Abstract
The orchids Ophrys sphegodes and O. exaltata are reproductively isolated from each other by the attraction of two different, highly specific pollinator species. For pollinator attraction, flowers chemically mimic the pollinators’ sex pheromones, the key components of which are alkenes with different double-bond positions. This study identifies genes likely involved in alkene biosynthesis, encoding stearoyl-acyl carrier protein (ACP) desaturase (SAD) homologs. The expression of two isoforms, SAD1 and SAD2, is flower-specific and broadly parallels alkene production during flower development. SAD2 shows a significant association with alkene production, and in vitro assays show that O. sphegodes SAD2 has activity both as an 18:0-ACP Δ9 and a 16:0-ACP Δ4 desaturase. Downstream metabolism of the SAD2 reaction products would give rise to alkenes with double-bonds at position 9 or position 12, matching double-bond positions observed in alkenes in the odor bouquet of O. sphegodes. SAD1 and SAD2 show evidence of purifying selection before, and positive or relaxed purifying selection after gene duplication. By contributing to the production of species-specific alkene bouquets, SAD2 is suggested to contribute to differential pollinator attraction and reproductive isolation among these species. Taken together, these data are consistent with the hypothesis that SAD2 is a florally expressed barrier gene of large phenotypic effect and, possibly, a genic target of pollinator-mediated selection.
Keywords: acyl-acyl carrier protein desaturase, isolation genes, pollination, speciation
Reproductive isolation is a central topic in the study of evolution, its origin and maintenance being critical for the process of speciation. This statement is especially true for ecological speciation, in which divergent selection pressures on key traits drive the establishment of reproductive isolation even in the absence of geographic barriers to gene flow (1). This process fits the genic view of speciation, in which only few loci of large effect may be responsible for species differentiation, whereas gene flow is possible throughout the rest of the genome (2, 3). In practice, the challenge in studying these processes is identifying the traits under divergent selection and their genetic basis (1). In plants with strong pollinator-mediated reproductive isolation (floral isolation), however, key floral traits are direct targets of selection (1, 4). By identifying the molecular mechanisms underlying these traits, genes directly involved in reproductive isolation (so-called “barrier” or “isolation” genes) or even speciation can be identified (3–5).
Strong floral isolation and high pollinator specificity make sexually deceptive orchids an excellent system for identifying barrier genes (4, 6). Rewardless orchids of the genus Ophrys attract male insects by sexual mimicry, inducing mating attempts of pollinators with flowers, whereby pollen is transferred. The key component to this system is the chemical mimicry of the pollinator female's sex pheromone (7, 8), a blend of substances consisting mostly of cuticular hydrocarbons, e.g., alkanes and alkenes. Alkenes (unsaturated hydrocarbons) are of special importance, and a different proportion of alkenes was found to be the major odor difference among two closely related Ophrys species attracting different pollinators (9). In Ophrys, speciation by pollinator shift has been hypothesized, and there is evidence both for pollinator-driven genetic differentiation and selection on floral hydrocarbon profiles (4, 6, 9, 10). In particular, specific pollinators mediate strong floral isolation among the coflowering closely related species O. sphegodes and O. exaltata by effectively preventing gene flow, whereas other reproductive barriers are largely absent (11). These species differ mainly in the double-bond position of their major alkenes (9), implying that the genes underlying this alkene difference may be barrier genes (6).
Although alkanes are common components of the wax layer covering the aerial parts of plants (12), alkenes are rare. Alkanes are synthesized from fatty acyl-coenzyme A (CoA) intermediates that undergo several rounds of chain elongation from the carboxyl terminus. These fatty acid (FA) intermediates undergo reduction to aldehydes and decarbonylation to form alkanes, mostly producing odd-numbered alkanes from even-numbered very-long-chain fatty acid (VLCFA) intermediates (12, 13). Alkenes are thought to follow the same synthesis scheme, except for the introduction of double-bonds in an additional desaturation step (6). Notably, biosynthesis of the alkenes in insect sex pheromones is likely very different from that in plants. Although insect acyl-CoA desaturases (which introduce the double-bond into alkene precursors) were identified as putative speciation genes (3, 5), plant acyl-acyl carrier protein (ACP) desaturases that are responsible for the conversion of saturated to unsaturated FAs are mostly unrelated to their animal counterparts (14). Specifically, plant homologs of the animal integral membrane acyl-CoA desaturases act mostly on acyl-lipid intermediates. In contrast, soluble, plastid-localized stearoyl-ACP desaturases (SAD) carry out the ubiquitous desaturation of 18:0 (saturated C18) to 18:1 (monounsaturated C18) FA intermediates (14).† Such SADs are candidates for the insertion of a double-bond into the precursors of alkenes in plants (6, 10). Double-bond insertion at position Δ9 of 18:0-ACP's carbon chain (counting from the substituted end) by a Δ9-SAD would yield 18:1Δ9-ACP. This product could be elongated, to e.g., 28:1Δ19-CoA (double-bond at position Δ19 = ω-9, with ω counting from the unsubstituted end), leading to the production of 27:1Δ9 alkenes upon decarbonylation. Therefore, species differences in alkene composition might result from changes in gene expression and/or enzyme activity of specific SAD-encoding genes among species, implying that such genes are candidate barrier genes in Ophrys orchids.
Here, we report the isolation of SAD homologs from O. sphegodes and O. exaltata and discuss their potential role as barrier genes. Specifically, we address the following questions: (i) are there any differences among species regarding SAD gene expression or protein structure, (ii) are such differences associated with alkene production, (iii) are SAD proteins functional desaturases, and (iv) is there any evidence for selection on these enzymes?
Results
Gene Cloning of Ophrys Stearoyl-ACP Desaturase Homologs.
Putative SAD-encoding transcripts were cloned by homology to Arabidopsis thaliana SSI2 (SUPPRESSOR OF SA-INSENSITIVITY2; At2g43710), the main Δ9-SAD-encoding gene of Arabidopsis. Three putative homologs, named SAD1–SAD3 (Fig. S1A), were identified from cDNA of Ophrys flower labella and their full coding sequence was obtained by RACE. SAD1 was identified only from O. sphegodes, whereas the SAD2 and SAD3 genes were cloned from both species. SAD3 showed only silent substitutions between the O. sphegodes and O. exaltata alleles (hereafter, denoted by Os and Oe prefixes). In contrast to SAD3, OsSAD2 and OeSAD2 differed at the amino acid level (Fig. S1A).
Evolutionary Analysis.
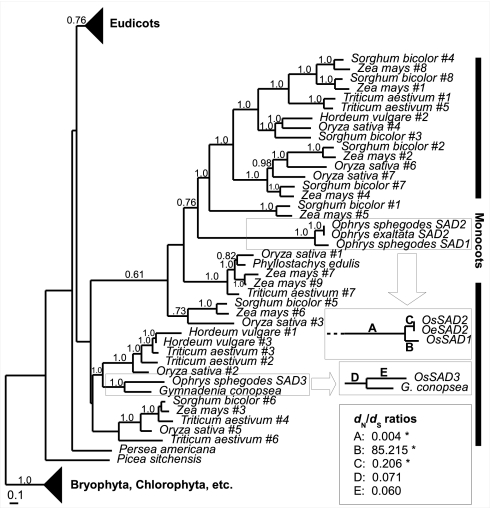
Homologs of A. thaliana SSI2 (Table S1) were identified in public sequence databases and used to construct a Bayesian inference phylogeny of plant acyl-ACP desaturases (Fig. 1 and Fig. S2A). There was only one group of monocot desaturases, with Ophrys SAD1 and SAD2 occupying a position separate from SAD3. This finding indicated that the gene duplication events associated with plant desaturase diversification occurred after the split of monocots and eudicots. Furthermore, the SAD1/SAD2 dichotomy is more recent than the split of proto-SAD1/2 and SAD3. To test for the signature of selection, a maximum likelihood-based analysis of synonymous mutations (dS; preserving the amino acid sequence) versus nonsynonymous mutations (dN; altering the amino acid sequence) was performed. This analysis revealed no indication of selection for SAD3. However, significant purifying selection (P = 0.002) was found on the SAD1/SAD2 clade before the split of SAD1 and SAD2, and significant positive or relaxed purifying selection (all P < 0.001) thereafter (Fig. 1, Fig. S2, and Tables S2–S4). A more conservative exact test of synonymous and nonsynonymous sites is consistent with this interpretation (Table S4).
Fig. 1.
Phylogenetic analysis of SAD homologs, showing monocot clade. Bayesian phylogeny with branch lengths from BaseML; numbers indicate posterior probabilities (where >0.5) next to branches. Selected branches for orchid desaturases are labeled, and the respective dN/dS ratios (from CodeML free-ratio model) are indicated in Inset. An asterisk marks branches A, B, and C, for which dN/dS ratios are significant (P < 0.01) among one- and two-ratio models (Tables S2–S3).
Cuticular Hydrocarbons and Gene Expression.
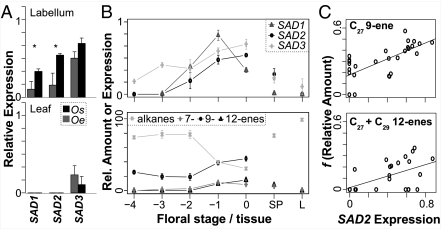
Because high levels of alkenes were found on flowers, but not on leaves of Ophrys (7), the occurrence of hydrocarbons and SAD expression in different O. sphegodes and O. exaltata tissues and floral developmental stages was investigated (Fig. 2, Fig. S3, and Fig. S4). Mature flowers of the two species differed significantly in the levels of different alkenes, with O. sphegodes producing high levels of 9-alkenes and 12-alkenes (strictly speaking, 11/12-alkenes; see Methods), and high levels of 7-alkenes in O. exaltata (Fig. S3 B and C). Expression of SAD1 and SAD2 (but not SAD3) differed among mature labella from the two species (Fig. 2A). Together with the finding that SAD3 was expressed in leaf tissue lacking alkenes, this suggests that SAD1 and/or SAD2 are involved in species-specific differences in alkene production. While alkanes were found in all tissues, most alkenes were barely detectable in leaves/bracts, sepals/petals, and labella from the smallest buds. The relative amount of alkenes, however, increased throughout flower development (Fig. 2B and Fig. S3 F–O). As judged by semiquantitative reverse transcriptase (RT)-PCR, SAD1, and SAD2 expression broadly paralleled alkene occurrence, but only SAD2 expression could significantly explain the presence of several 9- and 12-alkenes, which are different among species and detectable by pollinators (Fig. 2C and Fig. S3). Although SAD3 showed a significant association with one species-specific 9-alkene (C25; Fig. S3B), its lack of species-specific expression pattern makes it unlikely to be a causative factor.
Fig. 2.
SAD expression and hydrocarbons. (A) Mean relative expression of SAD1–SAD3 in O. sphegodes (Os) and O. exaltata (Oe), normalized to G3PDH control, in flower labella (Upper), and leaves (Lower). *P < 0.05 (one-way ANOVA). Error bars indicate SEM. (B) Normalized expression of SAD1–SAD3 (Upper) and relative amounts (%) of major hydrocarbon classes (Lower) in O. sphegodes flower labella of different developmental stages (−4, smallest bud; 0, flower at anthesis), mature sepals/petals (SP), and leaves (L). Error bars indicate SEM. (C) Correlation of normalized O. sphegodes SAD2 expression with relative alkene amount after f(x) = arcsin x0.5 transformation, for 27:1Δ9 alkene (Upper; adjusted R2 = 0.48, P = 2.8·10−5), and 27:1Δ12 + 29:1Δ12 alkenes (Lower; adjusted R2 = 0.32, P = 0.0009), showing regression lines.
Protein Predictions.
Because common stearoyl-ACP desaturases are plastid localized (14), we checked whether a plastid transit peptide was predicted for Ophrys SAD proteins. For SAD1 and SAD3 (but not SAD2), the presence of a transit peptide was predicted (Table S5). However, moderate prediction scores and N-terminal sequence divergence from the well-characterized Ricinus communis plant SAD (RcSAD) indicated that care is needed when postulating the subcellular localization of the Ophrys SADs. Using a crystal structure of RcSAD as a template, structural homology models were generated for OsSAD1, OsSAD2, OeSAD2, and OsSAD3 (which is identical in sequence to OeSAD3). These models were in good overall agreement, with differences among protein backbones localized mainly to one loop region (Fig. S1B). Geometry around the active site and substrate-binding pocket appeared to be mostly conserved among Ricinus and Ophrys proteins, and a canonical stearic acid (18:0) substrate modeled into RcSAD fitted into Ophrys structures similarly well (Fig. S1C). The most prominent difference between RcSAD and OsSAD2 is Leu123 at the aliphatic end of the hypothetical substrate-binding cavity (Fig. S1C). Between Ophrys SAD1 and SAD2s, there were several amino acid changes, mainly on the protein surface (Fig. S1D), and there was a marked difference in isoelectric point (Table S5). Hypothetically substrate-interacting regions were mostly similar among OsSAD2 and OeSAD2, but OsSAD1 showed some amino acid differences near the aliphatic end of the substrate-binding cavity (Fig. S1E). Overall, homology models suggested Ophrys proteins to be functional desaturases, although differences among the proteins indicated they might not be functionally equivalent.
SAD Functional Characterization.
Protein function of putative Ophrys desaturases OsSAD1, OsSAD2, and OeSAD2 was investigated in transgenic Arabidopsis and by in vitro assays of enzyme activity. The Ophrys SAD coding sequences were heterologously expressed in Arabidopsis under the control of the Cauliflower mosaic virus 35S RNA promoter. None of the transgenic plant lines complemented the dwarf phenotype of homozygous ssi2 mutants (SI Methods), indicating that orchid transgenes could not fully functionally replace the A. thaliana desaturase SSI2. However, the presence of the OsSAD2 transgene was significantly associated with changes in unsaturated C18 and C16 FA levels in Arabidopsis leaf lipids, suggesting that OsSAD2 has enzymatic activity in Arabidopsis (Fig. S5).
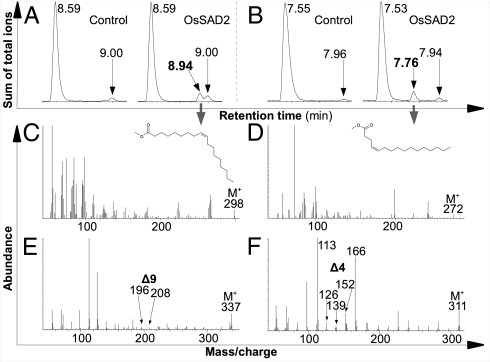
To uncover the specific reaction catalyzed by each Ophrys SAD, recombinant proteins were assayed for desaturase activity in vitro, using acyl-ACP from regiospecifically deuterated fatty acids. For OsSAD1, no product was detectable by gas chromatography coupled to mass spectroscopy (GC/MS), and lack of soluble OeSAD2 expression precluded its analysis. However, desaturase activity was observed for OsSAD2. Consistent with the lack of complementation of Arabidopsis ssi2 mutants, in vitro OsSAD2 activity was low. This low activity may reflect a requirement for specific ACP or ferredoxin proteins different from those present in enzyme assays or in Arabidopsis (cf. refs. 15 and 16). OsSAD2 was active both on 18:0 and 16:0 substrates, producing 18:1Δ9 and 16:1Δ4 products, respectively, as confirmed by MS of fatty acid methyl esters (FAMEs) of reaction products and their pyrrolidine derivatives (Fig. 3). Considering fatty acid elongation from the carboxyl end, these desaturation products would be expected to give rise to 9-alkenes and 12-alkenes, respectively.
Fig. 3.
GC/MS analysis of OsSAD2 desaturase assay. (A, C, and E) 12,12-2H2-18:0-ACP substrate. (B, D, and F) 7,7,8,8-2H4-16:0-ACP substrate. (A and B) GC trace showing assay without desaturase (control; Left) and with desaturase (Right), with retention times (minutes) indicated. Left peak, substrate; right peak, background FA; second peak (with desaturase only), specific reaction product. (C and D) MS fragmentation patterns of specific FAME peaks marked by a gray arrow in A and B, showing mass ion and depicting the molecular structure. (E and F) MS fragmentation patterns of pyrrolidine derivatives of FAMEs in C and D. Arrows indicate ions that are diagnostic for the double-bond positions inferred. This analysis confirms Δ9 and Δ4 double-bond positions for 18:1 and 16:1 reaction products, respectively.
Discussion
Reproductive isolation between O. sphegodes and O. exaltata depends on the attraction of two different, highly specific pollinator species by chemical mimicry of their sex pheromones (11). This specificity is due to the presence of alkenes with different double-bond positions (7–9). During development, these alkenes accumulate in the labella of Ophrys flowers. This accumulation is in marked contrast to the ubiquitous presence of alkanes on orchid surfaces, suggesting that alkene production is tissue- and stage-specific. Among the three putative orchid desaturases, SAD3 showed a relatively constant expression without obvious species difference, consistent with a function as a housekeeping desaturase rather than a factor linked to alkene production. By contrast, SAD1 and SAD2 expression broadly paralleled alkene production, and SAD2 showed a significant association with 9- and 12-alkene levels in O. sphegodes, supporting a functional link. SAD1 and SAD2 probably originated by gene duplication, forming a lineage distinct from SAD3. Purifying selection before this duplication event suggests a conserved role of the ancestral protein. The higher rate of amino acid change after duplication may indicate a partial release from functional constraints, although, considering that alkenes are likely under divergent selection (9), it is also possible that selection drove the divergence of protein function. Taken together, these results implicate Ophrys SAD2 as a desaturase-encoding gene associated with the biosynthesis of alkenes in the floral pseudopheromones.
OsSAD2 is a functional desaturase capable of producing 18:1Δ9 (ω-9) and 16:1Δ4 (ω-12) FA intermediates from which 9-alkenes and 12-alkenes could be synthesized (Fig. 4). However, because housekeeping desaturase activity should be ubiquitous and not restricted to alkene-producing tissues, other proteins must be involved to ensure that desaturation products enter the VLCFA elongation pathway in flowers. For example, changes in the activities of acyl-ACP thioesterase or acyl-CoA synthetase isoforms (12, 13) would be potential candidates. Several orchid genera related to Ophrys produce low levels of alkenes, which might have served as a preadaptation for sexual deception in Ophrys (17). If so, changes in the relevant proteins should be present in both Ophrys and related genera.
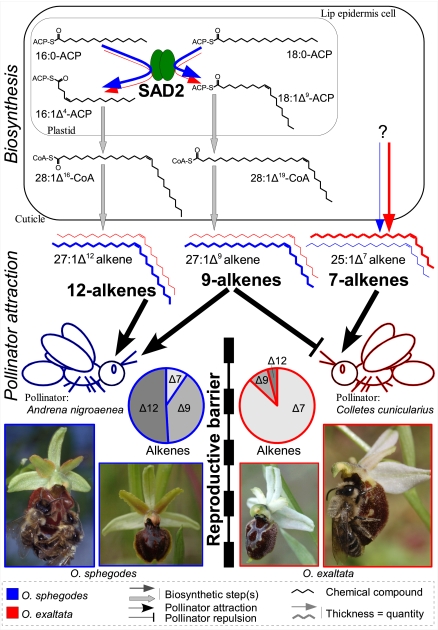
Fig. 4.
Model summarizing SAD2 involvement in floral isolation among O. sphegodes and O. exaltata. SAD2 activity is higher in O. sphegodes (blue arrows) than in O. exaltata (red arrows), due to expression (and possibly functional) differences. SAD2 reaction products are elongated and converted to 9- and 12-alkenes, the levels of which are higher in O. sphegodes than in O. exaltata. The exact source of high levels of 7-alkenes in O. exaltata is unknown. Floral alkenes are detected by pollinators, with 9- and 12-alkenes functioning as attractants to the bee Andrena nigroaenea (the pollinator of O. sphegodes). Conversely, the bee Colletes cunicularius (the pollinator of O. exaltata) is attracted by 7-alkenes, whereas 9-alkenes reduce this attraction. Overall, different alkene blends in the two species lead to differential pollinator attraction associated with reproductive isolation.
Different Ophrys species produce different alkenes, and double-bond differences will ultimately be due to desaturation reactions. Although several different mechanisms could potentially explain differences in desaturation among species, it appears that the higher expression of SAD2 in O. sphegodes contributes to higher 9- and 12-alkene levels in this species. Because OeSAD2 hardly differs from OsSAD2 around the active site and putative substrate-binding pocket (Fig. S1E), it is likely that both enzymes catalyze the same reaction. There are, however, amino acid changes on the surface of SAD2 (Fig. S1D), so that an additional activity change due to different interactions with reaction partners (e.g., specific ACP or ferredoxin isoforms) (15, 16) cannot be ruled out. Such a change may explain why only OsSAD2 affected unsaturated FA levels in transgenic Arabidopsis. SAD1 differs from SAD2 by both changes on the protein surface and changes in the substrate-binding pocket, especially where the aliphatic end of the substrate is expected to bind. However, two lines of evidence suggest that SAD1 is not a functional desaturase: First, SAD1 expression was not significantly associated with alkene production. Second, no evidence of SAD1 activity was detected in either in vitro assays or transgenic Arabidopsis.
The species-specific alkene differences associated with SAD2 are biologically relevant (Fig. 4). Electrophysiological studies with the two specific pollinators, the solitary bees Andrena nigroaenea (for O. sphegodes) and Colletes cunicularius (for O. exaltata), showed that both detect 9-alkenes (C23, C25, C27, C29) and some 12-alkenes (Andrena: C27, C29; Colletes: C29) (7, 8). Moreover, O. sphegodes alkene blends induced mating behavior in A. nigroaenea (7), whereas O. exaltata alkene blends containing 7- and 9-alkenes were less effective than only 7-alkenes for C. cunicularius (8), indicating that 9-alkenes may inhibit mating behavior in this pollinator. Taken together, these observations suggest that the alkenes linked to SAD2 activity are directly involved in the specificity of pollinator attraction and, thus, reproductive isolation among the two orchid species.
In conclusion, our data are consistent with the proposal that the SAD2 desaturase underlies the phenotypic difference in 9- and 12-alkenes among O. sphegodes and O. exaltata and, thereby, contributes to differential pollinator attraction and reproductive isolation among these species. SAD2 therefore represents a barrier gene of large phenotypic effect on pollinator attraction by orchid flowers.
Methods
Plant Material.
Plants of O. sphegodes Miller and O. exaltata Tenore subsp. archipelagi (Gölz & Reinhard) Del Prete were grown in a greenhouse at the Botanic Garden of the University of Zürich. For developmental stage-specific analysis of hydrocarbons and gene expression, inflorescences were taken on the first day of anthesis of the first flower of a given plant, flowers and buds were dissected, and the first open flower was used as a reference point.
Gene Cloning and Expression Analysis.
Total RNA was extracted from flash-frozen orchid tissue by using TRIzol reagent (Invitrogen) according to the manufacturer's instructions, followed by assessment of RNA quality and quantity by agarose gel electrophoresis and spectrophotometry using an ND-1000 (NanoDrop Technologies). Where necessary, RNA was further purified by LiCl precipitation (18). Total RNA was treated with DNase I (Fermentas) and reverse-transcribed into cDNA by using RevertAid M-MuLV H− Reverse Transcriptase (Fermentas), an anchored oligo-dT primer, and the supplier's protocol. Locus-specific and/or semiquantitative PCR was carried out by using RedTaq ReadyMix (Sigma), the supplier's protocol scaled to 10–20 μL with cDNA from 1 ng/μL total RNA as a template. For primers and cycling conditions, see Table S6 and SI Methods. Initial amplification of orchid SAD fragments used a nested degenerate primer approach. PCR products were cloned into pDRIVE (Qiagen), positive clones were identified, and they were Sanger-sequenced by using BigDye 3.1 and a 3130XL Genetic Analyzer (Applied Biosystems), as recommended by the manufacturers. Full-length coding sequence was isolated as detailed in SI Methods, deposited in GenBank (accession nos. FR688105–FR688110), and amplified essentially as before (but reactions also containing 0.015 units per μL Pfu DNA polymerase; Promega) with modified PCR primers (Table S6) to engineer flanking attB sequences during PCR, as recommended by Invitrogen. AttB-site containing PCR products of OsSAD1, OsSAD2, and OeSAD2 were cloned into pDONR207 by BP recombination (Invitrogen) to give pENTR207-SAD, followed by selection on LB agar containing 10 μg/mL gentamicin, plasmid isolation, and sequence confirmation as described before.
GC and GC/MS Analyses.
Cuticular hydrocarbons were extracted by washing plant tissue in 0.5 mL of n-hexane for 1 min, adding 100 ng of n-octadecane as an internal standard. GC was carried out as described (9), except for the use of a lower heating rate of 4 °C/min. Retention times were compared against those of synthetic hydrocarbon standards run with the same settings. The standards were: C19 and C21–C29 n-alkanes and odd-chain (Z)-7-C21–C25, (Z)-9-C21–C29, (Z)-11-C25– C29 and (Z)-12-C25–C27 n-alkenes. Several samples were reanalyzed on an Agilent 5975 GC/MS with the same oven and column settings. Discrimination of (Z)-11/12 alkenes is not possible with these parameters. However, double-bond positions have previously been determined: Both study species contain 11- and 12-alkenes, with 12-alkenes as the predominant isomer (19, 20). FAMEs extracted from Arabidopsis lines were analyzed by GC/MS using the same settings. FAMEs from desaturase assays were analyzed as in ref. 21.
Plant Expression of Desaturases and Biochemical Activity Assay.
To create 2×35S:SAD expression vectors, pENTR207-SAD entry clones were recombined with the binary plant expression vector pMDC32 (22) by LR recombination (Invitrogen) and selected on kanamycin. Plasmids were isolated, sequenced, and transformed into Agrobacterium tumefaciens strain LBA4404, which was, in turn, used to transform A. thaliana line SALK_036854 (23) by using the floral dip method (24). This line carries a T-DNA insertion in SSI2, associated with a recessive dwarf phenotype (SI Methods). Transgenic Arabidopsis plants were selected on MS (25) agar containing 0.05% Plant Preservative Mixture (Plant Cell Technology) and 25 μg/mL hygromycin. Selected independent transgenic lines in an ssi2/ssi2 background were tested for complementation: 35S:OsSAD1 (n = 2), 35S:OsSAD2 (n = 5), and 35S:OeSAD2 (n = 2). Transgene expression (Fig. S5B) and sequence were confirmed by RT-PCR and Sanger sequencing as described above. FAMEs were prepared by BCl3/methanol extraction (26).
Different constructs for protein expression in Escherichia coli were made and evaluated as detailed in SI Methods. Expression clones containing N-terminally modified orchid desaturases in the pET9d (Novagen) expression vector were chosen for functional analysis. In these clones, amino acids 2–5 (ELHL) were deleted to remove part of the putative chloroplast transit peptide. Proteins were purified and assayed as described (21), with minor modifications: only 7,7,8,8-2H4-16:0-ACP and 12,12-2H2-18:0-ACP substrates were used in assays containing 100 μg of desaturase, incubated for 2 h at 22 °C. FAMEs were suspended in 50 μL of hexane for GC/MS analysis.
Bioinformatic and Statistical Analyses.
Molecular mass and isoelectric point of proteins were predicted by using the ExPASy Server (27) and the presence of a chloroplast transit peptide predicted using the ChloroP 1.1 server (28). Homology modeling was performed by using the SWISS-MODEL server (29) and the 2.4-Å crystal structure 1OQ4 (chain A) (30) of RcSAD as a template. Validation and quality checking of the models were done by using the ProSA-web server (31) and Procheck software (32).
Homologs of the Arabidopsis SSI2 desaturase gene (Table S1) were extracted from public sequence databases as detailed in SI Methods and aligned based on amino acid sequence by using PRANK 0.91 (33). Poorly alignable regions were excluded from downstream analysis. The GTR+I+Γ nucleotide substitution model was selected by using MrModeltest 2.2 (34) and phylogenetic analysis conducted in MrBayes 3.1.2 (35), discarding results before apparent convergence of analysis chains (burn-in 1 million of 30 million generations). Branch lengths of the resulting consensus tree were optimized with BaseML and used as input for CodeML, both part of the PAML 4.3 (36) package. Different models of sequence evolution were calculated with CodeML and compared by likelihood ratio testing. Fisher's exact tests were done on (non)synonymous site counts (37) by using CodeML output. Statistical analyses were performed in Microsoft Excel and R 2.11.0 (38).
Supplementary Material
Acknowledgments
We thank A. Bolaños, A. Boyko, S. Cozzolino, M. Curtis, S. Kessler, M. and S. Schauer, and H. Zheng for providing laboratory materials, help, or source code, and M. Anisimova and M. and S. Schauer for discussions and comments. This work was supported by Austrian Science Fund Fellowship J2678-B16 (to P.M.S.), Swiss Federal Institute of Technology Zürich Grant TH 02 06-2 (to F.P.S.), the University of Zürich (U.G. and F.P.S.), and the Office of Basic Energy Sciences of the US Department of Energy (J.S. and E.J.W.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FR688105–FR688110).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013313108/-/DCSupplemental.
Shorthand notation for fatty acids and their derivatives is given in C:D form, where C specifies the number of carbon atoms and D the number of double-bonds; the position x of a cis double-bond in the carbon chain is indicated by Δx when counted from the substituted end (if applicable), or by ω-x when counting from the unsubstituted end.
References
- 1.Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- 2.Lexer C, Widmer A. The genic view of plant speciation: Recent progress and emerging questions. Philos Trans R Soc Lond B Biol Sci. 2008;363:3023–3036. doi: 10.1098/rstb.2008.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu C-I, Ting C-T. Genes and speciation. Nat Rev Genet. 2004;5:114–122. doi: 10.1038/nrg1269. [DOI] [PubMed] [Google Scholar]
- 4.Schiestl FP, Schlüter PM. Floral isolation, specialized pollination, and pollinator behavior in orchids. Annu Rev Entomol. 2009;54:425–446. doi: 10.1146/annurev.ento.54.110807.090603. [DOI] [PubMed] [Google Scholar]
- 5.Noor MAF, Feder JL. Speciation genetics: Evolving approaches. Nat Rev Genet. 2006;7:851–861. doi: 10.1038/nrg1968. [DOI] [PubMed] [Google Scholar]
- 6.Schlüter PM, Schiestl FP. Molecular mechanisms of floral mimicry in orchids. Trends Plant Sci. 2008;13:228–235. doi: 10.1016/j.tplants.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Schiestl FP, et al. Sex pheromone mimicry in the early spider orchid (Ophrys sphegodes): Patterns of hydrocarbons as the key mechanism for pollination by sexual deception. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2000;186:567–574. doi: 10.1007/s003590000112. [DOI] [PubMed] [Google Scholar]
- 8.Mant JG, et al. Cuticular hydrocarbons as sex pheromone of the bee Colletes cunicularius and the key to its mimicry by the sexually deceptive orchid, Ophrys exaltata. J Chem Ecol. 2005;31:1765–1787. doi: 10.1007/s10886-005-5926-5. [DOI] [PubMed] [Google Scholar]
- 9.Mant JG, Peakall R, Schiestl FP. Does selection on floral odor promote differentiation among populations and species of the sexually deceptive orchid genus Ophrys? Evolution. 2005;59:1449–1463. [PubMed] [Google Scholar]
- 10.Schlüter PM, et al. Genetic patterns and pollination in Ophrys iricolor and O. mesaritica (Orchidaceae): Sympatric evolution by pollinator shift. Bot J Linn Soc. 2009;159:583–598. [Google Scholar]
- 11.Xu S, et al. Floral isolation is the main reproductive barrier among closely related sexually deceptive orchids. Evolution. doi: 10.1111/j.1558-5646.2011.01323.x. in press. [DOI] [PubMed] [Google Scholar]
- 12.Samuels L, Kunst L, Jetter R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu Rev Plant Biol. 2008;59:683–707. doi: 10.1146/annurev.arplant.59.103006.093219. [DOI] [PubMed] [Google Scholar]
- 13.Jetter R, Kunst L. Plant surface lipid biosynthetic pathways and their utility for metabolic engineering of waxes and hydrocarbon biofuels. Plant J. 2008;54:670–683. doi: 10.1111/j.1365-313X.2008.03467.x. [DOI] [PubMed] [Google Scholar]
- 14.Shanklin J, Cahoon EB. Desaturation and related modifications of fatty acids. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:611–641. doi: 10.1146/annurev.arplant.49.1.611. [DOI] [PubMed] [Google Scholar]
- 15.Schultz DJ, Suh MC, Ohlrogge JB. Stearoyl-acyl carrier protein and unusual acyl-acyl carrier protein desaturase activities are differentially influenced by ferredoxin. Plant Physiol. 2000;124:681–692. doi: 10.1104/pp.124.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suh MC, Schultz DJ, Ohlrogge JB. Isoforms of acyl carrier protein involved in seed-specific fatty acid synthesis. Plant J. 1999;17:679–688. doi: 10.1046/j.1365-313x.1999.00418.x. [DOI] [PubMed] [Google Scholar]
- 17.Schiestl FP, Cozzolino S. Evolution of sexual mimicry in the orchid subtribe Orchidinae: The role of preadaptations in the attraction of male bees as pollinators. BMC Evol Biol. 2008;8:27. doi: 10.1186/1471-2148-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 19.Erdmann DH. Hamburg, Germany: Univ of Hamburg; 1996. Identification and synthesis of volatile signal compounds in insects and their host plants (Translated from German) PhD thesis. [Google Scholar]
- 20.Schulz CM. Hamburg, Germany: Univ of Hamburg; 2005. Chemical communication in insects: Identification and synthesis of volatile compounds (Translated from German) PhD thesis. [Google Scholar]
- 21.Whittle EJ, Tremblay AE, Buist PH, Shanklin J. Revealing the catalytic potential of an acyl-ACP desaturase: Tandem selective oxidation of saturated fatty acids. Proc Natl Acad Sci USA. 2008;105:14738–14743. doi: 10.1073/pnas.0805645105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtis MD, Grossniklaus U. A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 24.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 25.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 26.Cahoon EB, Shanklin J, Ohlrogge JB. Expression of a coriander desaturase results in petroselinic acid production in transgenic tobacco. Proc Natl Acad Sci USA. 1992;89:11184–11188. doi: 10.1073/pnas.89.23.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasteiger E, et al. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Press, Totowa, NJ: Humana; 2005. pp. 571–607. [Google Scholar]
- 28.Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moche M, Shanklin J, Ghoshal A, Lindqvist Y. Azide and acetate complexes plus two iron-depleted crystal structures of the di-iron enzyme Δ9 stearoyl-acyl carrier protein desaturase. Implications for oxygen activation and catalytic intermediates. J Biol Chem. 2003;278:25072–25080. doi: 10.1074/jbc.M301662200. [DOI] [PubMed] [Google Scholar]
- 31.Wiederstein M, Sippl MJ. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server issue):W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 33.Löytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;320:1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- 34.Nylander JAA. MrModeltest v2. Uppsala: Uppsala Univ; 2004. [Google Scholar]
- 35.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Kumar S, Nei M. Small-sample tests of episodic adaptive evolution: A case study of primate lysozymes. Mol Biol Evol. 1997;14:1335–1338. doi: 10.1093/oxfordjournals.molbev.a025743. [DOI] [PubMed] [Google Scholar]
- 38.R Development Core Team . Vienna: R Found for Stat Comput; 2010. R: A language and environment for statistical computing. 2.11.0. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.