Abstract
Chaperone-mediated autophagy is a selective mechanism for degradation of soluble cytosolic proteins in lysosomes that distinguishes itself from other autophagic pathways by the selectivity with which CMA substrates are targeted for degradation. The recent molecular dissection of this autophagic pathway and the development of experimental models with compromised CMA have unveiled the important contribution of this pathway to protein quality control. In fact, CMA activation seems to be a common mechanism of cellular defense against proteotoxicity.
Keywords: chaperones, lysosomes, oxidative stress, proteases, selective autophagy
Introduction to selective autophagy
Cells assure the renewal of their constituent proteins through a continuous process of synthesis and degradation that also allows for rapid modulation of the levels of specific proteins to accommodate to the changing extracellular environment [1]. Intracellular protein degradation is also essential for cellular quality control to eliminate damaged or altered proteins, thus preventing the toxicity associated with their accumulation inside cells. Two major proteolytic systems exert this cleaning function: the ubiquitin-proteasome system and the autophagic/lysosomal system [1]. In contrast to the rapid and selective degradation that characterizes the ubiquitin-proteasome system [2,3], lysosomal degradation or autophagy has been for years considered an “in bulk” non selective process. Recently, this lack of selectivity has been refuted as it has become evident that autophagy discriminates the intracellular components destined for degradation. Of the three types of autophagy described in mammals [4], the contribution to quality control of macroautophagy and microautophagy has been described in detail in other sections of this focused issue. We review here a third form of autophagy known as chaperone-mediated autophagy (CMA), that differs from the others in the mechanism for cargo selection and delivery to the lysosomal lumen for degradation. CMA substrate proteins are selectively targeted one-by-one to the lysosomes and are then translocated across the lysosomal membrane [5,6]. In this review, we briefly describe the steps and molecular components that participate in the degradation of cytosolic proteins via CMA and the most recent advances in our understanding of the physiological functions of this autophagic pathway, with particular emphasis on its role in protein quality control. We also comment on the consequences of CMA malfunctioning in the context of different human pathologies in which a primary defect in this pathway has been described.
Molecular characteristic of CMA
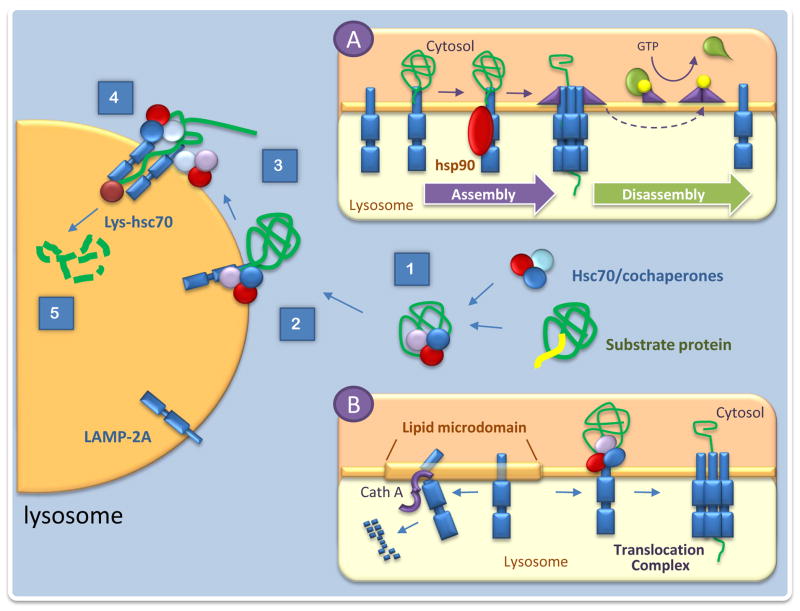
Selectivity in selection of CMA cargo is attained through the interaction of a cytosolic chaperone, the heat shock-cognate chaperone of 70kDa, hsc70, with a specific region in the amino acid sequence of the proteins destined for degradation. All CMA substrates contain in their sequence a consensus motif, biochemically related to the pentapeptide KFERQ, that when exposed (i.e. during protein misfolding or disassembly of protein complexes), is recognized by hsc70 and in a process modulated by the associated co-chaperones, leads to the delivery of the motif-bearing protein to lysosomes (Fig. 1) [7]. Lysosome-associated variant forms of hsc70, also contribute to later steps in CMA [8–10]. For example, it has been proposed that hsc70 along with the subset of co-chaperones associated to the lysosomal membrane contribute to the unfolding of the substrate protein, an essential requirement before translocation can occur [11].
Figure 1. Molecular components of chaperone-mediated autophagy.
Scheme of the sequential steps that mediate degradation of cytosolic proteins through chaperone-mediated autophagy (CMA): 1. Recognition of cytosolic proteins by hsc70 and its co-chaperones. 2. Binding of the chaperone/substrate complex to the receptor at the lysosomal membrane. 3. Unfolding of the substrate protein. 4. Translocation across the lysosomal membrane. 5. Degradation in the lysosomal lumen. Insets: A. Regulation of the stability of the CMA translocation complex B. Dynamics of the CMA receptor at the lysosomal membrane. .
Substrate proteins bind to the cytosolic tail of a constituent single span membrane protein the lysosome-membrane protein type 2A (LAMP-2A) and are then translocated into the lumen [5,6]. Complete translocation requires the presence of a form of hsc70 in the lysosomal lumen, proposed to drive internalization by a ratchet-like mechanism or at least prevent returning of substrate to the cytosol [8,10]. In contrast to other protein translocation systems, the formation of the CMA translocation complex at the lysosomal membrane is transient, and it persists only while the substrate is crossing the membrane (Fig. 1) [9]. LAMP-2A acts both as a receptor and also as essential component of the CMA translocation complex [12]. Binding of the substrate proteins to monomers of LAMP-2A at the lysosomal membrane promote its multimerization to form the complex required for substrate translocation. Membrane-associated molecules of hsc70 actively disassemble LAMP-2A into monomers to initiate a new cycle of binding and translocation (Fig. 1) [9].
Whereas cytosolic hsc70 is always in excess, binding of substrate proteins to the cytosolic tail of LAMP-2A is limiting for CMA [13]. In fact, levels of LAMP-2A at the lysosomal membrane are a direct determinant of cellular CMA activity, and changes in levels of this receptor are utilized by cells to modulate this autophagic pathway [14]. Activation of CMA in most cases does not require de novo synthesis of LAMP-2A and it is mediated instead by changes in the degradation, dynamics and organization of this receptor protein at the lysosomal membrane [9,13]. The mechanism that regulates the dynamics of LAMP-2A is still under investigation; but recent data supports that LAMP-2A distributes between fluid regions and lipid enriched microdomains at the lysosomal membrane where it can undergo degradation [15]. When high CMA activity is required, LAMP-2A is actively excluded from these domains and remains in the more fluid parts of the membrane where multimerization can take place [15]. Subcompartmentalization of LAMP-2A at the lysosomal membrane is tightly regulated by a lysosome-associated form of glial fibrillary acidic protein (GFAP) that stabilizes the CMA translocation complex in a GTP-dependent manner [16]. In the presence of GTP, a GFAP interacting partner, the elongation factor 1 alpha (EF1α), is released from the lysosomal membrane, leading to self-assembly of GFAP and the neutralization of its stabilizing function on the CMA translocation complex (Fig. 1).
In contrast to the comprehensive understanding of the regulation of the translocation mechanism, the intracellular signaling mechanisms that control activation/inhibition of CMA still remains unclear.
Physiological role of CMA: What crosses the lysosomal membrane and when
Sequence analysis has revealed that about 30% of cytosolic proteins contain in their sequence putative motifs for CMA targeting. Immunoprecipitation studies with an antibody that recognizes the structural characteristics of the CMA-tageting motif are in agreement with this estimation. The approximately fifty already validated CMA substrates fall in a broad range of protein categories, including, among others, glycolytic enzymes (GAPDH, aldolase, PGM) [17], transcription factors (c-fos, Pax, MEF2D) [18–20], inhibitors of transcription factors (IkBα) [21], calcium binding proteins (specific annexins) [22], lipid binding proteins (alpha-2-microglobulin) [23], proteins involved in vesicular trafficking (α-synuclein, Tau) [24,25] and even catalytic subunits of the proteasome [26]. Although many of these substrates organize as protein complexes in the cytosol, they only become amenable for CMA degradation when as single subunits. Organization of any of these proteins into irreversible oligomers or aggregates also prevents their degradation by this pathway. The inability of CMA to degrade protein inclusions explains why CMA is a successful mechanism for quality control of proteins while they remain soluble and “unfoldable”, but CMA cannot remove proteins severely altered which are no longer in solution.
Some level of basal CMA activity is detectable in almost all cells and it is responsible for some of the cell-type specific functions recently attributed to this pathway such as antigen presentation in professional antigen-presenting cells [27], preservation of neuronal viability by modulating levels of the transcription factor MEF2D [19] or control of renal tubular cells growth through the degradation of the Pax2 transcription factor [18]. However, CMA is maximally stimulated in response to stress, conferring onto CMA a role in protein quality control as well as an alternative source of amino acids. In fact, CMA is activated when cells are deprived of nutrients for relatively long periods of time (>10h) [28,29]. CMA supplies the cell with amino acids required to sustain protein synthesis under these conditions. In addition, CMA contributes to maintenance of the stability of the proteome during stress as it can mediate the selective removal from the cytoplasm of altered or damaged proteins without disturbing normally functioning nearby proteins. Different types of undesired posttranslational modifications (mild oxidation, partial unfolding, abnormal protein truncation, conjugation to toxins and formation of protein adducts with small molecules) have all been shown to enhance CMA of specific proteins [23,25,30–32]. The partial unfolding often associated with these modifications may facilitate CMA substrate recognition by hsc70 and translocation across the lysosomal membrane.
The physiological relevance of the selective removal of altered proteins by CMA has recently been confirmed by analyzing the cellular consequences of CMA malfunctioning [33]. Cultured cells with compromised CMA display unusual sensitivity to stress, that presumably originates from the higher content of oxidized and aggregated proteins observed in these cells [34]. In fact, upregulation of CMA has been observed upon exposure of cells to mild oxidative stress [32] and protein-damaging toxins [23], supporting that cells activate this pathway in their defense against proteoxicity.
CMA dysfunction in proteinopathies
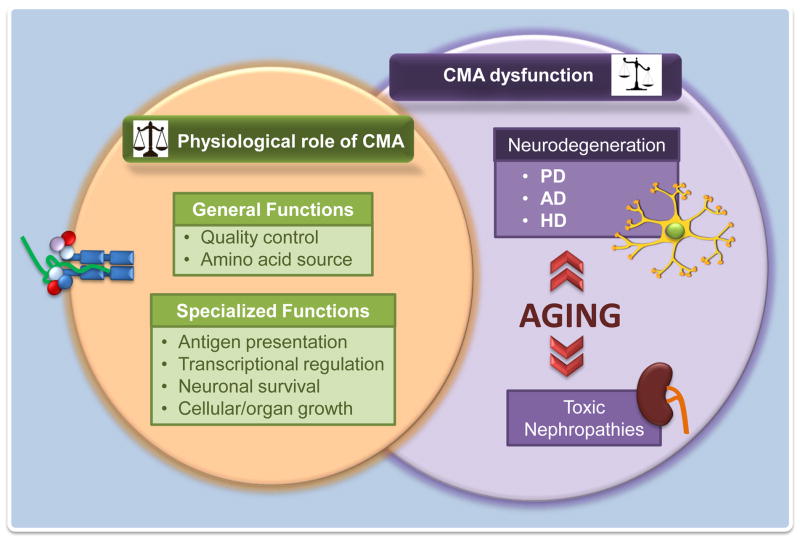
Dysfunction of CMA activity has been described in a growing number of pathologies (reviewed in [6,35,36]). Because of the interest for this focused issue, we describe here those pathologies related to alterations in protein homeostasis also known as proteinopathies or protein conformational disorders (Fig. 2).
Figure 2. CMA dysfunction in protein conformational disorders.
CMA contributes to protein quality control and maintenance of the stability of the proteome. Consequently, alterations in this autophagic pathway have been linked to different protein conformational disorders. This scheme displays some of these disorders in kidney and in the central nervous system, and illustrates the potential negative effect that changes with age in CMA activity have in the progression of these diseases.
The first connection between proteotoxicity and CMA in the context of disease was established with a type of nephropathy induced by exposure to gasoline derivatives, that upon interacting with the lipid binding protein alpha-2-microglobulin, promoted its accumulation into inclusions. Liver and kidney, the two tissues where this protein is detected, respond to the toxic modification of alpha-2-microgrobulin by upregulating CMA [23]. However, if the exposure persists, CMA capability is surpassed and protein aggregation occurs leading to proteotoxicity and disease [23].
The recent interest in autophagy malfunction in neurodegenerative diseases has revealed the contribution of primary defects in CMA to the pathogenesis of some of these protein conformational neuronal disorders (Fig. 2). Common to these pathologies is the accumulation of aberrant pathogenic proteins in the form of inclusions inside or outside neurons. Symptoms depend on the normal function of the pathogenic protein and the brain regions in which it is normally present.
A large number of these pathogenic proteins contain KFERQ motifs in their amino acid sequence making them putative candidates for CMA degradation (Fig. 2). In fact, CMA-targeting motifs have been identified in the Parkinson’s disease-related proteins α-synuclein, parkin, UCH-L1, pink-1 and DJ-1, in APP and Tau, associated to Alzheimer’s disease (AD) and in huntingtin, the protein mutated in Huntington’s disease (HD). Studies with in vitro reconstituted systems [25], in neuronal cells in culture [25,31,37] and more recently in vivo in the brain of PD mice [38] have demonstrated that a fraction of intracellular α-synuclein is normally degraded by CMA, but its pathogenic variants, although properly recognized by hsc70 and targeted to lysosomes, are unable to reach the lysosomal lumen [25]. The abnormally tight binding of these proteins to the lysosomal membrane and their organization into irreversible oligomeric complexes in this compartment is behind their autophagic failure [25,31]. Furthermore, this abnormal lysosomal binding of the pathogenic proteins blocks CMA of other cytosolic proteins. Impaired CMA could be behind the compromised response to stress of PD neurons. Abnormal interaction with CMA components in lysosomes has also been recently described for mutant forms of another PD-related protein, the ubiquitin carboxyl-terminal esterase L1 protein (UCHL-1) [39]. Contribution of other pathogenic PD proteins bearing CMA targeting motifs to CMA toxicity in PD requires further investigation.
Compromised translocation across the lysosomal membrane has also been described for some mutant forms of tau, a protein associated with AD pathogenesis and that accumulates in tangles in different tauopathies. Similar to pathogenic α-synuclein, mutant tau proteins are properly targeted to lysosomes, but in this case, translocation initiates but is halted at an early stage, leaving the N-terminal part of the protein accessible to the luminal proteases. Partial cleavage of the protein generates highly amyloidogenic peptides that organize into irreversible oligomers at the lysosomal membrane, interfering with CMA [24]. The extent to which CMA blockage contributes to pathogenesis of tauopathies or in the AD brain requires further investigation.
Despite the multiple CMA-targeting motifs identified in huntingtin, the protein that bears an abnormally expanded stretch of glutamines in HD patients favoring its aggregation in affected neurons, CMA degradation of the full size protein is negligible. A primary defect in macroautophagy seems responsible for proteotoxicity in HD and could explain the upregulation of CMA observed in this disease (Martinez-Vicente et al., submitted). In fact, previous studies have established the existence of reciprocal mechanisms of compensation between these two autophagic pathways [33,40]. Enhancers of CMA could thus have therapeutic potential in HD, as genetic upregulation of this pathway has been shown to improve cellular viability in HD brain slices in culture [30], and artificially enhanced targeting to CMA of mutant huntingtin has recently proven beneficial in HD mouse models [41].
Proteotoxicity associated with aging and CMA
Accumulation of protein damage is a common feature of most tissues in aging organisms. Compromised quality control with age contributes to altered protein homeostasis [42,43]. Age-dependent changes in the lysosomal system and the subsequent decline in autophagic activity were identified even before the molecular mechanisms that govern the different autophagic pathways were fully understood [42,43]. Both macroautophagy and CMA activity decrease with age in most organs in aging mammals. This reduction in CMA has been attributed to the lower LAMP-2A content in lysosomes from old cells due to problems with the stability of this receptor protein at the lysosomal membrane [44,45].
The contribution of reduced CMA activity to altered cellular homeostasis in old organisms has been recently confirmed using a transgenic mouse model with an inducible exogenous copy of LAMP-2A [46]. Activation of the transgene to compensate for the reduced levels of endogenous LAMP-2A and to preserve CMA function until late in life leads to reduced intracellular levels of oxidized and aggregated proteins in these animals, improves their response to stressors, and maintains better organ function. The age-dependent decline in CMA contributes, at least in part, to the aggravating effect of aging in the course of different protein conformational disorders with a primary compromise in CMA.
Concluding remarks
Selectivity in protein removal as part of cellular quality control has been traditionally attributed solely to the ubiquitin-proteasome system. However, the precise coordinated action of cytosolic chaperones and different protein components at the lysosomal membrane allows lysosomes to perform similar selective removal of proteins via CMA. This autophagic pathway reveals itself as an important component of the cellular response to those stressors damaging the soluble cellular proteome. The recently identified CMA compromise in a growing number of protein conformational disorders and the described decline in the activity of this selective pathway with age supports that interventions to modulate CMA activity could have therapeutic potential in these devastating disorders.
Acknowledgments
The authors want to express their profound appreciation for the late Fred Dice (“Paulo”) for his pioneering work on this pathway and for the great influence that he and his work have always had, and will continue to have, in this field. We thank Ms. Samantha Orenstein for critically reading the manuscript. Work in our laboratory is supported by NIH grants from NIA (AG021904, AG031782), NIDKK (DK041918), NINDS (NS038370), a Glenn Foundation Award and a Hirsch/Weill-Caulier Career Scientist Award. E.A. is a Fullbright Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest have been highlighted as * of special interest ** of outstanding interest
- 1.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 2.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navon A, Ciechanover A. The 26 S proteasome: from basic mechanisms to drug targeting. J Biol Chem. 2009;284:33713–33718. doi: 10.1074/jbc.R109.018481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. Basic review of functions of autophagy and connections to pathology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dice J. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 6.Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol Metab. 2010;21:142–150. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7**.Chiang H, Terlecky S, Plant C, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. First molecular component identified for CMA. [DOI] [PubMed] [Google Scholar]
- 8.Cuervo A, Dice J, Knecht E. A lysosomal population responsible for the hsc73-mediated degradation of cytosolic proteins in lysosomes. J Biol Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 9.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agarraberes F, Terlecky S, Dice J. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agarraberes F, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114:2491–2499. doi: 10.1242/jcs.114.13.2491. [DOI] [PubMed] [Google Scholar]
- 12**.Cuervo A, Dice J. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. Identification of the receptor for CMA substrates at the lysosomal membrane. [DOI] [PubMed] [Google Scholar]
- 13.Cuervo A, Dice J. Regulation of lamp2a levels in the lysosomal membrane. Traffic. 2000;1:570–583. doi: 10.1034/j.1600-0854.2000.010707.x. [DOI] [PubMed] [Google Scholar]
- 14.Cuervo A, Dice J. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000;113:4441–4450. doi: 10.1242/jcs.113.24.4441. [DOI] [PubMed] [Google Scholar]
- 15.Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Bandhyopadhyay U, Sridhar S, Kaushik S, Kiffin R, Cuervo AM. Identification of regulators of chaperone-mediated autophagy. Mol Cell. 2010;39:535–547. doi: 10.1016/j.molcel.2010.08.004. Evidence of a tight control of the lateral mobility of the CMA receptor at the lysosomal membrane and its conversion into a translocation complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuervo AM, Terlecky SR, Dice JF, Knecht E. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J Biol Chem. 1994;269:26374–26380. [PubMed] [Google Scholar]
- 18.Sooparb S, Price SR, Shaoguang J, Franch HA. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–2144. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 19.Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, Mao Z. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aniento F, Papavassiliou AG, Knecht E, Roche E. Selective uptake and degradation of c-fos and v-fos by rat liver lysosomes. FEBS Lett. 1996;390:47–49. doi: 10.1016/0014-5793(96)00625-4. [DOI] [PubMed] [Google Scholar]
- 21.Cuervo AM, Hu W, Lim B, Dice JF. IkappaB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell. 1998;9:1995–2010. doi: 10.1091/mbc.9.8.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuervo AM, Gomes AV, Barnes JA, Dice JF. Selective degradation of annexins by chaperone-mediated autophagy. Journal of Biological Chemistry. 2000;275:33329–33335. doi: 10.1074/jbc.M005655200. [DOI] [PubMed] [Google Scholar]
- 23.Cuervo A, Hildebrand H, Bomhard E, Dice J. Direct lysosomal uptake of alpha2-microglobulin contributes to chemically induced nephropathy. Kidney Int. 1999;55:529–545. doi: 10.1046/j.1523-1755.1999.00268.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Martinez-Vicente M, Kruger U, Kaushik S, Wong E, Mandelkow EM, Cuervo AM, Mandelkow E. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. First connection of CMA malfunctioning with a human disease. [DOI] [PubMed] [Google Scholar]
- 26.Cuervo AM, Palmer A, Rivett AJ, Knecht E. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem. 1995;227:792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhou D, Li P, Lin Y, Lott JM, Hislop AD, Canaday DH, Brutkiewicz RR, Blum JS. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Auteri J, Okada A, Bochaki V, Dice J. Regulation of intracellular protein degradation in IMR- 90 human diploid fibroblasts. J Cell Physiol. 1983;115:159–166. doi: 10.1002/jcp.1041150210. [DOI] [PubMed] [Google Scholar]
- 29.Cuervo A, Knecht E, Terlecky S, Dice J. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 30.Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, Martinez-Vincente M, Arrasate M, JGOS-R, Khashwji H, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Martinez-Vicente M, Talloczy Z, Kaushik S, Massey A, Mazzulli J, Mosharov E, Hodara R, Fredenburg R, Wu D, Follenzi A, et al. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. Evidence that undesired modifications in cytosolic proteins can exert a negative effect on CMA functioning. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiffin R, Christian C, Knecht E, Cuervo A. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Nat Acad Sci USA. 2006;103:5905–5910. doi: 10.1073/pnas.0507436103. First study of the consequences of CMA malfunctioning in cells, and evidence for the existence of a cross-talk among autophagic pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massey AC, Follenzi A, Kiffin R, Zhang C, Cuervo AM. Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy. 2008;4:442–456. doi: 10.4161/auto.5654. [DOI] [PubMed] [Google Scholar]
- 35.Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: Molecular mechanisms and physiological relevance. Semin Cell Dev Biol. 2010;21:719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kon M, Cuervo AM. Chaperone-mediated autophagy in health and disease. FEBS Lett. 2010;584:1399–1404. doi: 10.1016/j.febslet.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogiatzi T, Xilouri M, Vekrellis K, Stefanis L. Wild type alpha-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J Biol Chem. 2008;283:23542–23556. doi: 10.1074/jbc.M801992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mak SK, McCormack AL, Manning-Bog AB, Cuervo AM, Di Monte DA. Lysosomal degradation of alpha-synuclein in vivo. J Biol Chem. 2010;285:13621–13629. doi: 10.1074/jbc.M109.074617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabuta T, Wada K. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem. 2008;283:23731–22373. doi: 10.1074/jbc.M801918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaushik S, Massey A, Mizushima N, Cuervo AM. Constitutive Activation of Chaperone-mediated Autophagy in Cells with Impaired Macroautophagy. Mol Biol Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41**.Bauer PO, Goswami A, Wong HK, Okuno M, Kurosawa M, Yamada M, Miyazaki H, Matsumoto G, Kino Y, Nagai Y, et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol. 2010;28:256–263. doi: 10.1038/nbt.1608. Example of the therapeutic possibilities of the use of selective targeting of cytosolic proteins to lysosomes through CMA. [DOI] [PubMed] [Google Scholar]
- 42.Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morimoto RI, Cuervo AM. Protein homeostasis and aging: taking care of proteins from the cradle to the grave. J Gerontol A Biol Sci Med Sci. 2009;64:167–170. doi: 10.1093/gerona/gln071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiffin R, Kaushik S, Zeng M, Bandyopadhyay U, Zhang C, Massey AC, Martinez-Vicente M, Cuervo AM. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci. 2007;120:782–791. doi: 10.1242/jcs.001073. [DOI] [PubMed] [Google Scholar]
- 45.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 46**.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. Evidence that functional decline on CMA with age contributes to aging and that the age-related defect on this pathway can be corrected in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]