Abstract
OxLDL binding to CD36 is shown to result in macrophage activation and foam cell formation that have been implicated in atherosclerosis. However, CD36 has also been shown to induce inflammatory response to other ligands besides oxLDL. During the course of blocking CD36 oxLDL binding function using anti CD36 antibodies, we have identified a novel domain of CD36 that triggers inflammatory response-independent of oxLDL binding. OxLDL bound to the mouse reporter cell line RAW-Blue induced TNF-α and RANTES mRNA and protein expression. Pretreatment of RAW-Blue cells with an anti-mCD36 mAb, JC63.1, an activating mCD36 mAb, surprisingly did not inhibit oxLDL-induced response. Further, binding of this antibody to CD36 alone induced a pro-inflammatory cytokine response in RAW-Blue cells as well as primary mouse macrophages. The induction of cytokine response was specific only to this antibody and was CD36-dependent, since CD36−/− macrophages failed to induce a similar response. The interaction of the antibody to CD36 led to activation of NF-κB and MAP kinase. Notably, a CD36 peptide blocked oxLDL-induced foam cell formation and macrophage activation. However, the activating mCD36 mAb induced macrophage activation was not inhibited by CD36 peptide. Further, activating mCD36 mAb enhanced oxLDL- or TLR2- or TLR4-mediated inflammatory responses. Collectively, our data provide evidence that activating mCD36 mAb binds to a domain different from the oxLDL-binding domain on mouse CD36, and suggest that interaction at this domain may contribute to oxLDL-independent macrophage inflammatory responses that lead to chronic inflammatory diseases.
INTRODUCTION
CD36, one of the pattern recognition receptors, has been reported to bind with multiple ligands including oxLDL [1–3], thrombospondin-1 [4], free fatty acids [5], advanced glycation end products [6], β-amyloid [7,8], Plasmodium falciparum malaria-infected erythrocytes [9,10], apoptotic cells [11,12], non-opsonized bacteria [13] and FSL-1, a TLR2 ligand [14]. Due to its ability to bind to a broad range of ligands, CD36 has been shown to play a significant role in a number of physiological and pathological processes in vivo including atherogenesis, lipid sensing and metabolism, and innate immune response [15].
CD36 binding to oxidized-low density lipoprotein (oxLDL)3 has been shown to induce the pro-inflammatory cytokine responses in macrophages [16]. Further studies using macrophages from CD36−/− knockout mice have shown that oxLDL-induced foam cell formation is mediated by NF-κB and MAP kinase activation [3]. Though CD36−/− or SR-A−/− macrophages show reduced oxLDL-induced MAP kinase signaling and the formation of lipid-laden macrophages, there was no complete loss of oxLDL-induced foam cell formation and MAP kinase activation [3]. In vitro studies using CD36 knockout macrophages have shown reduced generation of foam cells, an early event in atherosclerosis [17,18]. However, in vivo studies using apolipoprotein E (apoE−/−) CD36−/− double knockout (apoE−/−CD36−/− DKO) mice have provided conflicting data [17,19–21]. Studies from one group showed apoE−/−CD36−/− DKO mice have attenuated atherosclerotic lesions [17,20], while the other group showed that loss of CD36 results in reduction of complexity of atherosclerotic lesions without reducing foam cell formation [19,21]. Though the reasons for the discrepancies are not clear, the later study has suggested that CD36-dependent and independent inflammatory response may be contributing to atherosclerosis [21,22].
Recent studies have suggested a broader role for CD36 in inflammatory cells besides oxLDL binding, which could exacerbate chronic inflammatory diseases [22]. For example, β-amyloid-mediated inflammatory response is dependent on CD36 expression [8,23]. Moreover, apolipoprotein C-III, that forms amyloid fibrils, induces TNF-α response also in a CD36 dependent manner [24]. CD36 has also been shown to play a pivotal role in bacterial infection. Hoebe et al [25] have shown CD36oblivious mice (that has a non-sense mutation in CD36) are more susceptible to staphylococcus aureus infection. Moreover, S. aureus-induced cytokine responses are significantly reduced in CD36−/− knockout mice [26]. Very recently, CD36-dependent inflammatory response, but not foam cell formation, has been implicated in non-alcoholic steatohepatitis [27]. Collectively, these findings suggest that oxLDL-independent CD36-mediated inflammatory response may be an important contributing factor to chronic inflammatory diseases.
In this report the effect of oxLDL on macrophage inflammatory response was determined in the presence of a blocking anti-mouse CD36 mAb, JC63.1 (referred as an activating mCD36 mAb) [28]. Surprisingly, the activating mCD36 mAb did not block oxLDL-induced macrophage activation. In fact, the mere binding of activating mCD36 to CD36 on macrophages induced pro-inflammatory cytokine response. This response was mediated via activation of NF-κB and MAP kinase signaling pathways. We also present data showing the binding of activating mCD36 mAb enhanced the cytokine response following co-operative interaction with oxLDL or TLR2. Our data provide evidence that activating mCD36 mAb binds to a domain different from the oxLDL-binding domain on mouse CD36. These findings suggest that interaction at this unknown domain may contribute to oxLDL-independent macrophage inflammatory responses that could contribute to chronic inflammatory diseases.
MATERIALS AND METHODS
Antibodies and chemicals
Details of anti-CD36 mAb and isotype control used in this study is presented in Table 1. Antibodies to phosphorylated forms of Erk1/2, SAPK/JNK, IκB, and their native forms; U0126, a MEK1/2 inhibitor (blocks Erk1/2 activation); and RIPA buffer were obtained from Cell Signaling (Beverly, MA). Mg-132, a proteasome inhibitor that blocks NF-κB activation, was purchased from Calbiochem (San Diego, CA). SP600125 (JNK inhibitor) and SB203580 (p38 MAP kinase inhibitor) were obtained from Alexis Biochemicals (Plymouth Meeting, PA).
TABLE 1.
Anti-mouse CD36 antibody used in this study
| Anti-CD36 mAb clone | Subtype | Vendors |
|---|---|---|
| JC63.1 | mIgA | Cayman Chemicals |
| JC63.1 | mIgA | Santa Cruz |
| 324216 | Rat IgG2b | RND |
| HM36 | Armenian Hamster IgG | Biolegend |
| Anti-mCD36 polyclonal | Goat IgG | RND |
| Anti-mCD36 polyclonal | Rabbit IgG | Cayman |
| Control antibody | ||
| M18–254 | mIgA | BD-Biosciences |
| mIgA | Santa Cruz | |
| 141945 | Rat IgG2b | RND |
| HTK888 | Armenian Hamster IgG | RND |
| Goat IgG | RND | |
| Rabbit IgG | RND |
Cell Culture
RAW-Blue cells (Invivogen, San Diego, CA) are derived from RAW264.7 macrophages with chromosomal integration of a secreted embryonic alkaline phosphatase (SEAP) reporter construct inducible by NF-κB and AP-1. RAW-Blue mouse macrophage cell lines were cultured in DMEM supplemented with 10% (v/v) FBS (Hyclone, Logan, UT) and zeocin (Invivogen, 200 μg/ml). RAW-Blue cells between 6–15 passages were used for the experiments. CHOK1-human CD36 transfectants (ATCC, CRL#2092, Manassas, VA) were cultured in RPMI-1640 supplemented with 10% FBS and G418 (1 mg/ml) and medium was replaced every 3 days. All cell culture reagents were purchased from Invitrogen (Carlsbad, CA).
Animals
ApoE−/− female mice bred onto a C57BL/6 background were purchased from Jackson Laboratory (Bar Harbor, ME). Dr. Maria Febbario (Cleveland Clinic Foundation, Cleveland, OH) kindly provided apoE−/−CD36−/− and apoE−/−SR-A−/− DKO mice [17]. Mice were housed in microisolator cages with filter tops and maintained on a 12-h light/dark cycle in a temperature-controlled room. Animal studies were conducted under guidelines and protocols approved by Institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences. Thioglycollate-elicited mouse peritoneal macrophages were obtained 4 days after injection of 4% thioglycollate (1 ml/mouse). The peritoneal cells were plated in RPMI-1640 supplemented with 10% (v/v) FBS, 2 mM L-glutamine, penicillin, streptomycin, and sodium pyruvate. Non-adherent cells were removed after 2 h and macrophages were used after 48 h.
Cell activation
RAW-Blue cells (1×105 cells/well) were cultured overnight and stimulated by the indicated reagents for 18 h or indicated time. The reagents used were oxLDL (5 μg/ml), activating mCD36 mAb (2 μg/ml) and indicated anti-CD36 mAb, and polyclonal antibodies (5 μg/ml). Cells treated with mIgA isotype control or LPS (Invivogen) were used as negative and positive controls, respectively. To determine signaling pathways, cells were pretreated with 100 nM of indicated inhibitors (Mg-132, SB203580, U0126, and SP600125) for 1 h followed by the addition of activating mCD36 mAb or oxLDL or LPS. All the reagents used in this study were tested for endotoxin contamination using endotoxin reporter cell line, HEK-BLUE (Invivogen), engineered to express TLR4 and SEAP under the control of NF-κB response element. The endotoxin levels were undetectable in the reagents used in this experiment. Polymyxin-B (10 μg/ml) was included in the indicated experiments to confirm that the effect is not due to undetectable levels of endotoxin.
For TLR ligand activation, RAW-Blue cells were incubated with mouse TLR ligands (Invivogen) in the presence or absence of the activating mCD36 mAb for 18 h at 37°C. Concentrations of TLR ligands were as follows: Pam3CSK4 (10 ng/ml), HKLM (108 cells/ml), LPS (100 ng/ml or indicated concentration), and FSL-1 (100 ng/ml). Supernatant was collected, and TNF-α and RANTES protein levels and SEAP enzyme activity were determined. Cells treated with TLR ligands alone, mIgA, and mIgA plus TLR ligands were used as controls.
CD36 cross-linking
RAW-Blue cells (5×105cells/well) were treated with activating mCD36 mAb (2 μg/ml) or isotype control mIgA (2 μg/ml) for 30 min on ice. Cells were washed twice with cold PBS and incubated with a goat anti-mIgA IgG (2 μg/mL, Southern Biotech, Birmingham, AL) for 30 min on ice followed by washing in cold PBS to cross-link CD36. Cells were further cultured in medium for 18 h at 37°C CO2 incubator. Cells treated with activating mCD36 mAb (2 μg/ml) or mIgA without cross-linking were used as controls. Supernatants collected after 18 h were used to analyze TNF-α, RANTES, and SEAP secretion.
Preparation of oxLDL
Native LDL (nLDL) and oxLDL were purchased from Academy Biomedials (Houston, TX). The degree of oxidation of nLDL and oxLDL was determined by measuring the amount of thiobarbituric acid-reactive substances (TBARS) as well as electrophoretic migration of LDL and oxLDL in TITAN-agarose gel (Helena Labs, Houston, TX) as described earlier [29]. nLDL had TBARS values of <1 nmol/mg. OxLDL had TBARS values of >10 and <30 nmol/mg and a relative electrophoretic mobility of 3.4 compared to LDL control. All lipoproteins were used for experiments within 3 weeks after preparation.
OxLDL binding assay
RAW-Blue cells (5×105) were incubated with corresponding mAbs or mIgA or mIgG at 4°C for 1 h, followed by incubation with oxLDL (10 μg/ml) at 4°C for 1 h. OxLDL binding was detected using rabbit anti-apoB IgG biotin (3 μg/ml) and saturating concentrations of streptavidin-PE (Jackson ImmunoResearch Inc., West grove, PA). Cells were washed in fixed in PRB/1% formalin and acquired in a FACSCalibur flow cytometry and analyzed using CellQuestPro software (BD-Biosciences, San Jose, CA).
NF-κB reporter assay
RAW-Blue cells (1×105 cells/well) were incubated with activating mCD36 mAb or indicated reagents for 18 h and SEAP secretion was determined. QUANTI-Blue™, an alkaline phosphatase substrate (Invivogen), was dissolved in endotoxin-free water and sterile filtered (0.22 μm). RAW-Blue cell supernatant (40 μl/well) was added to QUANTI-blue substrate (160 μl/well) and incubated at 37°C for 1 to 3 h. Absorbance was measured at 620 nm in a BMG Polarstar microplate reader.
Real-time RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions and treated with RNase-free DNase. Reverse transcription reaction and quantitative real-time PCR were described previously [30]. Real-time PCR primers (Integrated DNA Technologies, Inc. Coralville, IA) were as follows: β-actin (sense, ggctatgctctccctcacg; antisense, cgctcggtcaggatcttcat), TNF-α (sense, acaaggctgccccgactac; antisense, tggaagactcctcccaggtatatg), RANTES (sense, cgaaggaaccgccaagtg; antisense, ctagagcaagcgatgacagg). A two-step PCR with denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min for 40 cycles was conducted in iCycler (Bio-Rad, Richmond, CA) to determine the threshold cycle (Ct) value. Expression of TNF-α and RANTES was calculated using ΔΔCt method using threshold cycles for β-actin as normalization reference. All real-time PCR reactions were carried out at least twice from independent cDNA preparations. RNA without reverse transcriptase served as a negative control.
Cytokines ELISA
TNF-α and RANTES in the supernatant was determined by ELISA using Duoset ELISA kits (R&D, Minneapolis, MN) according to the manufacturer’s instructions. The optical density was determined using a BMG Polarstar microplate reader at 450 nm. A mean value of triplicate samples for each experiment and two or three separate experiments was used for analysis.
Western blot
RAW-Blue cells treated with indicated reagents were lysed in RIPA buffer with protease and phosphatase inhibitors (Cell Signaling Technologies Inc., Danvers, MA). Cell lysates were centrifuged at 10,000 rpm for 15 min to remove cell debris. Protein concentrations were determined by Bradford protein assay reagent (Bio-Rad). The lysates (10 μg protein) were separated by SDS-PAGE and transferred to nitrocellulose membrane. Membranes were blocked with 5% BSA/PBS containing 0.05% Tween-20 and were probed with antibodies against IκB, pIκB, SAPK/JNK (p54/p46), p-SAPK/JNK, Erk1/2 (p44/p42), p-Erk1/2, p-p38 and p38 MAP kinase. Bands were detected using ECL reagents (GE Healthcare, Piscataway, NJ) according to the manufacturer’s recommendations.
CD36 peptide binding
CD36 and scrambled control peptides [31] were synthesized and HPLC purified by Neo Bioscience (Cambridge, MA). Both peptides were about 99% pure as assessed by HPLC analysis by the company. Peptides were dissolved in 50% DMSO and stored in aliquots at −20°C. For foam cell assays, oxLDL was preincubated with CD36 or control peptides (at 4 μM) for 1 h followed by the addition to macrophages. The concentration of peptides used in these experiments were based on earlier report showing at 4 μM CD36 peptide inhibited oxLDL induced foam cell formation [31]. Cells were incubated for 24 h, fixed, and stained with Oil Red O and DAPI mount. To determine percent foam cells, total number of cells (DAPI+) and foam cells (Oil Red O+) cells were counted in five separate field using Olympus fluorescence microscope. Cells treated with oxLDL alone are used as positive control. Macrophages preincubated with the activating mCD36 mAb (JC63.1) at 5 μg/ml followed by the addition of oxLDL to determine the effect of anti-CD36 mAb on oxLDL induced foam cell formation.
To determine whether anti-CD36 mAb binds to different domain compared with oxLDL, anti-CD36 mAb binding to RAW-Blue cells were determined by cell ELISA. RAW-Blue cells (1 × 105/well) were plated and cultured overnight. OxLDL (10 μg/ml) or anti-mCD36 mAb (2 μg/ml) was preincubated with CD36- or control-peptides (4 μM) for 1 h at room temperature. Then the mixture was added to cells and incubated for 1 h at 4°C. Cells incubated with oxLDL or anti-CD36 mAb (mixed with DMSO at final concentration 0.2%) was used as positive controls. Cells incubated without any reagents were used as negative control. After washing the cells, cells were incubated with biotinylated anti-human apoB IgG (oxLDL binding) or anti-mIgA biotin (JC63.1 binding) for 1 h. After washing the cells in PBS/1% BSA, cells were incubated with SA-HRP followed by the addition of TMB-1, a HRP substrate. Color development was stopped by the addition of 2N sulfuric acid, and absorbance at 450 nm was read using a microplate reader (BMG Inc.).
Statistical analysis
Results are expressed as means ± SD. Data was analyzed by one-way ANOVA and a post hoc Tukey test was used as a multiple-comparison procedure. Differences were considered significant at P < 0.05. All analyses were performed using InStat 3.0a for Macintosh (Graphpad Software, San Diego, CA).
RESULTS
Binding of activating mCD36 mAb (JC63.1) to macrophage cells induces inflammatory cytokine response
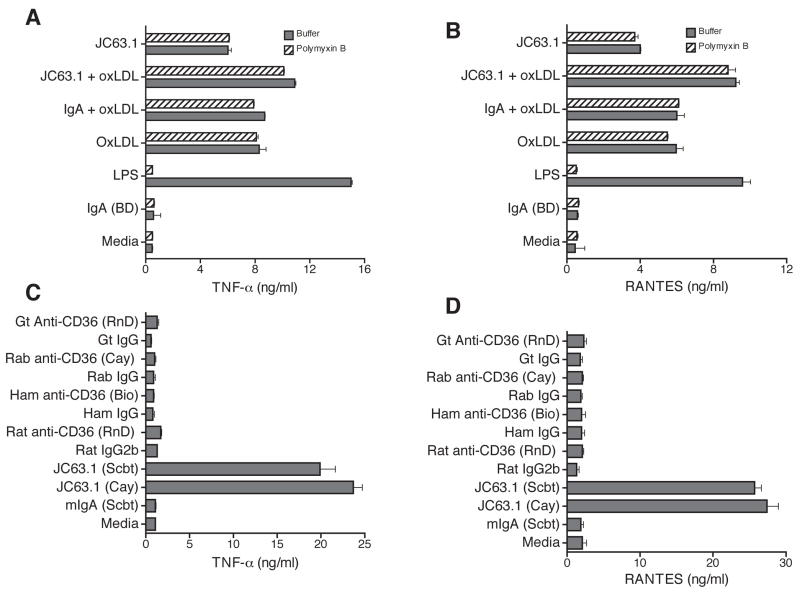
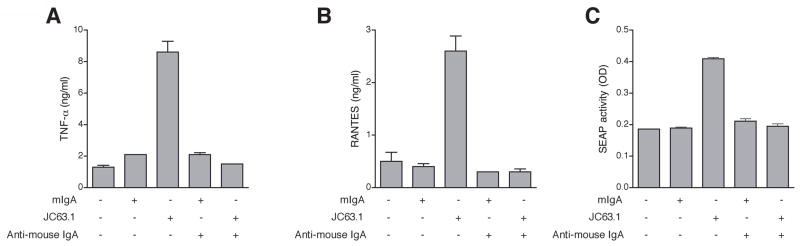
With an intention of searching for an alternate receptor for oxLDL besides CD36, blocking of CD36 receptor using different CD36 mAb was attempted. Mouse macrophage cell line, RAW-Blue, was pretreated with anti-mouse CD36 mAb (clone JC63.1) prior to the addition of oxLDL. OxLDL addition to RAW-Blue cells induced TNF-α and RANTES protein expression (Fig. 1A and B). An earlier report has shown that anti-mCD36 mAb (clone JC63.1) inhibited oxLDL uptake [28]. However, addition of anti-mCD36 mAb (JC63.1) did not block oxLDL-induced inflammatory cytokine responses. On the contrary, anti-mCD36 mAb (JC63.1) enhanced (P <0.001) oxLDL-induced TNF-α and RANTES expression (Fig. 1A and B). These findings raises the possibility that anti-mCD36 mAb (JC63.1) alone may be activating the macrophages to induce pro-inflammatory cytokine response. To address this possibility, RAW-Blue cells were incubated with anti-mCD36 mAb alone and cytokine response determined. Surprisingly, RAW-Blue cells incubated with anti-mCD36 mAb alone-induced TNF-α or RANTES secretion (Fig. 1A and B). To exclude the possibility that the anti-mCD36 mAb-induced TNF-α and RANTES is due to a low-level endotoxin contamination, polymyxin B was added during incubation. As expected LPS-induced cytokine response is completely blocked by polymyxin B, while addition of polymyxin-B did not show any difference in oxLDL- and/or anti-mCD36 mAb-induced TNF-α and RANTES expression (Fig. 1A and B). Under similar conditions isotype control mIgA from two different sources did not have effect. These findings suggest that the anti-mCD36 mAb, JC63.1 (hereafter referred as activating mCD36 mAb) binding to macrophage cell line results in macrophage activation and subsequent pro-inflammatory cytokine response.
Figure 1. Activating anti-mouse CD36 mAb alone induces inflammatory cytokine response.
RAW-Blue cells were pretreated with indicated reagents, and supernatant collected after 18 h was analyzed for TNF-α (A) and RANTES (B) secretion. Polymyxin B was included to exclude endotoxin contamination in antibody and oxLDL as the reason for the effect. RAW-Blue cells were treated with different commercially available antibody, activating mCD36 mAb (JC63.1), or other indicated anti-mCD36 mAb and polyclonal antibody and the control rabbit or goat IgG to determine the specificity of activating mCD36 mAb-induced TNF-α (C) and RANTES (D) secretion. Cells cultured in media, mIgA (from BD or Scbt), or rabbit IgG were used as controls. Error bars represent mean ± SD. Data are representative of three independent experiments.
Next, we addressed the specificity of activating mCD36 mAb induced inflammatory response. RAW-Blue cells were incubated with activating mCD36 mAb (from two different source) or other commercially available anti-mCD36 mAb and polyclonal antibody. As shown in Figure 1C, activating mCD36 mAb from two different source (Cayman and Santa Cruz biotech) induced TNF-α and RANTES expression, while other anti-mCD36 mAb and rabbit or goat anti-mCD36 polyclonal antibody did not induce inflammatory cytokine response (Fig. 1C and 1D). Further, neither the isotype control mIgA (from two different source) nor rabbit IgG induced responses (Fig. 1A-D). These findings suggest that the activating mCD36 mAb-induced macrophage inflammatory response is specific.
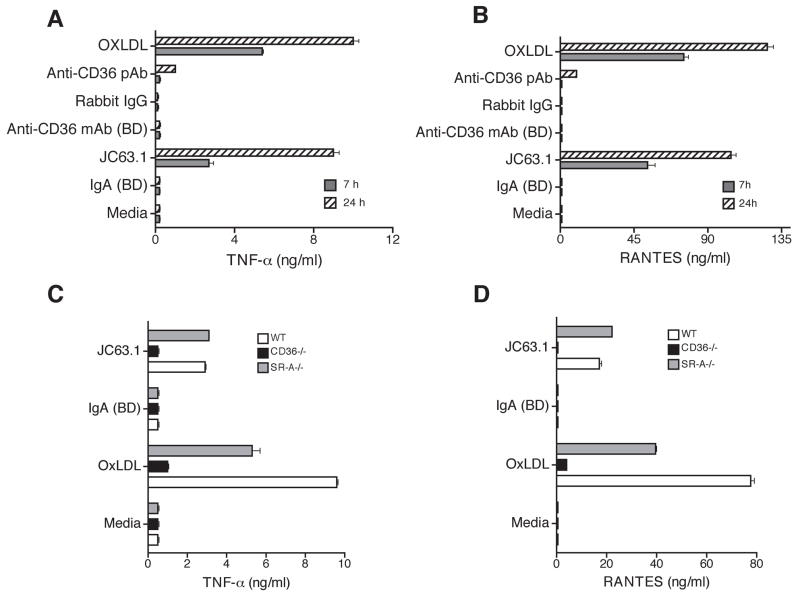
Activating mCD36 mAb induced inflammatory cytokine expression in murine peritoneal macrophages is dependent on CD36
To further confirm this unique phenomenon and to determine the cell-type specificity, experiments were repeated using primary mouse macrophages. Macrophage treated with the activating CD36 mAb-induced secretion of TNF-α (Fig. 2A) and RANTES (Fig. 2B). Under similar conditions other anti-mCD36 mAb, pAb, mIgA isotype control, or rabbit IgG alone did not induce TNF-α and RANTES release. To further confirm that the activating CD36 mAb induced inflammatory response is dependent on CD36 and not a non-specific interaction; peritoneal macrophages from apoE−/−, apoE−/−CD36−/− DKO, and apoE−/−SR-A−/− DKO mice were tested. Genotype analyses of macrophages from CD36−/− and SR-A−/− showed these macrophages indeed lack the expression of CD36 and SR-A (data not shown), while apoE−/− and apoE−/−SR-A−/− macrophages showed positive bands for CD36 gene. RT-PCR analyses also showed there is no CD36 mRNA expression in CD36−/− macrophages (data not shown). Treatment of apoE−/− and apoE−/−SR-A−/− macrophages with the activating CD36 mAb induced TNF-α and RANTES expression, while apoE−/−CD36−/− DKO macrophages did not show any detectable TNF-α and RANTES secretion (Fig. 2C and D). Under similar conditions LPS-induced TNF-α secretion was similar in both wild type and CD36-deficient macrophages (data not shown), as reported earlier [32]. Moreover, the oxLDL-induced TNF-α and RANTES were not completely inhibited in apoE−/−CD36−/− or apoE−/−SR-A−/− macrophages, suggesting that the deletion of CD36 or SR-A is not sufficient to decrease inflammatory response.
Figure 2. Activating mouse CD36 mAb induces inflammatory cytokine response in primary macrophages is dependent on CD36 expression.
Thioglycollate-elicited apoE−/− peritoneal macrophages were treated with indicated mouse anti-CD36 antibodies including the activating mCD36 mAb (JC63.1). Cells treated with mIgA, rabbit IgG, or oxLDL were used as controls. Supernatant collected after 7 or 24 h was used to determine TNF-α (A) and RANTES (B) secretion. Peritoneal macrophages collected from wild type (apoE−/−), CD36−/− (apoE−/−CD36−/− double deficient), and SRA−/− (apoE−/−SR-A−/− double deficient) were treated the activating mCD36 mAb. After 18 h, supernatant was collected and TNF-α (C) and RANTES (D) secretion were determined. Macrophages incubated with oxLDL and mIgA were used as positive and negative controls, respectively. Error bars represent mean ± SD. Data are representative of three independent experiments.
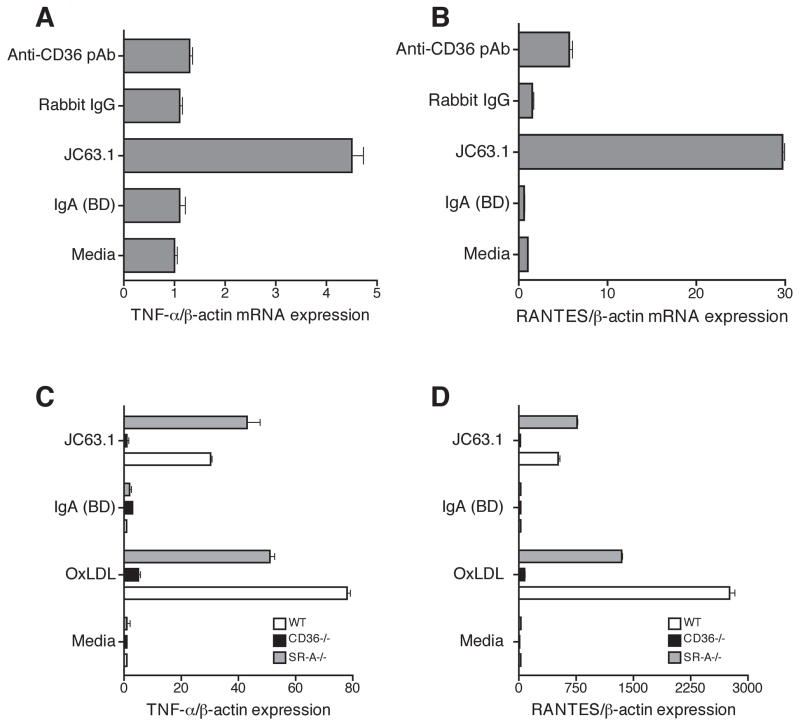
The activating mCD36 mAb-induced transcription of TNF-α and RANTES in macrophages
To further examine if activating mCD36 mAb-induced cytokine response at transcriptional level, quantitative RT-PCR analyses was performed. Macrophages treated with the activating CD36 mAb induced mRNA expression of TNF-α and RANTES on RAW-Blue cells, while other anti-mCD36 antibodies did not (Fig. 3A and B). Similarly, WT macrophages showed upregulation of TNF-α and RANTES mRNA expression in response to the activating mCD36 mAb as well as oxLDL (Fig. 3C and D). However, CD36-deficient macrophages did not respond to the activating mCD36 mAb-induced pro-inflammatory cytokine expression (Fig. 3C and D). These findings indicate that binding of activating mCD36 mAb induces pro-inflammatory cytokine responses at the level of transcription.
Figure 3. Activating mCD36 mAb induces TNF-α and RANTES mRNA expression.
RAW Blue cells and peritoneal macrophages were incubated with indicated reagents including activating mCD36 mAb for 18 h. Activating mCD36 mAb-induced TNF-α (C) and RANTES (D) mRNA expression in apoE−/− (WT), apoE−/−CD36−/− (CD36−/−), and apoE−/−SR-A (SR-A−/−) were determined by quantitative RT-PCR. Cells treated with media and oxLDL were used as controls. Error bars represent mean ± SD. Data are representative of three independent experiments.
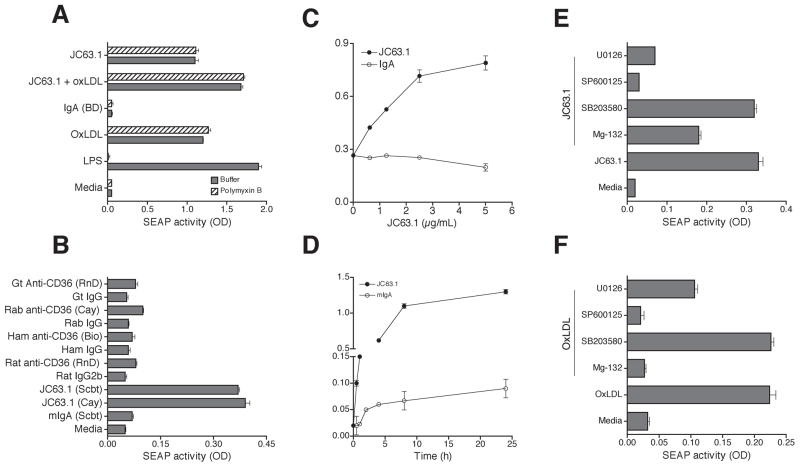
Signaling pathways involved in the activating CD36 mAb-induced inflammatory cytokine response
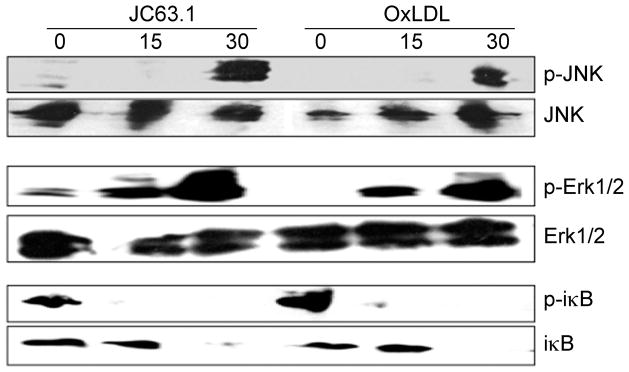
Earlier studies have shown that oxLDL binding to macrophages results in NF-κB and MAP kinase activation [3] and NF-κB and MAP kinase activation is necessary to promote foam cell formation [3]. Hence we investigated the signaling pathways regulating the inflammatory cytokine response initiated by the activating mCD36 mAb using NF-κB and AP-1 reporter RAW-Blue cells. OxLDL or LPS treatment of RAW-Blue cells induced SEAP secretion (Fig. 4A), while nLDL did not (data not shown). Interestingly, the activating anti-CD36 mAb induced SEAP activity while no response was observed with isotype control mIgA (Fig. 4A). Other anti-mCD36 mAb (mIgA) did not induce SEAP expression (Fig. 4B) suggesting the specificity of the NF-κB and AP-1 activation by the activating mCD36 mAb. Inclusion of polymyxin B in the assay conditions did not inhibit SEAP secretion by the activating mCD36 mAb or oxLDL. We then determined the kinetics and dose-dependent activation of NF-κB and AP-1 by the activating mCD36 mAb. The activating mCD36 mAb induced SEAP expression dose-dependently with a response is detected as low as 1 μg/ml (Fig. 4C). Kinetic analyses showed a time-dependent increase in SEAP secretion in RAW-Blue cells treated with the activating CD36 mAb with SEAP secretions detected as early as 30 min (Fig. 4D). Since SEAP reporter has both NF-κB and AP-1 response elements upstream to SEAP, we then determined the relative contribution of NF-κB and AP-1 using specific inhibitors. RAW-Blue cells were stimulated with the activating CD36 mAb in the presence of inhibitors of NF-κB and MAP kinase pathways. SPB00125, a selective inhibitor of JNK activation, strongly (>90%, P <0.001 from two independent experiments) inhibited SEAP secretion induced by the activating mCD36 mAb (Fig. 4E). MEK1/2 inhibitor (U0126) inhibited 75% (P <0.001 from two independent experiments) of SEAP secretion, while p38 kinase inhibitor (SB203580) did not inhibit SEAP secretion. NF-κB activation inhibitor blocked only 50% (P <0.001 from two independent experiments) of SEAP secretion induced by the activating mCD36 mAb. We then investigated whether oxLDL-induced signaling pathways differ from that of the activating mCD36 mAb. The activating mCD36 mAb-induced SEAP secretion is only partially (50%, P <0.001 from two independent experiments) inhibited by the NF-κB inhibitor (Fig. 4E). However, oxLDL-induced SEAP secretion is completely inhibited by the NF-κB inhibitor (Fig. 4F). Another striking difference is that the activating mCD36 mAb-induced response is completely abrogated by the MEK1/2 inhibitor (Fig. 4E), while oxLDL-induced response is inhibited about 40% (P <0.001 from two independent experiments) by MEK1/2 inhibitor (Fig. 4F). These findings suggest that the intracellular signaling pathways initiated by the activating mCD36 mAb are different from that of an oxLDL-induced response. We further confirmed these results by Western blot analysis. SAPK/JNK (p54/46) and Erk1/2 (p44/42) became phosphorylated transiently when cells were treated with the activating anti-CD36 mAb or oxLDL (Fig. 5). As seen with the inhibitor data, the activating anti-mCD36 mAb did not induce p38 phosphorylation (data not shown). Treatment of RAW-Blue cells with the activating anti-mCD36 mAb resulted in degradation of I-κB at 30 min and a concomitant increase in phospho-I-κB (Fig. 5), suggesting the activating anti-mCD36 mAb induced NF-κB activation. These findings indicate that NF-κB, JNK2, and Erk1/2 activation mediate the activating mCD36 mAb-induced pro-inflammatory cytokine responses. Moreover, the signaling cascade initiated by the activating mCD36 mAb is noticeably different from oxLDL-induced macrophage activation.
Figure 4. Activating mCD36 mAb-induced inflammatory cytokine response is dependent on NF-κB and MAP kinase activation.
To address signaling pathways regulating the inflammatory cytokine response initiated by the activating mCD36 mAb, RAW-Blue-SEAP reporter cells were used. RAW-Blue cells were pretreated with activating mCD36 mAb or other indicated reagents. Supernatant collected after 18 h was analyzed for SEAP reporter activity (A). RAW-Blue cells were incubated for 18 h with the activating mCD36 mAb or other anti-mCD36 mAb or pAb to determine the specificity of activating mCD36 mAb induced NF-κB and AP-1 activation (B). Cells treated with mIgA or cultured in medium were used as controls. RAW-Blue cells were treated with activating mCD36 mAb at different concentration (C) or different incubation time (D) to determine the dose-dependent and kinetics of macrophage activation. RAW-Blue cells were treated with activating mCD36 mAb (E) or oxLDL (F) in the absence or presence of Mg-132 (NF-κB inhibitor), SB203580 (p38 inhibitor), SP600125 (JNK inhibitor), or U0126 (Erk1/2 inhibitor). Cells cultured in media alone served as the control. Data are Mean ± SD of triplicates and representative of two independent experiments is presented.
Figure 5. Immunoblot analyses of MAP kinases and NF-κB induction by activating mCD36 mAb.
RAW-Blue cells were treated with activating mCD36 mAb (JC63.1, 5 μg/ml) or oxLDL (5 μg/ml) for 15 or 30 min. Cells were lyses, and Western blot analyses were carried out. Data represents two independent experiments.
CD36 cross-linking with the activating mCD36 mAb does not stimulate pro-inflammatory cytokine secretion
A recent report by Erdman et al. [32] showed that cross-linking mouse primary macrophages with anti-CD36 mAb (JC63.1) followed by anti-mIgA antibody did not induce TNF-α response. Hence the experiment was repeated to address the discrepancy in our data and the recently published article [32]. RAW-Blue cells treated with the activating anti-CD36 mAb followed by anti-mIgA secondary antibody did not induce cytokine response (Fig. 6A and B), and this finding is identical to the recent report [32]. However, RAW-Blue cells treated with the activating mCD36 mAb, without cross-linking (with no addition of cross-linking secondary antibody), induced TNF-α and RANTES protein expression (Fig. 6A and B). The increase in cytokine response also parallels the increase in SEAP activity (Fig. 6C). These findings suggest that internalization of CD36/anti-CD36 mAb complex does not result in macrophage inflammatory response. Moreover, incubating RAW-Blue cells at 4°C does not affect LPS-induced TNF-α secretion indicating that cells incubated at 4°C can respond to other inflammatory stimulus (data not shown) as reported using primary macrophages [32]. These findings suggest that the activating anti-CD36 mAb binding alone can activate macrophages and the subsequent pro-inflammatory cytokine response. However, internalization of cross-linked CD36, as shown in the recent report [32], cannot induce macrophage inflammatory responses.
Figure 6. CD36 cross-linking with activating mCD36 mAb does not induce pro-inflammatory response.
RAW-Blue cells were pretreated with activating mCD36 mAb (JC63.1) or IgA (isotype control), followed by the addition of anti mouse-IgA to induce CD36 cross-linking. Cells treated with media or mIgA or activating mCD36 mAb without cross-linking secondary antibody were used as controls. Supernatants were collected after 18 h, and secretion of TNF-α (A), RANTES (B), and SEAP activity (C) were determined.
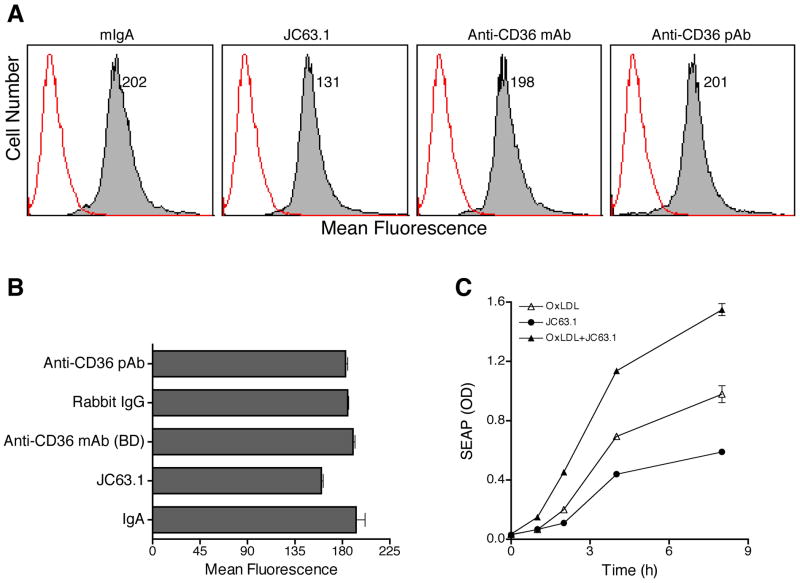
Activating mCD36 mAb binds to a domain different from the oxLDL-binding domain of CD36
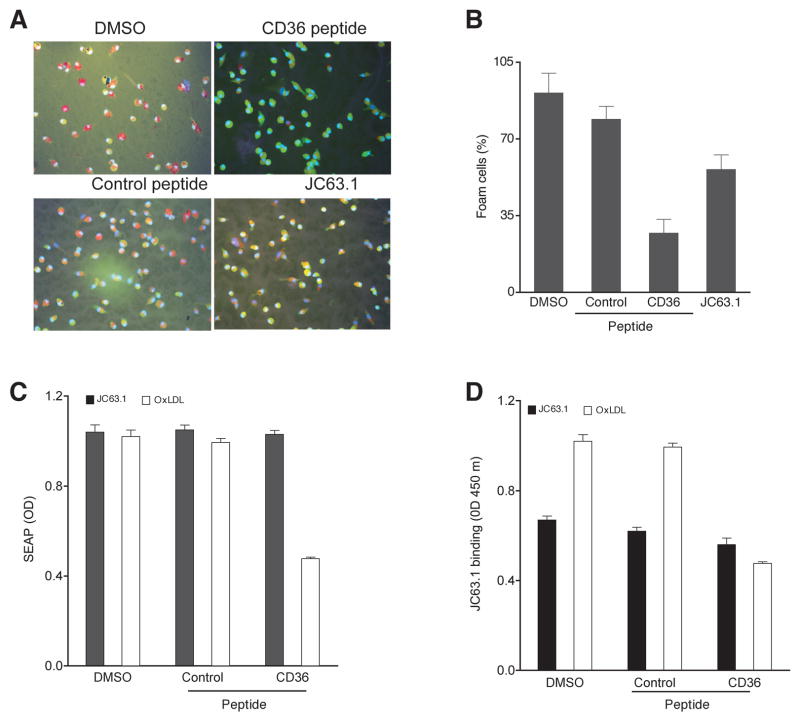
Thus far we showed activating mCD36 mAb induced pro-inflammatory cytokine responses by NF-κB and MAP kinase activation. However, earlier studies have shown that the activating mCD36 mAb (JC63.1) blocks oxLDL binding to macrophages [28]. Due to the unexpected finding, oxLDL binding to RAW-Blue cells in the presence of the activating anti-mCD36 mAb was determined. Binding assay was done at 4°C to prevent the internalization of oxLDL. FACS analysis showed that RAW-Blue cells bind to oxLDL (Fig. 7A) and this binding is only partially inhibited (25%–30%) by the activating anti-mCD36 mAb (Fig. 7B). However, oxLDL binding to RAW-Blue cells was not inhibited by other anti-mCD36 mAb or pAb (Fig. 7A and B). In addition to binding to mCD36, this activating mCD36 mAb can also recognize human CD36 (Cayman). Hence, we repeated the oxLDL-binding experiment using CHO-human CD36 transfected cells. OxLDL binding to CHO-human CD36 cells was also only partially inhibited (25%) by the activating mCD36 mAb (data not shown). These findings suggest that the activating mCD36 mAb and oxLDL binding domains on mCD36 are different. If oxLDL and the activating mCD36 mAb bind to different domains it raises a possibility that the activating mCD36 mAb would enhance oxLDL-induced inflammatory response. To address this possibility RAW-Blue cells were treated with activating mCD36 mAb and/or oxLDL and SEAP secretion was determined. OxLDL or the activating mCD36 mAb independently induced SEAP secretion (Fig. 7C). Interestingly, the combination of both the activating mCD36 mAb and oxLDL showed an enhanced SEAP activity (Fig. 7C) in a time-dependent manner. To further confirm the finding that the activating mCD36 mAb binds to a different domain, experiments were repeated using a CD36 peptide, which has been shown to block oxLDL induced foam cell formation in mouse macrophages [31]. Pre-incubation of oxLDL with CD36 peptide inhibited about 50% foam cell formation, while control scrambled peptide has no effect on foam cell formation (Fig 8A and B). After confirming the function of CD36 peptide, we then investigated whether the activating mCD36 mAb binds to the domain similar to oxLDL binding domain in CD36. CD36 peptide inhibited oxLDL-induced SEAP expression, a measure of RAW-Blue cell activation (Fig. 8C). However pre-incubation of CD36 peptide with the activating mCD36 mAb did not inhibit the activating mCD36-mAb induced RAW-Blue cell activation (Fig. 8C). We also determined the direct effect of CD36 peptide on anti-CD36 mAb binding to RAW-Blue cells using cell ELISA. Pre-incubation of oxLDL with the CD36 peptide inhibited oxLDL binding to RAW-blue cells (Fig. 8D), while control peptide did not. However, pre-incubation of anti-mCD36 mAb with the CD36 peptide only partially inhibited the activating mCD36 mAb binding to RAW-Blue cells. Under similar conditions, scrambled control peptide did not inhibit oxLDL or anti-CD36 mAb binding to RAW-Blue cells (Fig. 8D). These findings suggest that the activating mCD36 mAb may be binding to a novel domain, which may be in close proximity to oxLDL binding domain on mouse CD36.
Figure 7. Activating mCD36 mAb enhances oxLDL-induced macrophage activation.
OxLDL binding to RAW-Blue cells were determined by flow cytometry (A and B). Cells were treated with mIgA, activating mCD36 mAb (JC63.1), anti-mCD36 mAb, or anti-mCD36 pAb. OxLDL binding (closed histogram) was detected using goat anti apoB IgG-biotin followed by streptavidin-PE. Cells treated identically without oxLDL (open histogram) were used as a negative control. Number indicates mean fluorescence. C, Activating mCD36 mAb enhances oxLDL-induced macrophage inflammatory response. RAW-Blue cells were co-incubated with activating mCD36 mAb (2 μg/ml) and oxLDL (5 μg/ml). Cells treated with either one of the reagents were used to determine the basal activation. SEAP activity in the supernatant was analyzed to determine kinetics of inflammatory response.
Figure 8. Activating mCD36 mAb binds to a novel domain on mouse CD36.
Inhibition of OxLDL induced foam cell formation (A and B) by CD36 peptide was determined as described under ‘methods section’. CD36 peptide was preincubated with activating mCD36 mAb or oxLDL was added to RAW-Blue cells. Supernatant was collected to determine SEAP secretion (C). Effects of CD36 peptide on activating mCD36 mAb or oxLDL binding to RAW-Blue cells were determined by cell ELISA (D). Cells were treated with activating mCD36 mAb (JC63.1) or oxLDL preincubated with CD36- or control peptides were used. Cells treated with DMSO (at 0.2%, vehicle used to dissolve the peptides) were used as an additional control.
The activating mCD36 mAb enhanced TLR2 and TLR4-induced inflammatory responses
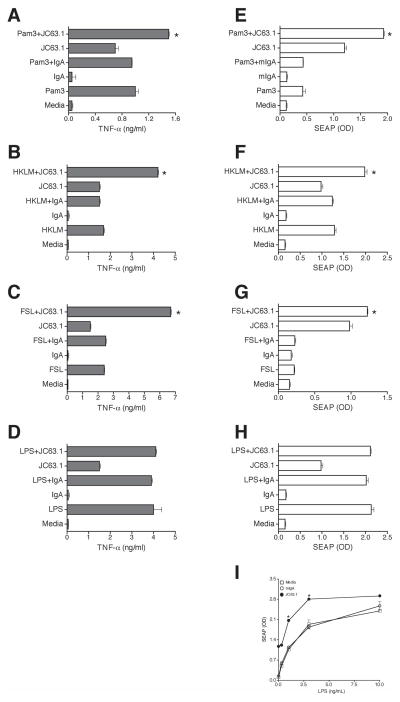
Data presented thus far demonstrated that the activating mCD36 mAb recognizing a novel domain on mCD36 induce pro-inflammatory cytokine responses. Next, we investigated the functional implications of the activating mCD36 mAb recognizing different domain on mCD36. CD36 has been shown to act as a co-receptor for TLR2 and TLR6 [33]. In fact, a very recent report shows CD36 co-association with TLR4 and TLR6 induced sterile inflammation [34]. We then determined whether the activating mCD36 mAb cooperatively interacts with TLR ligands to enhance inflammatory responses. To address such a possibility, RAW-Blue cell activation by the TLR ligand and/or the activating mCD36 mAb was determined by measuring TNF-α and SEAP expression. It should be noted that RAW-Blue cells express all TLRs including TLR2, TLR4, and TLR6 and excluding TLR5 (Invivogen). TLR2 ligands (Pam3CSK4, FSL1, and HKLM) or the activating mCD36 mAb alone induced TNF-α and SEAP expression (Fig. 9). Interestingly, the combination of the activating mCD36 mAb and TLR2 ligands, FSL-1 and HKLM, induced an approximately 2-fold higher (P <0.01 compared with TLR2 ligand or activating mCD36 mAb alone) expression of TNF-α (Fig. 9B-C) and SEAP (Fig. 9F-G), and Pam3CSK4 (at low dose 10 ng/ml) induced TNF-α (Fig. 9A) and SEAP secretion (Fig. 9E) is only marginal (1.3-fold). Under similar conditions, TLR4 ligand, LPS (Fig. 9D and H), and other TLR (TLR3, TLR5, TLR7 and TLR9) ligands did not show enhanced pro-inflammatory responses (data not shown). Recent studies have implicated TLR4/TLR6 heterodimer is involved in CD36-dependent macrophage activation and subsequent inflammatory responses [34]. Next we determined whether the lack of effect of LPS on JC63.1-induced additive response is due to the high dose of LPS used as a primary stimulus by titrating the LPS concentration. LPS at low concentration (0.1–10 ng/ml) dose-dependently induced SEAP activity with a maximum induction at 10 ng/ml (Fig. 9I). Notably, anti-CD36 mAb addition enhanced (approximately 2-fold higher expression) LPS-induced SEAP activity at lower dose of LPS (0.1 - 3 ng/ml) (Fig 9I). However at maximal stimulation of cells with high concentration of LPS (10 ng/ml) the JC63.1-mediated enhancement of inflammatory response is not apparent. These findings suggested that cooperative interaction between the activating mCD36 mAb and TLR2 and TLR4 ligands enhance macrophage activation and subsequent pro-inflammatory responses.
Figure 9. Activating mCD36 mAb enhanced TLR2-induced inflammatory response.
RAW-Blue cells were co-incubated with activating mCD36 mAb (JC63.1) and TLR2 (Pam3, FSL, and HKLM) or TLR4 (LPS, 100 ng/ml) ligands. Cells treated with either one of the reagents were used to determine the basal activation. Secretion of TNF-α (A-D) and SEAP activity (E-H) were determined. Inflammatory responses by TLR-2 ligands; Pam3 (A and E), FSL (B and F), HKLM (C and G), and TLR4 ligand LPS (D and H) are presented. Mean values between the combination of TLR2 ligands and activating mCD36 mAb is significantly different (*, p<0.01 by One-way ANOVA Tukey post-hoc multiple comparison test) from the response by TLR2 ligands or activating mCD36 mAb alone. I, Activating mCD36 enhanced TLR4-induced inflammatory response at low concentration of LPS. RAW-Blue cells were co-incubated with activating mCD36 mAb (JC63.1 at 2 μg/ml) and TLR4 (LPS) ligand at different concentration (0.1 - 10 ng/ml). Cells treated with either one of the reagents were used to determine the basal activation. Mean values between the combination of LPS and activating mCD36 mAb is significantly different (*, p<0.01 by One-way ANOVA Tukey post-hoc multiple comparison test) from the response by LPS or activating mCD36 mAb alone.
DISCUSSION
The principal finding in this report is identification of a novel domain on mouse CD36 that can induce macrophage inflammatory cytokine response independent of oxLDL- or TLR2- ligand binding function of CD36. We have identified this domain using an activating mCD36 mAb, which induced TNF-α and RANTES expression in mouse macrophage cell line and primary peritoneal macrophages. The inflammatory cytokine response initiated by this domain is mediated by MAP kinase and in part by NF-κB activation. Furthermore, we showed the binding of activating mCD36 mAb to this domain enhances TLR2- or TLR4- or oxLDL-mediated inflammatory cytokine response. Importantly, we present evidence that the activating mCD36 mAb binds to a domain different from oxLDL- or TLR2-ligand, FSL-1, binding domain on mouse CD36. The result of this study are relevant in the light of oxLDL-independent CD36-mediated inflammatory responses in chronic inflammatory diseases such as atherosclerosis [21,22], Alzheimer’s disease [8,23,24], S. aureus infection [25] and non-alcoholic steatohepatitis [27].
In this study using a mouse macrophage cell line, we demonstrated that the activating mCD36 mAb induced pro-inflammatory cytokines, TNF-α and RANTES in mouse RAW-Blue macrophage cell line and primary macrophages. We also presented evidence that the activating mCD36 mAb induced macrophage inflammatory responses is specific as the activating mCD36 mAb showed no effect on CD36−/− macrophages. We also showed activating mCD36 mAb (JC63.1) from two different commercial source induced TNF-α and RANTES response and the activating mCD36 mAb induced response is not inhibited by polymyxin B ruling out any possible endotoxin contamination which could non-specifically induce the inflammatory response. These findings indicate that the activating mCD36 mAb-induced response is specific. However, our finding is in contrast to a very recent report by Erdman et al. [32] showing that anti-CD36 mAb (JC63.1) did not induce TNF-α response in RAW264 macrophage cells. The difference between both studies is that in the study by Erdman et al. [32] RAW264 cells were treated with the activating mCD36 mAb followed by anti-mIgA antibody (secondary antibody) to induce CD36 cross-linking. In our study the activating mCD36 mAb was added without the cross-linking secondary antibody. Our finding showing addition of anti-CD36 Ab induced response is opposite to the finding showing cross-linking did not induce the response. We were surprised by our finding that activating mCD36 mAb from two different t sources (Cayman and Santa Cruz) induced macrophage activation, while control mIgA from Santa Cruz and BD-Biosciences did not. Moreover, all the experiments using the activating mCD36 mAb were conducted using polymyxin B. These findings rule out that the non-cross linking activation by the activating mCD36 mAb is not due to endotoxin contamination. Moreover, RAW-Blue cells kept at 4°C were responsive to other stimulus such as LPS, as reported earlier using primary macrophages in cross-linking experiment [32]. Together, these findings have indicated that CD36 internalization by antibody cross-linking does not induce inflammatory responses. Notably, these findings suggest that a putative ligand binding to the new domain on the extracellular region of mouse CD36 may be sufficient to induce inflammatory cytokine responses.
We investigated the signaling pathways contributing to the activating mCD36 mAb-induced inflammatory responses in macrophages. Using RAW-Blue, a NF-κB and AP-1 reporter cell line, we showed the activating mCD36 mAb-induced TNF-α and RANTES are mediated via activation of NF-κB and AP-1, a downstream target for MAP kinase pathway. Using pharmacological inhibitors and Western blot analysis, we showed JNK activation is contributing to the activating mCD36 mAb induced pro-inflammatory responses. However, blockade of p38 MAP kinase did not inhibit the response ruling out a role for p38 MAPK in this response. These findings suggest that the signaling cascade initiated by the activating mCD36 mAb for most part is similar to that reported for oxLDL [3]. However, comparison of the signaling cascade initiated by oxLDL and the activating mCD36 mAb showed two differences. OxLDL-induced NF-κB activation is completely inhibited by the NF-κB inhibitor; while only 50% inhibited the activating mCD36 mAb induced a response (Fig. 4E and F). Another striking difference is that the MKK1/2 inhibitor, specifically inhibiting Erk1/2 activation, completely inhibited the activating mCD36-induced inflammatory response, while partial inhibition was observed in the oxLDL-induced response (Fig. 4E and F). These findings suggest that the signaling cascade initiated by the activating mCD36 mAb binding to a new domain on mouse CD36 is different from the oxLDL-induced signaling pathways.
Our findings showing that the induction of inflammatory cytokine response by the activating mCD36 mAb is unexpected. One possible explanation for this response is that the activating mCD36 mAb may be binding to oxLDL binding domain on CD36 and act as a surrogate ligand. Earlier studies using this anti-mCD36 have shown that it inhibited oxLDL binding to primary macrophages [28] supports this possibility. However, oxLDL-binding studies using RAW-Blue cells showed only 30% inhibition was achieved by the activating mCD36 mAb (Fig. 7). On careful evaluation of the earlier report [28], it appears the authors have also achieved only partial inhibition of oxLDL binding, though the percent inhibition was not clearly stated. These findings raise a possibility that the activating mCD36 mAb may be binding to a novel domain that may be independent of oxLDL-binding domain of CD36, and the partial blocking of oxLDL-binding by the activating mCD36 mAb could be due to steric hindrance.
The next question is what is the functional domain recognized by the activating mCD36 mAb? Using GSH-human CD36 chimeric protein [35], as well as a panel of anti-human CD36 mAb, aa155–183 has been identified as oxLDL-binding domain on human CD36 [36]. A small CD36 peptide, aa160–168, inhibited oxLDL-mediated foam cell formation in mouse macrophages [31]. These findings have mapped the oxLDL-binding domain between aa155–183, and this domain is also conserved in human, mouse, and rat [31]. Earlier studies using mAb against aa155–183 of human CD36 has been shown to inhibit oxLDL binding [36], apoptotic neutrophils [37], and adherence of P. falciparum-infected RBC [38]. Similarly, β-amyloid, another CD36 ligand, inhibited mouse CD36-dependent oxLDL binding [7]. Based on these reports it is possible that multiple CD36 ligands such as oxLDL, apoptotic cells, β-amyloid and P. falciparum-infected RBC may be binding to a same domain on CD36. In the present study, we showed oxLDL binding is poorly inhibited by this activating mCD36 mAb (Fig. 7AB), ruling out a possibility that the activating mCD36 mAb may be binding to the oxLDL-binding domain on CD36. These findings suggest a novel domain on CD36 may initiate oxLDL-independent inflammatory response. These findings further suggest an interesting possibility that this novel domain on CD36 could be in close proximity to the oxLDL binding. Our data (Fig. 7C) showing enhanced inflammatory responses in RAW-Blue cells treated with the activating mCD36 mAb and oxLDL supports this possibility. We showed CD36 peptide blocked CD36-dependent foam cell formation, oxLDL binding to RAW-Blue cells and oxLDL-induced macrophage activation as determined by SEAP secretion (Fig. 8). Interestingly CD36 peptide did not block the activating CD36 mAb binding to RAW-Blue cells as well as the activating mCD36 mAb induced SEAP expression (Fig. 8). These findings suggest that the activating mCD36 mAb binding a domain different from oxLDL binding domain. In a very recent report Jimenez-Dalmaroni et al [14] have shown that soluble CD36 ectodomain binds to FSL-1 (a TLR2 ligand) and enhances TLR2-induced TNF-α secretion by macrophages. We showed the activating mCD36 mAb in combination with FSL-1 enhanced inflammatory responses in macrophages (Fig. 9). Based on this finding it is reasonable to predict that the activating mCD36 mAb binding site is different from FSL-1 binding site on mouse CD36. Based on earlier reports oxLDL, β-amyloid, apoptotic cells, P. falciparum-infected RBC, and FSL-1 may not be the potential ligands for this novel domain. More new ligands for CD36 are being identified, which include hexarelin, a hexapeptide member of the growth hormone releasing peptide [39], and serum amyloid antigen, which binds to human and rat CD36 [40]. Collectively, these findings suggest that the activating mCD36 mAb recognized a distinct domain, which is different from oxLDL- or TLR2-ligand binding domains. However, the nature of exogenous and/or endogenous ligand binding to this novel domain on mouse CD36 needs further investigation.
Recent studies have shown there is a cooperative interaction between TLRs, such as TLR2, TLR4, TLR6, and CD36 [33,34]. In this study we showed that activating mCD36 mAb binding to macrophages enhanced the TLR2-dependent inflammatory responses. Specifically, TLR2 and TLR4 ligands, Pam3, HKLM, and FSL-1 (TLR2 ligand) and at sub-saturating concentration of LPS (TLR4 ligand), induced TNF-α response and NF-κB and AP-1 activation was enhanced by the activating mCD36 mAb (Fig. 9). However, combining activating mCD36 mAb with TLR3, TLR5, TLR7, and TLR9 ligands did not show enhanced inflammatory responses suggesting the cooperative interaction of the activating mCD36 mAb is specific for TLR2. TLR2/TLR2, TLR2/TLR1 and TLR2/TLR6 dimerization has been implicated in inflammatory responses initiated by different TLR2 ligands [41]. TLR2/TLR2 homodimer is activated by Listeria-derived lipotechoic acid, a component of heat-killed Listeria monocytogenes, HKLM, Gram-positive bacteria [42,43]. Besides, TLR2 can also form heterodimers with other TLRs such as TLR1 and TLR6. TLR2/6 heterodimer is activated by the synthetic diacylated lipopeptide FSL-1 [44], while the synthetic triacylated lipopeptide Pam3CSK4 is recognized by TLR2/1 [45,46]. Since the activating mCD36 mAb enhanced all three TLR2 ligands, it is possible that the activating mCD36 mAb may be inducing responses initiated by TLR2/TLR2 homodimer as well as TLR2/TLR1 and TLR2/TLR6 heterodimers. Using HEK-293 cells expressing CD36 with TLR2, TLR4, or TLR6, recent studies have presented evidence that CD36 co-associates with TLR4 and TLR6 with minimal contribution from TLR2 when oxLDL was used as a ligand for CD36 [34]. These studies suggest that the activating mCD36 mAb binding to this new domain on CD36 may specifically result in co-association with TLR2 and/or TLR4. The activating mCD36 mAb provides an excellent research tool to decipher signaling pathways induced by CD36 interacting with co-receptors such as TLR2 homodimer, TLR/TLR1, TLR2/TLR6 and TLR4/TLR6 heterodimers.
In summary, we showed the activating mCD36 mAb (JC63.1) induced pro-inflammatory cytokine response with very minimal effect on oxLDL binding. Moreover, the activating mCD36 mAb enhanced the oxLDL-induced response. Analyses using a CD36 peptide showed CD36 peptide inhibited oxLDL induced macrophage activation, while no effect on the activating mCD36 mAb induced macrophage activation. Collectively, these findings suggest that the activating mCD36 mAb recognized a novel domain on mouse CD36. However an endogenous or exogenous ligand(s) binding to this domain need further investigation.
Acknowledgments
Supported by NIH grant R01HL86674 (SN) and in part by the USDA grant CRIS-6251-51000-005-02S to the Arkansas Children’s Nutrition Center.
We thank John Gregan for his help with manuscript preparation and Dr. Uma Nagarajan for the critical review of this article. We thank Ramona Burris, Amy Greenway, and Jessica Warden for their technical assistance.
Footnotes
Abbreviations used in this paper: oxLDL, oxidized low-density lipoprotein; SR-A, scavenger receptor A-I/II; apoE, apolipoprotein E; SEAP, secreted embryonic alkaline phosphatase; nLDL, native LDL.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Endemann G, Stanton LW, Madden KS, Bryant CM, White RT, Protter AA. CD36 is a receptor for oxidized low density lipoprotein. J Biol Chem. 1993;268:11811–11816. [PubMed] [Google Scholar]
- 2.Nicholson AC, Han J, Febbraio M, Silversterin RL, Hajjar DP. Role of CD36, the macrophage class B scavenger receptor, in atherosclerosis. Ann N Y Acad Sci. 2001;947:224–228. doi: 10.1111/j.1749-6632.2001.tb03944.x. [DOI] [PubMed] [Google Scholar]
- 3.Rahaman SO, Lennon DJ, Febbraio M, Podrez EA, Hazen SL, Silverstein RL. A CD36-dependent signaling cascade is necessary for macrophage foam cell formation. Cell Metab. 2006;4:211–221. doi: 10.1016/j.cmet.2006.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frieda S, Pearce A, Wu J, Silverstein RL. Recombinant GST/CD36 fusion proteins define a thrombospondin binding domain. Evidence for a single calcium-dependent binding site on CD36. J Biol Chem. 1995;270:2981–2986. doi: 10.1074/jbc.270.7.2981. [DOI] [PubMed] [Google Scholar]
- 5.Drover VA, Nguyen DV, Bastie CC, Darlington YF, Abumrad NA, Pessin JE, London E, Sahoo D, Phillips MC. CD36 mediates both cellular uptake of very long chain fatty acids and their intestinal absorption in mice. J Biol Chem. 2008;283:13108–13115. doi: 10.1074/jbc.M708086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohgami N, Nagai R, Ikemoto M, Arai H, Kuniyasu A, Horiuchi S, Nakayama H. Cd36, a member of the class b scavenger receptor family, as a receptor for advanced glycation end products. J Biol Chem. 2001;276:3195–3202. doi: 10.1074/jbc.M006545200. [DOI] [PubMed] [Google Scholar]
- 7.Kunjathoor VV, Tseng AA, Medeiros LA, Khan T, Moore KJ. beta-Amyloid promotes accumulation of lipid peroxides by inhibiting CD36-mediated clearance of oxidized lipoproteins. J Neuroinflammation. 2004;1:23. doi: 10.1186/1742-2094-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore KJ, El Khoury J, Medeiros LA, Terada K, Geula C, Luster AD, Freeman MW. A CD36-initiated signaling cascade mediates inflammatory effects of beta-amyloid. J Biol Chem. 2002;277:47373–47379. doi: 10.1074/jbc.M208788200. [DOI] [PubMed] [Google Scholar]
- 9.Crandall I, Guy RA, Maguire GF, Connelly PW, Kain KC. Plasmodium falciparum-infected erythrocytes and oxidized low-density lipoprotein bind to separate domains of CD36. J Infect Dis. 1999;180:473–479. doi: 10.1086/314897. [DOI] [PubMed] [Google Scholar]
- 10.Oquendo P, Hundt E, Lawler J, Seed B. CD36 directly mediates cytoadherence of Plasmodium falciparum parasitized erythrocytes. Cell. 1989;58:95–101. doi: 10.1016/0092-8674(89)90406-6. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg ME, Sun M, Zhang R, Febbraio M, Silverstein R, Hazen SL. Oxidized phosphatidylserine-CD36 interactions play an essential role in macrophage-dependent phagocytosis of apoptotic cells. J Exp Med. 2006;203:2613–2625. doi: 10.1084/jem.20060370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fadok VA, Warner ML, Bratton DL, Henson PM. CD36 is required for phagocytosis of apoptotic cells by human macrophages that use either a phosphatidylserine receptor or the vitronectin receptor (alpha v beta 3) J Immunol. 1998;161:6250–6257. [PubMed] [Google Scholar]
- 13.Baranova IN, Kurlander R, Bocharov AV, Vishnyakova TG, Chen Z, Remaley AT, Csako G, Patterson AP, Eggerman TL. Role of human CD36 in bacterial recognition, phagocytosis, and pathogen-induced JNK-mediated signaling. J Immunol. 2008;181:7147–7156. doi: 10.4049/jimmunol.181.10.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jimenez-Dalmaroni MJ, Xiao N, Corper AL, Verdino P, Ainge GD, Larsen DS, Painter GF, Rudd PM, Dwek RA, Hoebe K, Beutler B, Wilson IA. Soluble CD36 ectodomain binds negatively charged diacylglycerol ligands and acts as a co-receptor for TLR2. PLoS One. 2009;4:e7411. doi: 10.1371/journal.pone.0007411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janabi M, Yamashita S, Hirano K, Sakai N, Hiraoka H, Matsumoto K, Zhang Z, Nozaki S, Matsuzawa Y. Oxidized LDL-induced NF-kappa B activation and subsequent expression of proinflammatory genes are defective in monocyte-derived macrophages from CD36-deficient patients. Arterioscler Thromb Vasc Biol. 2000;20:1953–1960. doi: 10.1161/01.atv.20.8.1953. [DOI] [PubMed] [Google Scholar]
- 17.Febbraio M, Podrez EA, Smith JD, Hajjar DP, Hazen SL, Hoff HF, Sharma K, Silverstein RL. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J Clin Invest. 2000;105:1049–1056. doi: 10.1172/JCI9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunjathoor VV, Febbraio M, Podrez EA, Moore KJ, Andersson L, Koehn S, Rhee JS, Silverstein R, Hoff HF, Freeman MW. Scavenger receptors class A-I/II and CD36 are the principal receptors responsible for the uptake of modified low density lipoprotein leading to lipid loading in macrophages. J Biol Chem. 2002;277:49982–49988. doi: 10.1074/jbc.M209649200. [DOI] [PubMed] [Google Scholar]
- 19.Moore KJ, Kunjathoor VV, Koehn SL, Manning JJ, Tseng AA, Silver JM, McKee M, Freeman MW. Loss of receptor-mediated lipid uptake via scavenger receptor A or CD36 pathways does not ameliorate atherosclerosis in hyperlipidemic mice. J Clin Invest. 2005;115:2192–2201. doi: 10.1172/JCI24061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy DJ, Kuchibhotla SD, Guy E, Park YM, Nimako G, Vanegas D, Morton RE, Febbraio M. Dietary cholesterol plays a role in CD36-mediated atherogenesis in LDLR-knockout mice. Arterioscler Thromb Vasc Biol. 2009;29:1481–1487. doi: 10.1161/ATVBAHA.109.191940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manning-Tobin JJ, Moore KJ, Seimon TA, Bell SA, Sharuk M, Alvarez-Leite JI, de Winther MP, Tabas I, Freeman MW. Loss of SR-A and CD36 activity reduces atherosclerotic lesion complexity without abrogating foam cell formation in hyperlipidemic mice. Arterioscler Thromb Vasc Biol. 2009;29:19–26. doi: 10.1161/ATVBAHA.108.176644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore KJ, Freeman MW. Scavenger receptors in atherosclerosis: beyond lipid uptake. Arterioscler Thromb Vasc Biol. 2006;26:1702–1711. doi: 10.1161/01.ATV.0000229218.97976.43. [DOI] [PubMed] [Google Scholar]
- 23.El Khoury JB, Moore KJ, Means TK, Leung J, Terada K, Toft M, Freeman MW, Luster AD. CD36 mediates the innate host response to beta-amyloid. J Exp Med. 2003;197:1657–1666. doi: 10.1084/jem.20021546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medeiros LA, Khan T, El Khoury JB, Pham CL, Hatters DM, Howlett GJ, Lopez R, O’Brien KD, Moore KJ. Fibrillar amyloid protein present in atheroma activates CD36 signal transduction. J Biol Chem. 2004;279:10643–10648. doi: 10.1074/jbc.M311735200. [DOI] [PubMed] [Google Scholar]
- 25.Hoebe K, Georgel P, Rutschmann S, Du X, Mudd S, Crozat K, Sovath S, Shamel L, Hartung T, Zahringer U, Beutler B. CD36 is a sensor of diacylglycerides. Nature. 2005;433:523–527. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 26.Stuart LM, Deng J, Silver JM, Takahashi K, Tseng AA, Hennessy EJ, Ezekowitz RA, Moore KJ. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J Cell Biol. 2005;170:477–485. doi: 10.1083/jcb.200501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bieghs V, Wouters K, van Gorp PJ, Gijbels MJ, de Winther MP, Binder CJ, Lutjohann D, Febbraio M, Moore KJ, van Bilsen M, Hofker MH, Shiri-Sverdlov R. Role of Scavenger Receptor A and CD36 in diet-induced non-alcoholic steatohepatitis in hyperlipidemic mice. Gastroenterology. 2010;138:2477–2486. doi: 10.1053/j.gastro.2010.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang CP, Han S, Okamoto H, Carnemolla R, Tabas I, Accili D, Tall AR. Increased CD36 protein as a response to defective insulin signaling in macrophages. J Clin Invest. 2004;113:764–773. doi: 10.1172/JCI19528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart BW, Nagarajan S. Recombinant CD36 inhibits oxLDL-induced ICAM-1-dependent monocyte adhesion. Mol Immunol. 2006;43:255–267. doi: 10.1016/j.molimm.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 30.Thampi P, Stewart BW, Joseph L, Melnyk SB, Hennings LJ, Nagarajan S. Dietary homocysteine promotes atherosclerosis in apoE-deficient mice by inducing scavenger receptors expression. Atherosclerosis. 2008;197:620–629. doi: 10.1016/j.atherosclerosis.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 31.Kar NS, Ashraf MZ, Valiyaveettil M, Podrez EA. Mapping and characterization of the binding site for specific oxidized phospholipids and oxidized low density lipoprotein of scavenger receptor CD36. J Biol Chem. 2008;283:8765–8771. doi: 10.1074/jbc.M709195200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erdman LK, Cosio G, Helmers AJ, Gowda DC, Grinstein S, Kain KC. CD36 and TLR interactions in inflammation and phagocytosis: implications for malaria. J Immunol. 2009;183:6452–6459. doi: 10.4049/jimmunol.0901374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Triantafilou M, Gamper FG, Haston RM, Mouratis MA, Morath S, Hartung T, Triantafilou K. Membrane sorting of toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J Biol Chem. 2006;281:31002–31011. doi: 10.1074/jbc.M602794200. [DOI] [PubMed] [Google Scholar]
- 34.Stewart CR, Stuart LM, Wilkinson K, van Gils JM, Deng J, Halle A, Rayner KJ, Boyer L, Zhong R, Frazier WA, Lacy-Hulbert A, Khoury JE, Golenbock DT, Moore KJ. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat Immunol. 2010;11:155–161. doi: 10.1038/ni.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearce SF, Roy P, Nicholson AC, Hajjar DP, Febbraio M, Silverstein RL. Recombinant glutathione S-transferase/CD36 fusion proteins define an oxidized low density lipoprotein-binding domain. J Biol Chem. 1998;273:34875–34881. doi: 10.1074/jbc.273.52.34875. [DOI] [PubMed] [Google Scholar]
- 36.Navazo MDP, Daviet L, Ninio E, McGregor JL. Identification on human CD36 of a domain (155–183) implicated in binding low-density lipoproteins (Ox-LDL) Arterioscler Thromb Vasc Biol. 1996;16:1033–1039. doi: 10.1161/01.atv.16.8.1033. [DOI] [PubMed] [Google Scholar]
- 37.Navazo MD, Daviet L, Savill J, Ren Y, Leung LL, McGregor JL. Identification of a domain (155–183) on CD36 implicated in the phagocytosis of apoptotic neutrophils. J Biol Chem. 1996;271:15381–15385. doi: 10.1074/jbc.271.26.15381. [DOI] [PubMed] [Google Scholar]
- 38.Daviet L, Craig AG, McGregor L, Pinches R, Wild TF, Berendt AR, Newbold CI, McGregor JL. Characterization of two vaccinia CD36 recombinant-virus-generated monoclonal antibodies (10/5, 13/10): effects on malarial cytoadherence and platelet functions. Eur J Biochem. 1997;243:344–349. doi: 10.1111/j.1432-1033.1997.0344a.x. [DOI] [PubMed] [Google Scholar]
- 39.Bodart V, Febbraio M, Demers A, McNicoll N, Pohankova P, Perreault A, Sejlitz T, Escher E, Silverstein RL, Lamontagne D, Ong H. CD36 mediates the cardiovascular action of growth hormone-releasing peptides in the heart. Circ Res. 2002;90:844–849. doi: 10.1161/01.res.0000016164.02525.b4. [DOI] [PubMed] [Google Scholar]
- 40.Baranova IN, Bocharov AV, Vishnyakova TG, Kurlander R, Chen Z, Fu D, Arias IM, Csako G, Patterson AP, Eggerman TL. CD36 is a novel serum amyloid A (SAA) receptor mediating SAA binding and SAA-induced signaling in human and rodent cells. J Biol Chem. 2010;285:8492–8506. doi: 10.1074/jbc.M109.007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 42.Hauf N, Goebel W, Fiedler F, Sokolovic Z, Kuhn M. Listeria monocytogenes infection of P388D1 macrophages results in a biphasic NF-kappaB (RelA/p50) activation induced by lipoteichoic acid and bacterial phospholipases and mediated by IkappaBalpha and IkappaBbeta degradation. Proc Natl Acad Sci U S A. 1997;94:9394–9399. doi: 10.1073/pnas.94.17.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flo TH, Halaas O, Lien E, Ryan L, Teti G, Golenbock DT, Sundan A, Espevik T. Human toll-like receptor 2 mediates monocyte activation by Listeria monocytogenes, but not by group B streptococci or lipopolysaccharide. J Immunol. 2000;164:2064–2069. doi: 10.4049/jimmunol.164.4.2064. [DOI] [PubMed] [Google Scholar]
- 44.Okusawa T, Fujita M, Nakamura J, Into T, Yasuda M, Yoshimura A, Hara Y, Hasebe A, Golenbock DT, Morita M, Kuroki Y, Ogawa T, Shibata K. Relationship between structures and biological activities of mycoplasmal diacylated lipopeptides and their recognition by toll-like receptors 2 and 6. Infect Immun. 2004;72:1657–1665. doi: 10.1128/IAI.72.3.1657-1665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]