Abstract
The existence of “tumor-initiating cells” (TICs) has been a topic of heated debate for the last few years within the field of cancer biology. Their continuous characterization in a variety of solid tumors has led to an abundance of evidence supporting their existence. TICs are believed to be responsible for resistance against conventional treatment regimes of chemotherapy and radiation, ultimately leading to metastasis and patient demise. This review summarizes DNA repair mechanism(s) and their role in the maintenance and regulation of stem cells. There is evidence supporting the hypothesis that TICs, similar to embryonic stem (ES) cells and hematopoietic stem cells (HSCs), display an increase in their ability to survive genotoxic stress and injury. Mechanistically, the ability of ES cells, HSCs and TICs to survive under stressful conditions can be attributed to an increase in the efficiency at which these cells undergo DNA repair. Furthermore, the data presented in this review summarize the results found by our lab and others demonstrating that TICs have an increase in their genomic stability, which can allow for TIC survival under conditions such as anticancer treatments, while the bulk population of tumor cells dies. We believe that these data will greatly impact the development and design of future therapies being engineered to target and eradicate this highly aggressive cancer cell population.
Electronic supplementary material
The online version of this article (doi:10.1007/s10555-011-9277-0) contains supplementary material, which is available to authorized users.
Keywords: Tumor-initiating cells, DNA repair , Metastasis
Introduction
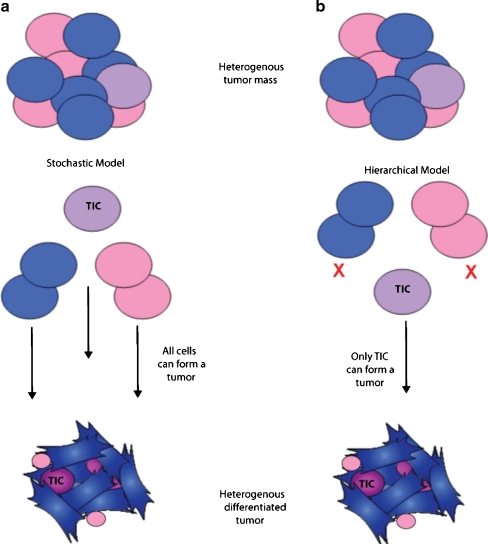
Cancer is defined as uncontrolled cell growth resulting from genetic mutations or exposure to environmental carcinogens that alter normal regulation. The uncontrolled cellular growth results in the formation of a mass of cells commonly referred to as a tumor. The majority of cells within a tumor share identical genetic and epigenetic gene signatures; however, there is a subset of cells that exist within the total population and carry unique signatures, thus demonstrating tumor heterogeneity. This process of tumor heterogeneity can be explained by two different models: the stochastic model and the hierarchy model (Fig. 1, adapted from [1]). The stochastic model hypothesizes that a tumor is biologically homogenous and both intrinsic and extrinsic factors affect cell behavior, leading to heterogeneous populations of cells. The hierarchy model hypothesizes that there are biologically distinct cells within the tumor, and it is this specific population of cells that is responsible for the initiation of tumor growth. The existence of these “tumor-initiating cells” (TICs) has been a topic of vigorous discussion for the last few years within the field of cancer biology. Their continuous characterization has led to an abundance of data supporting their existence; additionally, there is evidence suggesting that these cells are responsible for chemo- and radioresistance, hence serving as the foundation for metastasis and ultimately patient demise. Recently, our laboratory and others have observed that TICs have an increase in their genomic stability that allows them to have an enhanced ability to survive against treatment compared to the bulk population of cells. The following review will summarize evidence supporting the hypothesis that TICs, similar to embryonic stem (ES) cells and hematopoietic stem cells (HSCs), have an increase in their ability to deal with genotoxic stress and injury which appears to be due to an increase in the efficiency of DNA repair mechanism(s).
Fig. 1.
Process of tumor heterogeneity can be explained by two different models (adapted from [1]). a In the stochastic model, each cell from a heterogeneous tumor has the ability to form a new heterogeneously differentiated tumor. b In the hierarchical model, not all tumor cells have the ability to form a new heterogeneous differentiated tumor, and rather only the tumor-initiating cell or TIC does
Tumor-initiating cells
TICs represent a small population of cells that exist within a heterogeneous tumor based on the hierarchical theory of cancer development. Currently, TICs have been isolated from a variety of solid tumor types, including those from the bladder, brain, breast, colon, head/neck, liver, lung, ovary, pancreas, prostate, and skin [2–6]. Within the last few years, it has also been well documented that very low numbers of TICs (10–100) have the ability to form colonies in vitro or initiate tumor formation in vivo [7–12]. To achieve this same effect with total tumor cells, one to ten million cells are required. As early as 1994, Lapidot et al. [13] showed that “stem cells” could be isolated using fluorescence-activated cell sorting (FACS) based on the expression of the cell surface markers CD34 and CD38. The CD34+CD38− cells were identified as potential stem cells of acute myeloid leukemia. This “stem cell” phenotype was assigned since non-obese diabetes/severe combined immunodeficiency mice injected with low numbers of CD34+CD38− cells developed leukemia, whereas those injected with even larger numbers of more mature cells (CD34−CD38+) did not [13]. Overall, TICs are capable of undergoing the process of self-renewal and giving rise to differentiated tumor cells, while the bulk of tumor cells is highly differentiated, has limited proliferative potential, and is non-tumorigenic.
TICs have been isolated using several different methods, including FACS based on cell surface expression of CD44, CD133, α2β1 integrin for example, as well as flow cytometry-based “side populations” (reviewed by [14]). Additionally, TICs can be isolated by generating spheroids using specialized culture conditions and highly defined media called stem cell media [15–18]. Spheres generally express higher levels of stem cell genes and demonstrate higher tumorigenic potential in animals with similar levels to sorted TICs compared to total cells [17–21]. Our lab has also established a method to isolate these TICs based on the property of increased invasive ability [22]. Using an in vitro Matrigel assay and highly defined media [22, 23], we have isolated prostate TICs that have a stem-like phenotype [22], have undergone an epithelial-to-mesenchymal transition during the process of invasion, and are also highly tumorigenic when injected into mice. It is thought that these aggressive cells are also the most invasive cells and are able to survive and metastasize to other vital organs, leading to fatality in patients [24–26].
Recently, using this model of invasion to isolate pancreatic TICs, we observed significant increases in gene expression in a large number of genes related to DNA repair, specifically genes involved in BRCA1-mediated DNA repair (Mathews et al., unpublished). Additionally, various genes were identified as being upregulated as well and can be classified as members of hereditary breast cancer susceptibility signaling, including ATM, RAD50 and RAD51, PTEN, and a number of genes belonging to the Fanconi anemia family which have previously been linked to DNA repair mechanisms [27, 28]. Functionally, we also observed an increase in the ability of the TICs to repair DNA after challenge with gemcitabine compared to the total population of cells. These observations prompted us to explore the literature to determine whether other data supported a mechanism for increased DNA repair in stem cell populations.
Mechanisms of DNA repair
To understand the notion that stem cells display an increase in genomic stability, it is necessary that the different DNA repair mechanisms which cells use to maintain this stability are described (Table 1). These mechanisms include double-strand break repair (homologous recombination-mediated repair and non-homologous end-joining), mismatch repair, and nucleotide excision repair. A variety of the proteins involved in the process of DNA repair are defined as cancer susceptibility genes, and mutations or loss of expression in proteins such as ATM, BRCA1, BRCA2, p53, and a number of RAD proteins reveal that loss of heterozygosity at their genomic regions corresponds to the onset of sporadic breast carcinomas [29]. Our hypothesis is that in TICs, there is an increase in the expression of these DNA repair-related genes, therefore rendering the cells extremely stable to genetic insult and increasing their ability to survive and function as tumor initiators.
Table 1.
Main mechanisms of DNA repair and their contributing proteins
| Non-homologous end-joining (NHEJ) | KU70, KU80, DNA-PKc, XRCC4, and DNA ligase IV |
| Homologous recombination-mediated repair (HRR) | BRCA1, BRCA2, RAD50, RAD51, MRE11, NBS1 |
| Mismatch repair (MMR) | MutSα (MSH2/MSH6), MutSβ (MSH2/MSH3), MutLα (MLH1/PMS2), MutLβ (MLH1/MLH3), EXO1 |
| Nucleotide excision repair (NER) | DDB1, XPE, XPC-RAD23B, TFIIH, XPB, XPD, RPA |
| Base excision repair (BER) | AP-endonuclease, DNA polymerase-β, DNA ligase III, XRCC1 DNA polymerase-δ or -ε, PCNA, DNaseIV/FEN-1, DNA ligase I |
Double-strand break repair
The two major subdivisions of double-strand break repair (DSB) include homologous recombination-mediated repair (HRR) and non-homologous end-joining (NHEJ; reviewed in [30]). HRR uses thousands of bases of sequence homology either from a sister chromatid or a homologous chromosome during S/G2 phase and is the most error-free method of repair [30]. NHEJ, occurring during G1/S, requires little or no sequence homology for efficient repair and can be error-free or error-prone depending on the type of ends that are present at the site of the DSB. A key regulator in mediating which pathway a cell chooses to repair broken DNA is the multifunctional protein BRCA1 (as reviewed in [29]). BRCA1 preferentially channels DSB repair into HRR rather than NHEJ, and the process is started by a protein complex containing MRE11, RAD50, and NBS1, termed the MRN complex. A series of additional steps allows RAD51 to form a nucleoprotein filament and catalyze homologous pairing and strand exchange with the assistance of BRCA2. During NHEJ, an entirely different complex is formed using the KU70 and KU80 proteins, followed by recruitment and activation of the DNA protein kinase DNA-PKc, resulting in subsequent activation of XRCC4 and DNA ligase IV (LIG4) [31].
Mismatch repair
The mismatch repair (MMR) system repairs base–base mismatches that result in point mutations and insertion/deletion loops that can further result in frameshift mutations [32]. The MMR pathway, like other repair pathways, involves many proteins which act sequentially to repair the damaged DNA. The mismatch is first recognized by a protein called MutSα, which is a dimer of MSH2/MSH6 or MutSβ, containing a dimer of MSH2/MSH3 [32, 33]. Mismatch excision is initiated by the binding of MutLα, a dimer of MLH1/PMS2, or MutLβ, a dimer of MLH1/MLH3. Further recruitment of the exonuclease, EXO1, results in the sequential removal of nucleotides between an adjacent single-strand break up to and beyond the mismatch on the daughter strand. The DNA is then resynthesized by DNA polymerase-δ along with PCNA and RPA and ligated with DNA ligase [32, 33].
Nucleotide excision repair
The process of nucleotide excision repair (NER) is considered the most versatile form of DNA repair and operates on damaged/disruptive bases resulting from UV or oxidative damage [31]. Two different NER pathways exist and have substrate specificities depending on whether the damage is in the actively transcribed strand of a gene or elsewhere in the genome. The process global genomic NER (GG-NER) occurs if the damage is in the actively transcribed strand of a gene, and transcription-coupled repair (TCR) occurs if the lesions are directly associated with errors in transcription itself [31]. In GG-NER, two different heterodimeric proteins bind to the DNA, recognize the damage, and initiate repair. One of the complexes is named UV-damaged DNA-binding protein (UV-DDB) and consists of the proteins DDB1 and XPE. Binding of this complex then allows recruitment of XPC-RAD23B, a step which is not required in TCR, and permits the entry and binding of TFIIH which has ten different subunits. Two of the subunits, XPB and XPD, are helicases and are able to unwind the damaged DNA strand, permitting the binding of another protein called RPA. The gap repair proteins, RFC, PCNA, and DNA polymerase-δ are then able to carry out new DNA synthesis and seal the strand using DNA ligase I. TCR repairs damaged DNA more rapidly in the transcribed regions than in non-gene regions. In humans, TCR-NER requires all of the same proteins GG-NER does, expect for XPE, XPC, and RAD23B, thereby suggesting that an alternate mechanism is utilized [31].
Base excision repair
Base excision repair (BER) is most often used to protect cells from damage that is caused by cellular metabolism and by spontaneous depurinations [31]. There are two subdivisions of BER: “short-patch repair” and “long-patch repair.” Short-patch repair involves the repair of a single nucleotide, and long-patch repair involves repairing between 2 and 15 nucleotides. Both pathways include DNA glycosylases, endonucleases, and DNA polymerases [34]. During the short-patch repair, after base damage, recognition occurs by a DNA glycosylase/lyase and subsequent cleavage of the damaged base occurs by AP endonuclease. DNA polymerase-β will generate a repair patch and DNA ligase III will seal the DNA with the help of the XRCC1 protein. Long-patch repair utilizes proteins involved in DNA replication, including DNA polymerase-δ or -ε, PCNA, DNaseIV/FEN-1, and DNA ligase I.
The various mechanism(s) and pathways utilized by a cell to carry out DNA repair are extensive and complicated. DNA repair mechanism(s) are essential to the maintenance of a normal intact stable genome, and the hypothesis that TICs are capable of an enhanced efficiency of DNA repair is plausible. The role of DNA repair mechanism(s) in stem cells is a critical process necessary for stem cell regulation, and the hypothesis that DNA repair processes in TICs are more efficient is further investigated below (Table 2).
Table 2.
Cancer types, genes and DNA repair mechanisms
| Cells/cancer types | Genes | Mechanism | Reference |
|---|---|---|---|
| Embryonic stem cells | RAD51 | HRR | [38] |
| DNA-PKcs | DSB | [41] | |
| MSH2, MSH6 | MMR | [42] | |
| Hematopoietic stem cells (quiescent) | NHEJ | [45] | |
| Hematopoietic stem cells (proliferative) | HRR | [45] | |
| Normal multipotent stem cells from hair follicle bulge | Bcl-2, p53 | increase resistance DNA-damaged induced death | [46] |
| Keratinocyte stem cells | FGF2 | DBSR, SSBR | [47] |
| Glioma tumor-initiating cells | CHK1, CHK2 | Increase activation | [51] |
| BMI-1, DNA-PK, PARP-1 | Repair IR-induced DNA damage | [50] | |
| MGMT | DSB, NHEJ | [57] | |
| EZH2/HOX | Alkylating agent sensitivity | [61–63] | |
| PcG | [64, 65] | ||
| Breast cancer cell line MCF-7 | RAD51, APE1 | SSBR | [70] |
| AKT, WNT signaling | Increased DNA repair | [71, 72] | |
| p53-null Lin−CD29HCD24H | BRCA1 | Increased Expression | [73] |
| Metastatic malignant melanoma | Higher DNA repair | [67–69] | |
| Pancreatic cancer | BRCA1 | BRCA1-mediated repair | Unpublished, Mathews et al. |
| RAD51 | RAD51 DNA damage response | [74] |
DNA repair in embryonic and normal stem cells
Data obtained from ES cells suggest that they have robust mechanisms in place to preserve their genetic stability and, compared to somatic cells, display substantially lower mutation frequencies [30]. For example, compared to mouse embryonic fibroblasts (MEFs), ES cells have a 100-fold lower level of mutation in the Arpt gene [35, 36]. In a similar analysis, spontaneous mutation frequencies in the Hprt gene were not detectable in ES cells, whereas in MEFs, the rate was in the range of 105 [36]. Although suppression of mutagenesis is one mechanism that ES cells use to maintain their genomic stability, there is much more evidence suggesting that differences in this increased stability are due to increases in DNA repair [30]. The major pathway that ES cells utilize to repair damaged DNA is DSB repair via the HRR mechanism. This is thought to be the major pathway of repair since ES cells lack a G1 checkpoint, have short G1 and G2 cell cycle phases, and spend the majority of time in the S-phase [37]. Lastly, recent data demonstrate that RAD51 is a key player for HRR in ES cells. It has been shown that when RAD51 is deleted from ES cells, the cells are not viable; furthermore, cells isolated from RAD51 null blastocysts are unable to proliferate [38].
With regard to NHEJ, it is thought to play a minor role in repairing DNA in ES cells [30]; however, the majority of these data are from studies conducted using mouse cell lines. Recent data using human ES cells (hESCs) demonstrate that HRR is utilized extensively for repair, and this decreases throughout differentiation toward neural progenitor and astrocytes [39]. However, rapidly proliferating hESCs do utilize NHEJ in a process that is independent of ATM, DNA-PKcs, and PARP, but dependent on XRCC4. As these cells differentiate, the rate of NHEJ progressively increases while the fidelity of repair decreases [39, 40]. Overall, hESCs demonstrate an enhancement of DNA repair as a method to protect their genome and display higher levels of DNA-PKcs after irradiation, thus leading to a highly efficient DSB repair [41].
With regards to ES cells and MMR, there are significantly higher levels of MSH2 and MSH6 in ES cells compared to 3T3 cells [42]. When these cells are induced to differentiate with retinoic acid, the expression of MSH2 decreases as well. For the NER-based pathway, when ES cells are compared to either MEFs, CHO cells, or murine cardiomyocytes, low doses of UV radiation result in very similar levels of repair [43]. However, at higher doses, the ES cell repair machinery becomes saturated and the repair remains incomplete. This is contrary to previously published data suggesting that the NER is more efficient in ES cells compared to terminally differentiated cells and appears to be dependent on the cell cycle processes [44]. Although the results from a number of studies demonstrate opposing data, additional experiments need to be performed to determine the role of NER in stem cell populations.
Similar to HSCs, adult stem cells remain in a fairly quiescent state [45]. This state of quiescence is considered to be protective since it minimizes endogenous stress caused by cellular respiration and replication. Recent data demonstrate that quiescent HSCs use NHEJ while proliferating HSCs use the high-fidelity HRR mechanism [45]. It is speculated that the use of the NHEJ mechanism in quiescent cells renders the cells more susceptible to genomic instability associated with misrepaired DNA, leading to loss of HSC function and premalignant transformation. Alternatively, proliferating cells use the HRR mechanism to decrease any further risk of acquiring mutations. It has been proposed that it is logical to use HRR in long-lived quiescent HSCs in order to guard them against errors occurring during DNA replication and damage associated with oxidative stress. Furthermore, normal multipotent stem cells isolated from the hair follicle bulge have two important mechanisms for increasing their resistance to DNA damage-induced cell death: higher expression of the anti-apoptotic gene Bcl-2 and transient stabilization of p53 after DNA damage [46]. Investigation in keratinocyte stem cells showed increased levels of DSBR and single-strand break repair (SSBR) compared to progenitor cells, and further evidence shows that FGF2 is critical to mediate this repair [47]. These data further support that normal stem cells have evolved more efficient DNA repair mechanisms to help increase their overall survival compared to differentiated cells.
In a recent review by Frosina [48], the author summarized the extensive evidence supporting that hESCs, human and rat HSCs, and human bone marrow-derived mesenchymal stem cells have more efficient repair than differentiated cells [48]. These and other studies have led us to further investigate whether TICs are able to thrive due to an enhancement in their DNA repair mechanisms.
DNA repair in tumor-initiating cells
Frosina [49] also suggests that DNA repair is higher in both normal and cancer stem cells (CSCs or TICs) isolated from the central nervous system compared to differentiated and/or non-TICs. Bao et al. [50] have demonstrated that CD133+ glioma cells isolated from both human glioma xenografts and primary patient glioblastoma specimens preferentially activate the DNA damaged checkpoint in response to radiation and repair IR-induced DNA damage more effectively than the cells not expressing CD133. Ropolo et al. [51] have shown that glioma stem cells compared with non-stem cells have a significant increase in the population doubling time as well as an increase in the activation of CHK1 and CHK2 induced by IR in the CD133+ fractions. Although Ropolo et al. [51] did not find a significant change or enhancement of DNA repair in their glioma stem cell population, they speculate that it is a delay in cell cycle via an increase in cell cycle checkpoint kinases that allows for more time to repair DNA damage. Although there is a discrepancy between the studies peformed by Bao et al. [52] and Ropolo et al. [51], it is clear that the stem cell population utilizes a unique mechanism to ensure that there is efficient DNA repair; whether it may be an increase in DNA repair mechanism(s) or a delay in cell cycle is still under investigation.
To further elucidate this, McCord et al. [53] determined that clonogenic survival curves from six CD133+ glioblastoma tumor stem-like cell (TSC) lines were more radiosensitive compared to the parent lines. The authors state that the significance of these data is unclear since in vitro radiosensitivity does not predict in vivo tumor radioresponse; yet, the data do suggest that the molecular determinants of TSC radiosensitivity compared to total cells differ and could serve as important therapeutic targets. McCord et al. [53] demonstrate that CD133+ TSCs were able to activate the G2 checkpoint; however, they were deficient in activating the intra-S-phase arrest. The authors state that this could explain the relative radiosensitivity differences; however, the S-phase checkpoint plays a critical role in maintaining the genome, and this instability could actually be the driving force leading to tumor development and heterogeneity. Importantly, overall in glioblastoma TICs, there appears to be a universal requirement for more efficient DNA repair by the stem cell population compared to the non-stem cell population; further experiments will decipher their differential regulation.
Patient studies show that those diagnosed with malignant glioblastoma multiform (GBM) have a median survival of 5–8 months, and it is thought that the inefficient treatments available are due to resistance mechanisms acquired by aggressive TICs, such as the enhancement in DNA repair. Interestingly, there is evidence showing that post-irradiation in mice, the bulk GBM tumor responds and, as expected, the tumor shrinks [54]; however, CD133+ cells activate the checkpoint controls and repair damaged DNA more efficiently than the CD133− fractions, thus allowing for repopulation of the tumor. TICs may actually have similar DNA repair rates, but because they have lower levels of proliferation and constitutive activation of the checkpoint response, it allows for more time to repair breaks. We hypothesize that these highly aggressive cells could be targeted with DNA checkpoint inhibitors by increasing their sensitivity to radiation and allowing for their destruction. One such drug, AZD7762, produced by AstraZeneca, is an ATP-competitive checkpoint kinase inhibitor that assists DNA-damaging drugs by blocking the checkpoint response (reviewed in [55]). When the drug was administered in combination, it can block tumor formation in multiple xenograft models where DNA-damaging agents alone cannot [56]. The drug selectively blocks CHK1 and CHK2, abrogates S and G2 checkpoints, and enhances the efficacy of gemcitabine and topotecan, thus making it an attractive therapeutic for the treatment of cancers with a high stem cell fraction.
Recently, it was also demonstrated that the polycomb group (PcG) protein BMI-1 co-purifies with DNA DSB response and NHEJ proteins DNA-PK and PARP-1 in CD133+ GBM cells [57]. When BMI-1 is inactivated, recruitment of the DNA repair machinery is inhibited and the cells become increasingly radiosensitive. BMI-1 has been implicated as a regulator of stem cell maintenance in a number of cell lines and plays a significant role in maintaining TICs isolated from the prostate [58]. In addition to pharmacological inhibition of the DNA checkpoints, the targeted inhibition of BMI-1 combined with radiation could possibly lead to the destruction of GBM stem cells. Furthermore, another recent study demonstrated that when DNA-PKc levels were decreased with short hairpin RNA, glioma-initiating cells (GICs) were radiosensitized and underwent autophagy compared to cells expressing much higher levels of the enzyme [59]. Since DNA-PKc is a catalytic subunit, these data support the hypothesis that TICs in fact have more efficient DNA repair capabilities.
Alternatively, methylation of the O6-methylguanine DNA methyltransferase (MGMT) is the first predictive marker for benefit from alklyating agent therapy in the treatment of glioblastoma [60, 61]. This enzyme reverses alkylation at the O6 position of guanine, thus neutralizing the cytotoxic effect of alkylating agents such as temozolomide (TMZ). In this situation, if the DNA repair gene MGMT is actually methylated and thus not expressed, the patient responds to the therapy. Recent investigation determined that in MGMT methylated glioblastoma, promoter methylation is also highly enriched in GICs [61]. In contrast, another study demonstrated that compared to established glioma cell lines, neurosphere-forming GICs expressed higher levels of MGMT [62]; furthermore, when GBM-initiating cells were transduced with a shRNA to MGMT, the cells could be sensitized to TMZ treatment by decreasing both their ability to undergo DNA repair and efflux of the drug [63]. In the pediatric glioblastoma cell line KNS42, however, it was recently determined that these cells remain resistant to temozolmide treatment despite the absence of MGMT expression [60], thereby demonstrating that an MGMT-independent mechanism is able to regulate GIC survival. Although expression of MGMT leads to temozolmide resistance, regulation of this DNA repair pathway and maintenance of “stem-ness” is much more complex than once originally believed.
Further gene expression analysis determined that expression of HOX genes were significantly increased in a number of resistant cell lines and is associated with shorter survival in pediatric high-grade glioma patients [60]. Although HOX genes are known to play a role in the developing embryo and in the progression of adult glioblastoma, their role in regulating resistance to alkylating agents is unknown. HOX genes have recently been connected to the PcG proteins, such as EZH2 [64]. PcG proteins were originally identified in the fruit fly as repressors of the HOX genes; however, functional EZH2 expression is essential for the maintenance of glioblastoma cancer stem cells [65]. Furthermore, expression of EZH2 directly correlates with the progression of disease and is a member of the “death-from-cancer” signature, which is described in greater detail later in the review [66].
In human metastatic malignant melanoma, it has recently been shown that metastasis is associated with higher expression of DNA repair genes and that these aggressive cells are highly efficient at repairing damage caused by cytotoxic treatment regimes [67–69]. A majority of the repair genes overexpressed were in primary tumors with a poor prognosis; therefore, the authors speculate that primary melanoma cells are capable of undergoing metastasis and replication by using a fast and error-free method. A review published by the same authors stated a relatively new hypothesis similar to the one we are presenting here in relation to TICs: that the overexpression of DNA repair genes is associated with the onset of metastasis [69]. The authors stated that DNA repair is vital for normal life because defects in DNA repair activity are associated with a shorter life span as well as predispositions to cancer and/or aging. However, from a meta-analysis using the Gene Ontology database, they found that DNA repair pathways are overexpressed in a large set of primary tumors associated with a high risk of distant metastasis [61]. With regard to our hypothesis that DNA repair mechanisms are much more efficient in TICs, the meta-analysis performed supports this as there is evidence supporting the idea that TICs are responsible for metastasis.
The role of TICs in connection with enhanced DNA repair has also been demonstrated in other cancer types. In the breast cancer cell line MCF-7, it was recently shown that TICs isolated by mammosphere formation assays have a more active DNA SSBR pathway demonstrated by increased levels of RAD51 foci and expression of the APE1 protein compared to total cells [70]. Although no changes in DSBR were observed, the mammospheres appear to bypass the requirement to phosphorylate H2AX. The authors state that the mammospheres had a reduction in their propensity to undergo senescence due to an increase in telomerase activity and lower levels of p21 expression. Additionally, other groups have reported that activation of AKT and canonical WNT signaling in breast TICs results in an increased efficiency of DNA repair [71, 72].
The most significant data supporting TIC maintenance and DNA repair from the mammary gland are from a study conducted by Zhang et al. in 2008 [73]. The putative TICs isolated from the p53-null mouse mammary glands were Lin-CD29HCD24H, and compared to non-TICs, these expressed higher levels of genes related to the DNA damage response and repair, as well as genes involved in epigenetic regulation of self-renewal [73]. The upregulated genes included Brca1, which supports a recent finding in our laboratory where we have determined that both invasive cells and TICs have increased levels of a larger number of genes related to DNA repair in a pancreatic cancer cell model. Specifically, these DNA repair genes are involved in the BRCA1-mediated DNA repair pathway (Mathews et al., unpublished). Functionally, we observed an increase in the ability of pancreatic TICs to repair DNA after challenge with gemcitabine compared to the total population of cells. In line with our results, a previous investigation with different pancreatic cancer cell lines determined that an increased sensitivity to gemcitabine could be obtained by treating the cells with a CHK1 inhibitor, and this was accomplished by decreasing the RAD51 DNA damage response [74]. The results from this study and ours prompted us to investigate whether other forms of aggressive cancers also demonstrate an increase in expression of the DNA repair genes we identified.
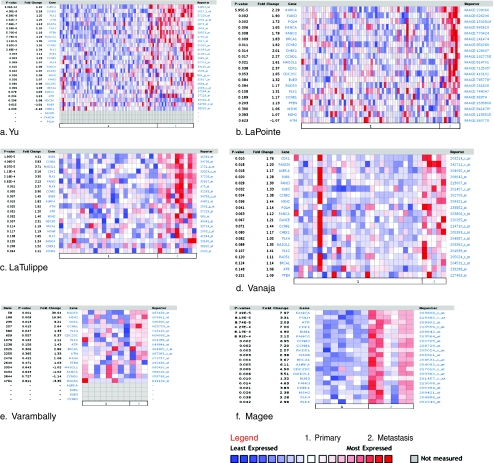
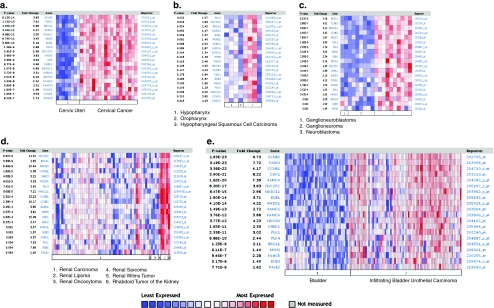
To further investigate the role of DNA repair genes in TICs, we took our gene list from the TICs derived from pancreatic cells and a “death-from-cancer” gene list [75]. This “death-from-cancer” gene list contains a number of DNA repair genes, and we utilized the Oncomine database to determine whether there was a similarity between the DNA repair genes from this list and TICs from pancreatic cells. The death-from-cancer signature contains elevated expression of genes related to DNA repair and cell cycle control in both metastatic breast and prostate cancers. In addition to displaying elevated expression of DNA repair and cell cycle genes, these aggressive cancer cells manifest a stem cell-like expression profile very closely related to a number of genes expressed in TICs from the prostate [58]. Expression of two “death-from-cancer” genes, EZH2 and BMI-1, were also found to be significantly increased in the mouse mammary gland TICs previously mentioned [73]. Thus, we selected DNA repair and cell cycle-related genes from the “death-from-cancer” gene list and observed significantly increased gene expression and/or DNA copy number of these DNA repair genes in aggressive pancreatic cancers [76] (Mathews et al., unpublished), and various cancer types as well (Figs. 2 and 3). We demonstrate a trend of increased expression of DNA repair genes in cancers from the prostate [66, 77–81] (Fig. 2a–f), the cervix [82], head and neck [83], brain [84], the kidney [85], and the bladder [86]. In Fig. 3a, we demonstrate that there are significant increases in DNA repair-related genes in cervical cancer compared to the cervix uteri. Figure 3b compares the expression between hypopharynx, oropharynx, and the more aggressive hypopharygenal squamous cell carcinoma. Figure 3c shows the expression of these genes in ganglioneuroblastoma, ganglioneuroma, and neuroblastoma. Figure 3d focuses on the increase of these genes in the aggressive renal Wilms’ tumors in comparison to renal carcinoma, renal lipoma, renal oncocytoma, renal sarcoma, and rhabdoid tumor of the kidney. Lastly, we show a significant increase of these genes in infiltrating bladder urothelial carcinoma compared to the bladder (Fig. 3e). Figures 2 and 3 demonstrate that there is a common trend of increased gene expression of DNA repair-related genes and cell cycle genes in various carcinomas, specifically cancers associated with aggressiveness. Although this connection between TICs and the aggressive nature of each tumor type listed has not been 100% elucidated, there is overwhelming evidence derived from Oncomine strongly suggesting an increased efficiency in DNA repair in these aggressive samples.
Fig. 2.
Oncomine analysis of DNA repair genes and “death-from-cancer” genes in metastatic prostate tissues compared to primary tumor tissues The heat maps represent raw data from the a number of studies comparing gene expression levels of DNA repair genes in 1 primary prostate tumor tissues to 2 metastatic tissues. The p value represents Student’s t tests comparing primary and metastatic expression. The fold change in expression, the gene name, and the reporter ID from the position on the array are provided as well. Genes demonstrating increases in metastatic tissues are shown in red. Studies were conducted by Yu et al. [66] (a), Lapointe [79] (b), LaTulippe et al. [80] (c), Vanaja et al. [78] (d), Varambally et al. [81] (e), and Magee et al. [77] (f)
Fig. 3.
Oncomine analysis of DNA repair genes and “death-from-cancer” genes in other cancers. The heat maps represent raw data from the a number of studies comparing gene expression levels of DNA repair genes in normal tissue and cancer tissues, as well as more aggressive forms of a number of the cancers shown. The p value represents Student’s t test comparing primary and metastatic expression. The fold change in expression, the gene name, and the reporter ID from the position on the array are provided as well. Genes demonstrating increases in cancerous tissues are shown in red. Studies were conducted by Pyeon et al. [82] in the cervix (a), Schlingemann et al. [83] in the head and neck (b), Albino et al. [84] in the brain (c), Yusenko et al. [85] in the kidney (d), and Sanchez-Carbayo et al. [86] in the bladder (e)
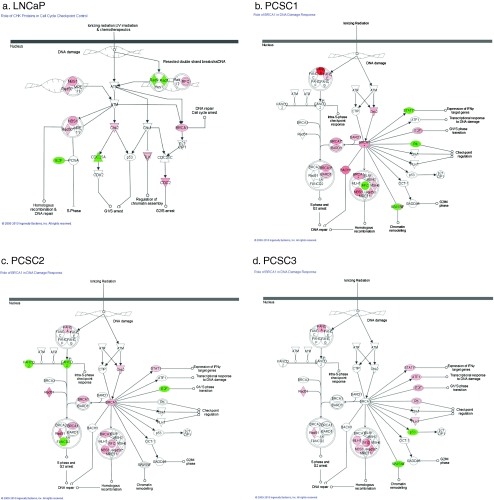
As seen in the data established from the Oncomine datasets, metastatic prostate cancer has a unique signature of increased DNA repair-related genes. We have previously shown that prostatospheres derived from both LNCaP and primary patient cell lines (PCSC1, PCSC2, and PCSC3) are representative of the TIC population and exhibit the ability to initiate tumors in vivo [19]. Upon further analysis of these prostatospheres using Agilent’s whole genome gene expression array and Ingenuity Pathway Analysis (IPA), we compared prostatospheres to the adherent population and discovered that a top function for significantly upregulated genes, at least ≥1.8-fold change, for all cell lines, was DNA replication, recombination, and repair. The molecules involved in these pathways which were significantly upregulated in our prostatospheres are organized by function in Electronic supplementary material Tables 1–4. We further used IPA to analyze the top canonical pathways used by the significantly upregulated genes. In the case of LNCaP prostatospheres and DNA repair mechanisms, IPA reports that the role of CHK proteins in cell cycle checkpoint control is a top canonical pathway (Fig. 4a). Interestingly, for all primary patient cell lines, PCSC1, PCSC2, and PCSC3, a top canonical pathway conserved in prostatospheres compared to the adherent population is the role of BRCA1 in DNA damage response, similar to what was seen in our pancreatic invasive and TIC cells (Fig. 4b, c). Thus, we see a conservation of increased DNA repair mediated by BRCA1 in both pancreatic and prostate TICs; hence, we propose that this be further examined in other cancer models.
Fig. 4.
Ingenuity analysis demonstrating significant changes in DNA replication, recombination, and repair pathways in prostatospheres. Genes highlighted in red demonstrate a significant increase in expression in prostatospheres compared to total cells. a LNCaP prostatospheres show an increase in CHK proteins in cell cycle checkpoint control. In PCSC1 (b), PCSC2 (c), and PCSC3 (d), prostatospheres have an increase in genes demonstrating a role of BRCA1 in DNA damage response
Conclusions
The existence of TICs has been a topic of heated debate for the last few years within the field of cancer biology. The data presented in this review summarize the results found by our lab and others demonstrating that TICs have an increase in their genomic stability. We hypothesize that this increase in DNA repair mechanisms could allow for TIC survival while the bulk population of tumor cells dies in response to treatment. Using the Oncomine database and IPA, we demonstrated that there is a trend in the increased expression of DNA repair-related and cell cycle genes in this specific population. TICs are defined by their ability to self-renew, differentiate, and initiate tumor formation. TICs are also associated with aggressiveness and are believed to be responsible for both metastasis and chemo- and radioresistance. The mechanism(s) by which TICs function to carry out these processes are under intense investigation. However, we speculate that the ability of a TIC to have enhanced and increased DNA repair efficiency is critical in the maintenance of TICs and may function in their ability to resist traditional anticancer treatments. We believe that these data will greatly impact the development of new therapies being designed to eradicate these highly aggressive cancers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 104 kb)
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Lesley A. Mathews and Stephanie M. Cabarcas contributed equally to this work.
References
- 1.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112(13):4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clinical Cancer Research. 2010;16(12):3113–3120. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 3.Sengupta A, Cancelas JA. Cancer stem cells: A stride towards cancer cure? Journal of Cellular Physiology. 2010;225(1):7–14. doi: 10.1002/jcp.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiman JM, Knutson KL, Radisky DC. Immune promotion of epithelial–mesenchymal transition and generation of breast cancer stem cells. Cancer Research. 2010;70(8):3005–3008. doi: 10.1158/0008-5472.CAN-09-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raimondi C, Gianni W, Cortesi E, et al. Cancer stem cells and epithelial–mesenchymal transition: Revisiting minimal residual disease. Current Cancer Drug Targets. 2010;10(5):496–508. doi: 10.2174/156800910791517154. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharyya S, Khanduja KL. New hope in the horizon: Cancer stem cells. Acta Biochimica et Biophysica Sinica (Shanghai) 2010;42(4):237–242. doi: 10.1093/abbs/gmq013. [DOI] [PubMed] [Google Scholar]
- 7.Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graziano A, d'Aquino R, Tirino V, et al. The stem cell hypothesis in head and neck cancer. Journal of Cellular Biochemistry. 2008;103(2):408–412. doi: 10.1002/jcb.21436. [DOI] [PubMed] [Google Scholar]
- 9.Cariati M, Purushotham AD. Stem cells and breast cancer. Histopathology. 2008;52(1):99–107. doi: 10.1111/j.1365-2559.2007.02895.x. [DOI] [PubMed] [Google Scholar]
- 10.Kasper S. Exploring the origins of the normal prostate and prostate cancer stem cell. Stem Cell Reviews. 2008;4(3):193–201. doi: 10.1007/s12015-008-9033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takaishi S, Okumura T, Wang TC. Gastric cancer stem cells. Journal of Clinical Oncology. 2008;26(17):2876–2882. doi: 10.1200/JCO.2007.15.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CJ, Dosch J, Simeone DM. Pancreatic cancer stem cells. Journal of Clinical Oncology. 2008;26(17):2806–2812. doi: 10.1200/JCO.2008.16.6702. [DOI] [PubMed] [Google Scholar]
- 13.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 14.Miki J, Rhim JS. Prostate cell cultures as in vitro models for the study of normal stem cells and cancer stem cells. Prostate Cancer and Prostatic Diseases. 2008;11(1):32–39. doi: 10.1038/sj.pcan.4501018. [DOI] [PubMed] [Google Scholar]
- 15.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes & Development. 2003;17(10):1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurt EM, Kawasaki BT, Klarmann GJ, et al. CD44+CD24(−) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. British Journal of Cancer. 2008;98(4):756–765. doi: 10.1038/sj.bjc.6604242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaviraghi M, Tunici P, Valensin S, et al. Pancreatic cancer spheres are more than just aggregates of stem marker positive cells. Bioscience Reports. 2010;31:45–55. doi: 10.1042/BSR20100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gou S, Liu T, Wang C, et al. Establishment of clonal colony-forming assay for propagation of pancreatic cancer cells with stem cell properties. Pancreas. 2007;34(4):429–435. doi: 10.1097/MPA.0b013e318033f9f4. [DOI] [PubMed] [Google Scholar]
- 19.Duhagon MA, Hurt EM, Sotelo-Silveira JR, et al. Genomic profiling of tumor initiating prostatospheres. BMC Genomics. 2010;11:324. doi: 10.1186/1471-2164-11-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherry MM, Reeves A, Wu JK, et al. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27(10):2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sansone P, Storci G, Tavolari S, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. Journal of Clinical Investigation. 2007;117(12):3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klarmann GJ, Hurt EM, Matthews LA, et al. Invasive prostate cancer cells are tumor initiating cells that have a stem cell-like genomic signature. Clinical & Experimental Metastasis. 2009;26(5):433–436. doi: 10.1007/s10585-009-9242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu SC, Bian XW. Enrichment of cancer stem cells based on heterogeneity of invasiveness. Stem Cell Reviews. 2009;5(1):66–71. doi: 10.1007/s12015-008-9047-8. [DOI] [PubMed] [Google Scholar]
- 24.Moss RA, Lee C. Current and emerging therapies for the treatment of pancreatic cancer. OncoTargets and Therapy. 2010;3:111–127. doi: 10.2147/OTT.S7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ischenko I, Seeliger H, Jauch KW, et al. Metastatic activity and chemotherapy resistance in human pancreatic cancer—Influence of cancer stem cells. Surgery. 2009;146(3):430–434. doi: 10.1016/j.surg.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Shah AN, Summy JM, Zhang J, et al. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Annals of Surgical Oncology. 2007;14(12):3629–3637. doi: 10.1245/s10434-007-9583-5. [DOI] [PubMed] [Google Scholar]
- 27.Thompson LH. Unraveling the Fanconi anemia–DNA repair connection. Nature Genetics. 2005;37(9):921–922. doi: 10.1038/ng0905-921. [DOI] [PubMed] [Google Scholar]
- 28.Thompson LH, Hinz JM, Yamada NA, et al. How Fanconi anemia proteins promote the four Rs: Replication, recombination, repair, and recovery. Environmental and Molecular Mutagenesis. 2005;45(2–3):128–142. doi: 10.1002/em.20109. [DOI] [PubMed] [Google Scholar]
- 29.Ralhan R, Kaur J, Kreienberg R, et al. Links between DNA double strand break repair and breast cancer: Accumulating evidence from both familial and nonfamilial cases. Cancer Letters. 2007;248(1):1–17. doi: 10.1016/j.canlet.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Tichy ED, Stambrook PJ. DNA repair in murine embryonic stem cells and differentiated cells. Experimental Cell Research. 2008;314(9):1929–1936. doi: 10.1016/j.yexcr.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sengupta S, Harris CC. p53: Traffic cop at the crossroads of DNA repair and recombination. Nature Reviews. Molecular Cell Biology. 2005;6(1):44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 32.Kinsella TJ. Coordination of DNA mismatch repair and base excision repair processing of chemotherapy and radiation damage for targeting resistant cancers. Clinical Cancer Research. 2009;15(6):1853–1859. doi: 10.1158/1078-0432.CCR-08-1307. [DOI] [PubMed] [Google Scholar]
- 33.Vaish M. Mismatch repair deficiencies transforming stem cells into cancer stem cells and therapeutic implications. Molecular Cancer. 2007;6:26. doi: 10.1186/1476-4598-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalhus B, Laerdahl JK, Backe PH, et al. DNA base repair—Recognition and initiation of catalysis. FEMS Microbiology Reviews. 2009;33(6):1044–1078. doi: 10.1111/j.1574-6976.2009.00188.x. [DOI] [PubMed] [Google Scholar]
- 35.Munroe RJ, Bergstrom RA, Zheng QY, et al. Mouse mutants from chemically mutagenized embryonic stem cells. Nature Genetics. 2000;24(3):318–321. doi: 10.1038/73563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas JW, LaMantia C, Magnuson T. X-ray-induced mutations in mouse embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(3):1114–1119. doi: 10.1073/pnas.95.3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savatier P, Lapillonne H, Jirmanova L, et al. Analysis of the cell cycle in mouse embryonic stem cells. Methods Mol Biol. 2002;185:27–33. doi: 10.1385/1-59259-241-4:27. [DOI] [PubMed] [Google Scholar]
- 38.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87(4):757–766. doi: 10.1016/S0092-8674(00)81394-X. [DOI] [PubMed] [Google Scholar]
- 39.Adams BR, Golding SE, Rao RR, et al. Dynamic dependence on ATR and ATM for double-strand break repair in human embryonic stem cells and neural descendants. PLoS ONE. 2010;5(4):e10001. doi: 10.1371/journal.pone.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams BR, Hawkins AJ, Povirk LF, et al. ATM-independent, high-fidelity nonhomologous end joining predominates in human embryonic stem cells. Aging (Albany NY) 2010;2(9):582–596. doi: 10.18632/aging.100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harfouche G, Martin MT. Response of normal stem cells to ionizing radiation: A balance between homeostasis and genomic stability. Mutation Research. 2010;704(1–3):167–174. doi: 10.1016/j.mrrev.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 42.Roos WP, Christmann M, Fraser St, et al. Mouse embryonic stem cells are hypersensitive to apoptosis triggered by the DNA damage O(6)-methylguanine due to high E2F1 regulated mismatch repair. Cell Death and Differentiation. 2007;14(8):1422–1432. doi: 10.1038/sj.cdd.4402136. [DOI] [PubMed] [Google Scholar]
- 43.Van Sloun PP, Jansen JG, Weeda G, et al. The role of nucleotide excision repair in protecting embryonic stem cells from genotoxic effects of UV-induced DNA damage. Nucleic Acids Research. 1999;27(16):3276–3282. doi: 10.1093/nar/27.16.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nouspikel T. DNA repair in differentiated cells: Some new answers to old questions. Neuroscience. 2007;145(4):1213–1221. doi: 10.1016/j.neuroscience.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 45.Mohrin M, Bourke E, Alexander D, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7(2):174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sotiropoulou PA, Candi A, Mascré G, et al. Bcl-2 and accelerated DNA repair mediates resistance of hair follicle bulge stem cells to DNA-damage-induced cell death. Nat Cell Biol. 2010;12(6):572–82. doi: 10.1038/ncb2059. [DOI] [PubMed] [Google Scholar]
- 47.Harfouche G, Vaigot P, Rachidi W, et al. Fibroblast growth factor type 2 signaling is critical for DNA repair in human keratinocyte stem cells. Stem Cells. 2010;28(9):1639–1648. doi: 10.1002/stem.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frosina G. The bright and the dark sides of DNA repair in stem cells. Journal of Biomedicine and Biotechnology. 2010;2010:845396. doi: 10.1155/2010/845396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frosina G. DNA repair and resistance of gliomas to chemotherapy and radiotherapy. Molecular Cancer Research. 2009;7(7):989–999. doi: 10.1158/1541-7786.MCR-09-0030. [DOI] [PubMed] [Google Scholar]
- 50.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 51.Ropolo M, Daga A, Griffero F, et al. Comparative analysis of DNA repair in stem and nonstem glioma cell cultures. Molecular Cancer Research. 2009;7(3):383–392. doi: 10.1158/1541-7786.MCR-08-0409. [DOI] [PubMed] [Google Scholar]
- 52.Bao S, Wu Q, Sathornsumetee S, et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Research. 2006;66(16):7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 53.McCord AM, Jamal M, Williams ES, et al. CD133+ glioblastoma stem-like cells are radiosensitive with a defective DNA damage response compared with established cell lines. Clinical Cancer Research. 2009;15(16):5145–5153. doi: 10.1158/1078-0432.CCR-09-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dirks PB. Cancer: Stem cells and brain tumours. Nature. 2006;444(7120):687–688. doi: 10.1038/444687a. [DOI] [PubMed] [Google Scholar]
- 55.Frosina G. DNA repair in normal and cancer stem cells, with special reference to the central nervous system. Current Medicinal Chemistry. 2009;16(7):854–866. doi: 10.2174/092986709787549253. [DOI] [PubMed] [Google Scholar]
- 56.Zabludoff SD, Deng C, Grondine MR, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Molecular Cancer Therapeutics. 2008;7(9):2955–2966. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 57.Facchino S, Abdouh M, Chatoo W, et al. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. The Journal of Neuroscience. 2010;30(30):10096–10111. doi: 10.1523/JNEUROSCI.1634-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crea, F., M.A. Duhagon Serrat, E.M. Hurt, et al. (2010). BMI1 silencing enhances docetaxel activity and impairs antioxidant response in prostate cancer. Int J Cancer. doi:10.1002/ijc.25522 [DOI] [PMC free article] [PubMed]
- 59.Zhuang, W., B. Li, L. Long, et al. (2011). Knockdown of the DNA-dependent protein kinase catalytic subunit radiosensitizes glioma-initiating cells by inducing autophagy. Brain Res, 1371, 7–15. [DOI] [PubMed]
- 60.Gaspar N, Marshall L, Perryman L, et al. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer Research. 2010;70(22):9243–9252. doi: 10.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sciuscio, D., A.C. Diserens, K. van Dommelen, et al. (2011). Extent and Patterns of MGMT Promoter Methylation in Glioblastoma and Respective Derived Spheres. Clin Cancer Res, 17(2), 255–266. [DOI] [PubMed]
- 62.Yuki K, Natsume A, Yokoyama H, et al. Induction of oligodendrogenesis in glioblastoma-initiating cells by IFN-mediated activation of STAT3 signaling. Cancer Letters. 2009;284(1):71–79. doi: 10.1016/j.canlet.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 63.Kato TN, Natsume A, Toda H, et al. Efficient delivery of liposome-mediated MGMT-siRNA reinforces the cytotoxity of temozolomide in GBM-initiating cells. Gene Therapy. 2010;17(11):1363–1371. doi: 10.1038/gt.2010.88. [DOI] [PubMed] [Google Scholar]
- 64.Korkaya H, Paulson A, Charafe-Jauffret E, et al. Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biology. 2009;7(6):e1000121. doi: 10.1371/journal.pbio.1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suva ML, Riggi N, Janiszewska M, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Research. 2009;69(24):9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 66.Yu J, Yu J, Rhodes DR, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Research. 2007;67(22):10657–10663. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- 67.Sarasin A, Dessen P. DNA repair pathways and human metastatic malignant melanoma. Current Molecular Medicine. 2010;10(4):413–418. doi: 10.2174/156652410791317011. [DOI] [PubMed] [Google Scholar]
- 68.Kauffmann A, Rosselli F, Lazar V, et al. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27(5):565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- 69.Sarasin A, Kauffmann A. Overexpression of DNA repair genes is associated with metastasis: A new hypothesis. Mutation Research. 2008;659(1–2):49–55. doi: 10.1016/j.mrrev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 70.Karimi-Busheri F, Rasouli-Nia A, Mackey JR, et al. Senescence evasion by MCF-7 human breast tumor-initiating cells. Breast Cancer Research. 2010;12(3):R31. doi: 10.1186/bcr2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woodward WA, Bristow RG. Radiosensitivity of cancer-initiating cells and normal stem cells (or what the Heisenberg uncertainly principle has to do with biology) Seminars in Radiation Oncology. 2009;19(2):87–95. doi: 10.1016/j.semradonc.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang M, Atkinson RL, Rosen JM. Selective targeting of radiation-resistant tumor-initiating cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(8):3522–3527. doi: 10.1073/pnas.0910179107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang M, Behbod F, Atkinson RL, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Research. 2008;68(12):4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parsels LA, Morgan MA, Tanska DM, et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Molecular Cancer Therapeutics. 2009;8(1):45–54. doi: 10.1158/1535-7163.MCT-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glinsky GV. Genomic models of metastatic cancer: Functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle. 2006;5(11):1208–1216. doi: 10.4161/cc.5.11.2796. [DOI] [PubMed] [Google Scholar]
- 76.Harada T, Chelala C, Bahkta V, et al. Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene. 2008;27(13):1951–1960. doi: 10.1038/sj.onc.1210832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Magee JA, Araki T, Patil S, et al. Expression profiling reveals hepsin overexpression in prostate cancer. Cancer Research. 2001;61(15):5692–5696. [PubMed] [Google Scholar]
- 78.Vanaja DK, Cheville JC, Iturria SJ, et al. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Research. 2003;63(14):3877–3882. [PubMed] [Google Scholar]
- 79.Lapointe J, Li C, Higgins JP, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(3):811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.LaTulippe E, Satagopan J, Smith A, et al. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Research. 2002;62(15):4499–4506. [PubMed] [Google Scholar]
- 81.Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8(5):393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 82.Pyeon D, Newton MA, Lambert PF, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Research. 2007;67(10):4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schlingemann J, Habtemichael N, Ittrich C, et al. Patient-based cross-platform comparison of oligonucleotide microarray expression profiles. Lab Invest. 2005;85(8):1024–1039. doi: 10.1038/labinvest.3700293. [DOI] [PubMed] [Google Scholar]
- 84.Albino D, Scaruffi P, Moretti S, et al. Identification of low intratumoral gene expression heterogeneity in neuroblastic tumors by genome-wide expression analysis and game theory. Cancer. 2008;113(6):1412–1422. doi: 10.1002/cncr.23720. [DOI] [PubMed] [Google Scholar]
- 85.Yusenko MV, Kuiper RP, Boethe T, et al. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer. 2009;9:152. doi: 10.1186/1471-2407-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sanchez-Carbayo M, Socci ND, Lozano J, et al. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. Journal of Clinical Oncology. 2006;24(5):778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 104 kb)