A mechanism of nuclear pore complex assembly into intact nuclear envelopes remains elusive. We explore roles of conserved inner nuclear membrane proteins, Heh1p and Heh2p, in this process. The data support the existence of a lumenal bridge between Heh1p and the nucleoporin Pom152p, which might facilitate early nuclear pore complex assembly events.
Abstract
Nuclear pore complexes (NPCs) provide a gateway for the selective transport of macromolecules across the nuclear envelope (NE). Although we have a solid understanding of NPC composition and structure, we do not have a clear grasp of the mechanism of NPC assembly. Here, we demonstrate specific defects in nucleoporin distribution in strains lacking Heh1p and Heh2p—two conserved members of the LEM (Lap2, emerin, MAN1) family of integral inner nuclear membrane proteins. These effects on nucleoporin localization are likely of functional importance as we have defined specific genetic interaction networks between HEH1 and HEH2, and genes encoding nucleoporins in the membrane, inner, and outer ring complexes of the NPC. Interestingly, expression of a domain of Heh1p that resides in the NE lumen is sufficient to suppress both the nucleoporin mislocalization and growth defects in heh1Δpom34Δ strains. We further demonstrate a specific physical interaction between the Heh1p lumenal domain and the massive cadherin-like lumenal domain of the membrane nucleoporin Pom152p. These findings support a role for Heh1p in the assembly or stability of the NPC, potentially through the formation of a lumenal bridge with Pom152p.
INTRODUCTION
The genome of eukaryotes is encapsulated by the nuclear envelope (NE)—a membrane system that is continuous with the endoplasmic reticulum (ER). The NE is composed of two parallel membranes, the inner nuclear membrane (INM) and the outer nuclear membrane (ONM) that are separated by an aqueous lumenal/perinuclear space (Hetzer et al., 2005). The surface of the NE is studded with ∼100-nm-diameter pores where the INM and ONM are connected to form a third highly curved membrane called the pore membrane (POM). The POM is filled with massive ∼50 MD protein assemblies, termed nuclear pore complexes (NPCs), which control the bidirectional flow of molecules across the NE (Strambio-De-Castillia et al., 2010).
The NPC is composed of ∼30 individual proteins, termed nucleoporins or nups, found in multiple copies such that an individual NPC is constructed of upwards of 500 individual subunits (Rout et al., 2000; Cronshaw et al., 2002; Alber et al., 2007b; D’Angelo and Hetzer, 2008). Nups are often isolated from cell extracts within distinct subcomplexes, contributing to the idea that the NPC is made up of several modular building blocks. These subcomplexes are assembled in multiples of eight into membrane, inner, and outer ring structures that form the core scaffold of the NPC (Alber et al., 2007a, 2007b). This scaffold surrounds and supports a transport channel that consists of nups with repetitive peptide motifs rich in phenylalanine-glycine (FG) amino acid residues. The FG-nups selectively bind nuclear transport factors to facilitate translocation of cargo molecules across the NPC (Wente and Rout, 2010).
The major nup subcomplex that contributes to the formation of the outer ring structures is thought to be the vertebrate (v) Nup107–160 complex or its yeast counterpart, the Nup84p-complex (Siniossoglou et al., 1996, 2000; Belgareh et al., 2001; Vasu et al., 2001; Alber et al., 2007b). The inner ring is composed of a number of nups, including vNup155, or its two yeast paralogues, Nup157p and Nup170p (Aitchison et al., 1995b; Nehrbass et al., 1996; Grandi et al., 1997; Marelli et al., 1998; Alber et al., 2007b). The inner ring directly interfaces with a membrane ring composed of three transmembrane nups called pore membrane proteins or poms. In vertebrates, the membrane ring is composed of vPom121 (Hallberg et al., 1993), vNdc1 (Stavru et al., 2006), and vgp210 (Greber et al., 1990). In budding yeast, a biochemical complex containing the three poms, Ndc1p (Chial et al., 1998), Pom34p (Rout et al., 2000), and Pom152p (Wozniak et al., 1994), has been isolated from cell extracts, and likely also helps form a lumenal ring that extends from the POM into the NE lumen (Alber et al., 2007b; Onischenko et al., 2009).
Although we have a solid grasp of the composition of the NPC, our understanding of the assembly of this elaborate machine is under intense scrutiny. NPC assembly occurs through two distinct mechanisms: one during postmitotic NE reformation that begins with the recruitment of the nup ELYS/Mel28 (Franz et al., 2007; Gillespie et al., 2007; Rasala et al., 2008) and the outer ring complex to chromatin (Belgareh et al., 2001; Harel et al., 2003b; Walther et al., 2003), followed by the stepwise recruitment of both membrane and inner ring complexes (Franz et al., 2005; Hawryluk-Gara et al., 2005; Mansfeld et al., 2006; Dultz et al., 2008; Hawryluk-Gara et al., 2008). The other mechanism occurs throughout interphase during which NPCs are inserted de novo into an intact NE (D’Angelo et al., 2006; Antonin et al., 2008). Many factors contribute to this process, including key regulators of nuclear transport like the GTPase Ran and karyopherins (a.k.a. importins) (Lusk et al., 2002; Ryan and Wente, 2002; Harel et al., 2003a; Ryan et al., 2003, 2007; D’Angelo et al., 2006), as well as specific nups (Mutvei et al., 1992; Zabel et al., 1996) and poms (Lau et al., 2004; Madrid et al., 2006; Miao et al., 2006; Doucet et al., 2010; Fichtman et al., 2010). Because the NE is intact during this process, nups must be recruited on both the cytoplasmic and nucleoplasmic sides of the NE to a site of NPC assembly. In addition, the insertion of nups into the NE must be coordinated with membrane-remodeling events that lead to the fusion of the INM and ONM, while maintaining NE integrity.
Recent work supports that vPom121 moves early to a site of interphase NPC assembly where it likely recruits additional assembly factors that might include membrane curvature–inducing proteins like the reticulons (Dawson et al., 2009; Doucet et al., 2010; Dultz and Ellenberg, 2010). Consistent with the idea that inducing membrane curvature at the NE plays a role in NPC assembly, a membrane curvature–sensing alkaline phosphatase S (ArfGAP1 lipid packing sensor) domain within vNup133 (Drin et al., 2007) plays a role in the recruitment of the outer ring complex (Doucet et al., 2010). Moreover, nups of both inner and outer rings structurally resemble vesicular coat complexes (Devos et al., 2004; Alber et al., 2007b; Hsia et al., 2007; Brohawn et al., 2008; Debler et al., 2008; Brohawn and Schwartz, 2009; Leksa et al., 2009; Whittle and Schwartz, 2009), supporting a general theme in which POM-proximal nups both sense and stabilize curved membranes.
It is likely that inner ring nups are recruited either before or perhaps simultaneously with the outer ring complex to a site of NPC assembly. Consistent with the latter idea, vPom121 has been shown to directly interact with components of both the inner (vNup155) and outer (vNup160) ring (Mitchell et al., 2010). By analogy in budding yeast, which uses only the interphase NPC assembly mechanism, direct interactions between Pom152p and Nup170p (Makio et al., 2009), as well as between Ndc1p and Nup53p or Nup59p (Onischenko et al., 2009), connect the membrane and inner ring complexes. These connections are critical for NPC assembly as misassembled nups accumulate in the cytoplasm (Makio et al., 2009; Onischenko et al., 2009; Flemming et al., 2009) and at the INM (Makio et al., 2009) when inner and/or membrane ring function has been disrupted. Taken together, these studies contribute to a model in which membrane and inner ring components contribute to a step in NPC assembly at or before the fusion of the INM and ONM.
We do not have a mechanistic understanding of INM/ONM fusion. Indeed, although proteins like the reticulons might impart curvature to the inner and outer membranes, it is unclear whether this induction of curvature would be sufficient to disrupt their lumenal leaflets to support fusion, and might predict a requirement for a lumenal fusogen. Consistent with this idea, the reticulon genes in Saccharomyces cerevisiae are not essential unless other components of either the membrane or outer ring complex are also simultaneously knocked out (Dawson et al., 2009). These data support the idea that there are additional membrane and/or lumenal proteins that could facilitate NPC assembly. A growing list of candidates includes components of the lipid synthesis machinery, including Apq12p (Scarcelli et al., 2007), Acc1p (Schneiter et al., 1996), and Brr6p (Hodge et al., 2010), in addition to a newly discovered POM-associated protein, Pom33p (Chadrin et al., 2010). None of these proteins, however, have significant domains that extend into the lumen. In contrast, a number of integral INM proteins have lumenal domains, such as the conserved LEM (Lap2, emerin, MAN1; Wagner and Krohne, 2007) and SUN (Sad1p, Unc84; Tzur et al., 2006) families, which could participate in making interactions across or within the NE lumen that might facilitate membrane fusion. Interestingly, recent work supports a potential lumenal connection between a SUN-family protein, SUN1, and NPCs that might be important for NPC distribution (Liu et al., 2007).
The interaction between SUN1 and NPCs highlights a growing physical and functional relationship between the INM and NPCs that is primarily mediated by interactions between the nuclear basket of the NPC and components of the INM and nuclear lamina (Strambio-De-Castillia et al., 2010). Whereas yeasts lack homologues of the nuclear lamins, proteins such as the myosin-like proteins (Mlp1p and Mlp2p) and Esc1p (establishes silent chromatin) provide an analogous structural and functional role for the yeast nucleus (Strambio-deCastillia et al., 1999; Hattier et al., 2007; Lewis et al., 2007). Yeast also have homologues of the LEM family of integral INM proteins that include Heh1p (also called Src1p) and Heh2p (King et al., 2006) that might also interact with NPCs. This idea is supported by synthetic genetic relationships between heh1Δ strains and strains containing deletions of genes encoding members of the THO-transcription export (TREX) complex (Grund et al., 2008). The function of Heh2p remains ill defined.
Here we further examine how Heh1p and Heh2p contribute to NE function. We describe a genetic interaction network that functionally links both Heh1p and Heh2p to the membrane, inner, and outer ring complexes of the NPC. The appearance of mislocalized nups in heh1Δ strains supports a role for Heh1p in NPC assembly and stability. Interestingly, we map the critical function of Heh1p in NPC assembly to its lumenal domain. Furthermore, we show evidence for a direct link between the lumenal domains of Heh1p and Pom152p. Together, our findings support the existence of a lumenal bridge that might directly contribute to early NPC assembly events.
RESULTS
Unique defects in nup distribution in heh1Δ and heh2Δ strains
To improve our understanding of the function of Heh1p and Heh2p, we examined whether their deletion impacted the subcellular distribution of key structural and functional components of the NE, including NPCs, Mlp1p, and Esc1p. As shown in Figure 1, heh1Δ, heh2Δ, and heh1Δheh2Δ strains did not display any marked differences in the distribution of Mlp1-GFP (green fluorescent protein) or Esc1-GFP. Both of these proteins were localized normally to the nuclear periphery in a punctate pattern as has been previously described (Strambio-deCastillia et al., 1999; Andrulis et al., 2002). In contrast, the deletion of HEH1 and HEH2 had a significant impact on the distribution of the nup GFP-Nup49p. GFP-Nup49 was seen distributed uniformly at the NE (maximum intensity projections of a z-series are shown in Figure 1) in wild-type (WT) cells, whereas a subset of heh1Δ cells (23%) showed a striking accumulation of brightly fluorescent cytoplasmic foci of GFP-Nup49p. These data suggest that pathways contributing to the assembly of new NPCs or the maintenance of the integrity of existing NPCs are disrupted in the absence of HEH1, whereas other elements of the INM are unperturbed. In contrast to heh1Δ cells, we did not observe significant numbers of cytoplasmic GFP-Nup49p foci in heh2Δ cells (Figure 1). Nonetheless, the distribution of NPCs at the NE was markedly changed, appearing clustered at the NE. In heh1Δheh2Δ cells, we observed both NPC clustering and nup cytoplasmic foci (Figure 1), suggesting an additive rather than a synergistic effect of HEH1 and HEH2 deletion on these unique NPC abnormalities. Together these data support a model in which Heh1p and Heh2p contribute to the normal distribution of NPCs, but by distinct mechanisms: Whereas Heh2p contributes to NPC distribution at the NE, Heh1p is required in a related pathway important for either the assembly of NPCs or their integrity.
FIGURE 1:
Specific defects in NPC distribution in heh1Δ and heh2Δ strains. Fluorescence micrographs (top panels) of deconvolved images of GFP-Nup49p, Mlp1-GFP, and Esc1-GFP in either heh1Δ, heh2Δ, or heh1Δheh2Δ strains. A merge between phase-contrast images and the fluorescent images are shown in bottom panels. To better visualize the cytoplasmic accumulation of GFP-Nup49 foci (arrows), a maximum intensity projection (MIP) is shown. Note that in heh2Δ cells there are few cytoplasmic GFP-Nup49p foci and the NPCs appear clustered at the NE.
Heh1p and Heh2p colocalize with a fraction of Nic96p
The specific NPC distribution defects observed in both heh1Δ and heh2Δ strains raised the possibility that Heh1p and/or Heh2p might directly interact with NPCs. To address this possibility, we examined the localization of both Heh1- and Heh2-GFP expressed at endogenous levels in WT cells. Expression of a Nic96-RFP (red fluorescent protein) fusion protein from the endogenous NIC96 chromosomal locus was used to colocalize NPCs (Figure 2). To achieve a high spatial resolution, we fixed cells to immobilize NPCs, and images were deconvolved using a stringent iterative algorithm. This approach allowed us to visualize distinct populations of both the Heh proteins and NPCs (Figure 2A). Both Heh1- and Heh2-GFP were localized in a punctate pattern that was remarkably similar to Nic96-RFP. When these images were merged (Figure 2A, merge), however, the Heh1-GFP and Heh2-GFP appeared intertwined but did not completely overlap with Nic96-RFP. Nonetheless, it is clear that there is an intimate relationship between the distributions of these subsets of proteins. Furthermore, as directed by the arrows in Figure 2A, we could clearly detect regions of the NE in which there was overlap between the Heh1- and Heh2-GFP fluorescence and that of Nic96-RFP, supporting the idea that Heh1p and Heh2p might associate directly with either a subset of NPCs or perhaps with nup subcomplexes not fully assembled into the NE.
FIGURE 2:
Heh proteins colocalize with a fraction of Nic96-RFP. The colocalization of endogenous levels of Heh1- and Heh2-GFP was determined with Nic96-RFP in WT (A) and nup133Δ strains where NPCs are clustered at the NE (B). To achieve a high spatial resolution, the cells were fixed and immobilized prior to imaging, and images were subsequently deconvolved. Arrows in merged images point to regions of overlap between the green and red channels. In (B), colocalization in nup133Δ cells was often only observed in one axial plane. The top two image series are identical cells separated by 0.2 μm in the z direction (Δz). (C) Quantitation of the percentage of nup133Δ cells with at least one region of colocalization of Heh1- or Heh2-GFP and Nic96-RFP.
To further evaluate the possibility that the Heh proteins interact with nups, we examined the distribution of Heh1- and Heh2-GFP in a nup133Δ strain. In this strain, NPCs are severely clustered at the NE, thus allowing one to more easily discriminate between nups and other components of the NE (Doye et al., 1994; Pemberton et al., 1995). Consistent with the idea that there is a relationship between the distribution of NPCs and the Heh proteins, both Heh1- and Heh2-GFP were no longer localized in a punctate pattern at the NE and appeared in a more uniform smooth distribution (Figure 2B). In addition, cells contained bright puncta that might indicate a clustering or aggregation of these proteins due to their mislocalization. We surmise that, because both Heh1p and Heh2p require functional NPCs to accumulate at the INM (King et al., 2006), disruption of NPC function in nup133Δ strains might also lead to their partial redistribution. Regardless, similar to the images in Figure 2A, we were able to detect regions of clear overlap between clusters of Nic96-RFP and both Heh1- and Heh2-GFP (Figure 2B, arrows). Often these regions colocalized at the edges of the clusters of NPCs and could be visualized in only one axial plane (compare the top two sets of panels in Figure 2B). We further quantified the number of nup133Δ cells in which we could observe at least one region of overlap between Heh1- or Heh2-GFP and Nic96-RFP. This analysis demonstrated that ∼70% of nup133Δ strains contained discernable colocalization between Heh1-GFP and Nic96-RFP (Figure 2C). Similarly, although within a smaller percentage of the population (∼33%), Heh2-GFP was observed colocalized with Nic96-RFP. These data support the notion that a fraction of Heh1p and Heh2p associate with nups in vivo.
Heh1p and Heh2p physically interact with nups
To further investigate the potential existence of a physical link between the Heh proteins and nups, we used an affinity pull-down approach. We used magnetic beads coupled to anti-GFP antibodies to affinity purify endogenously tagged Heh1-GFP or Heh2-GFP from cryolysates derived from equivalent amounts of cells expressing epitope-tagged nups. Proteins bound to Heh1-GFP and Heh2-GFP were evaluated by Western blot. We first tested for members of the membrane (Pom152p) and inner ring (Nup170p) complexes. As shown in Figure 3A, we could detect an HA (hemagglutinin)-tagged version of Pom152p and Nup170-myc bound to both Heh1-GFP and Heh2-GFP, suggesting that Heh1p and Heh2p associate with these nups in vivo. Although only a fraction of the total Nup170p or Pom152p present in the lysate was pulled down (Figure 3A, compare load [L] and unbound [UB] fractions, see figure legend for relative quantities), the interactions were nonetheless specific, as isolation of the anti-GFP beads from a cell extract lacking Heh1- or Heh2-GFP did not isolate these proteins (Figure 3B). Furthermore, we did not observe an interaction with histone H3 (Figure 3A) or an abundant ER-membrane protein, Sec63p. We also examined interactions with additional nups and were able to pull down detectable amounts of Nup85p with Heh2p, but we could not detect Nup188-myc, Nup60-myc, or the monoclonal antibody (mAb)414-reactive FxFG-motif containing nups (Davis and Blobel, 1986; Rout and Blobel, 1993). These data are consistent with a model in which Heh1p and Heh2p associate with specific nups that are localized at or near the POM (Alber et al., 2007b). The finding that only a portion of the total nups present are affinity purified is consistent with our observation that Heh1-GFP and Heh2-GFP associate with either a subpopulation of NPCs or a fraction of these nups that are not fully assembled into NPCs (Figure 2).
FIGURE 3:
Affinity purification of Heh1p and Heh2p demonstrates specific interactions with nups (A and B). Magnetic beads with α-GFP antibodies were used to pull out either Heh1-GFP or Heh2-GFP (or no GFP; B) from cell lysates derived from strains expressing HEH1-GFP or HEH2-GFP and the indicated c-myc- or HA-tagged nups. After washing, α-GFP beads were eluted with SDS–PAGE sample buffer. Proteins were separated by SDS–PAGE and Western blotted with the indicated antibodies (left) and were detected by HRP-conjugated secondary antibodies and enhanced chemiluminescence. Equivalent amounts of cells were used for each experiment. Equivalent amounts of load (L) and unbound (UB) fractions are shown in left panels and represent ∼0.5% of the total extract. Right panels show proteins in bound fractions to either Heh1-GFP (B1) or Heh2-GFP (B2), and represent 20% of bound proteins. For each row, the right and left panels can be directly compared, as they are the same membrane with identical exposure times.
Epistatic profiles of HEH1 and HEH2 with specific nup genes
We surmised that if Heh1p and Heh2p physically interacted with NPCs we would be able to detect genetic relationships that might help us to determine the potential function of this physical link. We therefore performed a detailed epistatic analysis by examining synthetic genetic relationships between nup genes and HEH1 and HEH2. We systematically crossed heh1Δheh2Δ strains to a battery of nup deletion strains and examined the viability and/or growth of the resulting double, triple, and, in some cases, quadruple gene deletion strain progeny. As described in Materials and Methods, interactions were classified after growth at 30°C as to whether there was no change (NC) in growth, synthetic lethality (SL), severe synthetic sickness (SSS), or synthetic sickness (SS).
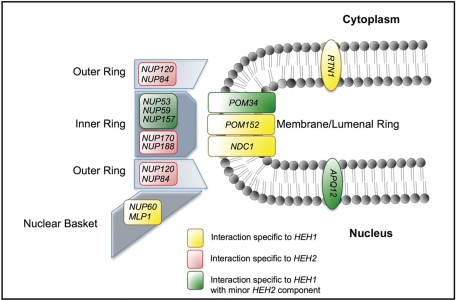
Strikingly, we observed specific synthetic genetic interactions between HEH1 and genes encoding components of the membrane ring. Most notably, we detected a complete loss of viability (SL) of heh1Δpom152Δ cells (Table 1 and Supplemental Figure S1). Because NDC1 is essential, we tested whether an NDC1-GFP allele expressed from the NDC1 chromosomal gene locus would affect the viability of heh1Δ and heh2Δ strains, and, as shown, we could detect a heh1Δ-specific SS interaction with this allele (Table 1). Consistent with the idea that the poms function together within a complex, we also detected a specific SSS interaction in the poor growth of heh1Δpom34Δ cells (Table 1 and Supplemental Figure S1).
TABLE 1:
Epistatic interaction profile of HEH1 and HEH2.
| Genotypes of crosses | heh1Δ | heh2Δ | heh1Δheh2Δ | |
|---|---|---|---|---|
| Membrane ring | ||||
| pom152Δ | SL | NC | SL | |
| pom34Δ | SSS | NC | SSS* | |
| NDC1-GFP† | SS | NC | n.d. | |
| Inner ring | ||||
| nup157Δ | SSS | NC | SL | |
| nup53Δ | SS | NC | SSS | |
| nup59Δ | SS | SS | SSS | |
| nup53Δnup59Δ | SS | SS | SL | |
| nup170Δ | NC | SSS | SSS | |
| nup188Δ | NC | SL | SL | |
| Outer ring | ||||
| nup120Δ | NC | SL | SL | |
| nup84Δ | NC | SL | SL | |
| Nuclear nups and associated protein | ||||
| nup60Δ | SS | NC | SS | |
| mlp1Δ | SS | NC | SS | |
| mlp2Δ | NC | NC | NC | |
| mlp1Δmlp2Δ | SS | NC | SS | |
| nup60Δ | SS | NC | SS | |
| esc1Δ | NC | NC | NC | |
| Membrane proteins | ||||
| apq12Δ | SSS | NC | SSS* | |
| rtn1Δ | SS | NC | SS | |
| yop1Δ | NC | NC | NC | |
| rtn1Δyop1Δ | SS | NC | SS | |
| pom33Δ | NC | NC | NC | |
| per33Δ | NC | NC | NC | |
| pom33Δper33Δ | NC | NC | NC |
NC, no change in growth; n.d., not done; SL, synthetic lethality; SS, synthetic sickness; SSS, severe synthetic sickness.
*Growth of these strains was slower than that of their heh1Δ counterpart. See Supplemental Figure S2.
†Because NDC1 is essential for viability, we tested whether expression of an NDC1-GFP allele was functional in heh1Δ and heh2Δ backgrounds.
Interestingly, the additional deletion of HEH2 further exacerbated the growth of heh1Δpom34Δ cells, suggesting that the viability of pom34Δ strains requires HEH1 and an additional shared functional element expressed by HEH2 (Supplemental Figure S2). Strikingly, this distinct epistatic profile was mirrored by strains with deletions of inner ring nup complex members. Specifically, we observed SS and SSS growth of heh1Δnup157Δ, heh1Δnup53Δ, and heh1Δnup59Δ strains, which was further affected by the deletion of HEH2. In these cases, heh1Δheh2Δnup53Δ and heh1Δheh2Δnup59Δ strains were now SSS, and we were unable to recover viable spores of either heh1Δheh2Δnup157Δ or heh1Δheh2Δnup53Δnup59Δ, suggesting that these deletion combinations resulted in synthetic lethality (Table 1). We hypothesize that these interactions reflect that Heh1p functions in a shared pathway with the inner and membrane ring complexes of the NPC. This pathway also requires the function of Heh2p. Interestingly, in contrast to the majority of the inner ring nup deletion strains, the deletion of NUP170 and NUP188 did not affect the growth of heh1Δ strains (Table 1). What was revealing, and unexpected, was that heh2Δnup170Δ was SSS, and heh2Δnup188Δ was SL, supporting the existence of a functional interaction between Heh2p and these inner ring nups that is independent of Heh1p.
We further examined epistatic relationships with other nup genes encoding two components of the outer ring complex, Nup120p and Nup84p. Remarkably, neither heh1Δnup120Δ nor heh1Δnup84Δ showed a significant interaction, whereas both heh2Δnup120Δ and heh2Δnup84Δ were SL (Table 1). These data support a model in which Heh1p and Heh2p have distinct functional relationships with the NPC, and might provide a rationale for the unique abnormalities in NPC distribution observed in heh1Δ and heh2Δ strains (Figure 1).
We next tested whether heh1Δ and heh2Δ strains affected the growth of strains lacking the nucleoplasmically oriented nups Nup60p and Mlp1p, or the INM-associated Esc1p. This analysis showed additional specific SS interactions exhibited by heh1Δmlp1Δ and heh1Δnup60Δ (Supplemental Figure S1), but not with heh1Δesc1Δ (Table 1). In the case of the heh1Δmlp1Δ strain, the additional deletion of MLP2 did not alter the growth of this strain. Therefore the HEH1-NUP60 and HEH1-MLP1 interactions were specific to HEH1, but, unlike interactions with inner and membrane ring nup genes, they conformed to a distinct epistatic profile where the interactions were weaker (i.e., SS vs. SSS or SL, Supplemental Figure S1) and they were not affected by the deletion of HEH2 (Table 1).
Interestingly, the deletion of APQ12, the gene encoding a membrane protein thought to affect membrane fluidity and NPC assembly (Scarcelli et al., 2007), exhibited a similar genetic interaction profile as the deletion of the inner ring nup genes. Specifically, heh1Δapq12Δ cells were SSS, and heh1Δheh2Δapq12Δ showed even slower growth (Table 1 and Supplemental Figure S2). This finding prompted us to also test whether we could detect interactions with additional membrane proteins known to affect NPC assembly or distribution. Consistent with this idea, we detected an SS interaction between HEH1 and the gene encoding the reticulon RTN1 (Table 1) (Dawson et al., 2009). This interaction was specific, as subsequent deletion of the functionally related YOP1, or HEH2, did not alter the growth of this strain (Table 1). Similarly, neither heh1Δ nor heh2Δ strains exhibited synthetic genetic relationships with pom33Δ, per33Δ, or pom33Δper33Δ strains (Chadrin et al., 2010). Taken together, our genetic analysis supports the existence of specific functional relationships between Heh1p and Heh2p with distinct complexes of the NPC, and membrane proteins that contribute to NPC assembly. A summary of these interactions can be found in Figure 4.
FIGURE 4:
Schematic of HEH1 and HEH2 interactions with nup genes. Genes encoding nuclear basket, membrane, inner, and outer ring complexes are shown in the context of the approximate location of their gene products relative to the POM. (Gray circles with tails represent phospholipids.) Blue-gray polygons are physical representations of the indicated nup subcomplexes. Interactions are colored as described in the key. The schematic is not intended to reflect the stoichiometry of these subcomplexes in the NPC.
Functional analysis of conserved domains of Heh1p and Heh2p
To investigate the functional determinants in both Heh1p and Heh2p that contribute to their specific interactions with nup subcomplexes, we generated truncation alleles of both HEH1 and HEH2, and tested whether they could complement the growth of synthetic genetic interactors (Figure 5A). Importantly, the INM targeting information of both Heh1p and Heh2p is encoded in their N-terminal domains (NTDs), such that the deletion of either the MAN1-C-terminal Homology Domain (MCHD) or their lumenal domains does not affect their INM localization (King et al., 2006; Grund et al., 2008). In addition, we confirmed that these alleles were expressed at or near levels of endogenous Heh1p and Heh2p (Supplemental Figure S3). As test cases for functional complementation, we chose the heh1Δpom152Δ and heh2Δnup170Δ strains. Our rationale was that, because heh1Δpom152Δ is SL, it represents a genetic background that is solely dependent on Heh1p function, whereas heh2Δnup170Δ depends solely on the function of Heh2p.
FIGURE 5:
Lumenal and nuclear domains of Heh1p and Heh2p contribute specificity to the functional interactions with the inner and membrane ring nups. (A) Schematic of N- and C-terminal deletion constructs of Heh1p and Heh2p. Heh1p and Heh2p share a similar domain architecture and topology: a helix-extension helix HEH/LEM domain and NTD, two transmembrane domains (TM), a lumenal domain (LUMD), and another nuclear domain (MCHD). Numbers are amino acid residues. (B–D) Colonies from tetrad dissections (in rows) of genetic crosses between the indicated strains on YPD or YPRG (for expression of GAL1 alleles). In (B) and (C), the underlined colony expresses an heh allele in the indicated deletion strain. In (D), underlined colonies represent, in descending order: heh1Δpom152Δ and heh1Δpom34Δ. Dashed underlined colonies are the double knockout strains expressing the heh2(heh1–442-735) allele. Note that heh2(heh1–442-735) rescues the growth and viability of both heh1Δpom34Δ and heh1Δpom152Δ strains.
We first crossed strains expressing heh2 alleles to a nup170Δ strain, then sporulated and dissected the tetrads into rows. As shown in Figure 5B, nup170Δ strains expressing heh2(1–570), an allele lacking the MCHD, grew slower than either heh2(1–570) or nup170Δ alone, suggesting that the MCHD is the critical functional element of Heh2p that contributes to the loss of viability of heh2Δnup170Δ strains. Consistent with this finding, further deletion of the Heh2p lumenal domain, heh2(1–345), was also unable to complement heh2Δnup170Δ.
In contrast to Heh2p, Heh1p lacking the MCHD [heh1(1–735)] was able to complement the lethality of heh1Δpom152Δ strains (Figure 5C). Strikingly, a subsequent deletion of the lumenal domain [heh1(1–480)] completely abolished the viability of the heh1(1–480)pom152Δ spore (Figure 5C), suggesting that the Heh1p lumenal domain was a critical determinant of the functionality of Heh1p in the absence of Pom152p. We further determined whether the lumenal domain was sufficient to complement heh1Δpom152Δ synthetic lethality by generating deletions of the Heh1p NTD, including GAL1::heh1(51–834), GAL1::heh1(304–834), GAL1::heh1(442–834), and the N-/C-terminal deletions of GAL1::heh1(442–735) and GAL1::heh1(442–703) (Figure 5A). This analysis cumulatively showed that the expression of the first transmembrane domain and lumenal domain was sufficient to restore viability of heh1Δpom152Δ strains (Figure 5C). Because heh1(442–703) could not be efficiently targeted to the INM due to the absence of the NTD, we generated an allele that would ensure the proper targeting of the Heh1p lumenal domain by attaching it to the Heh2p NTD: heh2(heh1–442-735) (Figure 5A). Consistent with the data that the lumenal domain was sufficient to rescue heh1Δpom152Δ lethality, heh2(heh1–442-735) also restored growth of heh1Δpom152Δ (Figure 5D). In addition, we found that heh2(heh1–442-735) could complement the SSS phenotype exhibited by heh1Δpom34Δ cells (Table 1 and Figure 5D). Together, these data suggest that the synthetic fitness defects of heh1Δpom152Δ and heh1Δpom34Δ cells reflect the loss of an essential lumenal function that can be performed by the Heh1p lumenal domain.
Disruption of Pom152p lumenal domain function
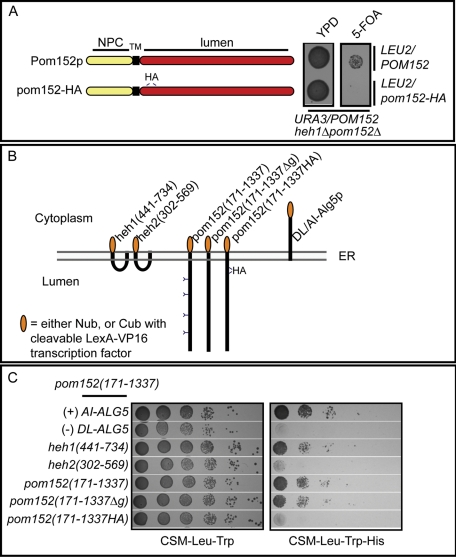
We were intrigued by the requirement and sufficiency of the Heh1p lumenal domain in rescuing the synthetic growth delays of heh1Δpom152Δ and heh1Δpom34Δ strains. These data point to a shared function of Heh1p and the poms within the NE lumen. We considered that, of the three poms, Pom152p has the largest lumenal domain composed of ∼1148 amino acid residues (Wozniak et al., 1994) and is thus the most obvious candidate for supporting this putative role. We hypothesized that, like Heh1p, disrupting the lumenal domain of Pom152p might lead to loss of a critical lumenal function. We therefore asked whether a previously published allele of POM152 (pom152-HA; Tcheperegine et al., 1999; Figure 6A) encoding a version of Pom152p with an in frame insertion of two HA epitopes in its lumenal domain, could complement the lethality of heh1Δpom152Δ. Importantly, this allele has previously been shown to functionally complement other pom152Δ synthetic lethal partners, including strains expressing nup170, nic96, and nup59 alleles (Tcheperegine et al., 1999), in addition to nup1Δ strains (Belanger et al., 2005). For this experiment, heh1Δpom152Δ strains with a URA3/POM152/CEN plasmid and either a LEU2/POM152/CEN or LEU2/pom152-HA/CEN plasmid were assessed for their ability to grow in the absence of URA3/POM152/CEN by plating on medium containing 5-fluoroorotic acid (5-FOA). As shown in Figure 6A, whereas heh1Δpom152Δ was viable with POM152, pom152-HA was unable to complement heh1Δpom152Δ lethality. Thus insertion of the lumenal HA epitopes in Pom152p disrupts a function required in the absence of Heh1p. Together, these data further reinforce the existence of a specific lumenal function for both Heh1p and Pom152p.
FIGURE 6:
Heh1p and Pom152p lumenal domains specifically interact. (A) Schematic showing both full-length Pom152p with NPC, TM, and lumenal (lumen) domains, in addition to the product of an allele of POM152 with two tandem HA peptides (pom152-HA) inserted into the lumenal domain. As shown at right, this allele is unable to support growth of heh1Δpom152Δ strains. An heh1Δpom152Δ strain covered with a URA3/POM152 plasmid and transformed with either a LEU2/POM152 or LEU2/pom152-HA plasmid was plated onto YPD or 5-FOA (to force the loss of URA3/POM152). Images were taken after 2 d at 30°C. (B) Schematic of lumenal domain constructs and topology in ER (gray lines are monolayers) expressed as fusions to both the N (Nub/prey/2μm/TRP1) or C terminus (Cub/bait/CEN/LEU2) of ubiquitin. Cub is fused to a cleavable LexA-VP16 transcription factor released upon Cub-Nub interaction that promotes transcription of the HIS3 gene in the two-hybrid query strain (NMY32). Positive interactions are thus assessed as growth on CSM-Leu-Trp-His. AI-Alg5 and DL-Alg5 are positive (+) and negative (–) controls for bait topology and self-activation, respectively. Numbers are amino acids. Y’s are glycosylation sites. HA is two tandem HA peptides inserted into the Pom152p lumenal domain. (C) NMY32 was simultaneously transformed with plasmids expressing the bait/Cub-fusion of pom152(171–1337) and the indicated prey/Nub fusions. Transformants were spotted in 10-fold serial dilutions on CSM-Leu-Trp plates and CSM-Leu-Trp-His plates and imaged after 3 d at 30°C.
A network of lumenal interactions
Because synthetic genetic relationships are often predictive of direct physical interactions, we wondered whether a lumenal contact exists between the Pom152p and Heh1p lumenal domains. To test this hypothesis, we used a split-ubiquitin–based membrane yeast two-hybrid system (Iyer et al., 2005). We expressed the transmembrane domain and lumenal domain of Heh1p, Heh2p, and Pom152p as fusions to the N (Nub) or C terminus (Cub) of ubiquitin in both single-copy (CEN/LEU2) “bait” and high-copy (2 μm, TRP1) “prey” vectors (Figure 6B). Lumenal domain interactions bring the Nub and Cub domains together allowing the reconstitution of ubiquitin. Ubiquitin is subsequently cleaved by endogenous proteases releasing the LexA-VP16 transcriptional activator that turns on HIS3 conferring growth on medium lacking histidine.
We first tested interactions using the Pom152p lumenal domain [pom152(171–1337)] as bait. We tested multiple control prey constructs that confirmed both bait expression and membrane topology (AI-ALG5), and an inability to self-activate (DL-ALG5) and produce a false-positive result (Figure 6C). We also confirmed the expression and stability of preys by Western blot (Supplemental Figure S4). Gratifyingly, when pom152(171–1337) was coexpressed with the Heh1p lumenal domain heh1(441–734), the strains were able to grow on medium lacking histidine, supporting our hypothesis that the lumenal domains of Heh1p and Pom152p were able to form a complex in vivo (Figure 6C). Importantly, consistent with our genetic analysis, the interaction between Pom152p and Heh1p lumenal domains was specific, as heh2(302–569) did not confer growth under these conditions.
It has been proposed that the oligomerization of the Pom152p lumenal domain could contribute to the formation of the lumenal ring of the NPC (Alber et al., 2007b). We therefore tested whether pom152(171–1337) could also interact with itself. As shown in Figure 6C, strains expressing pom152(171–1337) as both bait and prey were able to grow on medium lacking histidine, supporting the idea that the Pom152p lumenal domains are able to homo-oligomerize. We further demonstrated that the homo-oligomerization of pom152(171–1337) was not affected by mutating putative glycosylation sites in the Pom152p lumenal domain [pom152(171–1337Δg)] (Belanger et al., 2005). Strikingly, the insertion of two tandem HA peptides into the Pom152p lumenal domain (pom152(171–1337HA), at the same position as in pom152-HA (Figure 6A), prevented the interaction with pom152(171–1337). This finding suggests that pom152(171–1337HA) is a monomeric form of the Pom152p lumenal domain. Together, our data support a model in which homotypic interactions exist between Pom152p lumenal domains and they might be specifically required in a pathway shared with the Heh1p lumenal domain.
Nup mislocalization in heh1Δpom34Δ cells
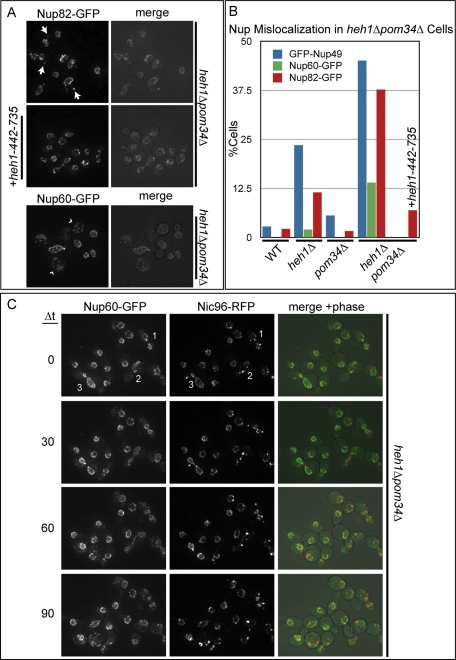
Having established a) a functional network of interactions between the Heh proteins and the NPC and b) the existence of a specific lumenal connection between Heh1p and Pom152p, we wished to evaluate how the disruption of the lumenal function of these proteins affected NPC assembly and/or stability. Since the synthetic growth delays of heh1Δpom34Δ strains could be complemented by the introduction of the Heh1p lumenal domain (Figure 5D), we used heh1Δpom34Δ cells as a proxy to examine the cellular effects of the loss of lumenal function. We first tested whether there were any disruptions to the localization of a number of nups beginning with the cytoplasmically oriented Nup82-GFP. As shown in Figure 7A, Nup82-GFP was dramatically mislocalized in heh1Δpom34Δ cells into cytoplasmic foci reminiscent of what we observed in heh1Δ strains (Figure 1) but present in a substantially higher proportion of the population (∼38% vs. ∼10%; Figure 7B). Consistent with the idea that this nup mislocalization was a result of the disruption of lumenal domain function, when we generated a heh1Δpom34Δ strain expressing the heh2(heh1–442-735) allele, we could restore NE localization of Nup82-GFP (Figure 7A). In this strain, we observed cytoplasmic Nup82-GFP foci in only ∼7% of cells (Figure 7B).
FIGURE 7:
The Heh1p lumenal domain can rescue nup mislocalization in heh1Δpom34Δ cells. (A) Maximum intensity projections of a deconvolved z-series showing the subcellular distribution of Nup82-GFP and Nup60-GFP in heh1Δpom34Δ cells. These images are merged with a phase-contrast image in right panels. Cytoplasmic nup foci are indicated by arrows. The mislocalization of Nup60-GFP is unique, and it appears in a reticular pattern (arrowheads). The mislocalization of Nup82-GFP can be rescued by the expression of the heh2(heh1–442-735) allele (+heh1–442-735). (B) Quantitation of the percentage of cells showing nup mislocalization in the indicated strains and rescue with the heh2(heh1–442-735) allele. (C) A time-lapse series (Δt is 30 min) showing the mislocalization of Nup60-GFP in heh1Δpom34Δ cells delayed in mitosis (see Supplemental Movie S1). Left and middle panels are maximum intensity projections of a z-series of images of Nup60-GFP and Nic96-RFP, respectively, and the right panels are a merge of green, red, and phase-contrast images. Cells marked as 1, 2, or 3 are all proceeding into mitosis. By 90 min, all three cells show a redistribution of both Nup60-GFP and Nic96-RFP, most strikingly in cell 3, where there is a mitotic catastrophe that results in a loss of NE organization.
We further examined the distribution of additional nups that once assembled into the NPC, exist either in the central channel (GFP-Nup49p) or at the nuclear basket (Nup60-GFP). GFP-Nup49 was mislocalized into cytoplasmic foci in proportions (43%) similar to Nup82-GFP. Interestingly, the nucleoplasmically facing Nup60-GFP did not accumulate within cytoplasmic foci. In ∼2% of heh1Δ cells and ∼11% of heh1Δpom34Δ cells (Figure 7B), however, we observed a striking mislocalization of Nup60-GFP (Figure 7A, arrowheads), whereby it redistributed into a cytoplasmic reticular pattern. In these cells, it was often no longer possible to determine the location of the NE from other cellular membranes, suggesting that there was a collapse of NE organization. Taken together, our data support that there is a general disruption of NPC integrity and/or a defect in the normal assembly of NPCs in heh1Δpom34Δ cells that is at least partially dependent on a lumenal function of Heh1p.
Mislocalization of Nup60-GFP occurs during mitotic delays
Because Nup60-GFP was mislocalized in only ∼11% of heh1Δpom34Δ cells, we wondered whether we could correlate its redistribution to a distinct phase of the cell cycle. Using time-lapse microscopy, we visualized the distribution of Nup60-GFP in heh1Δpom34Δ cells also expressing Nic96-RFP. Figure 7C shows a representative time-lapse of a subset of heh1Δpom34Δ cells at 30-min intervals (see Supplemental Movie S1 for complete time course). In the field shown there are three cells in which Nic96-RFP appears in foci in the cytoplasm (cells marked as 1, 2, and 3). All three cells had daughter buds that were similar in size to the mother, suggesting that they were delayed within the G2/M phase of the cell cycle. Consistent with a G2/M delay, cell 1 was unable to complete mitosis over the 90 min shown, and cells 2 and 3 take 60–90 min to complete anaphase (the average budding yeast cell cycle is ∼90 min). Interestingly, whereas the mislocalization of Nic96-RFP was immediately obvious in all three cells, there was also a gradual increase of Nup60-GFP in the cytoplasm alongside a concomitant decrease of its NE fluorescence intensity. Cell 2 was able to complete anaphase, but the resulting mother and daughter cells had significant perturbations to the distribution of both Nic96-RFP and Nup60-GFP. Cell 1 was not able to complete mitosis, and there was an eventual collapse of NE organization such that it was difficult to discern the location of the nucleus. More dramatically, the daughter of cell 3 appeared to undergo a similar loss of NE organization as both Nup60-GFP and Nic96-RFP were redistributed throughout the cell in what resembled a mitotic catastrophe, which occurred ∼15 min after anaphase completion (Supplemental Movie S1). We suspect that these cells ultimately senesce because neither cell 1 nor the daughter of cell 3 progress into another cell cycle over the next few hours (Supplemental Movie S1). These data support a model in which NPC integrity and/or assembly are disrupted in heh1Δpom34Δ cells during mitotic delays which might ultimately lead to the complete disruption of normal NE organization and a loss of cell viability.
DISCUSSION
We have uncovered an interaction network mediated by two conserved members of the LEM family of integral INM proteins, Heh1p and Heh2p, and the NPC. Despite the amino acid sequence-level similarity of these proteins (see Supplemental Figure S5), they display largely specific functional elements that uniquely impact NPC biology. Our data support a model in which Heh2p contributes to the normal distribution of NPCs at the NE, whereas Heh1p plays a role in a pathway that supports NPC assembly and/or integrity.
Our conclusion that Heh1p contributes to the assembly and/or integrity of NPCs is based primarily on the accumulation of a number of nups within the cytoplasm of heh1Δ strains, which is exacerbated in heh1Δpom34Δ cells (Figure 7). These nups include the cytoplasmically oriented Nup82p, the linker nup Nic96p, and the central channel FG-nup Nup49p. Other nups that we have investigated include Nup188p and Nup133p, both of which show a similar cytoplasmic distribution (unpublished data). Therefore in heh1Δpom34Δ cells there is a mislocalization of a number of the core scaffold and cytoplasmic nups of the NPC. The mislocalization of these specific subsets of nups is telling, because a similar complement of nups accumulates within analogous cytoplasmic structures in strains where the function of the membrane and inner ring nups is disrupted (Flemming et al., 2009; Makio et al., 2009; Onischenko et al., 2009). In addition, strains lacking Apq12p—a membrane protein that alters membrane fluidity to support NPC assembly—there is also a cytoplasmic accumulation of a similar subset of nups (Scarcelli et al., 2007). These shared phenotypes likely reflect the disruption of a common pathway or function, which is supported by our genetic analysis whereby HEH1 exhibits a distinct epistatic profile with inner and membrane ring nup genes, in addition to APQ12. Specifically, although these genetic relationships depend primarily on the function of Heh1p, they are at least partially dependent on Heh2p (Table 1 and Figure 4). Together, these data support a model in which Heh1p functions alongside Apq12p and components of the membrane and inner ring complex to ensure the proper assembly of cytoplasmic and core structures of the NPC.
Interestingly, although we do not observe the accumulation of cytoplasmic nup intermediates in heh2Δ strains, we observe NPC clustering at the NE similar to that seen in outer ring nup deletion strains (Figure 1; Doye et al., 1994; Aitchison et al., 1995a; Pemberton et al., 1995). This phenotypic relationship is again reflected in our genetic analysis showing specific interactions between HEH2 and two outer ring components encoded by NUP120 and NUP84 (Table 1 and Figure 4). These data raise the exciting possibility that Heh1p and Heh2p might both contribute to NPC assembly at discrete steps, one requiring the function of Heh1p with the inner/membrane ring complexes, the other with Heh2p and outer ring components. These data also predict the formation of a physical complex between Heh1p, Heh2p, and nups. Consistent with this idea, we can detect interactions between Heh1p and Heh2p and a fraction of the total cellular pool of Pom152p and Nup170p in our affinity pull-down analysis (Figure 3). Furthermore, by fluorescence microscopy, Heh1-GFP and Heh2-GFP colocalize with a fraction of Nic96-RFP (Figure 2). Together, these data suggest that Heh1p and Heh2p are not likely constitutive members of the NPC, but might exist with nups in a transient NPC assembly intermediate.
We envision a number of potential roles for Heh1p and Heh2p in the NPC assembly pathway. Because they have domains that extend into the nucleus and into the NE lumen, they are ideally positioned to interact with chromatin, recruit nuclear nups, and, by spanning the NE lumen, recruit membrane proteins/poms to the ONM. An obvious candidate that might interact with Heh1p across the NE lumen is Pom152p, because its lumenal domain is large (∼1148 amino acids), and it is predicted to form an extended conformation with repetitive β-rich domains similar to cadherin (Devos et al., 2006). In support of this model, we demonstrate a specific interaction between the Pom152p and Heh1p lumenal domains (Figure 6C). To our knowledge, this is the first example of a physical lumenal interaction for Pom152p, and it supports the existence of a novel NE lumenal bridge important for NE function.
We envision that a lumenal bridge between Heh1p and Pom152p could serve to coordinate both nuclear and cytoplasmic nups at a site of NPC assembly. We are also drawn to the possibility that this lumenal connection might directly function in INM/ONM fusion, perhaps by facilitating the disruption of the lumenal leaflets of the INM and ONM necessary for this event. Although our data do not directly address this possibility, they nonetheless point to a critical role for both the Heh1p and Pom152p lumenal domains in NPC assembly. A role for these lumenal domains in NPC assembly is best illustrated by the mislocalization of nups in heh1Δpom34Δ cells, which we link to the function of the Heh1p lumenal domain. Specifically, we can complement both heh1Δpom152Δ lethality and heh1Δpom34Δ synthetic growth delays by expressing the Heh1p lumenal domain (Figure 5D). Indeed, the lumenal domain is likely a key functional element that contributes to the specificity of the HEH1 genetic interactions with the membrane and inner ring complex (Figure 4). Most strikingly, the reintroduction of the lumenal domain of Heh1p in heh1Δpom34Δ strains rescues the mislocalization of Nup82-GFP, supporting the interpretation that the gain in viability under these conditions could be a result of a restoration of efficient NPC assembly (Figure 7). Furthermore, we find that the integrity of the Pom152p lumenal domain is essential to complementing heh1Δpom152Δ lethality as well (Figure 6A). Most interestingly, the allele that cannot restore viability of heh1Δpom152Δ likely encodes a version of Pom152p that is unable to homo-oligomerize (Figure 6C). These findings support a model in which the oligomerization of the Pom152p lumenal domain is required in early NPC assembly events.
Although much of this work focuses on the role of lumenal domains in NPC assembly, the lumenal domains of both Heh1p and Heh2p are directly connected to nuclear domains that have the capacity to influence transcriptional processes (Krogan et al., 2003; Grund et al., 2008), in addition to chromatin organization (Rodriguez-Navarro et al., 2002) and genome stability (Mekhail et al., 2008). We interpret the mitotic delays in heh1Δpom34Δ cells that lead to a dramatic loss of NE organization as a role for Heh1p (and perhaps NPCs) in these genome functions (Figure 7C). We are attracted to the possibility that chromatin binding by either Heh1p or Heh2p might directly influence NPC assembly. Consistent with this possibility, it has been demonstrated that disturbing chromatin organization can have a profound impact on the NE and NPCs (Titus et al., 2010). We imagine that Heh1p and Heh2p could transmit genomic information from the nucleus to the lumen and that this transmission could provide an input for the initiation of an NPC assembly event. Such a mechanism is plausible based on our ability to pinpoint the MCHD of Heh2p, a likely DNA-binding domain (Caputo et al., 2006), as the necessary element required for viability in the absence of Nup170p (Figure 5B). Dissecting the roles for Heh1p/Heh2p in chromatin organization and NPC assembly will be a key future challenge.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains (Supplemental Table S1) were grown at 30°C in YPD (1% yeast extract, 2% peptone, 2% dextrose), YPRG (1% yeast extract, 2% peptone, 1% raffinose, 1% galactose), or in complete synthetic medium (CSM) lacking the appropriate amino acid(s). Standard yeast manipulations were performed as described (Adams et al., 1997; Longtine et al., 1998). Plasmids are listed in Supplemental Table S2.
Epistatic analysis
Strains of heh1Δheh2Δ were systematically crossed with nup deletion strains, diploids were sporulated, and tetrads were dissected using standard yeast protocols and a Singer MSM 300 dissection microscope (Singer Instruments, Watchet, Somerset, UK). Synthetic genetic interactions were assessed after growing freshly dissected spores at 30°C on YPD plates for 48 h. A summary of interactions is presented in Table 1 and Figure 4. SL interactions were determined by an inability to recover viable double, triple, or quadruple deletion strain progeny (Supplemental Figure S1), whereas an SSS interaction was assessed by a clear difference in colony size (Supplemental Figure S1). In cases in which potential genetic interactions did not notably affect the growth of colonies on the dissection plate, strains were serially 10-fold diluted onto additional YPD plates and growth assessed after 24 h at 30°C. If there was a growth delay in these strains, they were assessed as SS (Supplemental Figure S1).
Microscopy
Cells were grown to early log-phase and immobilized on a 1.5% agarose pad containing CSM before imaging with an Applied Precision Deltavision wide-field deconvolution microscope (Applied Precision, Issaquah, WA) with a CoolSnap HQ2 camera (Photometrics, Tucson, AZ). For images shown in Figure 2, cells were collected by centrifugation and fixed in methanol for 10 min at 4°C. Cells were subsequently washed with phosphate-buffered saline, collected by brief centrifugation, and resuspended in Fluoromount-G (Electron Microscopy Sciences, Hatfield, PA), pipetted onto slides, and imaged.
Image manipulation and measurements
In all images shown, a z-series was deconvolved using the iterative algorithm in softWoRx (version 4.0.0; Applied Precision). For certain images (indicated in figure legends) maximum intensity projections were generated. Additional image manipulation and analysis were performed using ImageJ. In Figure 7, the contrast was linearly enhanced to saturate 0.4% of pixels to facilitate the visualization of nup mislocalization. To generate the chart in Figure 2C, dozens of nup133Δ cells expressing either Heh1-GFP or Heh2-GFP were examined for at least one region of colocalization with Nic96-RFP. These numbers were plotted as a percentage of total cells. Similarly, the presence of nup mislocalization was also quantified and presented as the percentage of cells in a given strain (Figure 7B).
Affinity purifications of Heh1- and Heh2-GFP
Cells were cryolysed using a ball mill (Retsch, Haan, Germany) as described (Alber et al., 2007a). For each experiment, ∼1 g of frozen grindate was resuspended in 5 ml of 50 mM HEPES, pH 7.4, 200 mM NaCl, 2 mM MgCl2, 0.1% Nonidet P-40, 1% Triton X-100. The lysate was homogenized using a polytron and centrifuged at 3000 × g for 10 min. Supernatants were incubated for 30 min with magnetic beads covalently coupled to anti-GFP antibodies (Miltenyi Biotech, Auburn, CA) at 4ºC. Beads were collected on a magnet then washed, and bound proteins were eluted with SDS–PAGE sample buffer. For Western blots, the following antibodies were used: mAb414 (Covance, Princeton, NJ), anti-GFP (gift from M. Rout), anti-Histone H3 (Millipore, Billerica, MA), anti-myc (9E10; Roche, Basel, Switzerland), and anti-HA (HA.11; Covance).
Membrane yeast two-hybrid system
Bait (CEN/LEU2) and prey (2μm/TRP1) plasmids encoding lumenal domains as N-terminal fusions of either the C or N regions of ubiquitin (Supplemental Table S2) were cotransformed into NMY32 (Supplemental Table S1) (Dualsystems Biotech, Schlieren Switzerland). After 3 d at 30°C, transformants were 10-fold serially diluted and plated onto CSM-Leu-Trp and CSM-Leu-Trp-His plates, and grown for 3 d at 30°C.
Supplementary Material
Acknowledgments
This work was initiated in the laboratory of G. Blobel and would not have been possible without his support. We thank S. Wente, R. Wozniak, M. Rout, and K. Belanger for reagents listed in the text and M. King and members of the Lusk lab for critical reading of the manuscript. We are also grateful to S. Osmani for sharing unpublished data. C.P.L is supported by the G. Harold and Leila Y. Mathers Charitable Foundation.
Abbreviations used:
- CSM
complete synthetic medium
- ER
endoplasmic reticulum
- Esc
establishes silent chromatin
- FG
phenylalanine-glycine
- GFP
green fluorescent protein
- HA
hemagglutinin
- INM
inner nuclear membrane
- LEM, Lap2, emerin
MAN1
- NC
no change
- MCHD
MAN1-C-terminal Homology Domain
- NE
nuclear envelope
- NPC
nuclear pore complex
- NTD
N-terminal domain
- ONM
outer nuclear membrane
- POM
pore membrane
- RFP
red fluorescent protein
- SL
synthetic lethality
- SS
synthetic sickness
- SSS
severe synthetic sickness
- SUN, Sad1p
Unc84
- TREX
transcription export
- WT
wild-type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-06-0554) on February 16, 2011.
REFERENCES
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol. 1995a;131:1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995b;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Zappulla DC, Ansari A, Perrod S, Laiosa CV, Gartenberg MR, Sternglanz R. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol Cell Biol. 2002;22:8292–8301. doi: 10.1128/MCB.22.23.8292-8301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Belanger KD, Gupta A, MacDonald KM, Ott CM, Hodge CA, Cole CM, Davis LI. Nuclear pore complex function in Saccharomyces cerevisiae is influenced by glycosylation of the transmembrane nucleoporin Pom152p. Genetics. 2005;171:935–947. doi: 10.1534/genetics.104.036319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Schwartz TU. Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice. Nat Struct Mol Biol. 2009;16:1173–1177. doi: 10.1038/nsmb.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo S, Couprie J, Duband-Goulet I, Konde E, Lin F, Braud S, Gondry M, Gilquin B, Worman HJ, Zinn-Justin S. The carboxyl-terminal nucleoplasmic region of MAN1 exhibits a DNA binding winged helix domain. J Biol Chem. 2006;281:18208–18215. doi: 10.1074/jbc.M601980200. [DOI] [PubMed] [Google Scholar]
- Chadrin A, Hess B, San Roman M, Gatti X, Lombard B, Loew D, Barral Y, Palancade B, Doye V. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J Cell Biol. 2010;189:795–811. doi: 10.1083/jcb.200910043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial HJ, Rout MP, Giddings TH, Winey M. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J Cell Biol. 1998;143:1789–1800. doi: 10.1083/jcb.143.7.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- D'Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- Dawson TR, Lazarus MD, Hetzer MW, Wente SR. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J Cell Biol. 2009;184:659–675. doi: 10.1083/jcb.200806174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt EC. A novel nuclear pore protein Nup133p with distinct roles in poly(A) +RNA transport and nuclear pore distribution. EMBO J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- Dultz E, Ellenberg J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol. 2010;191:15–22. doi: 10.1083/jcb.201007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtman B, Ramos C, Rasala B, Harel A, Forbes DJ. Inner/outer nuclear membrane fusion in nuclear pore assembly: biochemical demonstration and molecular analysis. Mol Biol Cell. 2010;21:4197–4211. doi: 10.1091/mbc.E10-04-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming D, Sarges P, Stelter P, Hellwig A, Bottcher B, Hurt E. Two structurally distinct domains of the nucleoporin Nup170 cooperate to tether a subset of nucleoporins to nuclear pores. J Cell Biol. 2009;185:387–395. doi: 10.1083/jcb.200810016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, de Marco A, Wilm M, Antony C, Mattaj IW. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J. 2005;24:3519–3531. doi: 10.1038/sj.emboj.7600825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8:165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol. 2007;17:1657–1662. doi: 10.1016/j.cub.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Dang T, Pane N, Shevchenko A, Mann M, Forbes D, Hurt E. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber UF, Senior A, Gerace L. A major glycoprotein of the nuclear pore complex is a membrane-spanning polypeptide with a large lumenal domain and a small cytoplasmic tail. EMBO J. 1990;9:1495–1502. doi: 10.1002/j.1460-2075.1990.tb08267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund SE, Fischer T, Cabal GG, Antunez O, Perez-Ortin JE, Hurt E. The inner nuclear membrane protein Src1 associates with subtelomeric genes and alters their regulated gene expression. J Cell Biol. 2008;182:897–910. doi: 10.1083/jcb.200803098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg E, Wozniak RW, Blobel G. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol. 1993;122:513–521. doi: 10.1083/jcb.122.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Chan RC, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes DJ. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell. 2003a;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003b;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hattier T, Andrulis ED, Tartakoff AM. Immobility, inheritance and plasticity of shape of the yeast nucleus. BMC Cell Biol. 2007;8:47. doi: 10.1186/1471-2121-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Platani M, Santarella R, Wozniak RW, Mattaj IW. Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol Biol Cell. 2008;19:1753–1762. doi: 10.1091/mbc.E07-08-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Shibuya EK, Wozniak RW. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell. 2005;16:2382–2394. doi: 10.1091/mbc.E04-10-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW, Walther TC, Mattaj IW. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Hodge CA, Choudhary V, Wolyniak MJ, Scarcelli JJ, Schneiter R, Cole CN. Integral membrane proteins Brr6 and Apq12 link assembly of the nuclear pore complex to lipid homeostasis in the endoplasmic reticulum. J Cell Sci. 2010;123:141–151. doi: 10.1242/jcs.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer K, Burkle L, Auerbach D, Thaminy S, Dinkel M, Engels K, Stagljar I. Utilizing the split-ubiquitin membrane yeast two-hybrid system to identify protein-protein interactions of integral membrane proteins. Sci STKE. 2005;2005:pl3. doi: 10.1126/stke.2752005pl3. [DOI] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Krogan NJ et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Lau CK, Giddings TH, Jr, Winey M. A novel allele of Saccharomyces cerevisiae NDC1 reveals a potential role for the spindle pole body component Ndc1p in nuclear pore assembly. Eukaryot Cell. 2004;3:447–458. doi: 10.1128/EC.3.2.447-458.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leksa NC, Brohawn SG, Schwartz TU. The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure. 2009;17:1082–1091. doi: 10.1016/j.str.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Felberbaum R, Hochstrasser M. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J Cell Biol. 2007;178:813–827. doi: 10.1083/jcb.200702154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Pante N, Misteli T, Elsagga M, Crisp M, Hodzic D, Burke B, Roux KJ. Functional association of Sun1 with nuclear pore complexes. J Cell Biol. 2007;178:785–798. doi: 10.1083/jcb.200704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lusk CP, Makhnevych T, Marelli M, Aitchison JD, Wozniak RW. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J Cell Biol. 2002;159:267–278. doi: 10.1083/jcb.200203079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid AS, Mancuso J, Cande WZ, Weis K. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J Cell Biol. 2006;173:361–371. doi: 10.1083/jcb.200506199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makio T, Stanton LH, Lin CC, Goldfarb DS, Weis K, Wozniak RW. The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J Cell Biol. 2009;185:459–473. doi: 10.1083/jcb.200810029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J, et al. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell. 2006;22:93–103. doi: 10.1016/j.molcel.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Marelli M, Aitchison JD, Wozniak RW. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J Cell Biol. 1998;143:1813–1830. doi: 10.1083/jcb.143.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Seebacher J, Gygi SP, Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M, Ryan KJ, Wente SR. The integral membrane protein Pom34p functionally links nucleoporin subcomplexes. Genetics. 2006;172:1441–1457. doi: 10.1534/genetics.105.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol. 2010;191:505–521. doi: 10.1083/jcb.201007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutvei A, Dihlmann S, Herth W, Hurt EC. NSP1 depletion in yeast affects nuclear pore formation and nuclear accumulation. Eur J Cell Biol. 1992;59:280–295. [PubMed] [Google Scholar]
- Nehrbass U, Rout MP, Maguire S, Blobel G, Wozniak RW. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J Cell Biol. 1996;133:1153–1162. doi: 10.1083/jcb.133.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E, Stanton LH, Madrid AS, Kieselbach T, Weis K. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J Cell Biol. 2009;185:475–491. doi: 10.1083/jcb.200810030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF, Rout MP, Blobel G. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:1187–1191. doi: 10.1073/pnas.92.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Ramos C, Harel A, Forbes DJ. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell. 2008;19:3982–3996. doi: 10.1091/mbc.E08-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Igual JC, Perez-Ortin JE. SRC1: an intron-containing yeast gene involved in sister chromatid segregation. Yeast. 2002;19:43–54. doi: 10.1002/yea.803. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KJ, McCaffery JM, Wente SR. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J Cell Biol. 2003;160:1041–1053. doi: 10.1083/jcb.200209116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KJ, Wente SR. Isolation and characterization of new Saccharomyces cerevisiae mutants perturbed in nuclear pore complex assembly. BMC Genet. 2002;3:17. doi: 10.1186/1471-2156-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KJ, Zhou Y, Wente SR. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol Biol Cell. 2007;18:886–898. doi: 10.1091/mbc.E06-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli JJ, Hodge CA, Cole CN. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J Cell Biol. 2007;178:799–812. doi: 10.1083/jcb.200702120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD, Tartakoff AM. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Stavru F, Hulsmann BB, Spang A, Hartmann E, Cordes VC, Gorlich D. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J Cell Biol. 2006;173:509–519. doi: 10.1083/jcb.200601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-deCastillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144:839–855. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- Tcheperegine SE, Marelli M, Wozniak RW. Topology and functional domains of the yeast pore membrane protein Pom152p. J Biol Chem. 1999;274:5252–5258. doi: 10.1074/jbc.274.8.5252. [DOI] [PubMed] [Google Scholar]
- Titus LC, Dawson TR, Rexer DJ, Ryan KJ, Wente SR. Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization. Mol Biol Cell. 2010;21:1072–1087. doi: 10.1091/mbc.E09-07-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur YB, Wilson KL, Gruenbaum Y. SUN-domain proteins: “Velcro” that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]
- Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner N, Krohne G. LEM-Domain proteins: new insights into lamin-interacting proteins. Int Rev Cytol. 2007;261:1–46. doi: 10.1016/S0074-7696(07)61001-8. [DOI] [PubMed] [Google Scholar]
- Walther TC, et al. The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2 doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Schwartz TU. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J Biol Chem. 2009;284:28442–28452. doi: 10.1074/jbc.M109.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RW, Blobel G, Rout MP. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J Cell Biol. 1994;125:31–42. doi: 10.1083/jcb.125.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel U, Doye V, Tekotte H, Wepf R, Grandi P, Hurt EC. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J Cell Biol. 1996;133:1141–1152. doi: 10.1083/jcb.133.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.