbors the sequence of the HIV-1 LTR and the CCR5 gene. Copy numbers obtained for each sample were extrapolated to the standard equation as described (Buzon et al., 2010). The concentrations obtained for LTR amplifications were then normalized to the CCR5 gene copy number as a measure of the number of cells present in each sample. The level of HIV-1 DNA into MOLT-infected cells was calculated from C34-containing cocultures.
Flow cytometry analysis
Permissive cells (CEM.NKR-CCR5 cells, primary CD4+ lymphocytes, or phosphate-buffered saline [PBS]/EDTA–detached permissive HeLa cells) were incubated with PE-labeled specific antibodies against HLA-A/B/C (previously incubated with a specific primary mAb) and CD4, CXCR4, or CCR5 in control cells or cells transfected with Arf6 constructs or siRNA oligonucleotides. Cells were then washed by ice-cold PBS, fixed in PBS with 1% paraformaldehyde, and analyzed by flow cytometry (XL-MCL system; Beckman-Coulter, CA), as described (Valenzuela-Fernandez et al., 2005; Barrero-Villar et al., 2008, 2009; Barroso-Gonzalez et al., 2009a).
Ligand-induced CXCR4 and CCR5 endocytosis
CEM-CCR5 cells (0.5 × 106 cells/ml) were incubated with 200 nM of SDF-1α or RANTES for 1 h at 37°C in free-serum RPMI 1640 medium, as described (Valenzuela-Fernandez et al., 2002; Barroso-Gonzalez et al., 2009a). Cells were then incubated for 3 min at 4ºC in an acidic buffer (50 mM glycine, pH 3) that stops receptor endocytosis and removes CXCR4-bound SDF-1α or CCR5-bound RANTES molecules that would mask cell-surface receptors recognition by specific mAbs. Cells were washed twice with ice-cold PBS and 0.1% bovine serum albumin (BSA), before incubation with specific anti-receptor PE-conjugated mAbs (1:50) for 1 h at 4°C, and then fixed in 1% paraformaldehyde in PBS. Samples were analyzed on a flow cytometer (XL-MCL system) by measuring cell-surface receptor labeling on EGFP+ cells, which express either Arf6-EGFP constructs or control- or siRNA-Arf6–pEGFP-N2-RNAi plasmids. Basal cell fluorescence intensity for cell-surface CXCR4 and CCR5 was determined using cells stained with a PE-conjugated IgG2a isotype control alone.
Immunofluorescence
Immunofluorescent lymphocytes or permissive HeLa cells (grown on glass coverslips, ∼50% confluent) were washed three times with PBS and fixed for 20 min in 2% paraformaldehyde with 1% sucrose in PBS. Cells were washed three times with PBS after fixation and, when indicated, permeabilized with 0.1% Triton X-100 in PBS. The cells were then washed with PBS after permeabilization and immunostained for 1 h at room temperature by Alexa 633–labeled phalloidin or Alexa 568–labeled goat anti–mouse against HLA-A/B/C previously incubated with a specific mAb and diluted in PBS with 0.1% BSA. For Arf6 silencing, cells were nucleofected with fluorescent siRNA-Arf6 oligonucleotides and analyzed for endogenous Arf6 knockdown by using specific primary antibody against Arf6 before incubation with Alexa 488 secondary antibody species specific, at 24 h posttransfection in permeabilized cells, as described earlier in this article. Coverslips were mounted in Mowiol-antifade (Dako) and imaged in x–y midsections in a FluoView FV1000 confocal microscope through a 1.35 numerical aperture (NA) objective (60×) (Olympus, Center Valley, PA) for high-resolution imaging of fixed cells. The final images and molecule codistributions were analyzed and quantified with MetaMorph software (Universal Imaging, Downington, PA).
Total internal reflection fluorescence microscopy and analysis of HIV-1 entry
We performed total internal reflection fluorescence microscopy (TIRFM) experiments in adherent, nonlymphoid permissive HeLa-derived cells stably expressing CD4, CCR5, and CXCR4 viral receptors, owing to the intrinsic difficulties of the EF microscopy approach in lymphoid cells. Permissive TZMbl cells, transiently expressing the different Arf6-HA or Arf6-EGFP constructs or fluorescent siRNA oligonucleotides (λEm 680 nm) together with PH-ECFP and CD4-DsRed molecules, or Arf6-ECFP constructs with CD4-DsRed, were imaged with an inverted microscope Zeiss 200M (Zeiss, Jena, Germany) through a 1.45 NA objective (αFluar, 100×, Zeiss), as described (Barroso-Gonzalez et al., 2009a, 2009b). Experiments were performed and imaged with cells in a Krebs-HEPES buffer containing 2 mM Ca2+ at 37°C. After transfection, TZMbl cells (1 × 105 cells/well) were grown overnight on coverslips placed on 12-well plates. Synchronous doses (multiplicity of infection 1.0) of nonreplicative HIV-1–Gag-EGFP virions were preincubated with permissive cells for 30 min at 4ºC to allow virus/CD4 binding and to prevent viral uptake. Cells were then washed with cold Krebs-HEPES buffer to remove unbound viral particles, and CD4-dependent HIV-1 uptake was studied at 37ºC by TIRFM. When T-20 (5 μM) was used, the anti-fusogenic peptide was added to all buffers and steps during all processes. The objective was coupled to the coverslip using an immersion fluid (n488 = 1.518, Zeiss). The expanded beam of an argon ion laser (LASOS Lasertechnik, Jena, Germany) was band-pass filtered and used to selectively excite different fluorescent proteins. The excitation light (around 488 nm) was directed to the objective by a dichroic mirror (500 nm) for EF illumination, and this was used to excite both the green fluorescent HIV-1–Gag-EGFP virions and the CD4-DsRed molecules at the plasma membrane. Green and red fluorescence was viewed through a DV2 image system (Photometrics, Tucson, AZ) that split red and green components of the image with a 565-nm dichroic mirror (565DCRX) and filtered them through D630/50-nm and D520/30 band-pass filters, respectively. The images were then projected side by side onto an EM-CCD digital camera (C9100-13, Hamamatsu Photonics, Hamamatsu City, Japan). The beam was focused on an off-axis position in the back focal plane of the objective. Light, after entering the coverslip, underwent total internal reflection as it struck the interface between the glass and the solution or cell at a glancing angle. Total internal reflection generates an EF that declines exponentially with increasing distance from the interface, depending on the angle at which light strikes the interface. The angle was measured using a hemicylinder, as described previously (Barroso-Gonzalez et al., 2009a, 2009b). Each cell was imaged using HC Image acquisition software (Hamamatsu Photonics) for up to 3 min with 0.25-s exposures at 1 Hz when illuminated under the EF. At the beginning of each cell acquisition, we took epifluorescence images using a Hg lamp that were projected onto a back-illuminated CCD camera (AxioCam MRm, Zeiss) through a dichroic and specific band-pass filter for fluorescent siRNA oligonucleotides. The splitter DV2 system was aligned in each recording session, and to do this we took an alignment image showing densely scattered 0.2-μm fluorescein isothiocyanate–conjugated beads (Molecular Probes). They were visible in both the green and red channels and thus provided markers in the x–y plane.
To determinate the overlap between HIV-1–Gag-EGFP and CD4-DsRed, EF images were taken. The EF images were background subtracted using MetaMorph. We plotted a small circle of 0.9-μm diameter and a big circle of 1.8-μm diameter around each structure expressing HIV-1–Gag-EGFP. The big circle was used to calculate the local background. The circles in the green image were transferred to the red image. Colocalization was scored positive when the fluorescence in the green channel was at least 10% of the local background. To study the entry of HIV-1–Gag-EGFP, we used only the ones that overlapped with CD4-DsRed and seemed likely to represent a single structure. Those structures that were >0.5 μm or showed extensive lateral movement were excluded from the analysis. To analyze entry events, EF stacks were searched for candidate HIV-1–Gag-EGFP spots that disappeared and seemed likely to represent single CD4-DsRed in that they were circular and of diffraction-limited size, as described (Merrifield et al., 2002; Barroso-Gonzalez et al., 2009b). A circle of 0.9-μm diameter was drawn around the center of each region, both in the red and green channels, and the average fluorescence intensity therein was calculated. The local background was the average fluorescence outside the region after excluding small regions where the intensity exceeded a threshold set by the user, one each for red and green channels. Time lapses showing CD4-DsRed–dependent HIV-1–Gag-EGFP entry or blockade were recorded under these technical conditions. Representative videos, shown in Supplemental Videos 1–6, were rated to 7 s, containing 15 frames/s. Total elapsed time is 105 s from the original stack.
Statistics
Data were compared using Student’s t test. One and two asterisks indicate p < 0.05 and p < 0.01, respectively.
Supplementary Material
Acknowledgments
This work and A.V.-F. are supported by SAF2008-01729 (Ministerio de Ciencia e Innovación, Spain), the European Regional Development Fund, 24661/07 and 24-0740-09 (Fundación para la Investigación y la Prevención del SIDA en España [FIPSE]), the HIVACAT Program, the FIS project PI08/1306, and the Spanish AIDS network (RD06/0006). L.G.-E. and J.B.-G. are supported by SAF2008-01729– and FIPSE-24-0740-09–associated fellowships, respectively. J.D.M. is supported by CONSOLIDER grant CSD-000005 and PI2007/017 (Gobierno de Canarias). J.B. is supported by the Instituto de Salud Carlos III and the Health Department (Generalitat de Catalunya). I.P. is supported by a predoctoral grant (Generalitat de Catalunya) and the European Social Fund. We thank the National Institutes of Health AIDS Research and Reference Reagent Program for providing HIV-1 plasmids and cells. The authors also thank María del Valle Croissier-Elías for critical reading of the manuscript and Patrick Dennis for his help in revising and editing the English content of the manuscript.
Abbreviations used:
- Arf6
ADP ribosylation factor 6
- BSA
bovine serum albumin
- CCP
clathrin-coated pit
- CCS
clathrin-coated structure
- CCV
clathrin-coated vesicle
- DIC
differential interference contrast
- ECFP
enhanced cyan fluorescent protein
- EF
evanescent field
- EGFP
enhanced green fluorescent protein
- FCS
fetal calf serum
- GDP
guanosine 5′-diphosphate
- GTP
guanosine 5′-triphosphate
- GTPase
guanosine 5′-triphosphatase
- HA
hemagglutinin
- HIV-1
human immunodeficiency virus type 1
- HLA
human leukocyte antigen
- HSV-1
herpes simplex virus 1
- IgG
immunoglobulin G
- LTR
long terminal repeat
- mAb
monoclonal antibody
- MHC
major histocompatibility complex
- NA
numerical aperture
- pAb
polyclonal antibody
- PBS
phosphate-buffered saline
- PE
phycoerythrin
- PH
pleckstrin homology
- PIP2
phosphatidylinositol-(4,5)-biphosphate
- PI4P5-K Iα
phosphatidylinositol-4-phosphate 5-kinase Iα
- PLCδ1
phospholipase C δ1
- PLD
phospholipase D
- RANTES
regulated on activation, normal T expressed and secreted
- RNAi
RNA interference
- SDF
stromal cell–derived factor
- SEM
standard error of the mean
- siRNA
small interfering RNA
- TIRFM
total internal reflection fluorescence microscopy
- VSV-G
vesicular stomatitis virus G
- WT
wild-type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-08-0722) on February 23, 2011.
The authors declare that they have no conflicting financial interests.
REFERENCES
- Aikawa Y, Martin TF. ADP-ribosylation factor 6 regulation of phosphatidylinositol-4,5-bisphosphate synthesis, endocytosis, and exocytosis. Methods Enzymol. 2005;404:422–431. doi: 10.1016/S0076-6879(05)04037-1. [DOI] [PubMed] [Google Scholar]
- Al-Awar O, Radhakrishna H, Powell NN, Donaldson JG. Separation of membrane trafficking and actin remodeling functions of ARF6 with an effector domain mutant. Mol Cell Biol. 2000;20:5998–6007. doi: 10.1128/mcb.20.16.5998-6007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson R, Riggs B, Sullivan W. Membrane traffic: a driving force in cytokinesis. Trends Cell Biol. 2005;15:92–101. doi: 10.1016/j.tcb.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Barral DC, Cavallari M, McCormick PJ, Garg S, Magee AI, Bonifacino JS, De Libero G, Brenner MB. CD1a and MHC class I follow a similar endocytic recycling pathway. Traffic. 2008;9:1446–1457. doi: 10.1111/j.1600-0854.2008.00781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero-Villar M, Barroso-Gonzalez J, Cabrero JR, Gordon-Alonso M, Alvarez-Losada S, Munoz-Fernandez MA, Sanchez-Madrid F, Valenzuela-Fernandez A. PI4P5-kinase Iα is required for efficient HIV-1 entry and infection of T-cells. J Immunol. 2008;181:6882–6888. doi: 10.4049/jimmunol.181.10.6882. [DOI] [PubMed] [Google Scholar]
- Barrero-Villar M, Cabrero JR, Gordon-Alonso M, Barroso-Gonzalez J, Alvarez-Losada S, Munoz-Fernandez MA, Sanchez-Madrid F, Valenzuela-Fernandez A. Moesin is required for HIV-1-induced CD4-CXCR4 interaction, F-actin redistribution, membrane fusion and viral infection in lymphocytes. J Cell Sci. 2009;122:103–113. doi: 10.1242/jcs.035873. [DOI] [PubMed] [Google Scholar]
- Barroso-Gonzalez J, El Jaber-Vazdekis N, Garcia-Exposito L, Machado JD, Zarate R, Ravelo AG, Estevez-Braun A, Valenzuela-Fernandez A. The lupane-type triterpene 30-oxo-calenduladiol is a CCR5 antagonist with anti-HIV-1 and anti-chemotactic activities. J Biol Chem. 2009a;284:16609–16620. doi: 10.1074/jbc.M109.005835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-Gonzalez J, Machado JD, Garcia-Exposito L, Valenzuela-Fernandez A. Moesin regulates the trafficking of nascent clathrin-coated vesicles. J Biol Chem. 2009b;284:2419–2434. doi: 10.1074/jbc.M805311200. [DOI] [PubMed] [Google Scholar]
- Begle A, Tryoen-Toth P, de Barry J, Bader MF, Vitale N. ARF6 regulates the synthesis of fusogenic lipids for calcium-regulated exocytosis in neuroendocrine cells. J Biol Chem. 2009;284:4836–4845. doi: 10.1074/jbc.M806894200. [DOI] [PubMed] [Google Scholar]
- Belov GA, Ehrenfeld E. Involvement of cellular membrane traffic proteins in poliovirus replication. Cell Cycle. 2007;6:36–38. doi: 10.4161/cc.6.1.3683. [DOI] [PubMed] [Google Scholar]
- Blagoveshchenskaya AD, Thomas L, Feliciangeli SF, Hung CH, Thomas G. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell. 2002;111:853–866. doi: 10.1016/s0092-8674(02)01162-5. [DOI] [PubMed] [Google Scholar]
- Blanco J, Bosch B, Fernandez-Figueras MT, Barretina J, Clotet B, Este JA. High level of coreceptor-independent HIV transfer induced by contacts between primary CD4 T-cells. J Biol Chem. 2004;279:51305–51314. doi: 10.1074/jbc.M408547200. [DOI] [PubMed] [Google Scholar]
- Borroni EM, Mantovani A, Locati M, Bonecchi R. Chemokine receptors intracellular trafficking. Pharmacol Ther. 2010;127:1–8. doi: 10.1016/j.pharmthera.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Brown FD, Rozelle AL, Yin HL, Balla T, Donaldson JG. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J Cell Biol. 2001;154:1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzon MJ, et al. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat Med. 2010;16:460–465. doi: 10.1038/nm.2111. [DOI] [PubMed] [Google Scholar]
- Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, Bonifacino JS. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2002;21:2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavenagh MM, Whitney JA, Carroll K, Zhang C, Boman AL, Rosenwald AG, Mellman I, Kahn RA. Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J Biol Chem. 1996;271:21767–21774. doi: 10.1074/jbc.271.36.21767. [DOI] [PubMed] [Google Scholar]
- Cavrois M, De Noronha C, Greene WC. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol. 2002;20:1151–1154. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- Chen P, Hubner W, Spinelli MA, Chen BK. Predominant mode of human immunodeficiency virus transfer between T-cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol. 2007;81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell. 2007;18:2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criss AK, Silva M, Casanova JE, McCormick BA. Regulation of Salmonella-induced neutrophil transmigration by epithelial ADP-ribosylation factor 6. J Biol Chem. 2001;276:48431–48439. doi: 10.1074/jbc.M106969200. [DOI] [PubMed] [Google Scholar]
- Daecke J, Fackler OT, Dittmar MT, Krausslich HG. Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J Virol. 2005;79:1581–1594. doi: 10.1128/JVI.79.3.1581-1594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit NM, Perelson AS. Multiplicity of human immunodeficiency virus infections in lymphoid tissue. J Virol. 2004;78:8942–8945. doi: 10.1128/JVI.78.16.8942-8945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem. 2003;278:41573–41576. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Honda A. Localization and function of Arf family GTPases. Biochem Soc Trans. 2005;33:639–642. doi: 10.1042/BST0330639. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Porat-Shliom N, Cohen LA. Clathrin-independent endocytosis: a unique platform for cell signaling and PM remodeling. Cell Signal. 2009;21:1–6. doi: 10.1016/j.cellsig.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Li G, Colombo MI, Stahl PD. A regulatory role for ARF6 in receptor-mediated endocytosis. Science. 1995;267:1175–1178. doi: 10.1126/science.7855600. [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C, van Donselaar E, Hsu VW, Yang C, Stahl PD, Peters PJ. ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J Cell Biol. 1998;140:603–616. doi: 10.1083/jcb.140.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earp LJ, Delos SE, Park HE, White JM. The many mechanisms of viral membrane fusion proteins. Curr Top Microbiol Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faurschou M, Borregaard N. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 2003;5:1317–1327. doi: 10.1016/j.micinf.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Folsch H, Mattila PE, Weisz OA. Taking the scenic route: biosynthetic traffic to the plasma membrane in polarized epithelial cells. Traffic. 2009;10:972–981. doi: 10.1111/j.1600-0854.2009.00927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M, Peters PJ, Boretto J, van Donselaar E, Neri A, D’Souza-Schorey C, Chavrier P. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- Gousset K, Ablan SD, Coren LV, Ono A, Soheilian F, Nagashima K, Ott DE, Freed EO. Real-time visualization of HIV-1 GAG trafficking in infected macrophages. PLoS Pathog. 2008;4:e1000015. doi: 10.1371/journal.ppat.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AT. Population biology of HIV-1 infection: viral and CD4+ T-cell demographics and dynamics in lymphatic tissues. Annu Rev Immunol. 1999;17:625–656. doi: 10.1146/annurev.immunol.17.1.625. [DOI] [PubMed] [Google Scholar]
- Ivanchenko S, Godinez WJ, Lampe M, Krausslich HG, Eils R, Rohr K, Brauchle C, Muller B, Lamb DC. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Hildreth JE, Schwartz DH. Actin-dependent receptor colocalization required for human immunodeficiency virus entry into host cells. J Virol. 1998;72:5251–5255. doi: 10.1128/jvi.72.6.5251-5255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Baranda S, et al. Filamin-A regulates actin-dependent clustering of HIV receptors. Nat Cell Biol. 2007;9:838–846. doi: 10.1038/ncb1610. [DOI] [PubMed] [Google Scholar]
- Johnson DC, Huber MT. Directed egress of animal viruses promotes cell-to-cell spread. J Virol. 2002;76:1–8. doi: 10.1128/JVI.76.1.1-8.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Kashefi K, Hollinshead M, Sattentau QJ. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J Exp Med. 2004;199:283–293. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Sattentau QJ. Retroviral spread by induction of virological synapses. Traffic. 2004;5:643–650. doi: 10.1111/j.1600-0854.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakkonen JP, et al. Clathrin-independent entry of baculovirus triggers uptake of E. coli in nonphagocytic human cells. PLoS One. 2009;4:e5093. doi: 10.1371/journal.pone.0005093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen JE, Massol RH, Nieland TJ, Kirchhausen T. HIV Nef-mediated major histocompatibility complex class I down-modulation is independent of Arf6 activity. Mol Biol Cell. 2004;15:323–331. doi: 10.1091/mbc.E03-08-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letinic K, Sebastian R, Toomre D, Rakic P. Exocyst is involved in polarized cell migration and cerebral cortical development. Proc Natl Acad Sci USA. 2009;106:11342–11347. doi: 10.1073/pnas.0904244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Belkina NV, Shaw S. HIV infection of T-cells: actin-in and actin-out. Sci Signal. 2009;2:pe23. doi: 10.1126/scisignal.266pe23. [DOI] [PubMed] [Google Scholar]
- Londono I, Marshansky V, Bourgoin S, Vinay P, Bendayan M. Expression and distribution of adenosine diphosphate-ribosylation factors in the rat kidney. Kidney Int. 1999;55:1407–1416. doi: 10.1046/j.1523-1755.1999.00365.x. [DOI] [PubMed] [Google Scholar]
- Macia E, Luton F, Partisani M, Cherfils J, Chardin P, Franco M. The GDP-bound form of Arf6 is located at the plasma membrane. J Cell Sci. 2004;117:2389–2398. doi: 10.1242/jcs.01090. [DOI] [PubMed] [Google Scholar]
- Malinowsky K, Luksza J, Dittmar MT. Susceptibility to virus-cell fusion at the plasma membrane is reduced through expression of HIV gp41 cytoplasmic domains. Virology. 2008;376:69–78. doi: 10.1016/j.virol.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Marchant D, Sall A, Si X, Abraham T, Wu W, Luo Z, Petersen T, Hegele RG, McManus BM. ERK MAP kinase-activated Arf6 trafficking directs coxsackievirus type B3 into an unproductive compartment during virus host-cell entry. J Gen Virol. 2009;90:854–862. doi: 10.1099/vir.0.005868-0. [DOI] [PubMed] [Google Scholar]
- Marechal V, Clavel F, Heard JM, Schwartz O. Cytosolic Gag p24 as an index of productive entry of human immunodeficiency virus type 1. J Virol. 1998;72:2208–2212. doi: 10.1128/jvi.72.3.2208-2212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal V, Prevost MC, Petit C, Perret E, Heard JM, Schwartz O. Human immunodeficiency virus type 1 entry into macrophages mediated by macropinocytosis. J Virol. 2001;75:11166–11177. doi: 10.1128/JVI.75.22.11166-11177.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Vicente C, Gomez-Fernandez JC, Corbalan-Garcia S. The ATP-dependent membrane localization of protein kinase Cα is regulated by Ca2+ influx and phosphatidylinositol 4,5-bisphosphate in differentiated PC12 cells. Mol Biol Cell. 2005;16:2848–2861. doi: 10.1091/mbc.E05-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh M, Helenius A. Virus entry: open sesame. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massol RH, Larsen JE, Kirchhausen T. Possible role of deep tubular invaginations of the plasma membrane in MHC-I trafficking. Exp Cell Res. 2005;306:142–149. doi: 10.1016/j.yexcr.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Matlin KS, Reggio H, Helenius A, Simons K. Pathway of vesicular stomatitis virus entry leading to infection. J Mol Biol. 1982;156:609–631. doi: 10.1016/0022-2836(82)90269-8. [DOI] [PubMed] [Google Scholar]
- McDonald D, Wu L, Bohks SM, KewalRamani VN, Unutmaz D, Hope TJ. Recruitment of HIV and its receptors to dendritic cell–T cell junctions. Science. 2003;300:1295–1297. doi: 10.1126/science.1084238. [DOI] [PubMed] [Google Scholar]
- Mellman I, Warren G. The road taken: past and future foundations of membrane traffic. Cell. 2000;100:99–112. doi: 10.1016/s0092-8674(00)81687-6. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudhakir D, Harashima H. Learning from the viral journey: how to enter cells and how to overcome intracellular barriers to reach the nucleus. AAPS J. 2009;11:65–77. doi: 10.1208/s12248-009-9080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N, Weigert R, Donaldson JG. Convergence of nonclathrin- and clathrin-derived endosomes involves Arf6 inactivation and changes in phosphoinositides. Mol Biol Cell. 2003;14:417–431. doi: 10.1091/mbc.02-04-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols BJ, Lippincott-Schwartz J. Endocytosis without clathrin coats. Trends Cell Biol. 2001;11:406–412. doi: 10.1016/s0962-8924(01)02107-9. [DOI] [PubMed] [Google Scholar]
- Nishi K, Saigo K. Cellular internalization of green fluorescent protein fused with herpes simplex virus protein VP22 via a lipid raft-mediated endocytic pathway independent of caveolae and Rho family GTPases but dependent on dynamin and Arf6. J Biol Chem. 2007;282:27503–27517. doi: 10.1074/jbc.M703810200. [DOI] [PubMed] [Google Scholar]
- Nordenfelt P, Winberg ME, Lonnbro P, Rasmusson B, Tapper H. Different requirements for early and late phases of azurophilic granule-phagosome fusion. Traffic. 2009;10:1881–1893. doi: 10.1111/j.1600-0854.2009.00986.x. [DOI] [PubMed] [Google Scholar]
- Pasqualato S, Menetrey J, Franco M, Cherfils J. The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2001;2:234–238. doi: 10.1093/embo-reports/kve043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauza CD, Price TM. Human immunodeficiency virus infection of T-cells and monocytes proceeds via receptor-mediated endocytosis. J Cell Biol. 1988;107:959–968. doi: 10.1083/jcb.107.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DM. The role of cell-to-cell transmission in HIV infection. Aids. 1994;8:719–731. doi: 10.1097/00002030-199406000-00001. [DOI] [PubMed] [Google Scholar]
- Pierini R, Cottam E, Roberts R, Wileman T. Modulation of membrane traffic between endoplasmic reticulum, ERGIC and Golgi to generate compartments for the replication of bacteria and viruses. Semin Cell Dev Biol. 2009;20:828–833. doi: 10.1016/j.semcdb.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- Puigdomenech I, Massanella M, Cabrera C, Clotet B, Blanco J. On the steps of cell-to-cell HIV transmission between CD4 T-cells. Retrovirology. 2009;6:89. doi: 10.1186/1742-4690-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigdomenech I, Massanella M, Izquierdo-Useros N, Ruiz-Hernandez R, Curriu M, Bofill M, Martinez-Picado J, Juan M, Clotet B, Blanco J. HIV transfer between CD4 T-cells does not require LFA-1 binding to ICAM-1 and is governed by the interaction of HIV envelope glycoprotein with CD4. Retrovirology. 2008;5:32. doi: 10.1186/1742-4690-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna H, Donaldson JG. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna H, Klausner RD, Donaldson JG. Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. J Cell Biol. 1996;134:935–947. doi: 10.1083/jcb.134.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabe H. Requirement for Arf6 in cell adhesion, migration, and cancer cell invasion. J Biochem. 2003;134:485–489. doi: 10.1093/jb/mvg181. [DOI] [PubMed] [Google Scholar]
- Schmoranzer J, Kreitzer G, Simon SM. Migrating fibroblasts perform polarized, microtubule-dependent exocytosis towards the leading edge. J Cell Sci. 2003;116:4513–4519. doi: 10.1242/jcs.00748. [DOI] [PubMed] [Google Scholar]
- Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard JM. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber BK, Pavlakis GN. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J Virol. 1992;66:7176–7182. doi: 10.1128/jvi.66.12.7176-7182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein BS, Gowda SD, Lifson JD, Penhallow RC, Bensch KG, Engleman EG. pH-independent HIV entry into CD4-positive T-cells via virus envelope fusion to the plasma membrane. Cell. 1987;49:659–668. doi: 10.1016/0092-8674(87)90542-3. [DOI] [PubMed] [Google Scholar]
- Sun X, Yau VK, Briggs BJ, Whittaker GR. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology. 2005;338:53–60. doi: 10.1016/j.virol.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Valenzuela-Fernandez A, et al. Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection. Mol Biol Cell. 2005;16:5445–5454. doi: 10.1091/mbc.E05-04-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela-Fernandez A, Palanche T, Amara A, Magerus A, Altmeyer R, Delaunay T, Virelizier JL, Baleux F, Galzi JL, Arenzana-Seisdedos F. Optimal inhibition of X4 HIV isolates by the CXC chemokine stromal cell-derived factor 1 alpha requires interaction with cell surface heparan sulfate proteoglycans. J Biol Chem. 2001;276:26550–26558. doi: 10.1074/jbc.M100411200. [DOI] [PubMed] [Google Scholar]
- Valenzuela-Fernandez A, et al. Leukocyte elastase negatively regulates Stromal cell-derived factor-1 (SDF-1)/CXCR4 binding and functions by amino-terminal processing of SDF-1 and CXCR4. J Biol Chem. 2002;277:15677–15689. doi: 10.1074/jbc.M111388200. [DOI] [PubMed] [Google Scholar]
- Vidricaire G, Tremblay MJ. Rab5 and Rab7, but not ARF6, govern the early events of HIV-1 infection in polarized human placental cells. J Immunol. 2005;175:6517–6530. doi: 10.4049/jimmunol.175.10.6517. [DOI] [PubMed] [Google Scholar]
- Vidricaire G, Tremblay MJ. A clathrin, caveolae, and dynamin-independent endocytic pathway requiring free membrane cholesterol drives HIV-1 internalization and infection in polarized trophoblastic cells. J Mol Biol. 2007;368:1267–1283. doi: 10.1016/j.jmb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Vitale N, Chasserot-Golaz S, Bailly Y, Morinaga N, Frohman MA, Bader MF. Calcium-regulated exocytosis of dense-core vesicles requires the activation of ADP-ribosylation factor (ARF)6 by ARF nucleotide binding site opener at the plasma membrane. J Cell Biol. 2002;159:79–89. doi: 10.1083/jcb.200203027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Rosales T, Rose JJ, Chowdhury B, Knutson JR, Venkatesan S. HIV-1 Nef binds a subpopulation of MHC-I throughout its trafficking itinerary and down-regulates MHC-I by perturbing both anterograde and retrograde trafficking. J Biol Chem. 2010;285:30884–30905. doi: 10.1074/jbc.M110.135947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder A, et al. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T-cells. Cell. 2008;134:782–792. doi: 10.1016/j.cell.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.