Abstract
LIMD1 is a tumour suppressor gene (TSG) down regulated in ∼80% of lung cancers with loss also demonstrated in breast and head and neck squamous cell carcinomas. LIMD1 is also a candidate TSG in childhood acute lymphoblastic leukaemia. Mechanistically, LIMD1 interacts with pRB, repressing E2F-driven transcription as well as being a critical component of microRNA-mediated gene silencing. In this study we show a CpG island within the LIMD1 promoter contains a conserved binding motif for the transcription factor PU.1. Mutation of the PU.1 consensus reduced promoter driven transcription by 90%. ChIP and EMSA analysis demonstrated that PU.1 specifically binds to the LIMD1 promoter. siRNA depletion of PU.1 significantly reduced endogenous LIMD1 expression, demonstrating that PU.1 is a major transcriptional activator of LIMD1.
Keywords: LIMD1, PU.1, Leukaemia, MicroRNA
1. Introduction
Lim domains containing protein 1 (LIMD1) is a bona fide tumour suppressor gene (TSG) encoded at chromosome 3p21.3, a region that commonly undergoes homozygous deletion, loss of heterozygosity and epigenetic silencing in many carcinomas, the most studied being lung [1–3]. Experimentally the ability of the A549 lung cancer cell line to form lung metastases in a mouse model is significantly reduced upon stable expression of LIMD1 [4]. Limd1−/− mice are predisposed to chemical-induced lung adenocarcinomas and genetic inactivation of Limd1 in mice heterozygous for oncogenic K-Ras (G12D) confers markedly increased tumour initiation, promotion, and mortality [5]. In corroboration with the mouse model, LIMD1 protein expression is reduced in 80% of lung squamous cell and adeno-carcinomas [5], and in 50% of head and neck squamous cell carcinomas (HNSCC) [6]. In childhood acute lymphoblastic leukaemias 20% of samples have chromosomal deletions at the 3p21.3 locus with LIMD1 loss flagged as a possible cause of tumour formation [7]. A reduction in LIMD1 expression is also correlated with poor prognosis and survival rates in breast cancer [8].
In the nucleus LIMD1 binds to the Retinoblastoma protein (pRb) and acts as a co-repressor of E2F-driven transcription [4]. More recently LIMD1 has been shown to be a critical effector protein of the miRNA-mediated gene silencing pathway. LIMD1 interacts simultaneously with eIF4E and core proteins of the microRNA induced silencing complex (miRISC) such as Ago1/2 in what is proposed to be a miRNA-induced inhibitory mRNA closed loop complex which may precede mRNA deadenylation and subsequent degradation [9]. It has been shown that several components of the miRNA pathway, including Ago2, TRBP and Dicer, are deleted or mutated in cancers [10–13] and LIMD1 loss may also ablate the tumour suppressive effects of this pathway.
Despite the validation of LIMD1 as a bona fide TSG, the processes controlling LIMD1 gene expression remain to be fully elucidated. Loss of heterozygosity, gene deletion and promoter methylation have been shown to cause decreased LIMD1 expression [5,14]. However currently there is no data on the transcriptional control of LIMD1. Preliminary characterisation of the LIMD1 promoter region identified a CpG island within which a 21 bp region was critical for transcription [5]. However, the identity of the controlling transcription factor(s) and the possibility of additional positive or negative regulatory elements within the entire CpG have not been examined.
PU.1, also referred to as spleen focus forming virus proviral integration protein (Spi1), was first identified as a putative oncogene in murine erythroleukaemias [15]. It is a member of the Ets family of transcription factors, of which there are 28 human members targeting over 200 genes including those involved in apoptosis, differentiation, transformation and development [16,17]. Constitutive PU.1 expression is essential for viability of haematopoietic stem cells (HSCs) [18], with a subsequent reduction in expression causing differentiation into megakaryocyte, B and T cell phenotypes and increased expression causing differentiation into macrophages [19]. A complete loss of PU.1 expression is indicative of committing pre-T cell to T-cell differentiation [20]. Pathologically PU.1 loss is associated with acute myeloid leukaemias with as little as a 20% reduction in expression resulting in an increase in pre-leukaemic haematopoietic cell number [21]. Reduced PU.1 expression and PU.1 dependent terminal differentiation markers are found in alveolar macrophages of patients suffering from pulmonary alveolar proteinosis (PAP) [22].
Herein we show that the LIMD1 promoter contains both positive and negative regulatory elements. Specifically, we identify a conserved binding motif for the Ets family transcription factor PU.1 and demonstrate that PU.1 specifically associates with the LIMD1 promoter at this binding motif. Mutations within the motif disrupt PU.1 binding and transcriptional activity, and in vivo depletion of endogenous PU.1 causes loss of LIMD1 protein expression. The implications of our findings with respect to LIMD1 regulation in haematopoietic derived malignancies are discussed.
2. Materials and methods
2.1. Promoter mapping analysis
As a point of reference the unconfirmed transcriptional start site (TSS) for LIMD1 was assigned according to the NCBI reference sequence NM_014240.2 (nucleotide 45636323 on the primary chromosome 3 ref assembly NC_000003.11), which is 49 bp upstream from the AUG (45636372). The LIMD1 promoter along with a series of ten 18–35 bp internal deletions (IΔ1–10) corresponding to internal regions 1–10 (IR1–10) (Supplementary data Fig. 1) were previously cloned as described [5] into a pGL4.10 [luc2] (Promega) firefly luciferase reporter vector. U2OS cells were co-transfected with 50 ng of promoter reporter and 5 ng of a Renilla luciferase vector (for normalisation). Luciferase activity was assayed using the Dual-Luciferase® Reporter Assay System (E1960, Promega) 24 h post-transfection.
2.2. Bioinformatic analysis
The LIMD1 promoter, encompassing 1990 bp upstream of the TSS, was scrutinised for putative transcription factor binding sites using MatInspector (http://www.genomatix.de/products/MatInspector/) with the Matrix Family Library Version 8.1 (Supplementary Table 1). The default thresholds were utilised, with a perfect and good matrix match scoring 1.00 and >0.80, respectively. For promoter homology/identity comparisons, genomic DNA sequences from mammalians that express PU.1 as identified using HomoloGene (http://www.ncbi.nlm.nih.gov/homologene/) were obtained using the Ensembl Genome Browser (www.ensembl.org) and aligned using ClustalW (www.ebi.ac.uk/clustalw) and BioEdit Sequence Alignment Editor©.
2.3. Point mutagenesis of IR5
The core putative PU.1 consensus binding sequence 5′-TTCC of the sense strand (5′-GGAA of the anti-sense strand) within IR5, which maps to −673 relative to the TSS was mutated from 5′-TTCC to 5′-TTTT using site directed mutagenesis (QuikChange XL Site-Directed Mutagenesis Kit, Stratagene #200517). Primers used were 5′ GCCTGGCGCACTCCTTTTGCGTCCCGCCGCCCTCCGG (forward) and 5′ CCGGAGGGCGGCGGGACGCAAAAGTGAGTGCGCCAGGC (reverse). As a control another C to T point mutation was introduced further downstream at −660 to the TSS, but still within the same internal region using 5′ GCCTGGCGCACTCACTTCCGCGTCCCGCTGCCCTCCGG (forward) and 5′ CCGGAGGGCAGCGGGACGCGGAAGTGAGTGCGCCAGGC (reverse). Both mutated plasmids were then reverse mutated back to the wild type plasmid as controls to ensure no other background mutations had been introduced through PCR.
2.4. siRNA targeted depletion and qRT-PCR analysis
siRNA targeted against PU.1, LIMD1, Ets-1 or a scrambled control sequence was electroporated into the human leukaemic monocytic lymphoma U937 cell line using the Amaxa Cell Line Nucleofector Kit V (Lonza VCA-1003) on a Nucleofector II electroporator (Lonza) utilising the U937 cell specific programme. siRNA targeted against LIMD1, PU.1, Elk-1 or a control scrambled sequence were transfected into U2OS using INTERFERin™ (Polyplus-transfection SA, Illkirch, France) as per manufacturer’s protocol. Gene knockdown was assayed 48 h post-transfection. RNA and protein were simultaneously extracted from U937 cells using an RNA/Protein Purification kit (Norgen Biotek Corp. 23000). RNA was extracted from U2OS using RNAqueous Micro Kit (Ambion AM1931). Protein knockdown was assayed by Western blot with rabbit monoclonal anti-PU.1 (Cell Signalling #2266) (for U937 cell knockdown only as the antibody could not detect the very low levels of endogenous PU.1 in U2OS cells), mouse monoclonal anti-LIMD1 [4], mouse monoclonal anti-Ets-1 (Transduction Laboratories E34620), rabbit polyclonal anti-Elk-1 (Santa Cruz SC-22804) and anti-β-actin (Sigma–Aldrich A3853) as a loading control. mRNA knockdown was quantified by qRT-PCR. cDNA was synthesised (Transcriptor High Fidelity cDNA Synthesis Kit, Roche Applied Science) and qRT-PCR performed using the primers PU.1 5′ CAGGGGATCTGACCGACTC (forward) and 5′ GCACCAGGTCTTCTGATGG (reverse) and the β-Tubulin housekeeper 5′ ATACCTTGAGGCGAGCAAAA (forward) and 5′ CTGATCACCTCCCAGAACTTG (reverse).
2.5. PU.1 subcloning
Human PU.1 cDNA was kindly provided by A. Rizzino (University of Nebraska Medical Centre). The cDNA was subcloned with the addition of 6N-terminal amino acids into a pcDNA4/HisMax TOPO vector (Invitrogen K86420) (further referred to as pcDNA4) using the primers 5′ CGCGAATTCCAGATGTTACAGGCGTGCAAAATGGAAGGGTTTCCCCTCGTCCCCCCTCCATC (forward) and 5′ GCGGGATCCTCAGTGGGGCGGGTGGCGCCGCTCGGCCAGGCCCCCGCGGCCCAGCACTTCGC (reverse). HA tagged PU.1 was generated by cut and pasting PU.1 cDNA from pcDNA4-PU.1 into a pCMV5-HA vector utilising the incorporated EcoR1 and BamH1 restriction sites.
2.6. Chromatin immunoprecipitation
Endogenous expression of PU.1 and LIMD1 in U937 cells was confirmed by Western blot using anti-PU.1 (Cell Signalling #2266) and anti-LIMD1 [4]. 8 × 106 U937 cells were serum starved overnight prior to 30 min stimulation with 20% FCS supplemented RPMI. Cells were fixed (1% paraformaldehyde, 10 min, 37 °C) and quenched with 2 × 10 ml 0.125 M glycine/PBS washes. Cells were resuspended in 1 ml harvesting buffer (0.125 M glycine, 1 mM EDTA, 1 mM PMSF in PBS), pelleted (3000 rpm, 4 °C, 10 min) and lysed in 100 μl lysis buffer (50 mM Tris–HCl, pH 8.0, 1% SDS, 10 mM EDTA). DNA was sheared to 200–600 bp by sonication then centrifuged (13 000 rpm, 4 °C, 10 min). Lysates were diluted 10-fold with dilution buffer (20 mM Tris–HCl, pH 8.0, 1% Triton-X-100, 2 mM EDTA, 150 mM NaCl) and pre-cleared for 1 h with IgG/Protein G before addition to 40 μl ExactaCruz™ E IP Matrix (Santa Cruz 45042) with either 10 μl anti-PU.1 (Cell Signalling #2266) or equalised isotype control antibody conjugated. After an overnight incubation IP beads were washed 4× for 10 min with wash buffer (10% lysis buffer/dilution buffer), 2× 10 min TE and eluted in 2× 75 μl elution buffer (1% SDS, 0.1 M NaHCO3). Crosslinks were reversed (0.2 M NaCl, 65 °C, 6 h), proteinase K treated and DNA purified (QIAquick PCR Purification Kit, Qiagen 28104). PCR was performed using the CD11b promoter as a positive control for PU.1 binding as previously described [23] and LIMD1 specific primers 5′ GCAGCAGGGACTGCGCCTGGCG (forward) and 5′ GGGGCTGGCGGCCCATTGTCCG (reverse), to amplify −751 to −650 of the promoter. To show specificity for the IΔ5 region a set of primers upstream (−1779 to −1630) of IΔ5 were included.
2.7. Production of recombinant Elk-1 and PU.1 Ets domain and nuclear extracts
The DNA-binding Ets domains of Elk-1 and PU.1 were subcloned into the pQE60 background, generating proteins that contained both His and HA epitope tags. These plasmids were expressed in the SG13009 strain of Escherichia coli and the resulting recombinant protein purified on nickel agarose beads (Qiagen). A total of 25 ng of recombinant Ets protein was used in subsequent electrophoretic mobility shift assays (EMSA). Nuclear extracts were prepared from HEK 293T cells either untransfected or transfected with HA-PU.1 or HA-Elk-1 using hypotonic and hypertonic lysis and flash frozen in liquid N2 as previously described [24].
2.8. EMSAs
Oligonucleotides representing the wild type IR5 sequence (WT: 5′ 32P-CTCACTTCCGCGTCCCGCCGC (forward) and 5′ GCGGCGGGACGCGGAAGTGAG (reverse) or point mutated (MT) IR5 sequence (MT1: 5′ CTCACTTTTGCGTCCCGCCGC (forward) and 5′ GCGGCGGGACGCAAAAGTGAG (reverse); MT2: 5′ CTCACGGCCGCGTCCCGCCGC (forward) and 5′ GCGGCGGGACGCGGCCGTGAG (reverse) were annealed. A mix of 4× binding buffer (140 mM KCl, 18 mM MgCl2, 12 mM spermidine), Poly dI/dC (26 μg/ml final concentration), sheared Herring sperm (100 μg/ml final concentration) and nuclear extract/recombinant protein was made up. The volume was made up with incubation buffer (10 mM HEPES, pH 7.8, 5% glycerol, 50 mM KCl, 1 mM EDTA, 1 mM DTT). Antibody or cold probe (either wt or containing point mutations) was added and the reaction incubated at room temperature for 1 h. Labelled probe was added and reactions incubated for a further 10 min at room temperature before loading onto a 5% polyacrylamide gel (0.5 × TBE). Gels were dried and developed using a Fuji-film LAS-3000 phosphor-imager.
3. Results
3.1. The CpG Island within the LIMD1 promoter contains both positive and negative regulatory elements
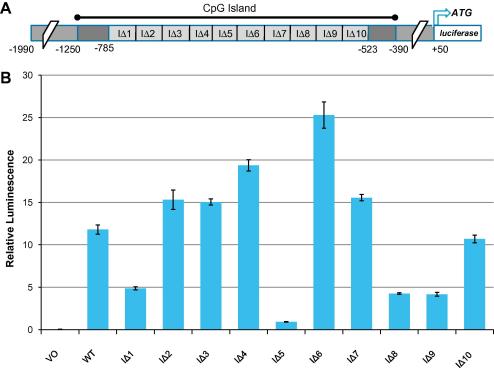
A series of 10 consecutive deletions within the CpG Island of the LIMD1 gene were created in order to identify positive or negative regulatory elements (Fig. 1A). The IΔ5 mutant (−678 to −658 relative to the transcriptional start site) reduced levels of promoter transcription by 90% (Fig. 1B), significantly greater than the IΔ1, 8 and 9 mutants which all exhibited a 65% reduction, implicating these regions of the promoter as containing positive regulatory elements (Fig. 1B). Conversely, IΔ4 and IΔ6 gave 50% and 100% increases in transcription, respectively, implicating these regions as negative regulatory elements (Fig. 1B). IΔ2, 3 and 7 showed approximately 25% increases in transcription and IΔ10 had no significant change in transcriptional activity (Fig. 1B). Comparable results were also obtained in the A549 lung cancer cell line with the IΔ5mutant (Supplementary data Fig. 2), however the decrease in transcriptional activity was not as great when compared to U2OS cells (60% compared to 90%) implying that tissue-specific factors may associate with this promoter region.
Fig. 1.
Identification of a transcriptionally crucial 21 bp region of the LIMD1 promoter by mutational analysis. (A) Schematic diagram of the LIMD1 promoter cloned into pGL4 firefly luciferase vector with the 10 internal deletions (IΔ) and previously identified CpG Island indicated. (B) U2OS cells were transfected with the internal promoter deletions as indicated, lysed 24 h post-transfection and firefly luciferase activity assayed with values normalised to renilla luciferase.
3.2. An Ets family consensus sequence is conserved between mammals within the LIMD1 promoter
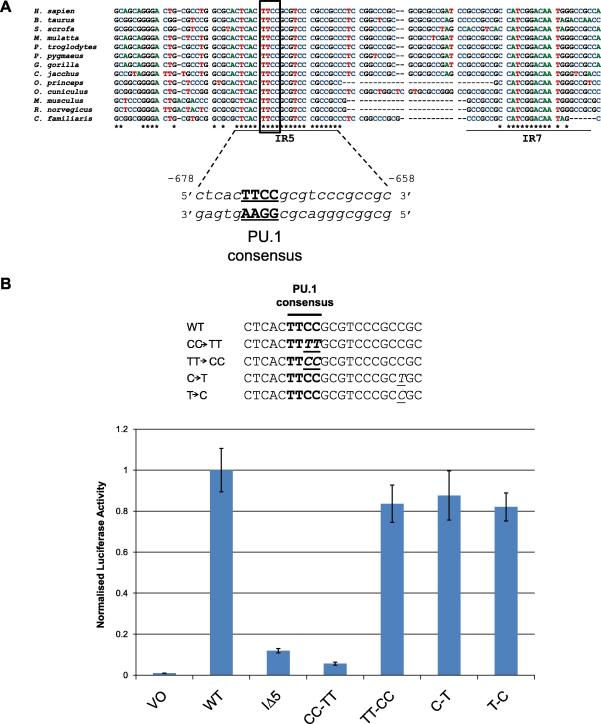
To determine why the internal region 5 (IR5) was critical for transcription, LIMD1 promoter sequences from different mammalians were scrutinised and found to be perfectly conserved across 13 species (Fig. 2A). The 3′ downstream IR7 was also conserved (Fig. 2A), however as its deletion did not cause a significant loss in transcription, no further investigations were performed on this region. To identify possible transcription factor binding sites within the critical IR5 of the LIMD1 promoter, the sequence was scrutinised using MatInspector. This analysis identified a consensus sequence for the Ets domain containing family member PU.1, with a Matrix score of 0.989 (1.0 is a perfect match and 0.8 is a good match), anchoring at position −673 to −670 relative to the TSS of the anti-sense strand 5′-GCGGAAGTG (Fig. 2A). No other potential transcription factor binding sites were identified within the core IR5 region.
Fig. 2.
IR5 contains a conserved PU.1 consensus that is required for transcription from the LIMD1 promoter. (A) LIMD1 promoter sequences from different LIMD1 expressing species was extracted from the Ensembl genome browser and aligned using ClustalW. Perfect homology is observed at the IR5 and within the IR7 consensus. Further scrutinisation of IR5 using MatInspector identified a putative PU.1 binding consensus sequence. (B) Point mutagenesis of the PU.1 binding consensus sequence compared to the WT consensus are shown (CC-TT and C-T). In addition we performed reverse mutagenesis to restore the PU.1 consensus (TT-CC) and reverse single conserved mutation outside of this consensus (T-C). All mutated promoter reporters together with WT and IΔ5 LIMD1 promoters were analysed in reporter assays as in Fig. 1B. Bold – PU.1 consensus; italics and underlined – mutated bases.
3.3. Mutation within the Ets binding domain consensus reduces transcription by 90%
To assess the requirement of the putative Ets family transcription factor consensus within IR5 for transcriptional control, the wild type LIMD1 promoter-luciferase construct was mutated within the binding consensus from CC to TT (Fig. 2B). This 2 base mutation resulted in a 90% decrease in promoter expression, similar to levels obtained with the 21 bp IΔ5 deletion (Fig. 2B). Reverse mutagenesis back to wild type sequence restored transcriptional activity to levels approaching that of wild type (Fig. 2B). As a control for consensus specificity, mutation of another cytosine within the IR5, but not part of the consensus motif, did not affect transcription. One way ANOVA statistical analysis confirmed values were not significantly different to that of WT (MS = 2.777779, F = 2.753897 and P-value = 0.088575).
3.4. ChIP analysis of IR5 within the LIMD1 promoter
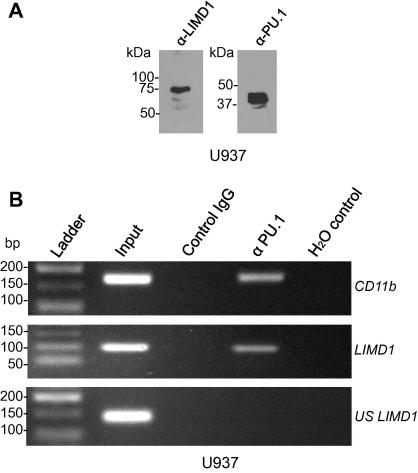
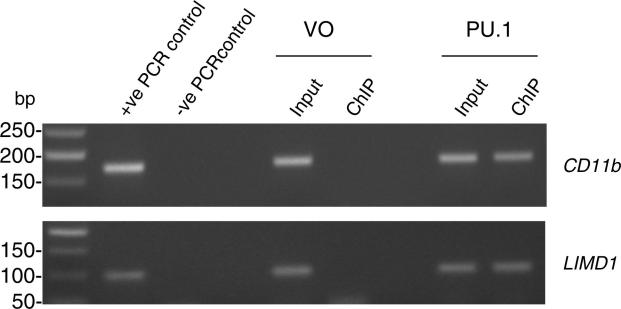
PU.1 is a well established transcription factor within the haematopoietic lineage, and so in order to establish the physiological role that PU.1 may have as a regulator of LIMD1 expression, we performed endogenous ChIP analyses using the U937 histiocytic lymphoma cell line. Endogenous expression of LIMD1 and PU.1 in this cell line was firstly confirmed by Western blot (Fig. 3A). In subsequent chromatin immunoprecipitation experiments, PU.1 co-immunoprecipitated the DNA region specifically spanning −751 to −650 of the LIMD1 promoter that includes IR5 (Fig. 3B). Primers were also included to amplify DNA further upstream of IR5 (but still within the promoter) however no amplicon was detected indicating the ChIP assay was specific for the PU.1 consensus containing region (Fig. 3B). As a positive control the primer set for the CD11b promoter (previously established to bind PU.1) was also included (Fig. 3B) [23,25]. Identical results with the LIMD1 and CD11b DNA elements were obtained using exogenously transfected pcDNA4 PU.1 in HEK 293T cells (Supplementary data Fig. 3).
Fig. 3.
ChIP analysis of PU.1 binding to the LIMD1 promoter. (A) Western blot analysis of U937 cell extracts for endogenous LIMD1 and PU.1 proteins as indicated. (B) Endogenous ChIP was performed with anti-PU.1 specific antibody (α-PU.1) or isotype IgG (control) using U937 nuclear extracts. Immunoprecipitated DNA was analysed via PCR utilising primers for the CD11b promoter (as a positive control), the LIMD1 promoter encompassing IR5, or primers targeted to a region within the LIMD1 promoter but upstream of IR5 (US LIMD1) as a negative control for binding. Water only was used for PCR control for each primer set (PCR control).
3.5. PU.1 and the PU.1 Ets domain alone bind to the IR5 DNA consensus in vitro
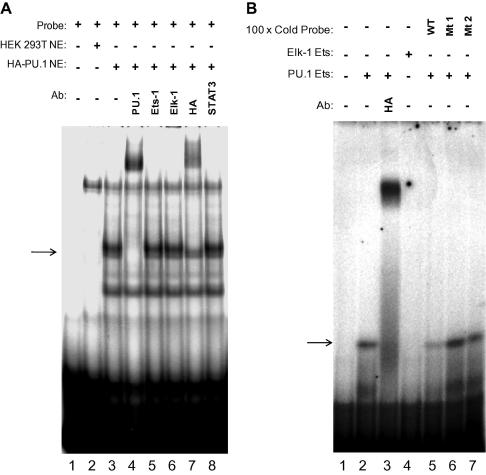
To support our findings from ChIP analyses we next performed electrophoretic mobility shift assays with nuclear extracts containing exogenous HA-tagged PU.1 and a 32P-labelled IR5 DNA consensus probe (Fig. 4A). A supershift was seen with both the αPU.1 and αHA antibodies but not with three control antibodies (αEts-1, Elk-1 and STAT3) (Fig. 4A). We then examined if the Ets domain of PU.1 alone could bind specifically to the IR5 sequence. Using recombinant HA-tagged PU.1 Ets domain in an EMSA assay we observed a supershift with an αHA antibody (Fig. 4B). The recombinant HA-tagged Ets domain of Elk-1 did not bind to this sequence (Fig. 4B, lane 4). Furthermore, in an EMSA assay with cold competitor DNA consensus probe, only the wild type consensus was able to compete, whereas two different PU.1 consensus point mutants (Mt1/Mt2) were unable to compete out labelled wild-type probe (Fig. 4B, lanes 5–7).
Fig. 4.
EMSA supershift analysis of the IR5 with PU.1 and competition with mutant oligos. (A) Nuclear extracts from untransfected HEK 293T cells (lane 2) or cells transfected with a vector for HA-PU.1 (lanes 3–8) were incubated with radiolabelled IR5 DNA alone or in the presence of antibodies against PU.1, Ets-1, Elk-1, the HA epitope and STAT3, as indicated. PU.1 complexes are indicated with an arrow. The partial supershift with the HA antibody likely reflects a degree of translation initiation at an internal AUG. (B) A recombinant HA-tagged Ets domain of PU.1 was incubated with IR5 DNA alone (lane 2), in the presence of anti-HA antibody (lane 3) or 100-fold excess of unlabelled wt or mutant IR5 DNA (lanes 5–7). In parallel, a recombinant HA-tagged Ets domain of Elk-1 did not form a complex with IR5 DNA (lane 4).
3.6. PU.1 siRNA targeted depletion results in loss of LIMD1 expression
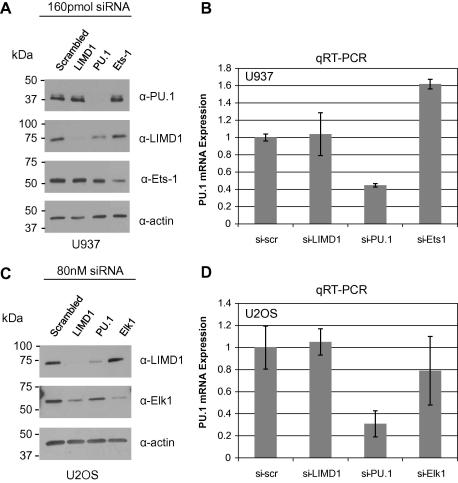
We next reasoned that if PU.1 was a major positive transcriptional activator of LIMD1 then depletion of PU.1 should result in down-regulation of LIMD1 mRNA and therefore protein levels. For continuity with the endogenous ChIP assay performed (Fig. 3B), we depleted endogenous PU.1 in U937 cells. siRNA depletion of endogenous PU.1 (Fig. 5A) significantly reduced LIMD1 protein levels. siRNA targeted depletion of (the Ets family member) Ets-1 was also performed. Depletion of Ets-1 did not alter LIMD1or PU.1 protein or PU.1 mRNA levels (Fig. 5A and B). As a control for specificity of siRNA depletion of PU.1, siRNA targeted against PU.1 did not affect Ets-1 protein levels (Fig. 5A), demonstrating that reduced LIMD1 expression was specific for PU.1 depletion.
Fig. 5.
PU.1 is required for the expression of LIMD1. (A) Western blot analysis of siRNA targeted depletion of PU.1 in U937 cells. Control scrambled LIMD1, PU.1 and Ets-1 targeting siRNA are indicated. (B) PU.1 mRNA expression was quantified by qRT-PCR. (C) Western blot analysis of siRNA targeted depletion of PU.1 in U2OS cells. Control scrambled, LIMD1, PU.1 and Elk-1 targeting siRNA are indicated (D) qRT-PCR was used to assay PU.1 knockdown as PU.1 levels in U2OS were not detectable by Western blot.
We also performed PU.1 knockdown in U2OS where levels were only detectable by qRT-PCR (Fig. 5D). As with U937 cells siRNA targeted depletion of PU.1 resulted in significantly reduced LIMD1 protein levels, as detected by Western blot (Fig. 5C). siRNA depletion of Elk-1 did not affect LIMD1 protein levels or cause a significant change in PU.1 mRNA levels. Of note is a decrease in the Elk-1 protein level upon knockdown of LIMD1, possibly implicating LIMD1 as a regulatory protein of Elk-1. This effect is also partially seen with PU.1 directed siRNA however this can most probably be attributed to the reduced LIMD1 protein levels as a result of PU.1 knockdown.
4. Discussion
The use of internal deletion mutants within the CpG Island has allowed us to identify core regions of promoter activity. Through reporter assays we observed areas of the promoter likely to contain positive and negative regulatory elements; however most notable was the IR5. Within this region we identified a conserved motif for the Ets transcription factor PU.1, and confirmed the functionality and physiological importance of PU.1 binding to this element both in vitro and in vivo. These analyses lead us to conclude that PU.1 is a major transcriptional activator of LIMD1.
PU.1 was initially identified as binding to the core sequence 5′-GAGGAA [26], however variations within the preceding and following bases around the GGAA motif have since been identified. Recent ChIP-Seq analysis of PU.1 binding in macrophages and B-cells revealed the core binding motif for PU.1 (GGAA) is commonly preceded by G/A/C then A/C/G, and followed by, in the majority of sequenced motifs, by GTG [27]. This correlates well with the IR5 identified sequence of GCGGAAGTG. Furthermore, other characterised macrophage and B lineage associated genes under PU.1 also have sequence homology to the IR5 sequence. Secretory interleukin-1 receptor antagonist (IL-1Ra) has the consensus GCGGAAATA [28], whilst Toll-like receptor 4 (TLR4) has the consensus GAGGAAGTG [29].
PU.1 has been extensively studied within the haematopoietic cell lineage with down regulation of this transcription factor leading to leukaemic transformation [21,30]. LIMD1 has not been studied in HSC lineage physiology or pathology, with the exception of one publication that flagged chromosomal loss that encompasses the LIMD1 gene as being possibly causative of childhood acute lymphoblastic leukaemia [7]. We have shown PU.1 binds to the LIMD1 promoter in the LIMD1 expressing U937 lymphoma cell line (Fig. 3B) and that PU.1 loss is causative of LIMD1 loss (Fig. 5). These experimental findings, in corroboration with the published tumour suppressive properties of LIMD1, could provide new insights into other HSC lineage derived malignancies. This may be applicable in leukaemias where PU.1 loss is already a well characterised phenomenon [21,30].
Published evidence suggests LIMD1 is silenced through genetic alterations that reduce expression levels rather than mutations to coding regions that affect activity [5]. Although a recent study has identified a small minority of head and neck squamous carcinoma patients with point mutations and one frame shift mutation [31], there is currently no reported evidence indicating such point mutations affect LIMD1 function. If the tumour suppressive effects of LIMD1 are applicable to HSC lineage related malignancies then identification of PU.1 as a major transcriptional activator of LIMD1 could lead to the development of innovative therapies to prevent or reactivate LIMD1 expression. Furthermore, as well as screening for LIMD1 gene loss as a pathological indicator bisulphite sequencing of the critical PU.1 consensus would also be indicative of LIMD1 expression (Fig. 2B).
PU.1 is an essential transcription factor in osteoclastogenesis, with PU.1 expression up-regulating miR-223 expression, which in turn down regulates NFI-A levels causing osteoclast differentiation [32,33]. A functional miRNA mediated gene silencing pathway is required for osteoclastogenesis: loss of the miRNA-silencing associated proteins DGCR8, Dicer1 and Ago2 impairs osteoclast differentiation and function [33]. LIMD1 binds Ago2 and is an important component of miRNA mediated silencing [9], so it is possible that reduced expression of LIMD1 may also impair osteoclast differentiation. During RANK-L mediated osteoclast differentiation, through TRAF6, PU.1 is upregulated [33,34]. Concurrent to this, Limd1 protein levels are also upregulated, where it interacts with TRAF6 to influence osteoclast differentiation [35]. In light of our findings it is feasible to link these two independent studies. Up-regulation of PU.1 induces expression of LIMD1, which may subsequently interact with TRAF6 to induce osteoclast differentiation. Furthermore, as increased LIMD1 expression increases the potency of miRNA silencing [9], down-regulation of NFI-A through mir-223 mediated miRNA silencing may be increased, further increasing osteoclastogenesis. Therefore in osteoclastogenesis we propose PU.1 may have a two pronged positive effect; firstly, as a transcriptional regulator of osteoclastogenesis specific miRNAs [36] and secondly, by increasing LIMD1 protein levels and its associated miRNA and TRAF6 interactions.
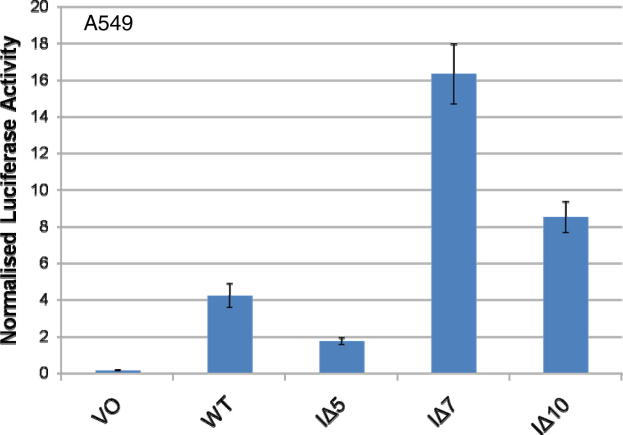
There is little published literature on the pathogenic role of deregulated PU.1 in non-HSC derived biology. Of note is an observation in the lung airway epithelia in allergic asthma where inhibition of microRNA-126 indirectly causes increased expression of PU.1, which consequently suppresses GATA3 expression and thus the GATA3 induced inflammatory response [37,38]. Our data also demonstrate a consistent reduction in transcriptional activity of LIMD1 with the IΔ5 mutant in the A549 lung cancer cell line which is thought to derive from alveolar basal epithelial cells (Supplementary data Fig. 2).
LIMD1 has reduced expression within 75–83% of lung carcinomas [4,5], with gene deletion, loss of heterozygosity and epigenetic silencing accounting for 70% of reductions in LIMD1 expression. If PU.1 is expressed in lung tissue, then the 5–10% shortfall in tumours with unexplained mechanisms for LIMD1 loss may be attributed to PU.1 gene silencing and/or regulation. Clearly, the siRNA mediated depletion of PU.1 and the consequent loss of LIMD1 expression (Fig. 5) supports this possibility.
As well as being a transcriptional activator, PU.1 has also been shown to associate with DNA methyl transferases 3a and b (Dnmt3a/b) and induce silencing of target genes with PU.1 binding sites through methylation, specifically exemplified by the p16INK4A tumour suppressor gene [39]. LIMD1 itself is epigenetically silenced through promoter methylation in the MDA-MB435 breast cancer cell line, with promoter methylation also evident in 26% of human lung tumours when compared to normal matched lung tissue [5,14]. In light of the dual function of PU.1, the role of PU.1 during LIMD1-loss associated transformation remains to be elucidated. In silico screening of the promoter identified a second putative Ets domain/PU.1 binding site at −120 relative to the TSS. As point mutations of the PU.1 motif within IR5 (Fig. 2B) resulted in a 90% decrease in transcription, it is likely that the second PU.1 site is not involved in transcriptional activation. However during transformation it could act as a site for PU.1 (coupled with Dnmt3a/b) binding, causing epigenetic silencing of LIMD1.
In conclusion we have shown that PU.1 is a major regulator of LIMD1 expression and thus it will be of interest to re-examine PU.1 related malignancies in light of these findings for deregulation of LIMD1 tumour suppressive functions such as miRNA-mediated silencing and pRB co-repression [4,9].
Acknowledgements
U937 cells were kindly donated by Dr. Andrew Bennett, University of Nottingham. We would like to thank Maureen Mee, Janice Saxton and Thomas Webb for their technical support and Li Li for guidance with ChIP assays. This work was supported by the Biotechnology and Biological Sciences Research Council [Grant BB/F006470/1 and BB/I007571/1] to Tyson V. Sharp and the British Heart Foundation [Studentship FS/05/103] to Peter E. Shaw.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.febslet.2011.03.013.
Appendix A. Supplementary data
Supplementary Fig. 1.
Sequence of the LIMD1 promoter utilised that spans to −1990 base pairs relative to the transcription start site as assigned according to the NCBI reference sequence NM_014240.2. The CpG Island is highlighted in bold with the 10 consecutive internal regions (IR1–10) indicated. The transcription start site is underlined and the start of the LIMD1 coding sequence is additionally italicised and in bold. The core PU.1 binding sequence within IR5 is double underlined.
Supplementary Fig. 2.
The A549 lung epithelia derived cell line was co-transfected with the wild type LIMD1 or indicated internal deletion mutant reporter vectors and resultant luciferase activity assayed 24 h post-transfection.
Supplementary Fig. 3.
Exogenous ChIP assay using HEK293-T cells was performed using transfected pcDNA4-PU.1 or pcDNA4-vector only. Immunoprecipitated DNA was analysed through PCR utilising primers for the CD11b promoter (previously described and thus used as a positive control) and LIMD1 promoter. +ve and −ve PCR controls were HEK293 genomic DNA and water (no DNA), respectively.
Raw data search results from the MatInspector scrutinisation of the LIMD1 promoter. As the promoter was scrutinised from 5′–3′ the start position of 1 corresponds to −1990 relative to the TSS.
References
- 1.Kok K., Naylor S.L., Buys C.H. Deletions of the short arm of chromosome 3 in solid tumors and the search for suppressor genes. Adv. Cancer Res. 1997;71:27–92. doi: 10.1016/s0065-230x(08)60096-2. [DOI] [PubMed] [Google Scholar]
- 2.Hesson L.B., Cooper W.N., Latif F. Evaluation of the 3p21.3 tumour-suppressor gene cluster. Oncogene. 2007;26:7283–7301. doi: 10.1038/sj.onc.1210547. [DOI] [PubMed] [Google Scholar]
- 3.Pfeifer G.P., Dammann R. Methylation of the tumor suppressor gene RASSF1A in human tumors. Biochemistry (Moscow) 2005;70:576–583. doi: 10.1007/s10541-005-0151-y. [DOI] [PubMed] [Google Scholar]
- 4.Sharp T.V., Munoz F., Bourboulia D., Presneau N., Darai E., Wang H.W., Cannon M., Butcher D.N., Nicholson A.G., Klein G., Imreh S., Boshoff C. LIM domains-containing protein 1 (LIMD1), a tumor suppressor encoded at chromosome 3p21.3, binds pRB and represses E2F-driven transcription. Proc. Natl. Acad. Sci. USA. 2004;101:16531–16536. doi: 10.1073/pnas.0407123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharp T.V., Al-Attar A., Foxler D.E., Ding L., Vallim T.Q. de A., Zhang Y., Nijmeh H.S., Webb T.M., Nicholson A.G., Zhang Q., Kraja A., Spendlove I., Osborne J., Mardis E., Longmore G.D. The chromosome 3p21.3-encoded gene, LIMD1, is a critical tumor suppressor involved in human lung cancer development. Proc. Natl. Acad. Sci. USA. 2008;105:19932–19937. doi: 10.1073/pnas.0805003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh S., Ghosh A., Maiti G.P., Alam N., Roy A., Roy B., Roychoudhury S., Panda C.K. Alterations of 3p21.31 tumor suppressor genes in head and neck squamous cell carcinoma: correlation with progression and prognosis. Int. J. Cancer. 2008;123:2594–2604. doi: 10.1002/ijc.23834. [DOI] [PubMed] [Google Scholar]
- 7.Tsuzuki S., Karnan S., Horibe K., Matsumoto K., Kato K., Inukai T., Goi K., Sugita K., Nakazawa S., Kasugai Y., Ueda R., Seto M. Genetic abnormalities involved in t(12;21) TEL-AML1 acute lymphoblastic leukemia: analysis by means of array-based comparative genomic hybridization. Cancer Sci. 2007;98:698–706. doi: 10.1111/j.1349-7006.2007.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spendlove I., Al-Attar A., Watherstone O., Webb T.M., Ellis I.O., Longmore G.D., Sharp T.V. Differential subcellular localisation of the tumour suppressor protein LIMD1 in breast cancer correlates with patient survival. Int. J. Cancer. 2008;123:2247–2253. doi: 10.1002/ijc.23851. [DOI] [PubMed] [Google Scholar]
- 9.James V., Zhang Y., Foxler D.E., de Moor C.H., Kong Y.W., Webb T.M., Self T.J., Feng Y., Lagos D., Chu C.Y., Rana T.M., Morley S.J., Longmore G.D., Bushell M., Sharp T.V. LIM-domain proteins, LIMD1, Ajuba, and WTIP are required for microRNA-mediated gene silencing. Proc. Natl. Acad. Sci. USA. 2010;107:12499–12504. doi: 10.1073/pnas.0914987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melo S.A., Ropero S., Moutinho C., Aaltonen L.A., Yamamoto H., Calin G.A., Rossi S., Fernandez A.F., Carneiro F., Oliveira C., Ferreira B., Liu C.G., Villanueva A., Capella G., Schwartz S., Jr., Shiekhattar R., Esteller M. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat. Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Wu J.F., Shen W., Liu N.Z., Zeng G.L., Yang M., Zuo G.Q., Gan X.N., Ren H., Tang K.F. Down-regulation of Dicer in hepatocellular carcinoma. Med. Oncol. 2010 doi: 10.1007/s12032-010-9520-5. [DOI] [PubMed] [Google Scholar]
- 12.Kumar M.S., Pester R.E., Chen C.Y., Lane K., Chin C., Lu J., Kirsch D.G., Golub T.R., Jacks T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim M.S., Oh J.E., Kim Y.R., Park S.W., Kang M.R., Kim S.S., Ahn C.H., Yoo N.J., Lee S.H. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. J. Pathol. 2010;221:139–146. doi: 10.1002/path.2683. [DOI] [PubMed] [Google Scholar]
- 14.Huggins C.J., Andrulis I.L. Cell cycle regulated phosphorylation of LIMD1 in cell lines and expression in human breast cancers. Cancer Lett. 2008;267:55–66. doi: 10.1016/j.canlet.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Moreau-Gachelin F., Tavitian A., Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 16.Hsu T., Trojanowska M., Watson D.K. Ets proteins in biological control and cancer. J. Cell. Biochem. 2004;91:896–903. doi: 10.1002/jcb.20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sementchenko V.I., Watson D.K. Ets target genes: past, present and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- 18.Iwasaki H., Somoza C., Shigematsu H., Duprez E.A., Iwasaki-Arai J., Mizuno S., Arinobu Y., Geary K., Zhang P., Dayaram T., Fenyus M.L., Elf S., Chan S., Kastner P., Huettner C.S., Murray R., Tenen D.G., Akashi K. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeKoter R.P., Kamath M.B., Houston I.B. Analysis of concentration-dependent functions of PU.1 in hematopoiesis using mouse models. Blood Cell Mol. Dis. 2007;39:316–320. doi: 10.1016/j.bcmd.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautreau L., Boudil A., Pasqualetto V., Skhiri L., Grandin L., Monteiro M., Jais J.P., Ezine S. Gene coexpression analysis in single cells indicates lymphomyeloid copriming in short-term hematopoietic stem cells and multipotent progenitors. J. Immunol. 2010;184:4907–4917. doi: 10.4049/jimmunol.0902184. [DOI] [PubMed] [Google Scholar]
- 21.Stirewalt D.L. Fine-tuning PU.1. Nat. Genet. 2004;36:550–551. doi: 10.1038/ng0604-550. [DOI] [PubMed] [Google Scholar]
- 22.Bonfield T.L., Raychaudhuri B., Malur A., Abraham S., Trapnell B.C., Kavuru M.S., Thomassen M.J. PU.1 regulation of human alveolar macrophage differentiation requires granulocyte-macrophage colony-stimulating factor. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003;285:L1132–L1136. doi: 10.1152/ajplung.00216.2003. [DOI] [PubMed] [Google Scholar]
- 23.Brugnoli F., Lambertini E., Varin-Blank N., Piva R., Marchisio M., Grassilli S., Miscia S., Capitani S., Bertagnolo V. Vav1 and PU.1 are recruited to the CD11b promoter in APL-derived promyelocytes: role of Vav1 in modulating PU.1-containing complexes during ATRA-induced differentiation. Exp. Cell Res. 2009 doi: 10.1016/j.yexcr.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Li L., Cheung S.H., Evans E.L., Shaw P.E. Modulation of gene expression and tumor cell growth by redox modification of STAT3. Cancer Res. 2010;70:8222–8232. doi: 10.1158/0008-5472.CAN-10-0894. [DOI] [PubMed] [Google Scholar]
- 25.Pahl H.L., Scheibe R.J., Zhang D.E., Chen H.M., Galson D.L., Maki R.A., Tenen D.G. The proto-oncogene PU.1 regulates expression of the myeloid-specific CD11b promoter. J. Biol. Chem. 1993;268:5014–5020. [PubMed] [Google Scholar]
- 26.Klemsz M.J., McKercher S.R., Celada A., Van B.C., Maki R.A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 27.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith M.F., Jr., Carl V.S., Lodie T., Fenton M.J. Secretory interleukin-1 receptor antagonist gene expression requires both a PU.1 and a novel composite NF-kappaB/PU.1/GA-binding protein binding site. J. Biol. Chem. 1998;273:24272–24279. doi: 10.1074/jbc.273.37.24272. [DOI] [PubMed] [Google Scholar]
- 29.Rehli M., Poltorak A., Schwarzfischer L., Krause S.W., Andreesen R., Beutler B. PU.1 and interferon consensus sequence-binding protein regulate the myeloid expression of the human Toll-like receptor 4 gene. J. Biol. Chem. 2000;275:9773–9781. doi: 10.1074/jbc.275.13.9773. [DOI] [PubMed] [Google Scholar]
- 30.Tatetsu H., Ueno S., Hata H., Yamada Y., Takeya M., Mitsuya H., Tenen D.G., Okuno Y. Down-regulation of PU.1 by methylation of distal regulatory elements and the promoter is required for myeloma cell growth. Cancer Res. 2007;67:5328–5336. doi: 10.1158/0008-5472.CAN-06-4265. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh S., Ghosh A., Maiti G.P., Mukherjee N., Dutta S., Roy A., Roychoudhury S., Panda C.K. LIMD1 is more frequently altered than RB1 in head and neck squamous cell carcinoma: clinical and prognostic implications. Mol. Cancer. 2010;9:58. doi: 10.1186/1476-4598-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugatani T., Hruska K.A. MicroRNA-223 is a key factor in osteoclast differentiation. J. Cell. Biochem. 2007;101:996–999. doi: 10.1002/jcb.21335. [DOI] [PubMed] [Google Scholar]
- 33.Sugatani T., Hruska K.A. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J. Biol. Chem. 2009;284:4667–4678. doi: 10.1074/jbc.M805777200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka S., Nakamura K., Takahasi N., Suda T. Role of RANKL in physiological and pathological bone resorption and therapeutics targeting the RANKL-RANK signaling system. Immunol. Rev. 2005;208:30–49. doi: 10.1111/j.0105-2896.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 35.Feng Y., Zhao H., Luderer H.F., Epple H., Faccio R., Ross F.P., Teitelbaum S.L., Longmore G.D. The LIM protein, Limd1, regulates AP-1 activation through an interaction with Traf6 to influence osteoclast development. J. Biol. Chem. 2007;282:39–48. doi: 10.1074/jbc.M607399200. [DOI] [PubMed] [Google Scholar]
- 36.Kong K.Y., Owens K.S., Rogers J.H., Mullenix J., Velu C.S., Grimes H.L., Dahl R. MIR-23A microRNA cluster inhibits B-cell development. Exp. Hematol. 2010 doi: 10.1016/j.exphem.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattes J., Collison A., Plank M., Phipps S., Foster P.S. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc. Natl. Acad. Sci. USA. 2009;106:18704–18709. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueno S., Tatetsu H., Hata H., Iino T., Niiro H., Akashi K., Tenen D.G., Mitsuya H., Okuno Y. PU.1 induces apoptosis in myeloma cells through direct transactivation of TRAIL. Oncogene. 2009;28:4116–4125. doi: 10.1038/onc.2009.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki M., Yamada T., Kihara-Negishi F., Sakurai T., Hara E., Tenen D.G., Hozumi N., Oikawa T. Site-specific DNA methylation by a complex of PU.1 and Dnmt3a/b. Oncogene. 2006;25:2477–2488. doi: 10.1038/sj.onc.1209272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data search results from the MatInspector scrutinisation of the LIMD1 promoter. As the promoter was scrutinised from 5′–3′ the start position of 1 corresponds to −1990 relative to the TSS.