A small G protein, RalA, was identified as a binding partner of PKCη. The binding led to activation of RalA and actin depolymerization associated with keratinocyte differentiation. These results provide new insight into the molecular mechanism of cytoskeletal regulation that leads to drastic change of cell shape.
Abstract
During differentiation, keratinocytes undergo a dramatic shape change from small and round to large and flat, in addition to production of proteins necessary for the formation of epidermis. It has been shown that protein kinase C (PKC) η is crucial for keratinocyte differentiation. However, its role in this process has yet to be fully elucidated. Here, we show that catalytic activity is not necessary for enlarged and flattened morphology of human keratinocytes induced by overexpression of PKCη, although it is important for gene expression of the marker proteins. In addition, we identify the small G protein RalA as a binding partner of PKCη, which binds to the C1 domain, an indispensable region for the morphological change. The binding led activation of RalA and actin depolymerization associated with keratinocyte differentiation. siRNA techniques proved that RalA is involved in not only the keratinocyte differentiation induced by PKCη overexpression but also normal keratinocyte differentiation induced by calcium and cholesterol sulfate. These results provide a new insight into the molecular mechanism of cytoskeletal regulation leading to drastic change of cell shape.
INTRODUCTION
For unicellular organisms, cell shape change associated with movement, cell division, and mating is fundamental for survival. In multicellular organisms, cell shape varies considerably in response to different cues. For example, neurons develop long neurites in response to several factors and hormones, including nerve growth factor (Wiesmann and de Vos, 2001; Tojima and Ito, 2004; Arimura and Kaibuchi, 2008), and lymphocytes chemotax to chemokines (Stephens et al., 2008). In most cases, dynamic morphological changes are triggered by environmental signals and involve cytoskeletal reorganization mediated by the small-molecular-weight G proteins. It is well known that RhoA, Rac1, and cdc42 induce stress fibers, membrane ruffles, and filopodia, respectively (Narumiya, 1996; Takai et al., 2001). Although how cells generate and maintain their shape and size has been well studied, the precise molecular mechanisms leading to dramatic morphological changes are not fully understood. The terminal differentiation of epidermal keratinocytes involves drastic and dynamic morphological changes, making them a model cell for studying drastic cytoskeletal rearrangement.
The epidermis consists of four layers: basal, spinous, granular, and cornified layers (Fuchs 1990; Koster and Roop, 2007). Once committed to differentiation, the basal cells lose their proliferative potential and are differentiated toward cornified cells becoming larger and flatter. Keratinocytes differentiation involves not only these morphological changes but also a shift in protein expression (Fuchs, 1990; Koster and Roop, 2007). When the basal cells are differentiated to the spinous cells, keratin expression shifts from K5/K14 to K1/K10. Differentiation from spinous to granular is marked by expression of keratinocyte-specific transglutaminase 1 (TGase1), and its substrates, including involucrin, filaggrin, and small proline rich protein (SPR), are expressed. TGase1 covalently cross-links the substrates, resulting in forming the cornified layer. Together the morphological and biochemical changes produce the epidermis, an effective function as a barrier to external surroundings.
Keratinocyte differentiation is partly regulated by protein kinase C (PKC), particularly the transition from spinous to granular cells (Denning, 2004; Dlugosz and Yaspa, 1994; Koster and Roop, 2007). Ten subtypes of PKC have been cloned and classified into three groups based on the structure of their regulatory domain (Nishizuka, 1992; Newton, 2006). Conventional PKCs (cPKC: α, βI, βII, and γ) have two common regions, diacylglycerol (DAG)-binding C1 domain and Ca2+-binding C2 domain, in the regulatory domain. Calcium, phosphatidylserine, and DAG are required for their activation. The novel PKCs (nPKC: δ, ε, η, and θ) are activated by DAG, but not by Ca2+, and atypical PKCs (aPKC: ι and ζ) are insensitive to both DAG and Ca2+. Of the 10 subtypes, keratinocytes express at least 5 PKC subtypes: α, δ, ε, η, and ζ (Denning, 2004). Of these, the role of PKCη in keratinocyte differentiation has been well studied (Osada et al., 1990, 1993; Ohba et al., 1998). Overexpression of PKCη induces the expression of TGase1 necessary for differentiation. Cholesterol sulfate (CS), an abundant lipid in the granular layer (Kagehara et al., 1994), specifically activates PKCη (Ikuta et al., 1994) and induces keratinocyte differentiation (Denning et al., 1995). However, the molecular mechanism by which this occurs is still unknown.
Therefore we performed a series of experiments to determine the molecular mechanism of PKCη-induced keratinocyte differentiation. Here, we show that the kinase activity of PKCη is not necessary for the morphological change, although it is very important for gene expression of the marker proteins. The small-molecular-weight G protein RalA binds to the C1 domain of PKCη and contributes to the morphological change during keratinocyte differentiation induced by not only PKCη but also calcium and CS. This is the first report to show RalA binds to C1 domain of PKCη and that this interaction results in RalA activation and actin depolymerization, necessary for keratinocyte differentiation. These findings provide information critical to our understanding of the mechanism regulating cell morphology.
RESULTS
PKCη is important for keratinocyte differentiation, but its kinase activity is not required for the morphological change
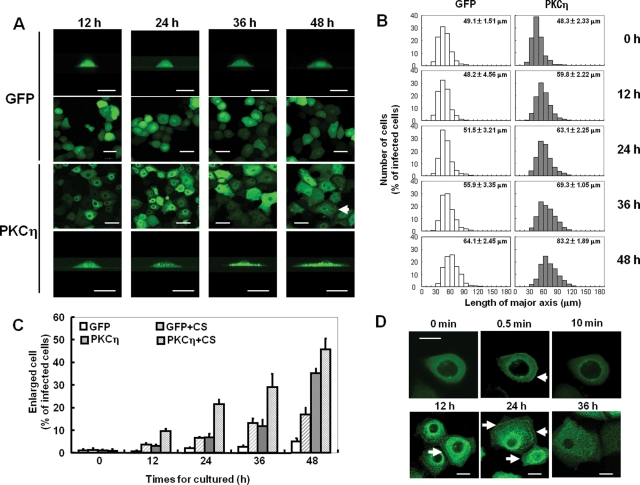
To elucidate the molecular mechanisms underlying the PKCη-associated keratinocyte differentiation, we first studied the effect of PKCη overexpression on cell height and area as morphological changes are a hallmark of it. Normal human epidermal keratinocytes (NHEK) expressing green fluorescent protein (GFP)-PKCη were flatter and more spread than cells expressing unconjugated GFP (Figure 1). Morphological changes were evident by 36 h and more pronounced by 48 h after adenoviral infection (Figure 1A). Before the infection with GFP-PKCη, the average diameter of NHEK was ∼48 μm; the diameter incresed to 69.3 and 83.2 μm at 36 and 48 h postinfection, respectively (Figure 1B). In contrast, NHEK cells expressing GFP alone were significantly less spread at both time points (55.9 and 64.1 μm at 36 and 48 h, respectively). To quantify enlargement, the number of the cells larger than 90 μm in diameter, which is almost twice as large as untreated cells, was counted and this percentage reported in Figure 1C. For cells expressing PKCη, the percentage of the enlarged cells was 15% at 36 h and 35% at 48 h, whereas that of the control cells was <10%, even at 48 h (Figure 1C). Interestingly, the distribution of PKCη-GFP changed from homogeneous at early time points to punctuate in the flattened cells (Figures 1A and 2A, arrows). This distribution of PKCη-GFP was also seen in CS-treated cells at 24 and 36 h (Figure 1D). Additionally, CS treatment induced the translocation of PKCη-GFP to the plasma membrane at 0.5 min, 12 h, and 24 h (Figure 1D, arrows). Finally, CS stimulation enhanced the effects, increasing the average size of PKCη-overexpressing cells from 69.3 μm to 79.7 μm at 36 h (Supplemental Figure S1), and the percentage of cells larger than 90 μm was ∼25% and 33% at 24 and 36 h, respectively (Figure 1C), more than double that of non–CS-treated PKCη-overexpressing cells. These results indicate that CS enhanced the effect of PKCη on the morphology, confirming that PKCη is involved in keratinocyte differentiation.
FIGURE 1:
Effect of PKCη overexpression on the morphology of keratinocytes. (A) Confocal images of NHEK cells expressing PKCη-GFP or GFP alone. NHEK cells were infected with adenoviruses encoding PKCη-GFP or GFP and fixed at indicated time points after the infection. The top and bottom rows show typical cross-section images. Arrow shows the enlarged and flattened cell having typical punctuate localization of GFP-PKCη. Bars are 50 μm. (B) Static analysis of the effect of PKCη overexpression on cell size. After being infected with the adenoviruses, NHEK cells were cultured for indicated time periods and fixed. The longest axis of 300 cells was measured using a soft wear of LSM501 and plotted as a percentage of the infected cells. Numbers in the graph represent average ± SE of the longest axis. (C) Effect of CS treatment on the PKCη-induced morphological change. NHEK cells expressing PKCη-GFP or GFP alone were cultured with or without 50 μM CS for indicated time periods. Longest axes of 100 cells were measured at each experiment, and average of the percentage of the cells having axis longer than 90 μm was indicated. Each data point indicates mean ± SE of three independent experiments. (D) Effect of CS treatment on the localization of PKCη in NHEK cells. NHEK cells expressing PKCη-GFP were cultured with 50 μM CS for indicated time periods and observed under confocal microscopy. Arrows indicate membrane localization of PKCη-GFP. Bars are 20 μm.
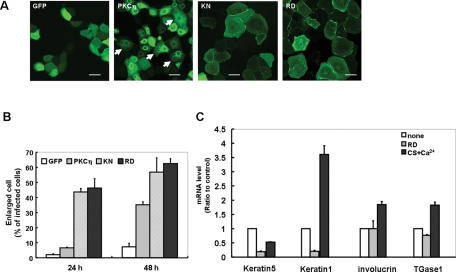
FIGURE 2:
Effect of PKCη activity on keratinocyte morphology and mRNA level of marker proteins. (A) Confocal images of NHEK cells expressing KN- or RD-PKCη. The images were taken 24 h after infection. Arrows indicate the enlarged and flattened cell having typical punctuate localization of GFP-PKCη. Bars are 50 μm. (B) Static analysis of the effect of KN- or RD-PKCη on cell size. NHEK cells expressing KN-GFP, RD-GFP, PKCη-GFP, or GFP alone were cultured for 24 or 48 h without any stimulation, followed by the same procedure described in Figure 1C. (C) Effect of overexpression of RD on mRNA level of the marker proteins. The mRNA was extracted from NHEK cells 48 h after the infection with RD or treated with or without 50 μM CS and 0.12 mM CaCl2 for 48 h. Data points represent mean ± SE of three independent experiments.
To determine whether PKCη activity is required for the observed morphological changes, we tested the effects of kinase-negative (KN)-PKCη and the PKCη regulatory domain (RD) on spreading. Compared with cells expressing wild-type PKCη, those expressing KN-PKCη and RD-PKCη were significantly larger at 24 h (Figure 2A). More than 40% of the cells expressing KN-PKCη or RD were larger than 90 μm by 24 h (Figure 2B), compared with <10% for wild-type PKCη (Figure 1, B and C). KN- and RD-PKCη showed punctuate localization in the cytoplasm in addition to the membrane localization at 24 h (Figure 2A). Similarly, overexpression of RD-PKCη enlarged and flattened HaCaT cells (see Figure 7, B and C, later in this article), suggesting that this phenomenon is not unique to NHEK cells. These results demonstrate that PKCη activity is not necessary for the enlargement and flattening of keratinocytes; RD is sufficient.
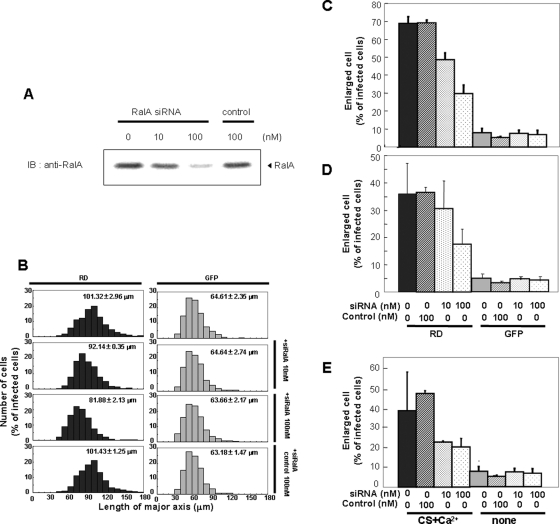
FIGURE 7:
Effect of RalA siRNA on the morphological change of keratinocytes. (A) Effect of siRNA on protein levels of endogenous RalA in HaCaT cells. After 48 h treatment with indicated concentration of RalA siRNA or control RNA (control), the cells were homogenized and applied to 10% SDS–PAGE followed by immunostaining using anti–RalA antibody. (B, C) Effect of siRNA on the RD-induced morphological change. After 24 h treatment with target (siRNA) or control RNA (control), HaCaT cells were infected with adenovirus for RD or GFP alone and cultured for an additional 24 h. The Longest axis of 300 cells in total was measured and plotted. Numbers in the graphs indicate average diameter of cells. The percentage of the cells having an axis longer than 90 μm was plotted in (C). (D) Effect on the RD-induced morphological change of NHEK. Same experiment was performed using NHEK cells. (E) Effect of RalA siRNA on the morphological change of NHEK induced by CS and calcium. After 24 h treatment with target or control RNA, NHEK cells were treated with or without 50 μM CS and 0.12 mM CaCl2 and cultured for an additional 24 h, followed by the same procedure as that described in (B) and (C).
However, keratinocyte differentiation involves both morphological and biochemical changes. We therefore used quantitative RT-PCR to determine the mRNA levels of marker proteins for keratinocyte differentiation. As reported previously, message levels for keratin 5 decreased in keratinocytes differentiated by calcium and CS, whereas those of keratin 1, involucrin, and TGase1 increased (Figure 2C). In contrast, the levels of these maker proteins did not increase upon expression of RD-PKCη. Notably, keratin 1 and 5 decreased in RD-expressing cells relative to control. These results indicate that overexpression of RD-PKCη induces the morphological, but not biochemical changes associated with differentiation. In other words, the two major processes in the differentiation, namely morphological changes and transcriptional regulation, can be distinguished by their sensitivity to PKCη. We therefore focused our study on defining the role of PKCη in the morphological changes associated with keratinocyte differentiation.
PS and C1 domains are important for RD-induced morphological change
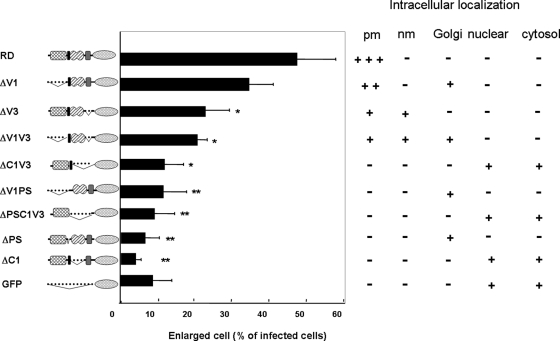
The RD of PKCη consists of several domains, including variable region 1 (V1), pseudosubstrate region (PS), conserved region 1 (C1), and variable region 3 (V3). To identify the domain(s) involved in the RD-induced morphological changes, RD mutants lacking each domain were produced and their effects on cell spreading determined. As shown in Figure 3, deletion of V1 domain did not significantly affect the RD-induced morphological changes; more than 30% of the cells overexpressing ΔV1 were flattened and enlarged. Further deletion of PS from ΔV1 (ΔV1PS), as well as deletion of C1 and V3 (ΔC1V3), abolished the shape change. Deletion of V3 alone (ΔV3), or in combination with V1 (ΔV1V3), decreased cell enlargement ∼50%. Notably, when either PS or C1 was removed (ΔPS and ΔC1), no morphological changes were seen. These results clearly show that PS, C1, and V3 are involved in the RD-induced morphological change; PS and C1 are particularly important. Interestingly, the mutants that did induce the morphological change (e.g., ΔV1 and ΔV3) localized to the plasma membrane like RD and KN-PKCη, but those mutants that did not induce cell enlagemnent tended to associate with nuclear and Golgi membranes or remain in the cytosol (Figure 3 and Supplemental Figure S2), demonstrating that plasma membrane localization is important to induce the morphological change.
FIGURE 3:
Localization of the PKCη mutant lacking domain(s) and their effects on the morphological changes. Twenty-four hours after the infection with adenovirus encoding PKCη mutants or GFP alone, NHEK cells were fixed following the same procedure described in Figure 1C. *, P < 0.05; **, P < 0.01 compared with RD. + or – represents localization of the mutants based on the results in Supplemental Figure 2. +++, strong localization; +, weaker localization; PM, plasma membrane; nm, nuclear membrane.
RalA binds to PKCη through C1 domain
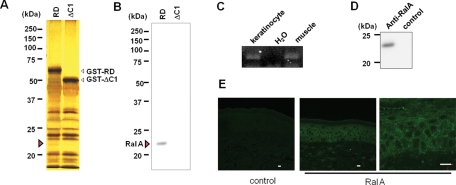
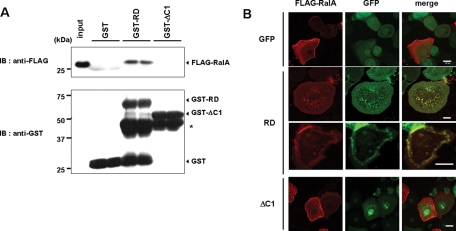
To identify candidate proteins that bind to the PKCη C1 domain and are involved in the RD-induced morphological change, because some C1 domains are known to be involved in the protein–protein interaction (Colon-Gonzalez and Kazanietz, 2006), we made glutathione S-transferase (GST) fusion proteins of RD or ΔC1 and immobilized them on glutathione–sepharose. Lysates of HaCaT cells were applied to the columns, and the bound proteins were eluted with glutathione, separated by SDS–PAGE, and compared by silver staining. A band of ∼23 kDa was present in the RD but not the ΔC1 lane (Figure 4A, red arrow). Mass spectrometric analysis revealed that RalA was present in this band, and immunoblotting for RalA confirmed its presence only in the RD lane (Figure 4B). RalA is a small GTP protein expressed in many tissues, but its expression in skin has not been reported. Therefore we determined the expression of RalA message and protein in keratinocytes. The results of RT-PCR and Western blotting revealed the presence of RalA in the cultured keratinocytes (Figure 4, C and D). Additionally, immunofluorescent staining revealed RalA expression throughout the epidermis. Specifically, its dotty localization on the plasma membrane was evident (Figure 4E, magnified image). Pull-down assays using purified FLAG-RalA and the GST fusion proteins confirmed that RalA directly bound to GST-RD but not to GST-ΔC1 or unconjugated GST (Figure 5A). Coincident with this, FLAG-RalA was colocalized with RD on the plasma membrane of unenlarged cells (Figure 5B, third row) and at dotty accumulation in the cytoplasm of enlarged cells (Figure 5B, second row). In contrast, there was no evidence of colocalization of RalA and ΔC1 (Figure 5B, bottom rows). Together these results indicate that RalA binds to PKCη via its C1 domain.
FIGURE 4:
RalA as binding protein of PKCη and its localization in the skin. (A) Detection of binding protein specific for RD-PKCη via its C1 domain. Silver staining of proteins in the HaCaT lysate eluted from the GST-RD or GST-ΔC1 column. (B) Presence of RalA in the eluate from GST-RD column. The same sample described in (A) was blotted onto PVDF membrane followed by immunoblotting using anti-RalA. (C) Detection of RalA mRNA in the keratinocytes by RT-PCR. mRNA was extracted from normally cultured NHEK cells. mRNA from mouse muscle was used for positive control. (D) Detection of endogenous RalA by mouse anti–RalA antibody in the cultured HNEK. For negative control, mouse anti–FLAG antibody was used. (E) Fluorescent immunostaining of endogenous RalA in the human epidermis. Bars are 20 μm.
FIGURE 5:
Importance of the C1 domain for the RalA binding. (A) Direct binding of FLAG-RalA to GST-RD, but not to GST-ΔC1. Pull-down assay using FLAG-RalA and GST-RD or GST-ΔC1 was performed as described in Materials and Methods. (B) Colocalization of FLAG-RalA with RD-GFP. NHEK cells expressing FLAG-RalA were subsequently infected with viruses coding RD-GFP (RD), ΔC1-GFP (ΔC1), or GFP alone. The cells were fixed 24 h after the infection, followed by immunocytochemistry using anti–FLAG antibody and Alexa 594–conjugated secondary antibody. Both red and green fluorescences were observed under confocal microscopy.
RD-PKCη specifically binds to RalA among several small-molecular-weight GTPases, and the binding is independent of activation state of RalA
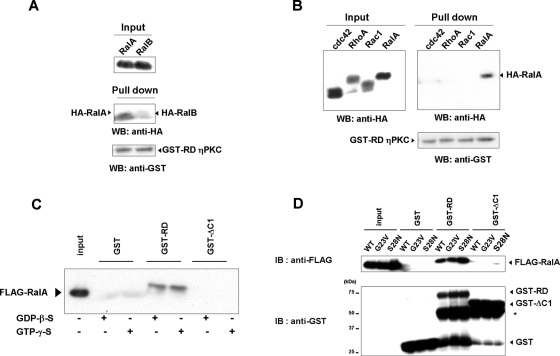
Ral is a member of the Ras family of small GTPases, and Ral proteins consist of two proteins, RalA and RalB, which are 85% identical. We therefore determined the specificity of the binding. Of the two, RD-PKCη preferentially associated with RalA (Figure 6A). In addition, RD-PKCη showed no binding to Rho family small GTPases RhoA, Rac1, and cdc42. These results suggest that binding of RD-PKCη to RalA is relatively specific.
FIGURE 6:
Characteristics of the binding between RalA and RD-PKCη. (A) Weaker binding of RD-PKCη to RalB than RalA. HA-tagged RalA or RalB was expressed in Cos-7 cells. The lysates were incubated with purified GST-RD or GST and pulled down by glutathione–sepharose 4B. The precipitates were applied to 10% SDS–PAGE followed by immunoblotting using HA or GST antibody. (B) No binding of RD-PKCη to Rho family GTPases. Pull-down assay using HA-tagged cdc42, RhoA, or RalA was performed as described in (A). (C) Effect of GDP-β-S or GTP-γ-S on the binding of GST-RD to FLAG-RalA. The binding assay using purified proteins was performed in the presence of 1 mM GDP-β-S or GTP-γ-S. (D) Binding of GST-RD to active form (G28V) and inactive form (G23V) of Ral A. *, degraded products of GST-RD and GST-ΔC1.
Like other small GTPases, RalA has two states: activated and inactivated. The former binds to GTP, whereas the later binds to GDP. To determine which state prefers to bind to RD-PKCη, effects of GTP-γ-S or GDP-β-S on the binding were determined. The binding of FLAG-RalA to RD-PKCη was independent of GTP-γ-S or GDP-β-S (Figure 6C). In addition, both mutants of RalA, G23V and S28N, mimicking active and inactive forms, bound to RD-PKCη (Figure 6D). These results demonstrate that the binding of RD-PKCη to RalA is independent of the nucleotide exchange.
RalA is involved not only in the RD-induced morphological change but also in the normal differentiation of keratinocytes induced by calcium and CS
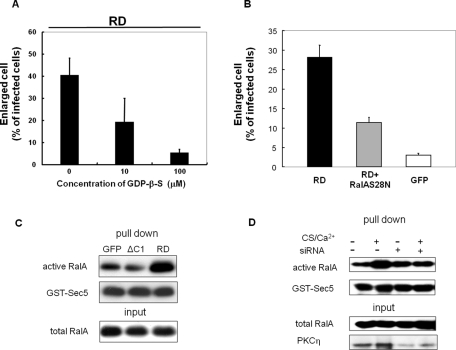
To determine whether RalA is really necessary for the RD-induced morphological changes, we used siRNA to down-regulate RalA in HaCaT cells. siRNA in the amount of 100 nM significantly decreased endogenous RalA expression in HaCaT cells and reduced the RD-dependent cell enlargement (Figure 7, A and B). The average diameter of the cells expressing RD was 101.32 μm, whereas 10 and 100 nM siRNA reduced the diameter to 92.14 and 81.88 μm, respectively (Figure 7B). The percentage of cells larger than 90 μm was reduced from 70–50% (10 nM) and 30% (100 nM), respectively (Figure 7C). siRNA treatment, however, did not affect the size of cells expressing GFP alone (Figure 7, B and C). Importantly, control siRNA, which has only four unmatched nucleotides, did not reduce the protein level of RalA, nor did it inhibit the RD-PKCη–induced morphological changes (Figure 7, A–C). Similar inhibitory effects of siRNA were obtained when NHEK cells were used (Figure 7D). These results demonstrate that RalA is involved in the RD-induced morphological changes. Consistent with this, treatment with GDP-β-S or overexpression of inactive form of RalA inhibited the RD-induced morphological change (Figure 8, A and B), suggesting that RalA is activated during the RD-induced morphological change. Indeed, activation of RalA was detected in the keratinocyte morphological change induced by RD but not ΔC1 (Figure 8C).
FIGURE 8:
Activation of RalA in the RD- and CS-induced morphological change. (A) Effect of GDP-β-S on the RD-induced morphological change. After infection with adenovirus for RD, NHEK cells were immediately cultured with or without GDP-β-S for 24 h. The percentage of the cells having an axis longer than 90 μm was plotted. Data points indicate mean ± SD of three independent experiments. (B) Effect of coexpression of inactive form of RalA (S28N) on the RD-induced morphological change. NHEK cells expressing RD, RD + FLAG-RalAS28N, or GFP alone were cultured for 24 h. After being fixed, the percentage of the cells having an axis longer than 90 μm was plotted. (C, D) Detection of active form of RalA by GST-Sec5. HaCaT cells after 24 h culture of the RD or ΔC1 transfection (C) and 48 h treatment with 50 μM CS and 0.12 mM CaCl2 with siRNA of PKCη or control (D). The cells were homogenized and applied to RalA activation assay as described in Materials and Methods.
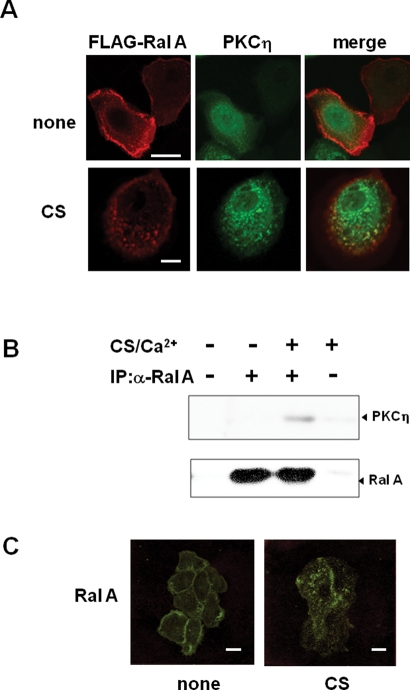
Furthermore, to determine whether RalA acts in morphological changes under more physiological conditions, we repeated the siRNA down-regulation in NHEK treated with calcium and CS. Down-regulation of RalA expression blocked the morphological changes induced by CS and calcium (Figure 7E). Similarly, siRNA of PKCη inhibited the morphological change induced by CS and calcium (Supplemental Figure S3). These results implicated that binding of endogenous RalA to endogenous PKCη is necessary for the normal keratinocyte differentiation. Interestingly, FLAG-RalA and full-length PKCη-GFP did not colocalize 24 h after infection (Figure 9A, top), when 90% of the cells are still smaller than 90 μm (Figure 1). Similarly to endogenous RalA, FLAG-RalA was localized on the plasma membrane, whereas full-length PKCη was detected throughout the cytoplasm (Figure 9A, top). In contrast, CS stimulation resulted in extensive colocalization of PKCη and RalA in puncta of enlarged and flattened cells (Figure 9A, bottom). This result is consistent with a model in which CS treatment enables full-length PKCη to bind to RalA, possibly by changing its conformation and targeting PKCη to the plasma membrane where RalA is localized, with following redistribution of RalA from the plasma membrane to the punctuate structure. Indeed, binding of endogenous RalA to PKCη was detected only in CS-treated cells (Figure 9B), under conditions in which endogenous RalA moved from the membrane to punctuate intracellular structure (Figure 9C). More importantly, activation of RalA was comfirmed in the CS-induced keratinocyte differentiation, and the activation was inhibited by siRNA of PKCη (Figure 8D). These results support that binding of RalA to PKCη and following activation of RalA contribute to the morphological changes during the normal differentiation of keratinocytes.
FIGURE 9:
CS-dependent interaction between RalA and full-length PKCη. (A) CS-dependent colocalization of full-length PKCη with FLAG-RalA. NHEK cells expressing FLAG-RalA (RalA) and PKCη-GFP (PKCη) were cultured for 24 h with or without 50 μM CS. After fixation, FLAG-RalA and PKCη-GFP were detected as described in (A). (B) Binding of endogenous RalA to endogenous PKCη. Immunoprecipitation using RalA antibody was performed as described in Materials and Methods. (C) CS-induced relocation of endogenous RalA. After 24 h incubation with or without 50 μM CS, NHEK cells were fixed followed by immnostaining with RalA antibody. Bars are 20 μm.
PKCη and RalA are colocalized with actin depolymerized during the differentiation of keratinocytes
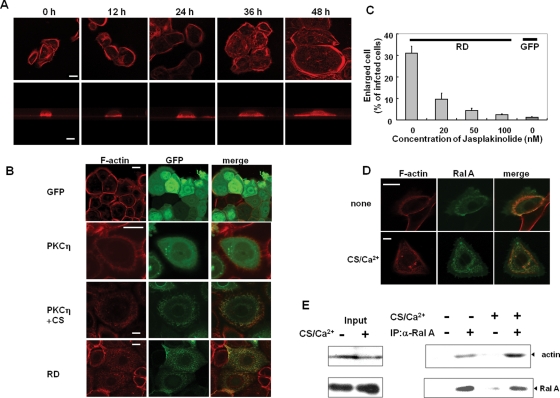
The morphological changes associated with keratinocyte differentiation require actin rearrangement. Thus the localization of actin and PKCη or RalA in response to CS or expression of RD was investigated. The cortical actin cytoskeleton was apparent in the untreated NHEK cells. In response to CS, F-actin reorganized from cortical bands to more fibrous and punctate structures as the cell spread (Figure 10A); PKCη colocalized with actin in these areas (Figure 10B, third row). In the absence of CS, neither unconjugated GFP nor PKCη-GFP colocalized with actin (Figure 10B, first and second rows). Similar F-actin reorganization and its colocalization with RD were confirmed in the case of RD expression; fibular actin was evident in the flattened cells by RD overexpression, whereas cortical actin was still observed in the small cells expressing no RD (Figure 10B, bottom row). These results suggest that actin depolymerization occurs during keratinocyte morphological changes induced by CS treatment or RD expression, and PKCη catalytic activity was not necessary for association with reorganized actin. Consistent with a requirement for actin depolymerization in the spreading, stabilization of actin with jasplakinolide inhibited the RD-induced morphological changes in a dose-dependent manner (Figure 10C).
FIGURE 10:
Functional interrelationships among actin, PKCη, and RalA. (A) Actin reorganization during morphological change induced by CS. After treatment with 50 μM CS for indicated time periods, HNEK cells were fixed and F-actin was visualized with rhodamin-phalloidine. The bottom row shows typical cross-section images All images were taken with the same magnification, and bar is 20 μm. (B) Colocalization of actin and PKCη. The NHEK cells expressing PKCη-GFP, RD-GFP, and GFP alone were cultured for 24 h with or without 50 μM CS. After being fixed, F-actin was visualized with rhodamin-phalloidine. Bars are 20 μm. (C) Effect of jasplakinolide on the RD-induced morphological change. Just after infection with the adenovirus, the NHEK cells were cultured in the presence and absence of jasplakinolide for 24 h. The percentage of the cells having an axis longer than 90 μm was indicated. (D) Colocalization of actin and RalA. The NHEK cells expressing FLAG-RalA were cultured with or without 50 μM CS and 0.12 mM CaCl2. After fixation, F-actin was visualized. Bars are 20 μm. (E) Association of RalA and actin. NHEK cells treated with or without 50 μM CS and 0.12 mM CaCl2 were immunoprecipitated with RalA antibody, followed by immunoblotting using actin.
Interestingly, in contrast to PKCη, RalA colocalized with actin in both resting and CS/Ca2+-treated cells; endogenous RalA was localized on the plasma membrane before the stimulation (Figure 10D). The presence of actin in RalA immunoprecipitates from control and CS/Ca2+-treated NHEK cells (Figure 10E) confirmed the association of endogenous RalA and actin in both the resting and differentiated cells, with more recovered following CS/Ca2+ treatments (Figure 10E). Notably, RalA immunoprecipitation from CS/Ca2+-treated cells also brought down PKCη (Figure 9B). These results are consistent with a model in which CS/Ca2+ increases RalA association with actin and PKCη, resulting in actin depolymerization with redistribution of the actin/RalA/PKCη complex, leading to cell spreading.
DISCUSSION
As previously reported (Ohba et al., 1998), overexpression of PKCη in human keratinocytes induced the enlarged and flattened morphology associated with differentiation. Interestingly, catalytic activity of PKCη was not necessary, and RD is sufficient for the induction of morphological change. Instead, binding of PKCη-RD to RalA was involved in the process. In other words, RD of PKCη not only prevents the kinase activity in the resting state but also functions as scaffold region in the active state. A similar additional function of RD of PKC was reported previously; RD of PKCε induces neurite outgrowth (Zeidman et al., 1999) and involvements of binding protein and/or lipids in this process are suggested (Zeidman et al., 2002; Ling et al., 2005; Shirai et al., 2007). Thus it may be common that regulatory domain of PKC has important roles in addition to “rid” of kinase domain. On the other hand, the kinase activity was necessary for the gene expression of the marker proteins, such as involucrin and TGase, necessary for differentiation. These results are very consistent with previous reports (Ueda et al., 1996; Ohba et al., 1998).
RalA is thought to be multifunctional in mammalian cells, such as regulating membrane transport, apoptosis, transcription cell migration, cell proliferation, and oncogenesis (Feig, 2003; Cammonis and White, 2005; van Dam and Robinson, 2006). However, before this study, there was no report to show its involvement in the morphological change during keratinocyte differentiation. Our novel findings provide a new idea, as shown in Figure 11, in the scenario that PKCη plays a crucial role in normal keratinocyte differentiation and CS is one of the triggers of the activation (Kashiwagi et al., 2002). In undifferentiated keratinocytes, PKCη is localized in the cytoplasm, whereas RalA and actin colocalized at the plasma membrane. Production of CS during the differentiation activates PKCη, resulting in up-regulation of marker proteins for differentiation. Coincidently, CS activation induces the membrane translocation of PKCη and enables its association with RalA. Alternatively, membrane localization of PKCη might require binding to RalA. In either case, the binding of PKCη to RalA appears to be a trigger of activation of RalA and actin depolymerization, leading to the morphological change. Finally, PKCη, RalA, and depolymerized actin are associated and colocalized in the cytoplasm of enlarged and flattened cells. In short, PKCη may function as scaffold proteins of RalA and actin, leading to actin depolymerization. The hypothesis is supported by the following findings. First, actin depolymerization occurs after PKCη translocation; actin depolymerization was evident at 36 and 48 h, whereas PKCη translocation was observed at 12 and 24 h (Figures 1 and 10). Simultaneous observation of PKCη and actin in the living keratinocytes proved the order more directly (data not shown). Second, overexpression of RD, which does not need any stimulator or time to induce both membrane translocation and the RalA binding, enhanced the rate of both the morphological change and the actin depolymerization (Figures 2, 5, and 10B). Third, only mutants that retained both plasma membrane localization and RalA binding induced the morphological changes and RalA activation (Figures 3, 5, and 8C). In addition, full-length PKCη cannot bind and colocalize with RalA without CS stimulation, but RD can (Figures 5B and 9). Furthermore, jasplakinolide inhibited the RD-induced morphological change (Figure 10C), whereas latrunculin A enhanced the morphological change (data not shown).
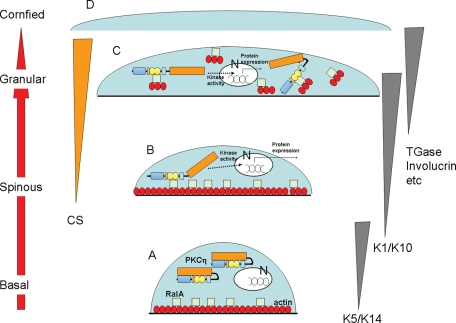
FIGURE 11:
Hypothetic roles of binding of PKCη and RalA during keratinocyte differentiation. (A) In basal cells, PKCη is localized in the cytoplasm, whereas RalA and actin present at the membrane. N, nucleus. (B) Production of CS during the differentiation activates PKCη, resulting in translocation to the membrane. The activated PKCη up-regulates marker proteins for differentiation (dotted lines) and binds to RalA. (C) Binding of PKCη to RalA somehow induces actin depolymerization, resulting in dottily colocalization of PKCη, RalA, and actin in the cytoplasm of enlarged cells. (D) Finally, the enlarged cells differentiated to cornified cells.
In addition to PKCη, many binding proteins have been identified, and the functions of RalA tightly depend on the protein–protein interactions. One of the first identified RalA binding partners was Ral binding protein 1, RalBP1 (Cantor et al., 1995), and the other binding proteins include PLD1, Sec 5, ZONAB, and PLCδ (van Dam and Robinson, 2006). Some of the interactions with binding proteins depend on RalA being activated, whereas others are constitutive. For example, RalBP1 binds to the active form of RalA, whereas PLD1 interacts with both forms. The former type of binding protein tends to bind to the switch region of RalA (van Dam and Robinson, 2006). The binding of RalA to PKCη was independent of the nucleotide exchange (Figure 6), suggesting that PKCη does not bind to the switch region. Although PKCη can bind to both forms, the active form of RalA seems to function in the morphological change because GDP-β-S and coexpression of inactive mutant (S28N) inhibited the RD-induced morphological change (Figure 8).
How RalA is activated during the differentiation is still unknown. Ral proteins can be activated by specific guanine nucleotide exchange factors (GEFs), including RalGDS (Feig, 2003; Cammonis and White, 2005; van Dam and Robinson, 2006). However, we failed to detect RalGDS associated with RalA or PKCη after the CS and calcium stimulation (data not shown), although PKCη is necessary for RalA activation (Figure 8D). RD-PKCη showed no GEF activity (data not shown). It has been recently reported that overexpression of novel class of GEF, RalGPS, causes morphological changes in HEK cells (Rebhum, 2000; Ceriani et al., 2007). Thus RalGPS is a potential candidate for activation of RalA during keratinocyte differentiation. Alternatively, additional production of RalA may be one of the activation mechanisms because the RD overexpression or CS treatment tended to increase mRNA of RalA (data not shown).
In addition, how the binding of RalA to PKCη induces morphological changes is still unclear. Overexpression of wild-type or active RalA alone did not induce the morphological change (data not shown), suggesting a requirement for other factors, possibly binding partner(s). One of the candidates is RalBP1, which interacts with Cdc42 and Rac1 (Cantor et al., 1995; Feig et al., 1996). Indeed, overexpression of inactive Rac1 inhibited the RD-induced morphological change (data not shown). However, we cannot exclude the possibility that inactive Rac1 prevented the adhesion of cells itself because Rac1 has been reported to be necessary for keratinocyte adhesion (Benitah et al., 2005; Tscharntke et al., 2007). The involvement of Rac1 in the morphological changes associated with keratinocyte differentiation is unlikely as it is highly expressed in the basal cells but not in their differentiated counterparts (Benitah et al., 2005), and reduction of Rac1 induces the keratinocyte differentiation (Nikolove et al., 2008). Rather, actin binding proteins are more likely candidates based on the fact that RalA targets filamin to induce filopodia (Ohta et al., 1999; Sugihira et al., 2001). An alternative possibility is the involvement of GTPase-activating proteins (GAP) because immunoprecipitates with RD-PKCη from differentiated keratinocytes, but not RD-PKCη itself, showed slight GAP activity (data not shown). These results suggest that both activation and inactivation of RalA may be involved in the morphological change, contributing to membrane trafficking to provide the additional plasma membrane needed for cell spreading. In fact, RalA is present on secretory vesicles in many cell types (Feig, 2003; van Dam and Robinson, 2006), and PLD, one of the RalA binding proteins, is involved in vesicle budding and transport (Jenkins and Frohman, 2005). More interestingly, PLD is also reported to regulate the differentiation of keratinocytes (Jjung et al., 1998, 1999). The speculation is very attractive, but further experiments are necessary.
In addition to filopodia formation and vesicle transportation described above, a link between RalA and regulation of cell morphology has been reported. Specifically, RalA promotes neurite branching (Lalli and Hall, 2005), and its activation has been implicated in cytoskeletal changes associated with cell stimulation by chemoattractant peptides (Bhattacharya et al., 2002). However, compared with Rho family small GTPases, the molecular mechanism by which RalA modulates the actin cytoskeleton is still enigmatic, and the biological function of RalA has not been fully elucidated. Our findings provide one possible model for how RalA and PKCη regulate the actin cytoskeleton and cellular morphology during keratinocyte differentiation.
MATERIALS AND METHODS
Materials
CS, GDP-β-S, GDP-γ-S were purchased from Sigma (St. Louis, MO). HuMedia-KG2 medium and DMEM were obtained from Kurabo Industries (Osaka, Japan) and Wako (Osaka, Japan). Both penicillin and streptomycin were products of Life Technologies (Carlsbad, CA), and fetal bovine serum (FBS) was from ICN Biomedicals (Irvine, CA). Anti-FLAG and RalA monoclonal antibodies were purchased from Sigma and BD Transduction Laboratories (Lexington, KY), respectively. Anti-GST was from Santa Cruz Biotechnology (Santa Cruz, CA). cDNA encoding FLAG or GST-fused RalA and hemagglutinin (HA)-tagged RalB were provided by T. Kataoka (Kobe University) and T. Sato (Kobe University), respectively. The plasmids encoding HA-tagged Rho family GTPases, Rac1, RhoA, and cdc42, were given by K. Kaibuchi (Nagaoya University).
Construction of adenovirus vector
The cDNA encoding PKCη and mutants were produced by a PCR with cDNA for mouse PKCη as the template. The sense and anti-sense primers used were as follows: for full-length PKCη, 5′-TTAGATCTGGCATGTCGTCCGGCACGATG-3′ and 5′-GGAGATCTCAGTTGCAATTGCAATTCCGGTGA-3′; for RD, 5′-TTAGATCTGGCATGTCGTCCGGCACGATG-3′ and 5′-GGAGATCTGATCCCATTTCCTTCTTCTTTGGA-3′; for V1 deletion (ΔV1), 5′-GGAGATCTATGCGCATCTTCAAGCATTTTA-3′ and 5′-GGAGATCTGATCCCATTTCCTTCTTTGGA-3′; for V1 and P1 deletion (ΔV1P1), 5′-TTAGATCTATGCATAAGTTCATGGCCACG-3′ and 5′-GGAGATCTGATCCCATTTCCTTCTTTGGA-3′; for V3 deletion (ΔV3), 5′-TTAGATCTGGCATGTCGTCCGGCACGATG-3′ and 5′-TTAGATCTCTCCACGGCATTCACCCCGCA-3′; for C1 and V3 deletion (ΔC1V3), 5′-TTAGATCTGGCATGTCGTCCGGCACGATG-3′ and 5′-TTAGATCTTCCGTTCACTTGATGGAC-3′; for PS, C1, and V3 deletion (ΔPSC1V3), 5′-TTAGATCTGGCATGTCGTCCGGCACGATG-3′ and 5′-GGAGATCTGGTCTCTCTGGAGAGTGGCTT -3′, respectively. The PCR products were first subcloned into pUC118 (Invitrogen, San Diego, CA).
In the case of ΔV1V3, ΔPS, and ΔC1, site-directed mutagenesis was performed according to manufacturer’s recommended protocol with ExSite PCR-based site-directed mutagenesis kit (Stratagene, La Jolla, CA). RD in pUC118 was used as a template, and the primers used were as follows: for ΔV1V3, 5′-GGAGATCTATGCGCATCTTCAAGCATTTTA-3′ and 5′-TTAGATCTCTCCACGGCATTCACCCCGCA-3′; for ΔPS, 5′-ATCACTCTGGAGAGTGGCTTC-3′ and 5′-CATAAGTTCATGGCCACGTAC-3′; for ΔC1, 5′-TCCGTTCACTTGATGGACTCG-3′ and 5′-GGGGTTAACGCCGTGGAGCTT-3′, respectively. Furthermore, to make adenovirus of KN mutants, a KN PKCη (Ohba et al., 1998) was used as a template and 5′-TTAGATCTGGCATGTCGTCCGGCACGATG-3′ and 5′-GGAGATCTCAGTTGCAATTGCAATTCCGGTGA-3′ for sense and anti-sense primers. All PCR products were verified by sequencing.
After being digested with BglII, the respective cDNA fragments and a cDNA encoding GFP, which was obtained from BS340 (Shirai et al., 1998) with HindIII digestion, were subcloned into the BglII and HindIII sites of pShuttle–CMV vector. After linearization by PmeI, the pShuttle vector having cDNA encoding fusion protein of GFP with PKCη or its mutant was coelectroporated with pAdEasy backbone vector into BJ5183 bacterial cells. The recombination was checked by PacI, cut, and the plasmids were purified by CsCl banding. After PacI digestion, 10 μg of the purified plasmids was lypofected into 50–70% confluent HEK293 cells plated on a 6-cm dish by Fugene (Roche, Indianapolis, IN). The cells were cultured in DMEM supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), and 10% horse serum (Life Technologies), scraped at 7 d posttransfection, and resuspended in 1 ml of phosphate-buffered saline (PBS) (−). After sonication and centrifugation, the supernatant was infected to 50–70% confluent HEK293 cells plated on a T75 flask. To amplify further, the infection of 30–50% of the viral supernatant to 50–70% confluent HEK293 cells plated on T75 flasks was repeated. Finally, the adenovirus was purified by CsCl banding and titrated.
Cell culture
NHEK were purchased from Kurabo Industries, and human HaCaT cells were kindly provided by Masahiro Oka (Kobe University). Keratinocytes were cultured in HuMedia-KG2 medium supplemented with insulin (10 μg/ml), epidermal growth factor (0.1 mg/ml), hydrocortizone (0.5 μg/ml), and gentamicin (50 μg/ml). HaCaT cells were cultured in DMEM supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), and 10% FBS. COS7 cells were purchased from Riken Cell Bank (Tsukuba, Japan) and cultured in DMEM supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), and 10% FBS. All cells were kept at 37°C in a humidified atmosphere containing 5% CO2.
Adenoviral infection and measurement of cell size
A total of 1 × 106 of the cells were seeded in a 3.5-cm, glass-bottom dish with 2 ml of the medium a day before the infection. High-titer virus solution (>MOI = 5) was added to the dish, and medium was changed to normal medium after 1 h. After culturing with or without reagents including CS, the cells were fixed at appropriate time points by 4% paraformaldehyde and 0.2% picric acid in 0.1 M phosphate buffer overnight at 4°C. The fixed cells were observed under confocal microscopy. The longest axis of 100 cells was measured using a soft wear of LSM501 for each experiment; three independent experiments were performed. Finally, the data were plotted as a percentage of the infected cells.
Construction of FLAG-tagged RalA mutants and HA-tagged RalA
The cDNA encoding FLAG-tagged RalA mutants were produced by site-directed mutagenesis according to manufacturer’s recommended protocol with QuickChange II XL site-directed mutagenesis kit (Stratagene). A cDNA for FLAG-tagged human RalA was used as the template. The sense and anti-sense primers used were as follows: for inactive form, S28N RalA, 5′-GGCAGTGGTGGCGTGGGCAAGAACGCGTTGACTCTACAGTTCATG-3′ and 5′-CATGAACTGTAGAGTCAACGCGTTCTTGCCCACGCCACCACTGCC-3′; for active form, G23V RalA, 5′-CATCATGGTGGGCAGTGTTGGCGTGGGCAAGAGTGCAATGACTCTACAGTTC-3′ and 5′-GAACTGTAGAGTCAGTGCACTCTTGCCCACGCCAACACTGCCCACCATGATG-3′.
To obtain a plasmid encoding HA-tagged RalA, a cDNA for human RalA obtained by BamHI cut from the plasmid encoding FLAG-tagged RalA was cloned into pEF-BOS-HA vector donated by K. Kaibuchi.
Immunocytochemistry
To compare localization of PKCη and its mutants with FLAG-RalA, NHEK cells were lipofected with FLAG-RalA by Fugene (Roche) as described previously (Shirai et al., 2007). Simultaneously, the cells were infected with adenoviruses and fixed as described. The fixed cells were permeabilized with 0.3% Triton X-100/PBS (PBS-T) for 30 min at room temperature and blocked for 1 h with 10% normal goat serum (NGS) in PBS. One-hour incubations with anti-FLAG or RalA monoclonal antibody (mAb) were performed at a 1:800 or 1:1000 dilution with 0.03% PBS-T, respectively. Alexa Fluor 594–conjugated anti–mouse immunoglobulin (Ig)G (Molecular Probes, Eugene, OR) was used as a secondary antibody.
To visualize F-actin, the fixed cells were sequentially treated with 0.3% PBS-T, 10% NGS, and diluted rhodamin–phalloidin (Molecular Probes) with PBS-T (1:250) for 1 h.
Human skin voluntarily provided was fixed with 4% paraformaldehyde in 0.1 M phosphate buffer. After dehydration, the fixed skin was blocked in paraffin wax. The blocked skin sample was sliced and transferred to slides coated with poly-l-lysine. The wax removed with Xylene, and the tissue sections were rehydrated through a graded series of ethanol before being placed in 10 mM PBS. The sections were preincubated with 0.3% H2O2 for 20 min to inactivate endogenous peroxidase activity, with 10% NGS for 1 h to block nonspecific binding sites, and then with 0.1% phenylhydrazine for 20 min to inactivate residential endogenous peroxidase activity. The sections were incubated with RalA antibody (1:2000) in PBS-T overnight at 4°C. After washing with PBS-T, the sections were incubated with Alexa 488–conjugated anti–mouse IgG secondary antibody (1:2000) for an additional 24 h. Finally, the fluorescence was observed under confocal microscopy after washing well.
Confocal microscopic analysis
The fluorescence of the GFP or Alexa 488 was monitored with a confocal laser scanning fluorescence microscope (LSM 510 invert; Carl Zeiss, Jena, Germany) at 488-nm argon excitation using a 515- to 535-nm band-pass barrier filter, whereas red fluorescence was monitored at 543-nm HeNe excitation using a 560-nm long-pass barrier filter.
Recombinant protein expression and purification
cDNA encoding RD or ΔC1 was digested with BglII from the pUC118-based plasmids described previously and subcloned into the BglII site of pGEX 6P-1 (GE Healthcare Bioscience, Uppsala, Sweden). For bacterial expression of GST-fusion proteins, BL21 (DE3) pLys cells were transformed with the expression plasmids. Expression of the recombinant proteins was induced by 0.1 mM isopropyl-1-thio-β-d-galactoside at 25°C for 16 h. The cells were harvested and lysed in the lysis buffer containing 20 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1 mM dithiothreitol (DTT), 5 mM MgCl2, 250 mM sucrose, 1% Triton X-100, 20 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride (PMSF) with handy sonic (Tomy Seiko, Tokyo, Japan). After ultracentrifugation at 10,000 × g for 30 min, fusion proteins were purified using Glutathione-Sepharose 4B (GE Healthcare Bioscience) as previously described (Yamaguchi et al., 2006).
Detection of protein(s) binding to C1 domain of PKCη
HaCaT cells harvested from 20 plates of 10-cm dishes were homogenized in 5 ml of the lysis buffer described above and stored on ice for 30 min. After ultracentrifugation at 10,000 × g for 30 min, the supernatant was applied onto the glutathione–sepharose 4B column immobilizing purified GST-RD or GST-ΔC1. After washing with TEDM buffer (20 mM Tris-HCl [pH8.0], 1 mM EDTA, 1 mM DTT, 5 mM MgCl2, 20 μg/ml leupeptin, and 1 mM PMSF), proteins bound to GST-RD or GST-ΔC1 were eluted with 10 mM reduced gluthathione and 0.1% SDS in 50 mM Tris-HCl buffer (pH 8.0). The eluted proteins were applied to SDS–PAGE followed by silver staining.
Mass spectrometric analysis
Proteins in SDS polyacrylamide gels were visualized by the reverse-staining method. The bands corresponding to proteins were excised, and then proteins in gels were reduced by incubating with 10 mM EDTA/10 mM DTT/100 mM ammonium bicarbonate for 1 h at 50°C and alkylated by treatment with 10 mM EDTA/40 mM iodoacetamide/100 mM ammonium bicarbonate for 30 min at room temperature. They were digested in gel with lysyl endopeptidase (LEP) from Achromobacter lyticus (Wako Pure Chemical Industries) in 100 mM Tris/HCl (pH 8.9) for 15 h at 37°C. Resulting peptide fragments were extracted from gels and then concentrated in vacuo. After desalting with ZipTipC18 (Millipore, Billerica, MA), peptide fragments were subjected to mass spectrometric analysis. Positive ion mass spectra were acquired in a Micromass Q-Tof2 mass spectrometer equipped with a nano electrospray ionization source. Tandem mass spectrometry (MS/MS) was performed by collision-induced dissociation using argon as the collision gas.
Pull-down assay
cDNA encoding FLAG-RalA or its mutant was transfected into COS7 cells by electroporation (Yamaguchi et al., 2006). After 48 h, cells were harvested and lysed in 200 μl of Tris-buffered saline (TBS)-T buffer (50 mM Tris-HCl [pH7.4], 150 mM NaCl, 1% Triton X-100, 1 mM PMSF, 20 μg/ml leupeptin). Total lysate of the COS7 cells expressing FLAG-RalA or its mutants was mixed with 2 μg of purified GST, GST-RD, or GST-ΔC1 in 1 ml of PBS(−) containing 1% bovine serum albumin and 50 μl of glutathione–sepharose 4B for 2 h at 4°C. To determine the effect of GTP or GDP on the binding, GDP-β-S or GTP-γ-S was added into the mixture. After centrifugation and washing with PBS(−) three times, the bound proteins were eluted with 50 μl of SDS buffer for 10% SDS–PAGE followed by immunoblotting using anti–GST, FLAG, or RalA antibody.
Immunoprecipitation
To detect interaction between endogenous RalA and PKCη, immunoprecipitation was performed. NHEK cells treated with or without calcium and CS were homogenized in the lysis buffer with sonication on ice and then centrifuged at 10,000 × g for 5 min. Cleared lysates were incubated with RalA mAb for 2 h at 4°C, and then 50 μl of protein G sepharose (GE Healthcare Bioscience) was added. After an additional 2 h of incubation at 4°C, reaction mixture was centrifuged and washed well with PBS(−). The pellet was resolved in 40 μl SDS buffer. The immunoprecipitants were separated on a 10% SDS–PAGE followed by immunoblotting using anti-PKCη (1:1000 dilution) and RalA antibody (1:10,000 dilution).
Immunoblotting
The separated proteins by SDS–PAGE were transferred to a polyvinylidene difluoride (PVDF) membrane and blocked with 5% skim milk in PBS-T. The membrane was immunostained with appropriate primary antibodies for 1 h at room temperature. After three rinses with PBS-T, the membrane was incubated with peroxidase-labeled anti–rabbit IgG or anti–mouse IgG (Jackson ImmunoResearch, West Grove, PA) for 1 h at room temperature. After extensively washing with PBS-T, the immunoreactive bands were visualized using an enhanced chemiluminescence detection kit (GE Healthcare Bioscience).
Qantative PCR
Total cellular RNA was extracted from NHEK using SV Total RNA Isolation System (Promega, Madison, WI) according to manufacturer’s recommended protocol and quantified spectrophotometically. A total of 200 ng of total RNA was reverse-transcribed into cDNA using ThermoScript RT-PCR System (Invitrogen) following standard protocol and was then applied to real-time PCR using CYBER green and an ABI Prism 7000 (Applied Biosystems, Foster City, CA). The primers used were as follows: for TGase1, 5′-TCTGGCACTCGAAGACCTGG-3′ and 5′-CTCGTCGTACTCATACTCGTCT-3′; for involucrin, 5′-ACCCATCAGGAGCAAATGAAA-3′ and 5′-GCTCGACAGGCACCTTCTG-3′; for keratin 5, 5′-ATCTCTGAGATGAACCGGATGATC-3′ and 5′-CAGATTGGCGCACTGTTTCTT-3′; for keratin 1, 5′-GATGAAATCAACAAGCGGACAA-3′ and 5′-TGGTAGAGTGCTGTAAGGAAATCAATT-3′. All samples were normalized to values of GAPDH, and the results are represented as fold changes of threshold cycle value relative to controls. Each sample was run in triplicate.
siRNA experiments
Target sequence for Ral A, CGCGGTGCAGATTCTTCTTAA (HsRALA), and control sequence, CGGAGCCGAAUCGUCAUAATT, were obtained from QIAGEN (Valencia, CA) and Invitrogen, respectively. siRNA for human PKCη (sc-44020) was obtained from Santa Cruz Biotechnology. Cells were transfected with dsRNA using lipofectamine 2000 (Invitrogen) and cultured for 48 h. After 24 h transfection of siRNA, the cells were infected with adenovirus for RD or GFP and cultured for an additional 24 h with or without CS and calcium.
RalA activation assay
The assay was essentially performed as described previously (Kawato et al., 2008). Briefly, treated or nontreated HaCaT cells were lysed in lysis buffer as described. After removal of insoluble materials, the lysate was incubated with glutathione–sepharose beads coated with 10 μg GST-Sec5-RBD at 4°C for 1 h. After washing, active RalA, which was pulled down with beads, was analyzed by immunoblotting using the anti–RalA mAb.
Supplementary Material
Abbreviations used:
- C1
conserved region 1
- CS
cholesterol sulfate
- GAP
GTPase-activating protein
- GDP
guanosine diphosphate
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- GTP
guanosine triphosphate
- NHEK
normal human epidermal keratinocyte
- PKC
protein kinase C
- PLC
phospholipase C
- PLD
phospholipase D
- PS
pseudosubstrate region
- RD
regulatory domain
- SPR
small
proline rich
- TGase 1
transglutaminase 1
- V1
variable region 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-09-0754) on February 23, 2011.
REFERENCES
- Arimura N, Kaibuchi K. Key regulators in neuronal polarity. Neuron. 2008;48:881–884. doi: 10.1016/j.neuron.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Benitah SA, Frye M, Glogauer M, Watt FM. Stem cell depletion through epidermal deletion of Rac1. Science. 2005;309:933–935. doi: 10.1126/science.1113579. [DOI] [PubMed] [Google Scholar]
- Bhattacharya M, Anborgh PH, Babwah AV, Dale LB, Dorbransky T, Benovic JL, Feldman RD, Verdi JM, Rylett RJ, Ferguson SSG. β-Arrestins regulate a Ral-GDS-Ral effector pathway that mediates cytoskeletal reorganization. Nat Cell Biol. 2002;4:547–555. doi: 10.1038/ncb821. [DOI] [PubMed] [Google Scholar]
- Cammonis JH, White MA. Ral GTPases: corrupting the exocyst in cancer cells. Trends Cell Biol. 2005;15:327–332. doi: 10.1016/j.tcb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Cantor SB, Urano T, Feig L. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani M, Scandiuzzi C, Amigoni L, Tisi R, Berruti G, Martegani E. Functional analysis of RalGPS2, a murine guanine nucleotide exchange factor for Ral A GTPase. Exp Cell Res. 2007;313:2293–2307. doi: 10.1016/j.yexcr.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Colon-Gonzalez F, Kazanietz MG. C1 domains exposed: from diacylglycerol binding to protein-protein interactions. Biochem Biophys Acta. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Denning MF. Epidermal keratinocytes: regulation of multiple cell phenotypes by multiple protein kinase C isoforms. Int J Biochem Cell Biol. 2004;36:1141–1146. doi: 10.1016/j.biocel.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Denning MF, Kazanietz M, Blumberg P, Yaspa H. Cholesterol sulfate activates multiple protein kinase C isoenzymes and induces granular cell differentiation in cultured murine keratinocytes. Cell Growth Differ. 1995;6:1619–1626. [PubMed] [Google Scholar]
- Dlugosz AA, Yaspa SH. Protein kinase C regulates keratinocytes transglutaminase (TGk) gene expression in cultured primary mouse epidermal keratinocytes induced to terminally differentiate by calcium. J Invest Dermatol. 1994;102:409–414. doi: 10.1111/1523-1747.ep12372171. [DOI] [PubMed] [Google Scholar]
- Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–425. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- Feig LA, Urano T, Cantor S. Evidence for a Ras/Ral signaling cascade. Trends Biochem Sci. 1996;21:438–450. doi: 10.1016/s0968-0004(96)10058-x. [DOI] [PubMed] [Google Scholar]
- Fuchs E. Epidermal differentiation: the bare essentials. J Cell Biol. 1990;111:2807–2814. doi: 10.1083/jcb.111.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta T, Chida K, Tajima O, Matsuura Y, Iwamori M, Ueda Y, Mizuno K, Ohno S, Kuroki T. Cholesterol sulfate, a novel activator for the η isoform of protein kinase C. Cell Growth Differ. 1994;5:943–947. [PubMed] [Google Scholar]
- Jenkins GM, Frohman MA. Phospholipase D: a lipid centric review. Cell Mol Life Sci. 2005;62:2305–2316. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jjung EM, Betancourt-Calle S, Mann-Blakeney R, Griner RD, Bollag WB. A potential role for ceramide in the regulation of mouse epidermal keratinocyte proliferation and differentiation. J Invest Dermatol. 1998;110:318–323. doi: 10.1046/j.1523-1747.1998.00137.x. [DOI] [PubMed] [Google Scholar]
- Jjung EM, Betancourt-Calle S, Mann-Blakeney R, Griner RD, Bollag WB. Sustained phospholipase D activation is associated with keratinocyte differentiation. Carcinogenesis. 1999;20:569–576. doi: 10.1093/carcin/20.4.569. [DOI] [PubMed] [Google Scholar]
- Kagehara M, Tachi M, Harii K, Iwamori M. Programmed expression of cholesterol sulfotransferase and transglutaminase during epidermal differentiation of murine skin development. Biochem Biophys Acta. 1994;1215:183–189. doi: 10.1016/0005-2760(94)90109-0. [DOI] [PubMed] [Google Scholar]
- Kashiwagi M, Ohba M, Chida K, Kuroki T. Protein kinase Cη (PKCη): its involvement in keratinocyte differentiation. J Biochem. 2002;132:853–857. doi: 10.1093/oxfordjournals.jbchem.a003297. [DOI] [PubMed] [Google Scholar]
- Kawato M, Shirakawa R, Kondo H, Higashi T, Ikedaq T, Okawa K, Fukai S, Nureki O, Kita T, Horiuchi H. Regulation of platelet dense granule secretion by Ral GTPase-exocyst pathway. J Biol Chem. 2008;283:166–174. doi: 10.1074/jbc.M705340200. [DOI] [PubMed] [Google Scholar]
- Koster MI, Roop DR. Mechanism regulating epithelial stratification. Annu Rev Cell Biol. 2007;23:93–113. doi: 10.1146/annurev.cellbio.23.090506.123357. [DOI] [PubMed] [Google Scholar]
- Lalli G, Hall A. Ral GTPases regulate neurite branching through GAP-43 and the exocyst complex. J Cell Biol. 2005;171:857–869. doi: 10.1083/jcb.200507061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingm M, Troller U, Zeidman R, Stensman H, Schultz A, Larssonm C. Identification of conserved amino acids N-terminal of the PKCεC1b domain crucial for protein kinase Cε-mediated induction of neurite outgrowth. J Cell Biol. 2005;280:17910–17919. doi: 10.1074/jbc.M412036200. [DOI] [PubMed] [Google Scholar]
- Narumiya S. The small GTPase Rho: cellular functions and signal transduction. J Biochem. 1996;120:215–228. doi: 10.1093/oxfordjournals.jbchem.a021401. [DOI] [PubMed] [Google Scholar]
- Newton AC. Newton, Totowa, NJ: Humana Press; 2006. The ins and outs of protein kinase C. In: Methods in Molecular Biology (vol. 233), ed. AC; pp. 3–7. [DOI] [PubMed] [Google Scholar]
- Nikolove E, Mitev V, Minner F, Deroanne CF, Poumay Y. The inhibition of the expression of the small Rho GTPase Rac 1 induces differentiation with no effect on cell proliferation in growing human adult keratinocytes. J Cell Biochem. 2008;103:857–864. doi: 10.1002/jcb.21455. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science. 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- Ohba M, Ishino K, Kashiwagi M, Kawanabe S, Chida K, Hou NH, Kuroki T. Induction of differentiation in normal human keratinocytes by adenovirus-mediated introduction of the η and δ isoforms of protein kinase C. Mol Cell Biol. 1998;18:5199–5207. doi: 10.1128/mcb.18.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase Ral A targets filamin to induce filopodia. Proc Natl Acad Sci USA. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada S, Mizuno K, Saido TC, Akita Y, Suzuki K, Kuroki T, Ohno S. A phorbol ester receptor/protein kinase, nPKCη, a new member of the protein kinase C family predominantly expressed in lung and skin. J Biol Chem. 1990;265:22434–22440. [PubMed] [Google Scholar]
- Osada S, et al. Predominant expression of nPKCη, a Ca2+-independent isoform of protein kinase C in epithelial tissues, in association with epithelial differentiation. Cell Growth Differ. 1993;4:167–175. [PubMed] [Google Scholar]
- Rebhum JF. Identification and characterization of a new family of guanine nucleotide exchange factors of the Ras-related GTPase Ral. J Biol Chem. 2000;275:13406–13410. doi: 10.1074/jbc.c000085200. [DOI] [PubMed] [Google Scholar]
- Shirai Y, Kashiwagi K, Yagi K, Sakai N, Saito N. Distinct effects of fatty acids on translocation of g- and e-subspecies of protein kinase C. J Cell Biol. 1998;143:511–521. doi: 10.1083/jcb.143.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai Y, Murakami T, Kuramasu M, Iijima L, Saito N. A novel PIP2 biding of εPKC and its contribution to the neurite induction ability. J Neurochem. 2007;102:1635–1644. doi: 10.1111/j.1471-4159.2007.04702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens L, Milne L, Hawkins P. Moving towards a better understanding of chemotaxis. Curr Biol. 2008;18:R485–R494. doi: 10.1016/j.cub.2008.04.048. [DOI] [PubMed] [Google Scholar]
- Sugihira K, Asano S, Tanaka K, Iwamatsu A, Okawa K, Ohta Y. The exocyst complex binds the small GTPase Ral A to mediate filopodia formation. Nat Cell Biol. 2001;4:73–78. doi: 10.1038/ncb720. [DOI] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Tojima T, Ito E. Signal transduction cascades underlying de novo protein synthesis required for neural morphogenesis in differentiating neurons. Prog Neurobiol. 2004;72:183–193. doi: 10.1016/j.pneurobio.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Tscharntke M, et al. Impaired epidermal wound healing in vivo upon inhibition or deletion of Rac1. J Cell Sci. 2007;120:1480–1490. doi: 10.1242/jcs.03426. [DOI] [PubMed] [Google Scholar]
- Ueda E, Ohno S, Kuroki T, Livneh E, Yamada K, Yamanishi K, Yasuno H. The η isoform of protein kinase C mediates transcriptional activation of the human transglutaminase 1 gene. J Biol Chem. 1996;271:9790–9794. doi: 10.1074/jbc.271.16.9790. [DOI] [PubMed] [Google Scholar]
- van Dam EM, Robinson PJ. Ral: mediator of membrane trafficking. Int J Biochem Cell Biol. 2006;38:1841–1847. doi: 10.1016/j.biocel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Wiesmann C, de Vos AM. Nerve growth factor: structure and function. Cell Mol Life Sci. 2001;58:748–759. doi: 10.1007/PL00000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Shirai Y, Matsubara T, Sanse K, Kuriyama M, Oshiro N, Yoshino K, Yonezawa K, Ono Y, Saito N. Phosphorylation and up-regulation of diacylglycerol kinaseγ via its interaction with protein kinase Cγ. J Biol Chem. 2006;182:73613–72613. doi: 10.1074/jbc.M606992200. [DOI] [PubMed] [Google Scholar]
- Zeidman R, Lofgren B, Pahlman S, Larsson C. PKCε, via its regulatory domain and independently of its catalytic domain, induces neurite-like processes in neuroblastoma cells. J Cell Biol. 1999;145:713–726. doi: 10.1083/jcb.145.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidman R, Troller U, Raghunath A, Pahlman S, Larsson C. Protein kinase C ε actin-binding site is important for neurite outgrowth during neuronal differentiation. Mol Biol Cell. 2002;13:12–24. doi: 10.1091/mbc.01-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.