Abstract
Background/Aims
In genetic studies of complex disease a consideration for the investigator is detection of joint effects. The Multifactor Dimensionality Reduction (MDR) algorithm searches for these effects with an exhaustive approach. Previously unknown aspects of MDR performance were the power to detect interactive effects given large numbers of non-model loci or varying degrees of heterogeneity among multiple epistatic disease models.
Methods
To address the performance with many non-model loci, datasets of 500 cases and 500 controls with 100 to 10,000 SNPs were simulated for two-locus models, and one hundred 500-case/500-control datasets with 100 and 500 SNPs were simulated for three-locus models. Multiple levels of locus heterogeneity were simulated in several sample sizes.
Results
These results show MDR is robust to locus heterogeneity when the definition of power is not as conservative as in previous simulation studies where all model loci were required to be found by the method. The results also indicate that MDR performance is related more strongly to broad-sense heritability than sample size and is not greatly affected by non-model loci.
Conclusions
A study in which a population with high heritability estimates is sampled predisposes the MDR study to success more than a larger ascertainment in a population with smaller estimates.
Key Words: Epistasis, MDR, Heterogeneity
Introduction
Detecting statistically epistatic relationships between genes or between genes and environmental factors requires a search through a very large space relative to that encountered when looking for main effects. Exhaustive searches through such spaces using traditional parametric methods encounter considerable multiple testing problems [1, 2]. The statistical corrections for such large numbers of tests are extreme and many nested non-independent models are examined. An impossible compromise must be struck between low statistical power due to conservative correction for many non-independent tests and burying a real signal in type I errors. In contrast, only testing models for which hypotheses exist a priori may fail to find real models with novel biological interpretations, of which there may be many.
Statistical epistasis describes an effect exceeding the combined individual effects of each genetic factor; this is due to the simultaneous presence of particular levels of two or more factors within individuals in a population [3]. These factors that participate in the departure from linear additivity [4] do not necessarily have a direct relationship biologically. Such genes might not physically interact; likewise an environmental effect may not directly influence the expression or otherwise perturb a gene product. The value of this discovery may thereby only be relevant to prediction of the trait, and not helpful to the biologist investigating a cryptic mechanism.
This is in contrast to biological epistasis [5], where variants of biomolecules physically interact in pathways to yield phenotypes in individuals. In an era of ever-growing candidate gene and whole genome association studies such epistatic models are of growing importance, with many reports in the literature of interactions and the development of methods to overcome the limitations of parametric methods to detect them.
Multifactor Dimensionality Reduction (MDR) finds models by scanning through all possible combinations of factors up to an order of interaction specified by the user [6, 7, 8, 9, 10, 11]. The MDR methodology has been successfully applied to detecting gene-environment and gene-gene interactions for several clinical phenotypes, including: Alzheimer disease [12], asthma [13, 14], atrial fibrillation [9, 15], myocardial infarction [16, 17], autism [18, 19], bladder cancer [20], familial amyloid polyneuropathy [21], hypertension [22, 23], multiple sclerosis [24], prostate cancer [25], schizophrenia [26], sporadic breast cancer [10], and type II diabetes [27].
MDR provides a means to find hypotheses for further testing for epistatic interactions in case-control data, using a permutation testing approach to adjust the significance level for the entire exhaustive search of the epistatic space. Previous reports from simulation studies have characterized the method's performance in the presence of missing data, phenocopy, 50% locus heterogeneity, various sample sizes, several epistatic models including two-locus through five-locus interactions [11], and multiple levels of class imbalance (in terms of the number of cases and controls in the dataset) [28]. In addition, the algorithm has been refined and improved with the addition of the balanced accuracy [28], NMI fitness metrics [29], cross-validation optimization [30], continuous outcomes[31], and extensions to family data with the development of the MDR Pedigree Disequilibrium Test (MDR-PDT) [12].
Locus heterogeneity or model heterogeneity, where multiple independent epistatic models may predispose an individual to become a case, has been demonstrated to be difficult for MDR and other methods [11]. However, recent innovations in interpretation of MDR results have improved MDR performance in this regard [32]. Best models from an MDR analysis are the largest, most consistent signals observed in the data. Since no explicit test of effect modification is conducted in the MDR algorithm, this signal may be due to main effects, main effects and interactions, pure interactions, noise, or any combination. As a result, MDR may find some but not all loci contributing to prevalence in complex diseases. We considered this to also be a success of the method, since these results contain some true positive results, and some false positives, as with any statistical procedure; and so we have modified the scoring of power to account for the correct discovery of either epistatic model or any locus from either epistatic model.
The goal of this work is to further characterize the performance of MDR given large-scale datasets or locus heterogeneity. To investigate MDR performance in the presence of genetic heterogeneity, we examine two purely epistatic models acting on various proportions of cases within a population in case-control data and across three levels of genetic heterogeneity. This is an expansion of the heterogeneity study from [11] and [32]. These heterogeneity investigations survey all combinations of ten genetic models where minor allele frequency, broad-sense heritability, prevalence among cases, and effect size are varied.
This study also characterizes the performance of MDR when analyzing large datasets of up to 10,000 SNPs. The large-scale data experiments survey 28 genetic models where number of loci, minor allele frequency, broad-sense heritability, effect size, and the number of independent noise loci are varied. Results suggest that while MDR may not solve the intractable issue of heterogeneity confronting genetic epidemiology, it does have some power and is a valuable tool for approaching epidemiological problems involving complex genetic architectures.
Materials and Methods
Multifactor Dimensionality Reduction
The MDR algorithm has been described in detail [10, 11, 28]. Briefly, the steps of MDR follow:
-
(1)
MDR randomly splits the data into k portions, for use in k-fold cross-validation. Cross validation (CV) functions optimally between 5 and 10 intervals, with lower values of k optimized for computation time.
-
(2)
In k – 1/k of the data, the ratios of cases/controls at all multilocus genotypes within a combination of loci are established.
-
(3)
The multilocus genotype combinations are combined to form one binary variable summarizing risk for each multilocus comparison; such that all high risk genotypes are one group and low risk genotypes form the second group. Balanced Accuracy (BA) or (sensitivity + specificity)/2, where sensitivity is true positives/total sample size and specificity is true negatives/total sample size, is computed and used to select models from each order of comparison, or number of loci, for testing. The model with the highest BA is tested in the remaining 1/k of the data to determine the model's ability to predict outcomes in independent datasets. For k CV intervals, k models will be tested in test sets.
-
(4)
This procedure is repeated k times. Maximized average predicted BA and maximized cross-validation consistency over the k-fold cross-validation procedure are used to select the final model. Among all models with the highest observed CV consistency, the highest BA is the tiebreaker. If these two criteria support different models, then the model with fewest loci is selected, according to the principle of statistical parsimony.
To estimate the statistical significance of the result, permutation testing is employed. To obtain an estimate of the empirical null distribution of results, affection status is randomized according to the original proportions in the dataset. This disrupts associations that may exist between predictor and outcome variables. The MDR procedure is performed as above on the permuted data. This procedure of generating permuted data and subsequent analysis is repeated at least 1,000 times. The actual result is compared to the distribution of ordered results from the permutations to determine significance. A significant result suggests a main or joint effect on risk of genotypes at tested loci. The type I error rate of this procedure is always nominal, as long as the search performed for each permutated dataset is the same size as the search performed in the original data (unpublished results). For the experiments in this study, a C++ version of the MDR algorithm was used. However, a Java MDR software package with a graphical user interface and many additional features for data analysis is freely available for download and use from www.epistasis.org.
Genetic Data Simulation
Simulation Software
The simulation software packages genomeSIM [33] and simpen [modified from [34]] were used to generate genetic models and simulate data. Genetic models were generated by simpen, a software package that uses a genetic algorithm to evolve purely epistatic penetrance models using a multiobjective fitness function specifying a target number of model loci, minor allele frequency (MAF), marginal penetrance variance, odds ratio, and heritability. The odds ratio in this case is the average ratio of odds of disease given an exposure to a high-risk genotype relative to exposure to a low-risk genotype, assuming no loss of cases to follow-up due to death or other causes. To calculate this quantity, the product of the penetrance of each multilocus genotype and the frequency of that genotype are found, estimating the expected prevalence of cases in the population for each genotype. Each genotype prevalence is then divided by the sum of the prevalence values for all multilocus genotypes for the model, providing the proportion of cases expected under random sampling at each multilocus genotype. This procedure is also conducted for 1 minus each penetrance, providing the expected proportion of controls per genotype. Where the expected proportion of cases equals or exceeds that of controls, a genotype is denoted high risk; otherwise it is low risk. High risk cells are those genotypes for which the penetrance equals or exceeds the model prevalence. Thus the expected number of high risk cases, low risk cases, high risk controls and low risk controls are found and this is used to calculate the odds ratio for the interaction from a 2 × 2 table. All 28 penetrance models for this study were purely epistatic and contained no main effects larger than relative risk 1.001. The parameters for each simulation are detailed in table 1. All loci in the simulations were independent to provide conservative estimates of power due to increased data noise. It is expected that data with extensive correlation (linkage disequilibrium) among non-model loci would provide fewer spurious signals relative to independent loci since correlated loci would tend to behave similarly to one another, thereby effectively reducing the number of independent non-model variables. This is analogous to the principles underlying the multiple testing correction method of Nyholt [35]. If MDR model loci are in strong LD with nearby loci, mapping fidelity is decreased, as the signals may arise at the correlated non-model locus, but the genomic region is mapped with similar power (unpublished results).
Table 1.
Simulation parameters for MDR power studies
| Model | Loci | Minor allele frequency | Heritability | Odds ratio |
|---|---|---|---|---|
| 1 | 2 | 0.2 | 0.005 | 1.10 |
| 2 | 2 | 0.2 | 0.010 | 1.26 |
| 3 | 2 | 0.2 | 0.050 | 1.79 |
| 4 | 2 | 0.2 | 0.100 | 3.00 |
| 5 | 2 | 0.2 | 0.150 | 4.50 |
| 6 | 2 | 0.2 | 0.200 | 6.00 |
| 7 | 2 | 0.2 | 0.250 | 7.00 |
| 8 | 2 | 0.4 | 0.005 | 1.15 |
| 9 | 2 | 0.4 | 0.010 | 1.28 |
| 10 | 2 | 0.4 | 0.050 | 1.79 |
| 11 | 2 | 0.4 | 0.100 | 2.85 |
| 12 | 2 | 0.4 | 0.150 | 3.49 |
| 13 | 2 | 0.4 | 0.200 | 6.00 |
| 14 | 2 | 0.4 | 0.250 | 7.00 |
| 15 | 3 | 0.2 | 0.005 | 1.14 |
| 16 | 3 | 0.2 | 0.010 | 1.30 |
| 17 | 3 | 0.2 | 0.050 | 1.78 |
| 18 | 3 | 0.2 | 0.100 | 2.55 |
| 19 | 3 | 0.2 | 0.150 | 4.20 |
| 20 | 3 | 0.2 | 0.20 | 6.00 |
| 21 | 3 | 0.2 | 0.250 | 7.00 |
| 22 | 3 | 0.4 | 0.005 | 1.12 |
| 23 | 3 | 0.4 | 0.010 | 1.21 |
| 24 | 3 | 0.4 | 0.050 | 1.83 |
| 25 | 3 | 0.4 | 0.100 | 3.27 |
| 26 | 3 | 0.4 | 0.150 | 4.35 |
| 27 | 3 | 0.4 | 0.200 | 6.00 |
| 28 | 3 | 0.4 | 0.250 | 7.00 |
Large-Scale Simulations
One objective was to characterize the performance of MDR as the search space grows across several genetic scenarios. Two and three locus epistatic models were simulated in datasets of various sizes (table 1). MDR was then run on all datasets and the proportion of the time that the correct best model was found by MDR is reported as the power. Permutation tests were not performed here due to computational infeasibility to evaluate hundreds of large datasets. Datasets with 500 cases and 500 controls and 100, 500, 1,000, 5,000, and 10,000 markers were simulated for two-locus models and 500 cases and 500 controls with 100 and 500 markers were simulated for three-locus models. Fewer markers were simulated for the 3-locus models due to the much longer computation times necessary to exhaustively search for such models. This is due to the property of exhaustive searches where for a 2-locus search in 500 loci, 1.25 × 105 models must be examined, while a 3-locus search in 500 loci consists of 2.07 × 107 models. Comparatively, for a 2-locus search in 10,000 loci, 4.99 × 107 models are evaluated, while in a 3-locus search in 1,000 loci, 1.66 × 108 models are evaluated, and in 10,000 loci there are 1.67 × 1011 3-locus models. With regard to whole-genome association studies, in 500,000 markers 1.26 × 1011 2- locus models are searched. A single whole-genome 2-locus search with MDR takes approximately 4 days to complete using 160 2-gigahertz processors with at least 2 gigabytes of RAM each. Smaller searches scale linearly with the size of the analysis.
Genetic Heterogeneity Simulations
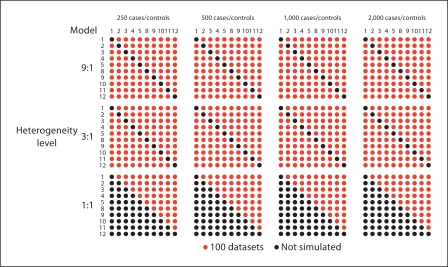
To model genetic heterogeneity, all possible non-redundant two-way combinations of ten selected two-locus models (a subset of the 14 two-locus models in table 1) were considered (table 2; fig. 1). These models were the most subtle effects from our large-scale simulations, as we anticipated looking at power in large sample sizes. Also, some subset of models had to be selected for computational feasibility. Our approach for simulating multiple purely epistatic disease models was similar to that taken in previous studies [11]. Three levels of genetic heterogeneity were examined, where a two-locus model specified the risk profile of a proportion of cases and another model specified the rest of the cases. The ratios of models simulated in cases were 9:1 (i.e. 9/10 cases had one model and 1/10 of the cases had another model), 3:1, and 1:1. There were 90 unique combinations of two-locus models in various proportions for each of the 9:1 and 3:1 levels of heterogeneity, and 45 unique combinations of models for the 1:1 level, for a total of 225 unique simulations, each with 100 datasets (fig. 1). This is because for the 1:1 ratio of models, the combination model 1: model 2 is equivalent to the combination model 2:model 1, whereas for a 3:1 or 9:1 ratio of models, model 1:model 2 is not equivalent to model 2:model 1. All datasets for the heterogeneity study contained 20 total SNPs (16 independent noise markers with random MAF and two 2-locus epistatic models). This number of markers was selected based on the observations from the large scale simulation which indicated that noise markers do not have a strong effect on the sensitivity of MDR (table 3). Four sample sizes for all 225 scenarios were simulated with 250, 500, 1,000, and 2,000 cases and controls. The effect of unequal numbers of cases and controls on performance was also considered as in [28]. For each heterogeneity scenario, data were also simulated with two and four times the controls as there were cases (online supplementary materials, www.karger.com/doi/10.1159/000181157). This approach yielded 225 scenarios in 4 sample sizes and 3 levels of data imbalance, for a total of 2,700 simulations of 100 datasets each.
Table 2.
Simulation II: simulation parameters for heterogeneity MDR power study for two epistatic loci
| Model | Minor allele frequency | Heritability | Heteroeneity combinations | Proportion of cases with a given model |
|---|---|---|---|---|
| 1 | 0.2 | 0.005 | ||
| 2 | 0.2 | 0.01 | ||
| 3 | 0.2 | 0.05 | ||
| 4 | 0.2 | 0.1 | all non-redundant | 0.10/0.90 |
| 5 | 0.2 | 0.15 | pairwise | 0.25/0.75 |
| 8 | 0.4 | 0.005 | combinations | 0.50/0.50 |
| 9 | 0.4 | 0.01 | of the 10 models | |
| 10 | 0.4 | 0.05 | ||
| 11 | 0.4 | 0.1 | ||
| 12 | 0.4 | 0.15 |
A total of 2,700 sets of parameters were simulated. 225 groups of 100 datasets were simulated for all possible non-redundant combinations of ten two-locus models at the three levels of heterogeneity. This was repeated in four sample sizes and three levels of data imbalance. In addition, three scoring schemes were applied (any correct locus, either correct two-locus model, both two-locus models), for a total of 8,100 power results on 100 datasets each.
Fig. 1.
Design for Simulation II. Nonredundant combinations of models 1–5 and 8–12 were simulated for three levels of heterogeneity. To obtain average power for a heterogeneity scenario for a particular model against the other nine models, the average power of a row or column is calculated within the 9: 1 or 3: 1 levels of heterogeneity, for 900 total datasets. For the 1: 1 level the transpose of the upper diagonal matrix is copied into the lower diagonal and averages are calculated in the same way. This design was also performed with 1: 2 and 1: 4 case:control ratios (see online supplementary materials).
Table 3.
Simulation I results
| Model | Number of loci |
||||
|---|---|---|---|---|---|
| 100 | 500 | 1,000 | 5,000 | 10,000 | |
| MDR power with large scale datasets for two-locus models | |||||
| 1 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 0 | 0 |
| 3 | 100 | 100 | 100 | 100 | 100 |
| 4 | 100 | 100 | 100 | 100 | 100 |
| 5 | 100 | 100 | 100 | 100 | 100 |
| 6 | 100 | 100 | 100 | 100 | 100 |
| 7 | 100 | 100 | 100 | 100 | 100 |
| 8 | 0 | 0 | 0 | 0 | 0 |
| 9 | 1 | 0 | 0 | 0 | 0 |
| 10 | 100 | 98 | 98 | 95 | 94 |
| 11 | 100 | 100 | 100 | 100 | 100 |
| 12 | 100 | 100 | 100 | 100 | 100 |
| 13 | 100 | 100 | 100 | 100 | 100 |
| 14 | 100 | 100 | 100 | 100 | 100 |
| MDR power with large-scale datasets for three-locus models | |||||
| 15 | 0 | 0 | |||
| 16 | 0 | 0 | |||
| 17 | 85 | 62 | |||
| 18 | 100 | 100 | |||
| 19 | 96 | 85 | |||
| 20 | 100 | 100 | |||
| 21 | 100 | 100 | |||
| 22 | 0 | 0 | |||
| 23 | 0 | 0 | |||
| 24 | 100 | 99 | |||
| 25 | 100 | 99 | |||
| 26 | 100 | 100 | |||
| 27 | 100 | 100 | |||
| 28 | 100 | 100 | |||
Model Evaluations
Power was calculated for each set of 100 datasets generated in each of the models. Power was estimated as the number of times MDR correctly identified the functional loci from each set of 100 datasets for each of three criteria: (1) finding any functional locus; (2) either two-locus model, or (3) both two-locus models (data not shown). The results for the strictest definition of power (3 above) were very low values with no scenario exceeding 10% power. Average power across nine simulations of 100 datasets each where a given model was simulated as one proportion of the cases and the other nine models were the remainder was calculated for each model for each level of heterogeneity. Groups of models with high and low broad-sense heritabilities were also averaged in this way.
Results
Large-Scale Dataset Simulations
The results of the power studies in large-scale data indicated that in studies with thousands of markers, two-locus searches with MDR are a powerful means of finding epistatic effects (table 3). Where effect sizes are small, MDR lacks power due to the dimensionality and subtlety of signals. Here, MDR has no power to correctly select the correct two loci as the best model when the odds ratio of the interactive effect was 1.3 or less. This most likely would gradually be remedied by much larger samples. Two-locus models with odds ratios larger than 1.75 and broad-sense heritability of 0.1 or larger had very good power with estimates ranging from 94 to 100% for model 10. The rest of the two-locus models simulated either had 0% power or 100% power at all levels of noise. There was not a strong trend of power loss when independent markers were added to the data.
For three-locus models, the power was less than that for two-locus models; although high power was observed for some of the models. This set of three-locus models was only investigated in 100- and 500-locus datasets due to computational restrictions. Models 17 and 19 showed some attenuation of power as noise loci were added to the data. The power for model 17 with 100 markers in the data was 85%, and with 500 markers was 62%. The power for model 19 with 100 markers was 96% and with 500 markers was 85%.
Genetic Heterogeneity Simulations
The results of the heterogeneity studies varied depending on the different definition of power used to score the MDR analysis outcomes. The most conservative definition where all 4 loci from both models must be found had almost no power for any combination of models or level of heterogeneity (online supplementary materials). However, when the definition of power was more liberal, allowing the correct discovery of either epistatic model or any locus from any epistatic model, large increases in power were observed. This phenomenon had been previously explored in [32], where the data from [11] was revisited using these new definitions of power. However, those simulations were based on a 1:1 ratio of cases to controls simulated with pairs of 6 purely epistatic models simulated without regard to the model odds ratio. These results also showed that the presence of a model with high broad sense heritability greatly enhanced the power of the analysis beyond that observed when the sample size was large.
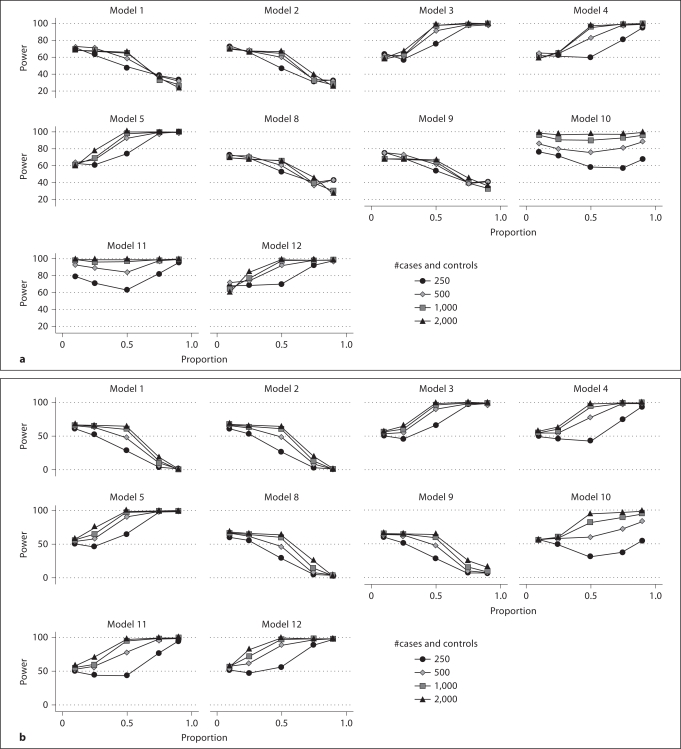
For these results, the average power was calculated for each model at each heterogeneity ratio and at each sample size (fig. 2a, b). Here it is apparent that as low heritability models become predominant, power decreases. The power under these simulation parameters is also not extremely sensitive to sample size at the extremes of heterogeneity, while in models that are simulated at a ratio of 1:1, sample size is most influential. An exception to that is model 10, where large differences in performance across sample sizes are observed at the 9:1 level of heterogeneity, where 90% of cases are simulated on model 10.
Fig. 2.
Power in the presence of heterogeneity. a MDR power to detect any correct locus from either two-locus model. b MDR power to detect either correct two-locus model. The x-axis represents the proportion of cases simulated with the labeled model. The y-axis represents the average power of 9 independent simulations of 100 datasets each where the labeled model is the proportion of cases on the x-axis and [1 – proportion of cases] are simulated with the other 9 models. When x = 0 or 1, there is no heterogeneity. Thus, each point represents the average rate of success in 900 total datasets. a The ability of MDR to find any correct model locus from either two-locus model. b The ability of MDR to find either two-locus model, but not both.
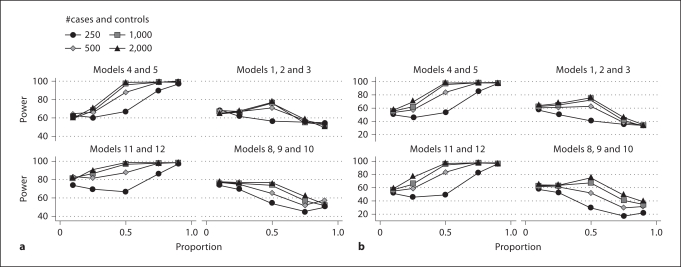
Also explored was the average power when weak models are plotted versus strong models (fig. 3a, b). These results show that the power of MDR is more related to the signal from the data than the sample size collected. For instance, at the 1:1 level of heterogeneity, all but the 250 cases/250 controls sample had more than 80% power for the high heritability models for both definitions of power, while for the low heritability models, even the 2,000 cases/2,000 controls data did not reach 80% average power. The slightly lower average power at the 1:9 level of heterogeneity of the strong models from these figures can be attributed to the majority of the signal being due to the average performance of all other models, where the low heritability models determine most of the cases.
Fig. 3.
Extreme ranges of heritability. a MDR power to detect any correct locus from either two-locus model for high and low broadsense heritability and allele frequencies. b MDR power to detect either correct two-locus model for high and low broad-sense heritabilities and allele frequencies. Models 4 and 5 (0.2 minor allele frequency) and 11 and 12 (0.4 minor allele frequency) have broadsense heritabilities of 0.1 and 0.15 respectively (high heritability), while models 1, 2 and 3 (0.2 minor allele frequency) and models 8, 9 and 10 (0.4 minor allele frequency) have heritabilities of 0.005, 0.01 and 0.05 respectively (low heritability). The x-axis represents the proportion of cases simulated with the labeled models. When x = 0 or 1, there is no heterogeneity. The y-axis represents the average power of 18 (high heritability) or 27 (low heritability) independent simulations of 100 datasets each where the labeled model is the proportion of cases on the x-axis and [1 – proportion of cases] are simulated with the other 8 (high heritability) or 7 (low heritability) models. Each point represents the average rate of success in 1,800 datasets (2 × 900) for the left two graphs (high heritability) and 2,700 datasets (3 × 900) for the right two graphs (low heritability). a The ability of MDR to find any correct model locus from either two-locus model. b The ability of MDR to find either two-locus model, but not both.
Also simulated were 1:2 and 1:4 levels of data imbalance across all parameters presented here. Those results resembled the results presented here and are presented in online supplementary materials.
Discussion
The results of these experiments show that MDR can find epistatic models in the presence of many non-model loci. The number of markers present in the data will however affect the type of analysis performed. Due to the computation time of about 15,000 h to perform a single 2-locus search in a dataset containing 500,000 markers, while the analysis of one dataset is feasible, the permutation testing is infeasible, as this would take 15,000,000 h for 1,000 permutations and 1.5 × 1010 h for 1 million permutations. Instead, for very large searches, we recommend a 2-way split of the data, where one half of the samples are used for searching for 2-locus epistatic models with MDR and the other half is used for independent hypothesis tests for a few top models with logistic regression. Bonferroni corrections for significance should be applied to the results of such regression tests. Otherwise where feasible given the user's access to computer power, the permutation test can be performed.
Heterogeneity can negatively impact MDR performance, especially when power is judged as the ability to detect all model loci in two epistatic models functioning independently. Here we focused on relatively small datasets with 20 markers in light of our observations in the large-scale dataset study above and considering the large number of scenarios we wanted to investigate. However, we observed that under these circumstances, MDR performs well when used to detect either of two susceptibility models, or a single locus from any of the models. These studies show that the results from [11] may lead to undue pessimism with regard to the use of MDR as a tool to detect true associations in data collected for complex traits. This topic was revisited in [32] using the scoring rules we apply here. Here we have simulated many more scenarios than those presented previously by varying heterogeneity level, sample size and case:control ratio. This approach has provided detailed observations about MDR performance in complicated analysis situations. In addition, MDR has also been compared to some other methods for searching for interactions in genetic data [36]. In that manuscript, the authors show that MDR can have superior sensitivity compared to some machine learning methods and regression-based methods that condition analyses of interactions on main effects. This behavior is thought to be due to the exhaustive search performed by MDR, and is more pronounced in higher order epistatic models.
The results of the current study illustrate a property of MDR that is similar to what has been seen elsewhere for parametric statistics. This is that effect size influences performance more than sample size. Heritability estimates can vary across populations for a trait. Thus, the selection of a study population where the heritability estimates for a trait are high will certainly predispose the study to success more than a vigorous and successful ascertainment [37]. While the overall effect size may range within a given trait heritability, in purely epistatic models, these quantities are related, where higher heritability corresponds with higher odds ratios, and so the analogy with parametric statistics holds [38]. Here, we have simulated various heritabilities in our data, but this could emulate the choice an investigator might make; whether to pursue a more convenient ascertainment in a population with a lower estimate of heritability, where environment explains more trait variance, or go in search of a study population where the trait is more genetic, and heritability estimates are higher.
For example, the power to detect a mutation at a single locus with an odds ratio of 1.25 at an α = 0.05 with a χ2 test where 30% of controls are exposed with 500 cases and 500 controls are completely genotyped is 38%. The power to detect the same mutation in 1,000 cases and 1,000 controls is 65%. The power to detect a mutation with an odds ratio of 1.5 in 500 cases and 500 controls with an exposure frequency of 30% in controls is 99%. While not a novel concept in statistics, this property of MDR has not previously been demonstrated. The heterogeneity experiment results show that the average power with a strong primary model is larger than the power with a relatively weak primary model with higher sample sizes.
Additionally, the performance of MDR is not extremely attenuated by noise encountered in larger searches with larger datasets. This is encouraging in the whole genome association era, and although these experiments are not on the scale of whole-genome association studies, these results show that MDR use is appropriate for very large candidate gene studies where epistasis models might be encountered (and even two-locus searches in whole genome association data).
The approach to interpreting multi-locus models from MDR has been under recent development. MDR screens a search space and ranks the signals according to the fitness metric employed. Here we applied balanced accuracy, but any fitness function could be used. Since this procedure does not explicitly look at effect modification across genotypes, some other method, such as regression, must be employed post hoc to determine the nature of the multi-locus model. Recent experiments investigating the Type I error of regression after a search through all possible interactions suggest that this procedure is not valid, and that the means used to look at effect modification must be inherently corrected for the search conducted to find the model to conduct a valid test. Extensions to MDR and MDR-PDT are in development that will allow this to be accomplished. However, in general it is our opinion that a multilocus model may contain real signals and noise markers due to the manner in which MDR examines data searching for the largest signal. For example, when a large main effect, such as APOE in Alzheimer's disease, is present in the data, all multilocus models containing APOE might be significant by the permutation test. Therefore, we advocate considering nested models from the MDR output as the source of the signal. Replication datasets should be designed with this in mind, and significant MDR results should not be interpreted literally as effect modification across several loci.
In summary, MDR is statistically efficient in data with many variables. Each variable from an MDR model should be considered a potential association or member of an interaction. Most importantly, MDR performance is more sensitive to effect size and the selection of study populations with high heritability estimates than the sample size collected.
Supplementary Material
Supplementary materials
References
- 1.Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 2.Sabatti C, Service S, Freimer N. False discovery rate in linkage and association genome screens for complex disorders. Genetics. 2003;164:829–833. doi: 10.1093/genetics/164.2.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore JH, Williams SM. Traversing the conceptual divide between biological and statistical epistasis: systems biology and a more modern synthesis. Bioessays. 2005;27:637–646. doi: 10.1002/bies.20236. [DOI] [PubMed] [Google Scholar]
- 4.Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Trans R Soc Edinburgh. 1918;52:399–433. [Google Scholar]
- 5.Bateson W. Mendel's Principles of Heredity. Cambridge: Cambridge University Press; 1909. [Google Scholar]
- 6.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- 7.Hahn LW, Moore JH. Ideal discrimination of discrete clinical endpoints using multilocus genotypes. In Silico Biol. 2004;4:183–194. [PubMed] [Google Scholar]
- 8.Moore JH. Computational analysis of gene-gene interactions using multifactor dimensionality reduction. Expert Rev Mol Diagn. 2004;4:795–803. doi: 10.1586/14737159.4.6.795. [DOI] [PubMed] [Google Scholar]
- 9.Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden T, Barney N, White BC. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol. 2006;241:252–261. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 10.Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24:150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- 12.Martin ER, Ritchie MD, Hahn L, Kang S, Moore JH. A novel method to identify gene-gene effects in nuclear families: the MDR-PDT. Genet Epidemiol. 2006;30:111–123. doi: 10.1002/gepi.20128. [DOI] [PubMed] [Google Scholar]
- 13.Chan IH, Leung TF, Tang NL, Li CY, Sung YM, Wong GW, Wong CK, Lam CW. Gene-gene interactions for asthma and plasma total IgE concentration in Chinese children. J Allergy Clin Immunol. 2006;117:127–133. doi: 10.1016/j.jaci.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 14.Millstein J, Conti DV, Gilliland FD, Gauderman WJ. A testing framework for identifying susceptibility genes in the presence of epistasis. Am J Hum Genet. 2006;78:15–27. doi: 10.1086/498850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai CT, Lai LP, Lin JL, Chiang FT, Hwang JJ, Ritchie MD, Moore JH, Hsu KL, Tseng CD, Liau CS, Tseng YZ. Renin-angiotensin system gene polymorphisms and atrial fibrillation. Circulation. 2004;109:1640–1646. doi: 10.1161/01.CIR.0000124487.36586.26. [DOI] [PubMed] [Google Scholar]
- 16.Coffey CS, Hebert PR, Krumholz HM, Morgan TM, Williams SM, Moore JH. Reporting of model validation procedures in human studies of genetic interactions. Nutrition. 2004;20:69–73. doi: 10.1016/j.nut.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Mannila MN, Eriksson P, Ericsson CG, Hamsten A, Silveira A. Epistatic and pleiotropic effects of polymorphisms in the fibrinogen and coagulation factor XIII genes on plasma fibrinogen concentration, fibrin gel structure and risk of myocardial infarction. Thromb Haemost. 2006;95:420–427. doi: 10.1160/TH05-11-0777. [DOI] [PubMed] [Google Scholar]
- 18.Ashley-Koch AE, Mei H, Jaworski J, Ma DQ, Ritchie MD, Menold MM, Delong GR, Abramson RK, Wright HH, Hussman JP, Cuccaro ML, Gilbert JR, Martin ER, Pericak-Vance MA. An analysis paradigm for investigating multi-locus effects in complex disease: examination of three GABA receptor subunit genes on 15q11-q13 as risk factors for autistic disorder. Ann Hum Genet. 2006;70:281–292. doi: 10.1111/j.1469-1809.2006.00253.x. [DOI] [PubMed] [Google Scholar]
- 19.Ma DQ, Whitehead PL, Menold MM, Martin ER, Ashley-Koch AE, Mei H, Ritchie MD, Delong GR, Abramson RK, Wright HH, Cuccaro ML, Hussman JP, Gilbert JR, Pericak-Vance MA. Identification of significant association and gene-gene interaction of GABA receptor subunit genes in autism. Am J Hum Genet. 2005;77:377–388. doi: 10.1086/433195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrew AS, Nelson HH, Kelsey KT, Moore JH, Meng AC, Casella DP, Tosteson TD, Schned AR, Karagas MR. Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking and bladder cancer susceptibility. Carcinogenesis. 2006;27:1030–1037. doi: 10.1093/carcin/bgi284. [DOI] [PubMed] [Google Scholar]
- 21.Soares ML, Coelho T, Sousa A, Batalov S, Conceicao I, Sales-Luis ML, Ritchie MD, Williams SM, Nievergelt CM, Schork NJ, Saraiva MJ, Buxbaum JN. Susceptibility and modifier genes in Portuguese transthyretin V30M amyloid polyneuropathy: complexity in a single-gene disease. Hum Mol Genet. 2005;14:543–553. doi: 10.1093/hmg/ddi051. [DOI] [PubMed] [Google Scholar]
- 22.Sanada H, Yatabe J, Midorikawa S, Hashimoto S, Watanabe T, Moore JH, Ritchie MD, Williams SM, Pezzullo JC, Sasaki M, Eisner GM, Jose PA, Felder RA. Single-nucleotide polymorphisms for diagnosis of salt-sensitive hypertension. Clin Chem. 2006;52:352–360. doi: 10.1373/clinchem.2005.059139. [DOI] [PubMed] [Google Scholar]
- 23.Williams SM, Ritchie MD, Phillips JA, III, Dawson E, Prince M, Dzhura E, Willis A, Semenya A, Summar M, White BC, Addy JH, Kpodonu J, Wong LJ, Felder RA, Jose PA, Moore JH. Multilocus analysis of hypertension: a hierarchical approach. Hum Hered. 2004;57:28–38. doi: 10.1159/000077387. [DOI] [PubMed] [Google Scholar]
- 24.Brassat D, Motsinger AA, Caillier SJ, Erlich HA, Walker K, Steiner LL, Cree BA, Barcellos LF, Pericak-Vance MA, Schmidt S, Gregory S, Hauser SL, Haines JL, Oksenberg JR, Ritchie MD. Multifactor dimensionality reduction reveals gene-gene interactions associated with multiple sclerosis susceptibility in African Americans. Genes Immun. 2006;7:310–315. doi: 10.1038/sj.gene.6364299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J, Lowey J, Wiklund F, Sun J, Lindmark F, Hsu FC, Dimitrov L, Chang B, Turner AR, Liu W, Adami HO, Suh E, Moore JH, Zheng SL, Isaacs WB, Trent JM, Gronberg H. The interaction of four genes in the inflammation pathway significantly predicts prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:2563–2568. doi: 10.1158/1055-9965.EPI-05-0356. [DOI] [PubMed] [Google Scholar]
- 26.Qin S, Zhao X, Pan Y, Liu J, Feng G, Fu J, Bao J, Zhang Z, He L. An association study of the N-methyl-D-aspartate receptor NR1 subunit gene (GRIN1) and NR2B subunit gene (GRIN2B) in schizophrenia with universal DNA microarray. Eur J Hum Genet. 2005;13:807–814. doi: 10.1038/sj.ejhg.5201418. [DOI] [PubMed] [Google Scholar]
- 27.Cho YM, Ritchie MD, Moore JH, Park JY, Lee KU, Shin HD, Lee HK, Park KS. Multifactor-dimensionality reduction shows a two-locus interaction associated with Type 2 diabetes mellitus. Diabetologia. 2004;47:549–554. doi: 10.1007/s00125-003-1321-3. [DOI] [PubMed] [Google Scholar]
- 28.Velez DR, White BC, Motsinger AA, Bush WS, Ritchie MD, Williams SM, Moore JH. A balanced accuracy function for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction. Genet Epidemiol. 2007;31:306–315. doi: 10.1002/gepi.20211. [DOI] [PubMed] [Google Scholar]
- 29.Bush WS, Edwards TL, Dudek SM, Mc Kinney BA, Ritchie MD. Alternate contingency table measures improve the power and detection of multifactor dimensionality reduction. BMC Bioinformatics. 2008;9:238. doi: 10.1186/1471-2105-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motsinger AA, Ritchie MD. The effect of reduction in cross-validation intervals on the performance of multifactor dimensionality reduction. Genet Epidemiol. 2006;30:546–555. doi: 10.1002/gepi.20166. [DOI] [PubMed] [Google Scholar]
- 31.Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet. 2007;80:1125–1137. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie MD, Edwards TL, Fanelli TJ, Motsinger AA. Genetic heterogeneity is not as threatening as you might think. Genet Epidemiol. 2007;31:797–800. doi: 10.1002/gepi.20256. [DOI] [PubMed] [Google Scholar]
- 33.Dudek SM, Motsinger AA, Velez DR, Williams SM, Ritchie MD. Data simulation software for whole-genome association and other studies in human genetics. Pac Symp Biocomput. 2006:499–510. [PubMed] [Google Scholar]
- 34.Moore JH, Hahn LW, Ritchie MD, Thornton TA, White B. Routine discovery of high-order epistasis models for computational studies in human genetics. Appl Soft Comput. 2004;4:79–86. doi: 10.1016/j.asoc.2003.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Motsinger AA, Reif DM, Fanelli TJ, Ritchie MD. A comparison of analytical methods for genetic association studies. Genet Epidemiol. 2008 doi: 10.1002/gepi.20345. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 37.Terwilliger JD, Weiss KM. Confounding, ascertainment bias, and the blind quest for a genetic ‘fountain of youth’. Ann Med. 2003;35:532–544. doi: 10.1080/07853890310015181. [DOI] [PubMed] [Google Scholar]
- 38.Culverhouse R, Suarez BK, Lin J, Reich T. A perspective on epistasis: limits of models displaying no main effect. Am J Hum Genet. 2002;70:461–471. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials