Abstract
Polycomb group proteins (PcG) such as Enhancer of zeste homolog 2 (EZH2) are epigenetic transcriptional repressors that function through recognition and modification of histone methylation and chromatin structure. Targets of PcG include cell cycle regulatory proteins which govern cell cycle progression and cellular senescence. Senescence is a characteristic of melanocytic nevi, benign melanocytic proliferations that can be precursors of malignant melanoma. In this study, we report that EZH2, which we find absent in melanocytic nevi but expressed in many or most metastatic melanoma cells, functionally suppresses the senescent state in human melanoma cells. EZH2 depletion in melanoma cells inhibits cell proliferation, restores features of a cellular senescence phenotype, and inhibits growth of melanoma xenografts in vivo. p21/CDKN1A is activated upon EZH2 knockdown in a p53-independent manner and contributes substantially to cell cycle arrest and induction of a senescence phenotype. EZH2 depletion removes histone deacetylase 1 (HDAC1) from the CDKN1A transcriptional start site and downstream region, enhancing histone 3 acetylation globally and at CDKN1A. This results in recruitment of RNA polymerase II, leading to p21/CDKN1A activation. Depletion of EZH2 synergistically activates p21/CDKN1A expression in combination with the HDAC inhibitor trichostatin A. Since melanomas often retain wild-type p53 function activating p21, our findings describe a novel mechanism whereby EZH2 activation during tumor progression represses p21, leading to suppression of cellular senescence and enhanced tumorigenicity.
Keywords: melanoma, polycomb, senescence, EZH2, p21
INTRODUCTION
Senescence is a cellular stress response resulting in a blockade of cellular proliferation, characteristic morphological changes, and the expression of characteristic molecular markers such as senescence-associated β-galactosidase (SA-β-gal) (1–3). Overexpression of activated oncogenes such as HRASG12V in primary cells triggers oncogene-induced senescence, featuring a cellular phenotype indistinguishable from that of cells entering senescence following extending in vitro replication (4). Overexpression of melanoma oncogenes such as BRAFV600E (5) and NRASQ61K (6) provokes oncogene-induced senescence in melanocytes. BRAFV600E and activating mutations at codon 61 in NRAS, both found frequently in melanoma (7–8), are also associated with acquired (9) and congenital (10) melanocytic nevi respectively. Both types of nevi are benign precursors to melanoma and exhibit characteristics of senescent cells (5, 11). Progression to melanoma is associated with (5, 12–13) loss of the tumor suppressor p16INK4A, encoded by the CDKN2A locus, through mutation, deletion, or promoter DNA methylation (14). Since senescence is a distinct mechanism of tumor suppression (2, 15–16), determining how melanoma cells escape senescence and devising methods to restore senescence to these cells may be important for developing additional therapeutic options for this malignancy.
Polycomb group (PcG) proteins are global repressors of gene expression effecting transcriptional suppression epigenetically through the formation of polycomb repressor complexes (PRC), including PRC1 and PRC2 (17–18). Enhancer of Zeste Homologue 2 (EZH2), the catalytic subunit of PRC2, has a histone methyltransferase activity for histone 3 lysine 27 trimethylation (H3K27me3) (19). Generation of the H3K27me3 mark by PRC2 establishes a strong repressive signal for gene expression (20). PcG proteins suppress CDKN2A expression (21). In senescent cells, PcG proteins are downregulated or dissociated from the CDKN2A locus leading to a decreased level of histone H3K27me3 and reactivation of INK4/ARF genes (22). Hence, activation of expression of PcG proteins in senescent cells, including those of pre-malignant benign tumors resulting from oncogene activation, could contribute to the loss of tumor suppressor gene activity and drive malignant progression. PcG proteins are overexpressed in various types of human cancer including prostate cancer and breast cancer and are functionally important for maintaining the malignant phenotypes of those cells (23–24). However, the function of EZH2 in progression from the senescent state and its mechanisms of action other than CDKN2A suppression remain unclear.
In this study, we explored the role of EZH2 in senescence and tumorigenicity in nevi and melanoma cells. We provide evidence that EZH2 is important to maintain resistance to the senescent state in human melanoma cells. We observed a striking difference, on a cell-by-cell basis, between the level of EZH2 expressed in acquired melanocytic nevi, where it is absent from all or nearly all benign nevus cells, and samples from metastatic melanomas, where it is detected in a majority of melanoma cells in most samples studied. Depletion of EZH2 in melanoma cells resulted in a senescent phenotype that was significantly dependent upon activation of p21/CDKN1A expression in a p53-independent manner. Chromatin immunoprecipitation (ChIP) analysis shows that EZH2 maintains HDAC1 at the CDKN1A transcriptional start site and downstream regions of the gene. EZH2 depletion removes HDAC1 from these regions, triggering histone acetylation and recruitment of RNA polymerase II, resulting in p21/CDKN1A activation. Our findings are consistent with a model for senescence bypass in melanoma development whereby EZH2 expression overcomes p53-dependent senescence to promote malignant progression.
MATERIALS AND METHODS
Identification and quantification of EZH2-expressing melanocytic cells in vivo
Human samples were collected under the auspices of approved human clinical protocols passed by the Institutional Review Board of the Intramural Research Program of the National Cancer Institute. Relative expression levels of EZH2 were determined in melanocytes in normal human skin, benign melanocytic nevi, and metastatic melanoma specimens using two-color immunofluorescence. For immunofluorescence studies, mouse monoclonal anti-EZH2 (Cell Signaling Technology, #AC22) and rabbit polyclonal anti-Melan-A (Santa Cruz #sc-28871) were used each at a 1:50 dilution following fixation in methanol for 20 min at −20 C° and 4% paraformaldehyde in PBS. Secondary antibodies used were Alexa Fluor 488, F(ab′)2 fragment of goat anti-rabbit IgG (H + L) 1:1000 (Molecular Probes A-21425) and Alexa Fluor 555 F(ab′)2 fragment of goat anti-mouse IgG (H + L) 1:1000 (Molecular Probes A-11070).
Cell culture and growth measurement
UCD-Mel-N cells were a kind gift from Dr. Estela Medrano (Baylor College of Medicine). SK-Mel-30, SK-Mel-103, SK-Mel-119, SK-Mel-147, and SK-Mel-173 cells were graciously provided by Dr. Norman Sharpless (Univ. of North Carolina) with the permission of Dr. Alan Houghton (Memorial Sloan-Kettering). Cells were grown in DMEM (Invitrogen) supplemented with 10% fetal calf serum (FCS) and were maintained in a humidified incubator at 37°C and 5% CO2. To determine the cell growth rate, cells were plated in 24-well plates (1×104 cells/well) and transfected either with a control siRNA or an siRNA targeting EZH2 (see below). After 24 h and each successive day for 5 consecutive days, cells were trypsinized briefly with 0.05% trypsin/0.02% EDTA and cell number counted. For treatment of cells with the histone deacetylase inhibitor Trichostatin A (TSA, Sigma), TSA was dissolved in dimethyl sulfoxide (DMSO) (Sigma). Cells were treated with 1 μM TSA or an equal volume of DMSO as a control for 24 h. For the combined EZH2 siRNA/1 μM TSA treatment, cells were harvested 3 days following transfection with EZH2 siRNA, and TSA was added only for the last 24 h before harvesting.
RNA interference
Cells were seeded and incubated at 37°C for 24h and transfected with EZH2 siRNA (25) or nonspecific control siRNA (Qiagen, Inc) using LipofectRNAiMax (Invitrogen) according to the manufacturer’s protocol. For detection of markers of senescence other than SA-β-Gal, a second transfection was performed 3 days after the first transfection prior to fluorescence or immunofluorescence detection of SAHF, α-tubulin distribution, or H3K9me3. To account for any off-target effects of the siRNA targeting EZH2, key experiments were repeated with a pool of 4 siRNAs (ON-TARGETplus SMARTpool L-004218-00-0005, Millipore) targeting different regions of the EZH2 transcript than the original siRNA used. siRNA sequences are described in the Supplementary Information. For sustained suppression of EZH2 expression, SMARTvector lentiviral particles (Thermo Scientific) carrying short hairpin RNA (shRNA) targeting EZH2 and a non-specific shRNA control, shN1 (Thermo Scientific, #S-005000-01) were used to transduce melanoma cells.
For stable, RNAi-mediated suppression of CDKN1A expression, four shRNA-expressing lentiviral expression constructs (V2LHS-202469 (shp21-1), V2LHS-268120 (shp21-2), V2LHS-230118 (shp21-3), and V2LHS-230370 (shp21-4)) (Thermo Scientific) targeting CDKN1A (NM_000389) were utilized. For stable, RNAi-mediated suppression of TP53 expression, two constructs (V2LHS-93613 (shp53-1) and V2LHS-93615 (shp53-2)) (Thermo Scientific) targeting TP53 (NM_001126113), were utilized. A human GIPZ lentiviral shRNA control (shN2) (Thermo Scientific, #RHS4346) was obtained for use with the above vectors.
Immunofluorescence staining
Cells grown on coverslips were fixed with 4% paraformaldehyde in PBS for 5 min, then permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature. Cells were blocked with 5% nonfat dry milk (Carnation)/0.2% Tween-20 in PBS pH 7.4 for 1 h and incubated with primary antibodies anti-EZH2 (1:500), anti-H3K27me3 (1:1000), anti-H3K9me3 (Abcam #ab-8898, 1:1000), or anti-α-tubulin (Sigma #T-6199, 1:5000) for 1 h. Cells were incubated with fluorescent secondary antibodies for 30 min, washed three times with PBS, and stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclear DNA.
Flow cytometric analysis of cell cycle distribution. Cells were collected and fixed with ice-cold 70% ethanol for 1 h. Cells were washed twice in PBS and incubated with 25 μg/ml propidiumiodide (PI, Sigma) with 100 μg/ml RNase A in PBS. The PI-stained cells were analyzed on a FACScan. The distribution of cells in different cell cycle phases was calculated using ModFit LT (BD Biosciences).
Western blot analysis
Cells were lysed in RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% Nonidet-P40, 1% sodium deoxycholate, 0.1% SDS) containing a protease inhibitor cocktail (Sigma). For histone detection, an acid extract procedure was used according to the protocol recommended by the manufacturer (Millipore). Following determination of protein concentration, cell lysates were subjected to SDS-PAGE, transferred to PVDF membranes (Invitrogen), and used for Western blotting.
Senescence-associated β-galactosidase staining
Cells were fixed by incubation in 2% formaldehyde/0.2% glutaraldehyde/PBS for 5 minutes at room temperature and stained for senescence-associated β-galactosidase activity in X-gal solution (1 mg/ml X-gal, 0.12 mM K3Fe[CN]6, 0.12 mM K4Fe[CN]6, 1 mM MgCl2 in PBS at pH 6.0) at 37°C (3).
Quantitative real-time PCR
RNA was isolated with TRIzol (Invitrogen). Reverse transcription and quantitative real-time PCR was performed with SYBR Green one-step reagent (Bio-Rad) on a Chromo4 Optical Detection System (MJ Research). The transcript levels were measured in a triplicate set of reaction for each gene and calculated as fold change relative to nonspecific siRNA transfected cells. Relativeβ2-microglobulin (β2M) or GAPDH levels were used for normalization. PCR primers for CDKN1A, GAPDH, and β2M are listed in Supplementary Table 1A.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed on chromatin isolated from cells 72h after transfection with an siRNA targeting EZH2 or with control siRNA using the Chromatin Immunoprecipitation Assay Kit (Millipore, 17-295), according to the manufacturer’s instructions. Details are described in the Supplementary Methods.
Anchorage-independent growth and xenograft studies of EZH2-depleted human melanoma cells
To determine the role of EZH2 on anchorage-independent growth, cells were resuspended in medium supplemented with agar at a final concentration of 0.35% as the top layer and 0.6% agar as base layer and plated at a density of 2.5 × 104 cells per well in 6-well plates in triplicate. Colonies were counted following a defined period of growth. For in vivo tumorigenicity studies, UCD-Mel-N cells were transduced with lentiviruses encoding shRNAs targeting EZH2 and control shRNA as described above. 48h after infection, cells were selected with 1 μg/ml puromycin for 4 days, then allowed to recover. To generate xenografts from these modified cells, 2 × 105 transduced UCD-Mel-N cells were resuspended in 100 μL of a DMEM/Matrigel (Basement Membrane Extract; R&D Systems) mixture (3:1 v/v) and injected subcutaneously into the right flank of 6-week-old NOD/SCID mice. Tumor dimensions were measured every 3 days and volume calculated using the formula 1/2×(length)×(width)2. All animal experiments were conducted in accord with a protocol approved by the National Cancer Institute’s Animal Care and Use Committee.
RESULTS
Expression of EZH2 in vivo in melanocytes in human skin, nevi, and metastatic melanomas
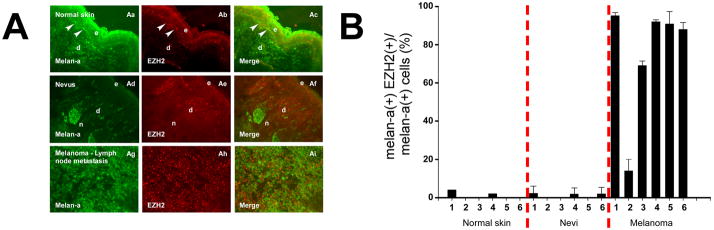
Using immunofluorescence with tissue specimens, we found EZH2 was rarely expressed in melanocytes in specimens of normal human skin (Figure 1Aa–c), with nuclear expression of EZH2 detectable in melanocytes from only 2/6 specimens studied and in < 5% of the cells examined in these 2 samples (Figure 1B). EZH2 was also rarely detectable in melanocytes in 6/6 specimens of melanocytic nevi examined (< 1% of cells; Figure 1Ad–f, Supplementary Figure 1). In contrast, EZH2 was expressed in 14–95% of melanoma cells in 6 different specimens of metastatic melanomas (Figures 1Ag–i, 1B, Supplementary Figure 2), with expression in the majority of melanoma cells in 5/6 specimens examined. These results extend results of other studies describing EZH2 expression in nevi and melanoma cells (26–27) by quantifying expression in these lesions and in normal melanocytes in vivo on a cell-by-cell basis in clinically correlated tumor specimens.
Figure 1.
Expression of EZH2 in skin, melanocytic nevi, and metastatic melanomas. (A) In vivo melanocytic expression of EZH2. Aa–Ac depict expression of Melan-a (Aa), EZH2 (Ab), and merged image (Ac) in normal human skin. Arrowheads point to Melan-a-expressing melanocytes in basal layer (Aa), which do not express EZH2 (Ab). Epidermis (e) and dermis (d) are delineated. Ad–Af show expression of Melan-a (Ad), EZH2 (Ae), and merged image (Af) in an example of a compound melanocytic nevus Nests (n) of adherent nevus cells in the dermis are depicted. EZH2 expression is absent in these and in individual melanocytes (Ae). Ag–Ai demonstrate expression of Melan-a (Ag), EZH2 (Ah) in tissue from a melanoma lymph node metastasis. Merged image (Ai) shows that most cells express both Melan-a and EZH2. (B) Proportions of Melan-a-expressing cells in normal human skin, melanocytic nevi, and metastatic melanomas that express EZH2. Nevi and melanoma specimens correspond to specimens presented in Figure 1A (#1) and to specimens presented sequentially (#2–6) in Supplementary Figures 1 and 2. Error bars represent standard deviations from the mean.
Suppression of EZH2 expression induces senescence features in human melanoma cells
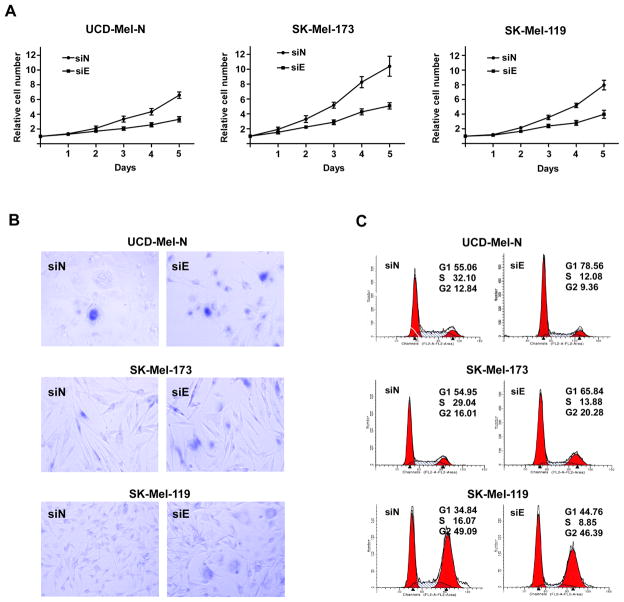
Because of the absence of EZH2 expression in nevi, we hypothesized that EZH2 suppresses senescence in melanoma cells, thereby promoting their proliferation and tumorigenicity. EZH2 was depleted from a set of human melanoma cell lines using siRNA and proliferation, cell cycle progression, and senescence characteristics measured. Depletion of EZH2 was accompanied by a global decrease of H3K27me3 in UCD-Mel-N cells (Supplementary Figure 4A), indicating that maintenance of the repressive H3K27me3 mark in these cells is dependent upon the expression of EZH2 as described previously (18–19, 28–29). Growth of EZH2 siRNA cells was significantly decreased compared to control-transfected cells (Figure 2A, Supplementary Figure 3B). EZH2 siRNA cells displayed a flattened, enlarged morphology and expressed SA-β-Gal (Figure 2B, Supplementary Figure 3C). In addition to SA-β-Gal expression, additional senescence markers following extended EZH2 depletion (6 days) include senescence-associated heterochromatic foci (SAHFs) and the heterochromatin-associated H3K9me3 modification (1) (Supplementary Figure 4C) as well as an increase in cell size (30), as evaluated by tubulin immunofluorescence (Supplementary Figure 4B). Flow cytometric analysis demonstrated that the proportion of cells in the S phase decreased in EZH2 siRNA cells with a concomitant increase of cells in the G1 phase (Figure 2C, Supplementary Figure 3D), a significant difference (p < 0.01, Supplementary Figure 3E) in all 3 cell lines tested. In SK-Mel-173 cells, we also noticed a slight increase in the proportion of cells in the G2 phase (Figure 2C). Apoptosis was not observed in the EZH2 siRNA cells as shown by absence of a sub-G1 fraction (Figure 2C).
Figure 2.
EZH2 depletion induces a cellular senescence phenotype in melanoma cells. Cell proliferation rate, SA-β-gal expression, and cell cycle distribution were determined following EZH2 depletion with a small-interfering RNA. (A) Growth curves of EZH2 siRNA (siE)- and control siRNA (siN)-treated UCD-Mel-N, SK-Mel-173, and SK-Mel-119 cells. Each data point represents the average of three independent measurements. Error bars indicate the standard deviation. (B) SA-β-gal staining in melanoma cells following EZH2 depletion (siE) or control siRNA transfection (siN). (C) Cell cycle analysis of melanoma cells with EZH2 depletion. Cell phase distribution from 1/3 representative experiments performed for control siRNA (siN) and siEZH2 (siE)-treated cells.
Effect of EZH2 depletion on melanoma growth in vivo
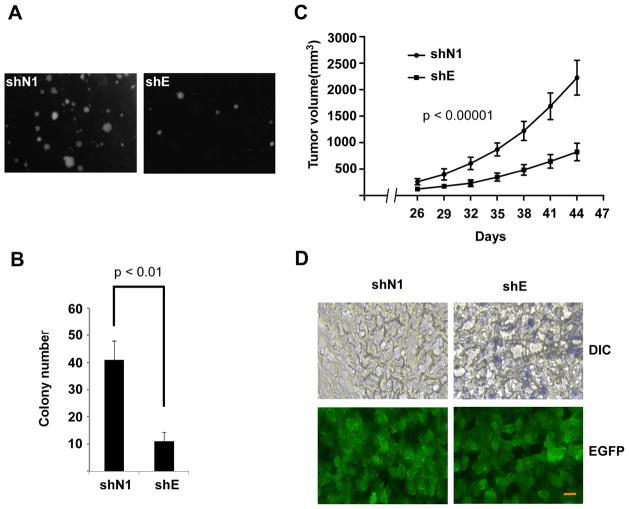
We asked whether depletion of EZH2 would inhibit melanoma growth in vivo using a xenograft tumor model. To obtain sustained depletion of EZH2, we identified a lentiviral shRNA particle construct, shEZH2-3, with the best knockdown efficiency which was selected for further experiments (Supplementary Figure 5A). Both EZH2 shRNA and control shRNA UCD-Mel-N cells were able to form colonies in soft agar during the 2-week observation period (Figure 3A), but the number of colonies was markedly reduced by EZH2 knockdown (Figure 3B). To compare the rate of tumor formation in vivo, EZH2 shRNA or control shRNA cells in Matrigel were subcutaneously injected into NOD/SCID mice. A substantial inhibition of tumor growth compared with control cells was evident in cells stably depleted of EZH2 (Figure 3C). Analysis of tumor xenografts revealed frequent SA-β-gal-expressing cells in the xenografts from EZH2-depleted cells, but not in the shNeg xenografts (Figure 3D), whereas both tumors maintained expression of EGFP indicating continued expression of the integrated lentiviral construct.
Figure 3.
Stable depletion of EZH2 reduces tumorigenicity of human melanoma cells. (A) Reduced colony formation following EZH2 depletion. Representative photomicrographs shown of anchorage-independent colonies 20 days after initiating growth of EZH2 shRNA cells (shE) and control shRNA cells (shN1) in soft agar. (B) Mean colonies per field. Error bars represent standard deviation of 3 independent platings. p < 0.01 (**) for the difference between shN1 and shE by Students’s t-test. (C) Reduced tumor volumes in NOD/SCID mice injected with EZH2 shRNA cells compared to control shRNA cells. Each data point represents the mean volume (±SEM) of subcutaneous tumor xenografts developing in 12 mice per experimental group. p < 0.00001 for the difference between shE and shN1 tumor growth curves by repeated measures analysis of variance. (D) Expression of SA-β-gal is observed in xenograft tumor specimen from EZH2 shRNA tumor (upper panel, right), but not from control tumor (upper panel, left). EGFP expression (lower panels) confirms continued expression of the shRNA construct in the xenograft cells. Results are representative of two tumors analyzed. Scale bar,50 μM.
EZH2 depletion activates p21 expression in melanoma cells
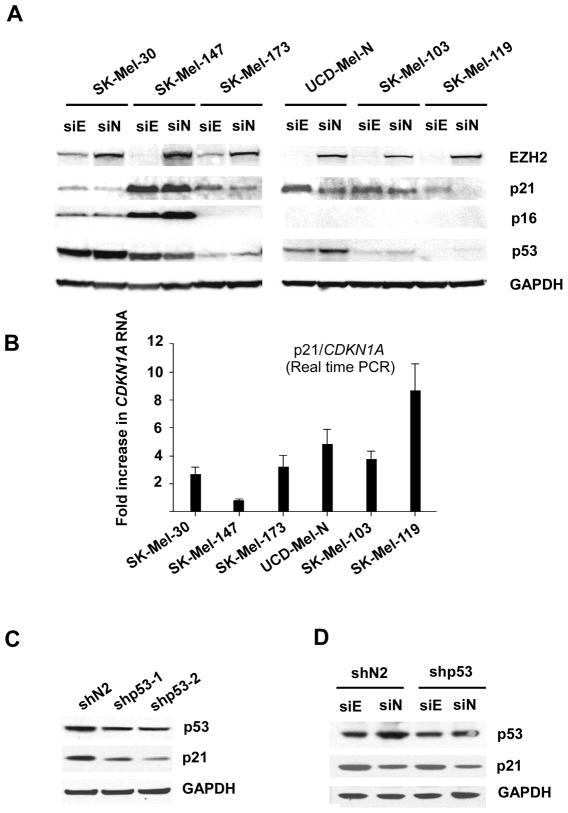
Since cell cycle regulatory pathways mediated by cyclin-dependent kinase inhibitors play a critical role in senescence induction (31), we examined their expression following EZH2 depletion. As shown in Figure 4A, EZH2 siRNA efficiently suppressed EZH2 expression in all six cell lines examined. p16 is undetectable in UCD-Mel-N, SK-Mel-103, SK-Mel-119, and SK-Mel-173 cells, due to 9p21 loss (32). Thus p16 expression was not reactivated in those lines after EZH2 depletion. In SK-Mel-30 and SK-Mel-147 cells where p16 is mutated and expressed at a high level (32), depletion of EZH2 did not change p16 expression. However, depletion of EZH2 elevated expression of p21 in 5/6 cell lines. The effect of loss of EZH2 upon induction of p21 expression was transcriptional, since transcripts of CDKN1A were upregulated in the 5/6 cell lines that exhibited increases in p21 at the protein level (Figure 4B). p53 transcriptionally activates p21, binding to two distinct sites in the p21/CDKN1A promoter. However, in cell lines exhibiting an increase in p21, p53 was decreased, inversely correlating to p21 expression.
Figure 4.
Western blot analysis of cell cycle regulatory protein expression in human melanoma cells with EZH2 depletion. (A) Relative expression of cell cycle regulatory proteins p16, p21, and p53 were determined upon EZH2 depletion (siE) or control siRNA transfection (siN) in human melanoma cell lines. GAPDH serves as loading control. (B) Quantitative real-time PCR measurement of CDKN1A induction in human melanoma cells lines following EZH2 depletion. The fold induction of CDKN1A expression is depicted for each of the cell lines studied. Error bars represent standard deviation from the mean. (C) Western blot analysis of p53 depletion by p53 shRNA (upper panel). Two distinct stable shp53-expressing cell lines of UCD-Mel-N cells (shp53-1 and shp53-2) were generated along with a control cell line expressing an irrelevant shRNA (shN1). p21 expression in indicated stable cell lines with depletion of p53 (middle panel). GAPDH (lower panel) was used as a loading control. (D) p53 depletion does not attenuate EZH2 siRNA-mediated upregulation of p21 expression. An siRNA targeting EZH2 or a control siRNA was transfected into stable shp53-expressing or control shRNA vector-expressing UCD-Mel-N cells. Relative levels of p53 (upper panel), p21 (middle panel), and GAPDH as loading control (lower panel) are depicted.
The inverse relationship between p53 and p21 expression suggested that the activation of p21 observed was largely independent of changes in p53 activity. To test this independence more directly, we targeted p53 with shRNA in UCD-Mel-N cells. p53 lentiviral-shRNA transduction moderately suppressed p53 expression in UCD-Mel-N cells (Figure 4C). The p53 level decreased to a level comparable with that in EZH2 siRNA treated cells (Figure 4A). However, in contrast to the upregulation of p21 expression noted with EZH2 depletion (Figure 4A), depletion of p53 diminished p21 expression (Figure 4C), demonstrating that p53 regulates constitutive p21 expression in UCD-Mel-N cells, but is not responsible for EZH2 siRNA-mediated p21 activation. Activation of p21 by EZH2 depletion was not blocked by a reduction in p53 (Figure 4D). These results indicate that p21 induction occurs through a mechanism other than p53 activation.
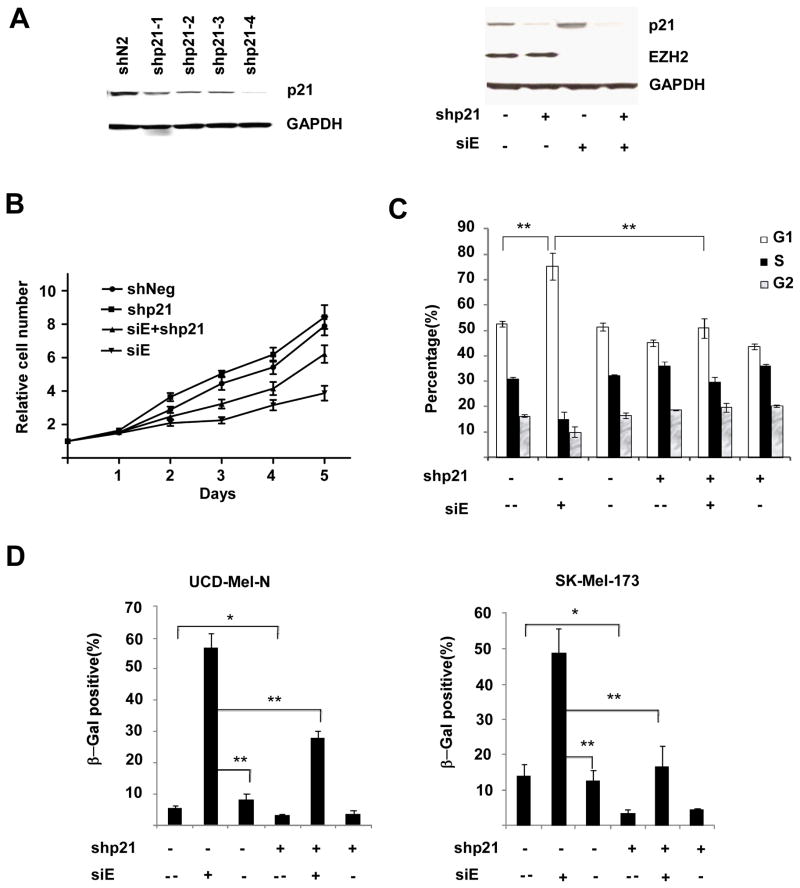
Suppression of p21 expression blocks cellular senescence
To determine whether the upregulation of p21 induced by EZH2 depletion contributes to cellular senescence and growth arrest, we utilized RNA interference to suppress p21/CDKN1A induction. p21/CDKN1A was depleted efficiently with a lentivirally-delivered shRNA (shp21-4) (Figure 5A), which was used to generate shp21 UCD-Mel-N cells and a control cell line (Figure 5A). In shp21 UCD-Mel-N cells, there was a slight but consistent decrease in the G1 phase population when compared with control cells (from 54% to 45%), with an increase of S phase cells (from 30% to 38%), indicating that the presence of p21 at its constitutive levels exerts a modest inhibitory effect regulating the G1/S transition in UCD-Mel-N cells (Figure 5C). This effect was corroborated by a trend toward increased cell proliferation in p21-depleted UCD-Mel-N cells (Figure 5B). We then depleted EZH2 in these stable cell lines and showed that p21 shRNA could suppress p21 induction under these conditions (Figure 5A). While in control cells EZH2 depletion resulted in an increase of the G1 phase population from 54% to 71% (p < 0.01, Figure 5C), in shp21 cells the G1 population only increased from its baseline value of 45% to 56%. The difference between the percentage of EZH2-depleted cells with (71%) and without (56%) p21 was significant (p < 0.01, Figure 5C). Together, these data indicate that suppression of p21 partially inhibits the G1 cell cycle arrest caused by EZH2 depletion. This finding of partial inhibition was supported by the higher rate of proliferation observed in cells with both EZH2 and p21 suppressed compared to cells with only EZH2 suppressed (Figure 5B). Hence, repression of p21 by EZH2 promotes cell cycle progression.
Figure 5.
Suppression of p21 inhibits the senescence phenotype induced by depletion of EZH2. (A) Left panel, suppression of p21 in UCD-Mel-N cells by shRNAs targeting p21/CDKN1A (shp21-1,2,3,4) or an irrelevant control shRNA (shN1). p21 and GAPDH (as loading control) levels are shown by Western blotting. Right panel, maintenance of p21 suppression in UCD-Mel-N cells stably transduced with either p21 shRNA (shp21 +) or a control shRNA (shp21 −) following transfection with EZH2 siRNA (siE +) or control siRNA (siE −). Western blot of p21 and EZH2 levels are shown with GAPDH as loading control. (B) Relative rates of cell proliferation of UCD-Mel-N cells stably transduced with either control shRNA or shRNA targeting p21 with or without transient depletion of EZH2. Cells were counted at the indicated number of days after transfection with control siRNA or siRNA targeting EZH2 (siE). Experiment was performed in triplicate with error bars representing standard deviation from the mean. (C) G1 cell cycle arrest induced by EZH2 siRNA can be partially inhibited by p21 suppression. UCD-Mel-N cells stably expressing shp21 (shp21 +) or shN1 (shp21 −) were either not treated (siE --), transfected with an siRNA targeting EZH2 (siE +), or transfected with a control siRNA (siE −) and cell cycle distribution measured. Error bars represent standard deviation of experiments performed in triplicate. ** indicates p < 0.01 for comparison by Student’s t-test. (D) SA-β-gal expression in UCD-Mel-N (left panel) and SK-Mel-173 (right panel) cells transfected with EZH2 siRNA (siE +) or control siRNA (siE −), or untreated (siE --), with or without stable p21 suppression (shp21 +, shp21 − respectively). Cells were stained, photographed under microscopy, and counted to determine the percentage of SA-β-gal-expressing cells. At least 1000 cells were counted in random fields in each of the duplicate wells. The results are presented as mean ± SD of three independent experiments. Differences were analyzed by Student’s t test, and differences with p value < 0.05 (*) and p < 0.01 (**) noted.
We also determined whether p21 repression by EZH2 suppressed senescence as measured by SA-β-gal expression. We noticed that the cell lines utilized to assess changes in SA-β-Gal expression with EZH2 depletion, UCD-Mel-N, SK-Mel-173, and SK-Mel-119 (Figure 2B), displayed a small fraction of SA-β-Gal-expressing cells. This was quantified in control and shp21 UCD-Mel-N and SK-Mel-173 cells, where p21 suppression was found to have a small but statistically significant effect reducing the proportion of these cells expressing SA-β-gal (Figure 5D). The percentage of SA-β-gal-expressing cells decreased from 5% to 3% in UCD-Mel-N cells, and from 12% to 3% in SK-Mel-173 cells. Depletion of EZH2 with siRNA in each of the transduced cells increased the percentage of cells expressing SA-β-gal, but this increase was significantly blunted in cells with p21 suppression. 57% of control transduced UCD-Mel-N cells expressed SA-β-gal with EZH2 depletion, whereas only 28% expressed SA-β-gal when p21 was also suppressed. Likewise, in SK-Mel-173 cells, the 49% of cells expressing SA-β-gal was reduced to 16% with p21 suppression (Figure 5D).
EZH2 depletion increases histone acetylation
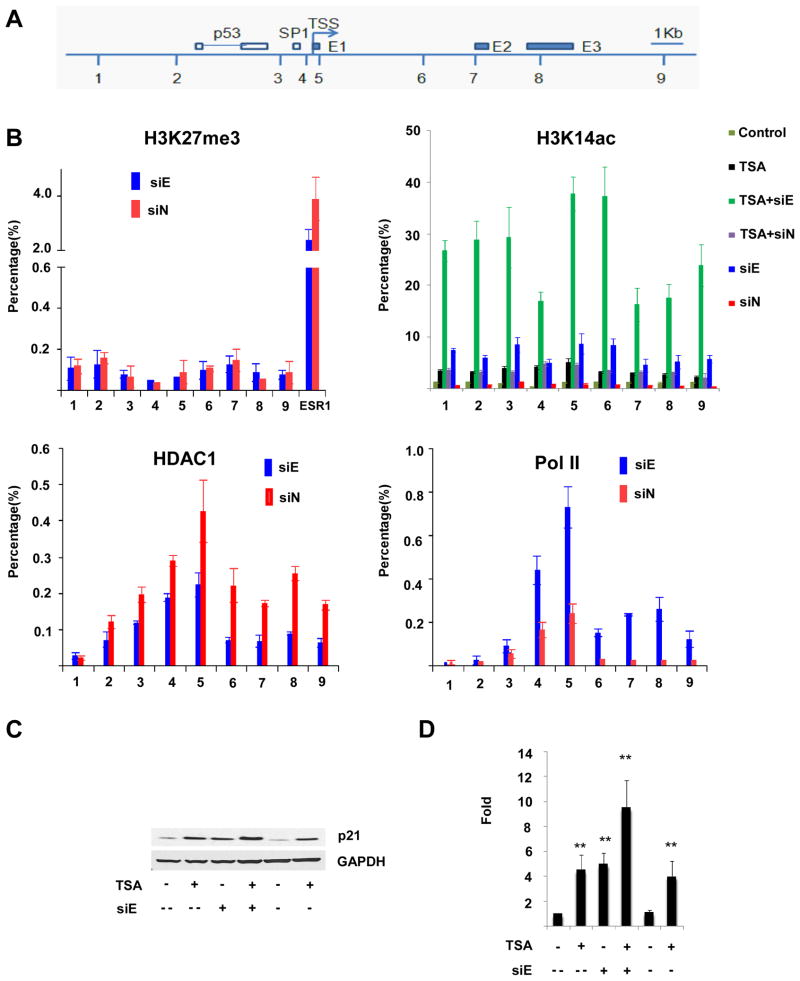
EZH2 has a specific H3K27 methyltransferase activity (18, 33) associated with gene repression (20, 29). EZH2 also associates with HDAC1 (34), recruiting it to the DAB2IP promoter (35). Histone deacetylation is a requirement for EZH2 repression at the E-cadherin promoter (36). To determine whether p21 activation in UCD-Mel-N cells is associated with changes in CDKN1A H3K27 trimethylation or histone acetylation, ChIP was used to characterize the relative density of repressive H3K27me3 marks and activating (37) H3K14Ac marks at the CDKN1A genomic locus, a ~20-kb region upstream and downstream of the transcription start site (Figure 6A).
Figure 6.
Effect of EZH2 depletion on histone 3 interactions and modifications at CDKN1A. (A) Schematic drawing of CDKN1A locus depicting transcription start site (TSS), p53 and SP1 transcription factor binding sites, and the location of the three exons (E1,E2,E3). Positions of primer pairs used for chromatin immunoprecipitation (ChIP) are indicated (1–9). (B). ChIP analyses of CDKN1A using antibodies recognizing H3K27me3, H3K14ac, HDAC1, and RNA polymerase II were performed in UCD-Mel-N cells with EZH2 depletion (siE) or transfection with a control siRNA (siN). The precipitated DNA was amplified by real-time quantitative PCR (qPCR) using primers 1–9 from distinct regions of CDKN1A. Each experiment was repeated independently at least twice. The results were normalized to IgG control. The data are expressed as percentage of immunoprecipitated DNA relative to input DNA. Primers to the ESR1 gene were used as a positive control. Histone acetylation at CDKN1A was determined not only under the above conditions (siE and siN), but also in the above conditions with cells treated with 1 μM TSA (TSA+siE, TSA+siN), in cells treated with TSA alone (TSA), and in untreated or untransfected cells (Control). Error bars represent standard deviation from the mean. (C,D) Combined effects of HDAC inhibition and EZH2 depletion upon p21/CDKN1A transcript and protein expression. An additive effect of HDAC inhibition with trichostatin A (TSA) and EZH2 depletion (siE) on the upregulation of p21 protein expression as measured by Western blotting (C) and of CDKN1A mRNA expression as measured by quantitative real-time PCR (D) is observed relative to cells treated with TSA (TSA +) or EZH2 siRNA (siE +), respectively, alone. siE −, control siRNA cells; siE --, untreated cells; ** signifies p < 0.01 for indicated comparisons.
H3K27me3 was present throughout the CDKN1A genomic region at a very low, but detectable level, which did not change following depletion of EZH2. The gene ESR1, a frequent polycomb target in cancer cell lines (38), was analyzed as a positive control (Figure 6B, Supplementary Figures 5A, 5B). In contrast, EZH2 depletion significantly increased H3K14ac at CDKN1A. This increase was comparable or slightly greater than the increase in H3K14ac observed after treatment of the cells with 1 μM TSA. Treatment with a combination of EZH2 siRNA and 1 μM TSA synergistically increased H3K14ac compared to treatment with EZH2 siRNA or TSA alone (Figure 6B). The effect of EZH2 depletion upon H3K14ac at the CDKN1A locus is not likely limited to this gene, since EZH2 depletion resulted in an increase in global H3K14ac acetylation in UCD-Mel-N, SK-Mel-173, and SK-Mel-119 cells corresponding with a global decrease in H3K27me3 (Supplementary Figure 5D).
EZH2 depletion reduced HDAC1 localization at CDKN1A, indicating that EZH2 is important for maintaining HDAC1 at CDKN1A. Reduction in HDAC1 corresponded to the recruitment of RNA polymerase II to the CDKN1A transcription start site and downstream region (Figure 6B), resulting in transcriptional activation (Figure 6D). Treatment with 1μM TSA and EZH2 depletion resulted in approximately 4-fold and 5-fold inductions of CDKN1A expression, respectively. Combined treatment with TSA and EZH2 siRNA resulted in an approximately 10-fold increase of CDKN1A expression, correlating with the synergistic increase in histone acetylation (Figure 6D). The combined depletion of EZH2 and histone deacetylase inhibition also resulted in enhanced expression of p21 as detected by Western blotting (Figure 6C).
Neither EZH2 (Supplementary Figure 5A) nor BMI1 (Supplementary Figure 5B), a component of PRC1, which recognizes and binds H3K27me3 (39) were significantly enhanced at CDKN1A. The CDKN1A promoter was not significantly methylated in UCD-Mel-N, SK-Mel-119, and SK-Mel-173 cells. Methylation status was not significantly altered following depletion of EZH2, indicating that the activation of p21 expression following EZH2 depletion does not result from a change in CDKN1A promoter methylation (Supplementary Figure 6).
DISCUSSION
EZH2 activity and function in melanoma cells and in melanocyte senescence
We describe EZH2-dependent repression of p21/CDKN1A that suppresses senescence in melanoma cells. Senescence in melanocytes is governed by both the p16INK4A/Rb and p53/p21 pathways (Supplementary Figure 7A). Although neither pathway is absolutely required, inhibition of both pathways together abrogates the oncogene-induced senescence response (40). p16INK4A expression is induced following overexpression of BRAFV600E (5) or NRASQ61K (6) in primary human melanocytes and is also observed in acquired (11) and congenital (5) melanocytic nevi that preferentially harbor BRAFV600E (9) or activating NRAS (10) mutations, respectively. However, expression of p16INK4A in benign melanocytic nevus cells is heterogeneous (5, 11), implying that its expression in every cell is not required for maintaining senescence, and patients lacking functional p16 develop melanocytic nevi (41). In vitro, melanocytes with p16INK4A expression suppressed undergo a delayed, p53/p21-mediated senescence, suggesting that the p16INK4A/Rb pathway is the dominant senescence pathway in these cells (40). Nonetheless, melanocytes obtained from patients lacking functional p16 also undergo p53-mediated senescence, including upregulation of the p53 target p21 (41). Accordingly, as p16 expression is lost during the transition of benign compound nevi to dysplastic nevi and radial growth phase melanoma, p21 begins to be detected more frequently and at higher levels (11). These findings suggest that following loss of p16/RB activity in human melanocytes, including loss that occurs in vivo as melanocytic tumors progress from the benign nevus stage (42), an alternative mechanism of senescence that includes induction of p53/p21-dependent is activated (Supplementary Figure 7B).
Our findings are consistent with a model whereby EZH2 repression of p21, through maintenance of HDAC1 at CDKN1A resulting in low histone acetylation, bypasses an otherwise intact p53-dependent p21 activity to overcome p53-mediated senescence (Supplementary Figure 7C). This mechanism could promote tumor progression via an alternative to somatic mutation of p53, which remains wild-type in most primary and metastatic melanomas (43)(44). Our observations and those of others (26–27) suggest that EZH2 becomes activated in subpopulations of tumor cells during the transition from benign nevus to dysplastic nevus and radial growth phase melanoma. EZH2-expressing cells may preferentially expand to yield the high proportions of EZH2-positive cells we found in metastatic specimens. We do not currently understand how EZH2 expression is upregulated during the transition from benign nevus to malignant melanoma cell. In other cell types, loss of microRNA expression has been associated with the induction of EZH2 expression in both normal development and malignancy (45–48), and may be a potential mechanism to consider for EZH2 activation in melanocytic neoplasia.
p21 as an epigenetic target of EZH2 in melanoma
p21 is a common epigenetic target in cancer, with HDAC inhibitor-mediated de-repression of p21/CDKN1A occurring in cancer cells in a p53-independent manner. Our results show that EZH2-dependent regulation of p21 accounts for a substantial component of the effects of EZH2 on cell cycle and senescence properties in melanoma cells. p21 remains functional in the melanoma cell lines examined because its suppression alone lowers the G1/S phase ratio (Figure 5C) and reduces the proportion of cells expressing SA-β-Gal. The G1 cell cycle arrest and induction of SA-β-Gal expression observed with EZH2 depletion are significantly attenuated under conditions of p21 suppression (Figure 5C,D), indicating that the upregulation of p21 that occurs upon knockdown of EZH2 is a major effector of these cellular changes.
Activation of p21/CDKN1A occurring upon EZH2 suppression appears p53-independent. We observed a decrease in p53 levels upon EZH2 depletion in those cell lines exhibiting p21 activation (Figure 4A). Stable depletion of p53 did not block p21 induction caused by EZH2 knockdown, even though depletion of p53 was accompanied by a reduction in p21 level (Figure 4B). Hence p53 retains p21-activating activity in melanoma cells, consistent with previously reported findings that, in melanoma cells, p21 can mediate p53-dependent responses (49–50).
Loss of EZH2 substantially increased histone acetylation at CDKN1A in the absence of changes in H3K27 trimethylation, PcG occupancy, or promoter DNA methylation. The increase in histone acetylation is due to loss of HDAC1 at CDKN1A, favoring the recruitment of RNA polymerase II and transcription. We show for the first time that modulating the level of cellular EZH2 affects HDAC1 occupancy of a critical gene regulating cell senescence. How EZH2 functions noncanonically to maintain HDAC1 at CDKN1A in melanoma cells, thereby suppressing senescence, is worthy of future investigation. EZH2, in addition to its enzymatic function as a histone methyltransferase, may have a more general, structural role stabilizing HDACs in chromatin remodeling complexes. EZH2 occupancy of H3K27me3 sites in melanoma cell chromatin might facilitate guiding HDACs to indirectly regulated sites such as CDKN1A via mechanisms such as noncoding RNA (51) or chromatin looping interactions (52). In this regard, current thinking suggests that HDACs are not core members of PRC2 but instead engage in more transient interactions with its components to effect epigenetic regulation of gene expression (53). The precise mechanism to determine how EZH2 interacts with CDKN1A will require further investigation using techniques that enable both characterization of other chromatin-modifying proteins at CDKN1A in melanoma cells and exploration of long-range chromatin interactions.
In closing, we demonstrate that EZH2 suppresses senescence in melanoma cells by repressing p21/CDKN1A expression independent of p16 expression or p53 function. We describe a novel mechanism for EZH2 regulation of p21/CDKN1A occurring through changes in histone acetylation rather than changes in H3K27me3. This mechanism is in direct contrast to polycomb regulation of the p16/INK4A gene, which is highly H3K27 trimethylated and bound directly by polycomb proteins to establish repression (22). Because of the potential relevance of senescence to the outcome of cancer therapy(15) in melanoma, inhibition of EZH2 may be a promising approach for melanoma treatment. Our observation that EZH2 depletion in synergy with HDAC inhibition activates p21 expression further through enhancing acetylation at CDKN1A suggests a potential combination approach to melanoma therapy.
Supplementary Material
Acknowledgments
We thank Doug Lowy and Kathy Kelly for a critical reading of the manuscript, and John Wunderlich and Steve Rosenberg of the NCI Surgery Branch for providing valuable tissue specimens. We acknowledge David Venzon and David J. Liewehr for their contributions to the statistical analysis. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This paper is dedicated to the memory of Leonard S. Kim, Princeton Class of 1985.
Financial support: This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research
References
- 1.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–40. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 2.Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10:51–7. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A Biomarker that Identifies Senescent Human Cells in Culture and in Aging Skin in vivo. Proc Natl Acad Sci. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras Provokes Premature Cell Senescence Associated with Accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 5.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–4. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 6.Haferkamp S, Scurr LL, Becker TM, Frausto M, Kefford RF, Rizos H. Oncogene-induced senescence does not require the p16(INK4a) or p14ARF melanoma tumor suppressors. J Invest Derm. 2009;129:1983–91. doi: 10.1038/jid.2009.5. [DOI] [PubMed] [Google Scholar]
- 7.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 8.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 9.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nature Genetics. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 10.Bauer J, Curtin JA, Pinkel D, Bastian BC. Congenital Melanocytic Nevi Frequently Harbor NRAS Mutations but no BRAF Mutations. J Invest Derm. 2006;127:179–82. doi: 10.1038/sj.jid.5700490. [DOI] [PubMed] [Google Scholar]
- 11.Gray-Schopfer VC, Cheong SC, Chong H, Chow J, Moss T, Abdel-Malek ZA, et al. Cellular senescence in naevi and immortalisation in melanoma: a role for p16? Br J Cancer. 2006;95:496–505. doi: 10.1038/sj.bjc.6603283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sparrow LE, Eldon MJ, English DR, Heenan PJ. p16 and p21WAF1 protein expression in melanocytic tumors by immunohistochemistry. Am J Dermatopathol. 1998;20:255–61. doi: 10.1097/00000372-199806000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Wang YL, Uhara H, Yamazaki Y, Nikaido T, Saida T. Immunohistochemical detection of CDK4 and p16INK4 proteins in cutaneous malignant melanoma. Br J Dermatol. 1996;134:269–75. [PubMed] [Google Scholar]
- 14.Walker GJ, Flores JF, Glendening JM, Lin AH, Markl ID, Fountain JW. Virtually 100% of melanoma cell lines harbor alterations at the DNA level within CDKN2A, CDKN2B, or one of their downstream targets. Genes Chromosomes Cancer. 1998;22:157–63. doi: 10.1002/(sici)1098-2264(199806)22:2<157::aid-gcc11>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002;109:335–46. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 16.Wu CH, van Riggelen J, Yetil A, Fan AC, Bachireddy P, Felsher DW. Cellular senescence is an important mechanism of tumor regression upon c-Myc inactivation. Proc Natl Acad Sci. 2007;104:13028–33. doi: 10.1073/pnas.0701953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alkema MJ, Bronk M, Verhoeven E, Otte A, van’t Veer LJ, Berns A, et al. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian polycomb complex. Genes Devel. 1997;11:226–40. doi: 10.1101/gad.11.2.226. [DOI] [PubMed] [Google Scholar]
- 18.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Devel. 2002;22:2893–905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs JJL, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–8. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 22.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Devel. 2007;21:525–30. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 24.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol. 2006;24:268–73. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 27.McHugh JB, Fullen DR, Ma L, Kleer CG, Su LD. Expression of polycomb group protein EZH2 in nevi and melanoma. J Cutan Path. 2007;34:597–600. doi: 10.1111/j.1600-0560.2006.00678.x. [DOI] [PubMed] [Google Scholar]
- 28.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, et al. Histone Methyltransferase Activity of a Drosophila Polycomb Group Repressor Complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 29.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandeck C, Mooi WJ. Oncogene-induced cellular senescence. Adv Anat Pathol. 2010;17:42–8. doi: 10.1097/PAP.0b013e3181c66f4e. [DOI] [PubMed] [Google Scholar]
- 31.Lundberg AS, Hahn WC, Gupta P, Weinberg RA. Genes involved in senescence and immortalization. Curr Opin Cell Biol. 2000;12:705–9. doi: 10.1016/s0955-0674(00)00155-1. [DOI] [PubMed] [Google Scholar]
- 32.Shields JM, Thomas NE, Cregger M, Berger AJ, Leslie M, Torrice C, et al. Lack of Extracellular Signal-Regulated Kinase Mitogen-Activated Protein Kinase Signaling Shows a New Type of Melanoma. Cancer Res. 2007;67:1502–12. doi: 10.1158/0008-5472.CAN-06-3311. [DOI] [PubMed] [Google Scholar]
- 33.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–96. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 34.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–8. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 35.Chen H, Tu S-w, Hsieh J-T. Down-regulation of Human DAB2IP Gene Expression Mediated by Polycomb Ezh2 Complex and Histone Deacetylase in Prostate Cancer. J Biol Chem. 2005;280:22437–44. doi: 10.1074/jbc.M501379200. [DOI] [PubMed] [Google Scholar]
- 36.Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–84. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–50. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 39.Min J, Zhang Y, Xu R-M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Devel. 2003;17:1823–8. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haferkamp S, Tran SL, Becker TM, Scurr LL, Kefford RF, Rizos H. The relative contributions of the p53 and pRb pathways in oncogene-induced melanocyte senescence. Aging (Albany NY) 2009;1:542–56. doi: 10.18632/aging.100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sviderskaya EV, Gray-Schopfer VC, Hill SP, Smit NP, Evans-Whipp TJ, Bond J, et al. p16/cyclin-dependent kinase inhibitor 2A deficiency in human melanocyte senescence, apoptosis, and immortalization: possible implications for melanoma progression. J Natl Cancer Inst. 2003;95:723–32. doi: 10.1093/jnci/95.10.723. [DOI] [PubMed] [Google Scholar]
- 42.Bennett DC. Human melanocyte senescence and melanoma susceptibility genes. Oncogene. 2003;22:3063–9. doi: 10.1038/sj.onc.1206446. [DOI] [PubMed] [Google Scholar]
- 43.Hartmann A, Blaszyk H, Cunningham JS, McGovern RM, Schroeder JS, Helander SD, et al. Overexpression and mutations of p53 in metastatic malignant melanomas. Int Journal Cancer. 1996;67:313–7. doi: 10.1002/(SICI)1097-0215(19960729)67:3<313::AID-IJC1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 44.Haapajarvi T, Pitkanen K, Laiho M. Human melanoma cell line UV responses show independency of p53 function. Cell Growth Differ. 1999;10:163–71. [PubMed] [Google Scholar]
- 45.Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–9. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 46.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–12. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 48.Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–43. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 49.Gorospe M, Cirielli C, Wang X, Seth P, Capogrossi MC, Holbrook NJ. p21(Waf1/Cip1) protects against p53-mediated apoptosis of human melanoma cells. Oncogene. 1997;14:929–35. doi: 10.1038/sj.onc.1200897. [DOI] [PubMed] [Google Scholar]
- 50.Bae I, Smith ML, Sheikh MS, Zhan Q, Scudiero DA, Friend SH, et al. An abnormality in the p53 pathway following gamma-irradiation in many wild-type p53 human melanoma lines. Cancer Res. 1996;56:840–7. [PubMed] [Google Scholar]
- 51.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, et al. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–27. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–9. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.