Abstract
Myosin VI is a molecular motor implicated in many processes, and it likely associates with a variety of cargoes that specify its functions. Although it is critical to Drosophila development, little is known about its cellular roles. To reveal its involvement in specific pathways, we sought to identify the binding partners of Drosophila myosin VI. We used affinity chromatography and mass spectrometry to discover interacting proteins, which we tested for direct binding. Using this approach, we found that the microtubule-associated protein Cornetto bound myosin VI, and we demonstrated a role for both in secretion of the lipidated morphogen Hedgehog. We also identified a number of other binding proteins, and further characterization of their interactions with myosin VI will advance our understanding of the roles of these complexes in cellular and developmental processes. Thus, our method has provided us the means to gain valuable insight into the multifaceted roles of a motor protein in vivo.
Keywords: unconventional myosins, cytoskeleton, trafficking
Myosins comprise a superfamily of actin-based motor proteins involved in a variety of processes (1). On the molecular scale, significant progress has been made in recent years toward understanding their mechanical properties (2, 3). On the organismal scale, it is also clear that they are important for the development and function of tissues and organs (4–6). Yet for many of these proteins, there is little data explaining their roles on the cellular level. As functions of motor proteins are thought to be determined by the types of cargoes they transport (7), an important step in furthering our understanding of motors’ functions is identifying the proteins and organelles with which they interact (8).
Toward this goal, we have focused on myosin VI, which participates in a wide range of processes such as vesicle trafficking and maintenance of stereocilia for mammalian hearing (9). Its broad utilization in higher organisms is perhaps due to its unique directionality toward the minus ends of actin filaments, unlike all other studied myosins (reviewed in ref. 10 and others). Although yeast two-hybrid screens and other approaches have made progress toward identifying some myosin VI cargoes (11), less is known about the molecular details of how myosin VI complexes function in vivo. Such information is particularly lacking about this protein in Drosophila, despite its significance in fly development.
Depletion of Drosophila myosin VI (Jaguar; referred to as M6 throughout) protein levels, expression of a dominant negative M6 truncation, or injection of a function-blocking M6 antibody produces a variety of phenotypes that depend on the stage and tissue targeted (12). These data have revealed roles for M6 in pseudocleavage furrow formation in the syncytium (13), dorsal closure later in embryogenesis (14), and spermatogenesis in the adult male (15), among other processes (16).
Though its importance is evident, it is unclear what M6 contributes as a motor protein to developmental events, because very few binding partners are known. Recent data have revealed that M6 transports Miranda to the basal region of dividing neuroblasts (17) and cooperates with Echinoid in dorsal closure (18). And although M6 coimmunoprecipitates and colocalizes with the microtubule-binding protein CLIP-190 in the embryonic nervous system (19), the function of this complex is not known. The many processes perturbed upon M6 disruption, however, in addition to its broad expression pattern (20), suggest a diversity of functions outside of these known interactions. Therefore, we chose a proteomics-based approach to identify M6 cargoes in Drosophila, anticipating that their identities might begin to reveal its cellular roles. From our screen, we have found a number of proteins that interact directly with M6, and we chose to further characterize the binding of M6 to Cornetto. We also demonstrate a role for both proteins in secretion of lipid-modified Hedgehog, and we expect that future work on the remainder will continue to shed light on M6’s many cellular functions.
Results
Many Proteins Are Retained by Immobilized Myosin VI.
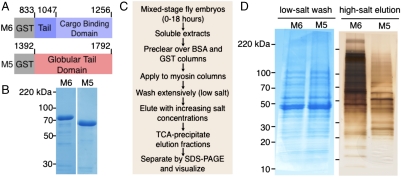
We set out to identify binding partners of M6 using affinity chromatography (21), given that affinity techniques are among the most accurate and comprehensive of the large-scale methods to discover interactions (22). To control for specificity, we chose to compare proteins bound to M6 to those that bound to the type-V Drosophila myosin, another actin-based motor with fewer noted functions in embryogenesis (23). We constructed columns from purified cargo-binding domains of M6 and myosin V (Didum; referred to as M5 throughout) (Fig. 1 A and B), applied Drosophila embryonic extract to the columns, and eluted proteins with increasing salt concentrations (Fig. 1C). Though the protein content of the low-salt washes from the M5 and M6 columns appeared very similar (Fig. 1D, Left), there were numerous differences between the subsequent high-salt elutions (Fig. 1D, Right), which we subjected to mass spectrometry analysis. The sensitivity of our method enabled the identification of a large number (> 1,000) of proteins that associate, directly or indirectly, with M6 or M5.
Fig. 1.
Affinity column construction and analysis. (A) Schematic of protein constructs used to make affinity columns with residue numbers indicated. (B) Coomassie-stained SDS-PAGE of purified proteins used for column construction after affinity and ion exchange chromatography. (C) Extract preparation, column application, and elution sample handling workflow. Elutions included sequential steps of 250 mM, 500 mM, and 1 M KCl. (D) SDS-PAGE of TCA-precipitated fractions. (Left) Coomassie-stained 100 mM KCl washes. (Right) Silver-stained 1 M KCl elutions.
To narrow our list of candidates, we devised a stringent specificity criterion and selected only those with at least five times as many unique peptides in the M6 sample as in the M5 elution; this filtering still left us with hundreds of proteins. Noting the wide range of unique peptide counts (number of unique sequences obtained for each protein), we further refined our specificity criterion by normalizing to the predicted size of each parent protein. We divided the number of unique peptides obtained from any given protein by its calculated molecular weight in kilodaltons (kDa), yielding a parameter we refer to as the unique peptide ratio (UPR). We ranked the candidates by UPR, which all fell in the range from 0–1. Considering that Miranda, a protein known to interact directly with M6 (17), had a UPR greater than 0.2, whereas most proteins did not, we focused on the subset of proteins that ranked above this cutoff.
A Number of Highly Ranked Candidates Bind Directly to Myosin VI.
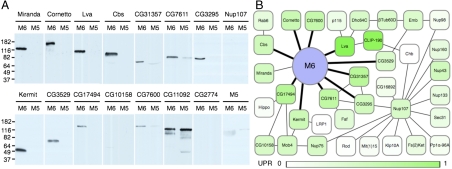
Although there were still many proteins in this category, the stringency of our washes (at salt concentrations chosen to screen nonspecific protein interactions) and the data filtering made it likely that many of the candidates bound directly to M6, with the remainder potentially interacting indirectly as part of larger complexes. To test for direct binding partners, we transcribed and translated a number of highly ranked candidate proteins in vitro and compared binding to immobilized M6 and M5 (Fig. 2A). Miranda, one of the few proteins known to directly associate with Drosophila M6 (17), served as our positive control and showed high specificity for M6 binding over M5 (Fig. 2A). Though it is possible that these interactions depend on an intermediate from the lysates used for translation, they are likely to be direct; we will refer to them as such for clarity, noting that further tests could confirm these assignments.
Fig. 2.
M6 binds novel cargoes and is likely present in larger complexes. (A) Anti-myc Western blots of tagged candidate proteins retained by immobilized M6 or M5 after washing with 300 mM salt. Candidate names are indicated above each set of comparisons. Molecular weight markers and their sizes in kilodaltons are indicated on the left of each set. Lava lamp is listed as Lva, and only a construct containing the N-terminus was translated. CG11092 and CG2774 were chosen as specificity controls and were present in both M6 and M5 elution samples. Experiments were repeated several times each, and consistent results were obtained. (B) M6 interaction map constructed in Cytoscape (52). Included are novel (bold lines) and known (thin lines) physical interactions, culled from the DroID database (31) and primary sources (17, 25); all were present in and specific to the M6 elution. Those proteins with the highest UPR are colored the darkest shade of green, fading to white for the lowest-ranked. Not all interactions are shown for some very highly connected proteins (Emb, βTub60D, Nup107), and CG7671 is represented as Nup43. Kermit has been linked genetically to M6 (28), but their direct binding has not been previously demonstrated.
Cornetto, a presumed microtubule-binding protein (24) that asymmetrically localizes in neuroblasts, specifically bound to M6. Other M6 interactions include Golgi proteins, namely Lava lamp (25, 26) and Centrosomin’s beautiful sister (Cbs), the latter of which has also been linked to the centrosome cycle (27). Kermit, the orthologue of the vesicle-associated protein GIPC (28), which is known to bind myosin VI (29), also bound directly in our assay. Another protein that bound specifically and may be involved in vesicle trafficking is CG3529, which is similar to human Tom1 and Tom1L2, two proteins that associate with clathrin (30). From data on their mammalian orthologues, CG31357, CG3295, and CG7611 are predicted to interact with each other (31); we have found that all three proteins also bind M6 directly, suggesting they function together in a complex with yet-unknown function.
We tested both CG17494 and CG10158 for direct binding, two proteins that interact with each other in a yeast two-hybrid assay (32). Of these two, only CG17494, one of our highest-ranking candidates (Table S1), associated reproducibly with M6. Although these and another direct binder, CG7600, have no known functions in flies, CG17494 may be involved in muscle development (33). Not all proteins tested bound specifically to M6. Some, like CG11092, also bound to M5, and others, like Nup107 and CG2774, bound directly to neither myosin in vitro. Notably, M5 does not seem to interact directly with M6, despite the presence of many M5 peptides in the high-salt M6 elution (Table S1).
Incorporating information from our binding data, database-curated interactions, and primary references, we constructed an interaction map for M6 (Fig. 2B). Listed in Table S1 and Fig. 2B are the proteins that bound directly to M6, as well as other proteins from our M6 elution sample that are known to associate with them. Of the proteins that likely associate in complexes, those with higher UPRs (darker green squares, Fig. 2B) tended to be direct binding partners. Many of the interactions are with proteins with unknown cellular roles and no previous connection to M6 function. Nearly half of the candidates tested showed reproducible association with M6 (those shown in Fig. 2A), implying our stringent criteria were useful in leading us toward direct binders. Although the remainder did not bind well in our assay, it is possible that they require posttranslational modifications or other nonprotein factors not present in our in vitro system. Otherwise, they may be indirectly associated with M6 through larger complexes.
Having found a number of interacting proteins, we began searching for evidence of cooperation between our candidates and M6 in an in vivo context. Given that M6 shows a striking enrichment in the leading edge cells of Drosophila embryos during dorsal closure (14, 34) (Fig. S1A), a disruption in that localization pattern could be readily discernible. When we examined the localization of M6 in embryos mutant for or expressing RNAi against several of its binding proteins, we observed some qualitative differences in M6 staining in stage 13–14 compared to wild-type, such as a reduction of M6 staining at the leading edge (Fig. S1, arrowheads). Although this screen facilitates the identification of embryos with obvious defects, a negative result does not necessarily correlate with a lack of effect on embryogenesis. Additionally, due to the difficulty in interpreting these images, the limitations imposed by the use of uncharacterized mutant lines, and the unknown extent of RNAi-mediated knockdown, we focused instead on one interacting protein to validate our binding data and test its in vivo relevance with a quantitative assay.
A Cornetto Fragment Is Sufficient for Binding with Micromolar Affinity to the Myosin VI Tail.
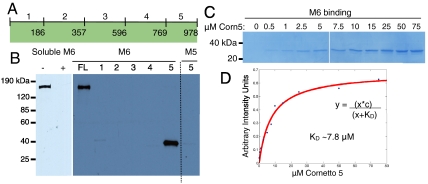
Cornetto, one of our highest-ranked hits, is a protein with no reported function, though it has regions predicted to form coiled-coils and localizes to the apical cortex of dividing neuroblasts during anaphase and telophase through an unknown mechanism (24). Interestingly, although M6 itself does not show an asymmetric localization in these cells, one of its few reported binding partners, Miranda, depends on M6 for its asymmetric translocation, albeit to a basal crescent (17). To further characterize the interaction between M6 and Cornetto, we translated five similarly sized fragments spanning the Cornetto protein (Fig. 3A), attempting to preserve predicted secondary and tertiary protein structure. The C-terminal fragment, Cornetto 5 (Fig. 3A), was necessary and sufficient for the interaction (Fig. 3B, Right). In addition, soluble M6 cargo-binding domain was able to effectively compete with immobilized M6 for Cornetto binding, indicating that the interaction is specific (Fig. 3B, Left).
Fig. 3.
Biochemical analysis of Cornetto binding to M6. (A) Schematic of full-length Cornetto and constructs used to make five fragments spanning the protein, with residue numbers indicated. (B) Anti-myc Western blots showing the binding of myc-tagged Cornetto, or Cornetto fragments, to M6. (Left) Full-length Cornetto binds to immobilized M6 (lane 1), and is effectively competed off the resin upon addition of an excess of soluble M6 (lane 2). (Right) A binding assay with each of the five fragments of Cornetto to M6. Only fragment 5 retains full binding to M6, and it does not bind to M5 (last lane). (C) Coomassie-stained SDS-PAGE showing an in vitro binding assay with recombinant purified Cornetto 5 and M6. M6 immobilized on resin (approximately 5 μM as a bead suspension) was incubated with a range of Cornetto 5 concentrations (from 0–75 μM), and the protein remaining bound to the resin after washing was loaded on the gel. (D) Bands from the binding assay were quantified in ImageJ, and values were plotted in Matlab and fit to a curve with the equation indicated. Based on the fit, the estimated dissociation constant (KD) is between 5 and 10 μM.
In early embryos, M6 is reported to have an intracellular concentration of about 0.5 μM (35). The concentration of Cornetto in cells is not known; however, in general, motor-cargo interactions in vivo form complexes with affinities in the micromolar range (36, 37). This range is likely conducive for the dynamic nature of these interactions, as motors must release their cargo at specific cellular locations. To estimate the dissociation constant of the interaction between the Cornetto C-terminal region and the M6 cargo-binding domain, we expressed and purified both recombinant proteins. After ensuring that binding of Cornetto 5 to immobilized M6 had reached saturation, we estimated a dissociation constant of about 8 μM from the binding curve (Fig. 3 C and D).
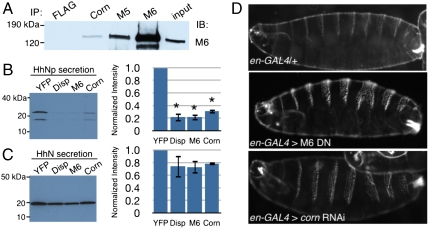
We sought to confirm that Cornetto and M6 form a complex in Drosophila embryonic cells. After generating and affinity-purifying antibodies against M6, M5 (Fig. S2) and Cornetto, we immunoprecipitated each protein from total cell lysates and detected M6 by Western blotting. M6 coimmunoprecipitated with Cornetto, indicating that they do indeed interact in vivo (Fig. 4A). As expected from our mass spectrometry results, M6 and M5 likely reside on some of the same subcellular compartments, as M6 also coimmunoprecipitated with M5 (Fig. 4A). However, our in vitro binding experiments indicate that only the M6–Cornetto association is direct and specific.
Fig. 4.
M6 and Cornetto are involved in the secretion of processed Hedgehog. (A) Immunoprecipitations from S2R+ cell lysates with the antibodies indicated. Anti-M6, anti-Cornetto, and anti-M5 antibodies all coimmunoprecipitate M6, whereas a nonspecific antibody (anti-FLAG) does not. The input lane represents 1% of each total sample. Similar results were obtained from immunoprecipitations of whole fly embryo lysates. (B) Representative anti-Hedgehog Western blot of conditioned media from cells transfected with full-length Hedgehog and each of the dsRNAs as indicated: YFP (nonspecific control), Dispatched (Disp), M6, or Cornetto (Corn). Bands were quantitated and normalized to the signal in the YFP sample. Three independent experiments were averaged, and results are graphed on the Right (± SEM). ∗P < 0.01, very significant compared with nonspecific control, as determined by t test. (C) Representative anti-Hedgehog Western blots of conditioned media from cells transfected with a truncated version of Hedgehog (HhN) and each of the dsRNAs as before. Bands were quantitated and normalized to the signal in the YFP sample as before, and the averages are shown on the Right (± SEM, n≥2). (D) Cuticles prepared from crosses between engrailed-GAL4 and control (y1w67c23), M6 dominant negative (UAS-jaguar tail-CBD), or Cornetto RNAi (UAS-cornetto RNAi) lines. Examples are representative of the normal pattern (control) and the 5–10% fraction affected (M6 DN and corn RNAi).
Myosin VI and Cornetto Are Involved in Protein Secretion, and Disruption of Either Protein Leads to Functional Consequences in Fly Development.
Both M6 and Cornetto are expressed in epidermal cells (14, 24, 34), and the mammalian orthologue of M6 has been reported to play roles in Golgi organization and protein secretion (38, 39, and references within). These observations led us to examine the roles of each in Hedgehog (Hh) export, encouraged by the appearance of M6 as a hit in a yet-unpublished screen for genes involved in Hh secretion (http://www.flyrnai.org/cgi-bin/RNAi_public_screen.pl?project_id=66). The export of cleaved, processed Hedgehog (HhNp) is preceded by several processing steps, and the lipid modification and cleavage reactions that occur in Hh-producing cells are crucial for achieving its distribution pattern in embryos (reviewed in ref. 40). As expected, the dsRNA-mediated knockdown of Dispatched (Disp) protein, which is required for the release of HhNp from Hedgehog-producing cells (41), resulted in a significant reduction (approximately 80%) of HhNp secreted by S2R+ cells exogenously expressing full-length Hh (Fig. 4B). Interestingly, the introduction of dsRNA targeting either M6 (Fig. S3) or Cornetto resulted in a similarly drastic reduction (70–80%) of HhNp secretion. The levels in the medium of a truncated form of Hedgehog that lacks the cholesterol modification (HhN), and does not require Disp for its release by cells (41) (Fig. 4C), were not significantly affected by the knockdown of M6 or Cornetto (Fig. 4C).
Next, we wanted to determine whether the role M6 and Cornetto play in HhNp export is important for pattern formation during embryogenesis. Late in embryonic development, epidermal cells begin secreting the larval cuticle, a chitinous exoskeleton deposited in a segmented pattern; this patterning depends on signals from morphogens such as Hedgehog (reviewed in ref. 42). We examined the cuticles of late embryos and larvae expressing either an M6 truncation or cornetto RNAi specifically in the cells that secrete Hh (42). The M6 truncation was used because large amounts of M6 protein are maternally contributed and persist throughout early stages of embryogenesis (14). Overexpression of a dominant negative, as our protein construct is expected to act (14), would better inhibit this pool of protein than induction of RNAi targeting M6. Upon examination of the embryos and larvae by darkfield microscopy, we found that a portion (5–10%) consistently display segmentation defects that are rarely found in controls (≤ 1%) (Fig. 4D). The segment fusions observed are similar to those occasionally found in embryos with hypomorphic hedgehog mutations (43), and thus our data are consistent with a role for M6 and Cornetto in Hh export.
Discussion
Myosin VI May Be a Part of Many Functional Complexes.
The wide variety of phenotypes that emerge upon disruption of M6 during fly development led us to hypothesize that the motor might have multiple binding partners. This was indeed borne out by the number of specific hits identified from the mass spectrometry screen. We suspect that M6 is a component of many different networks, as suggested by Fig. 2B, and that its association with any one complex or cargo depends on their spatial and temporal expression patterns. In addition, some cargoes are likely shared with M5, because although they do not bind directly (Fig. 2A), much endogenous M5 protein was retained on our M6 column (Table S1). Though this current study focused on M6, we did find that many proteins were common to both samples, and these may represent the overlap between the two motors’ functions in trafficking or other processes.
Our work in identifying many M6 binding partners has provided headway into the understanding of the cellular functions of this motor protein. For example, M6 was recently revealed as a modifier of Notch signaling (44, 45), but its function in this important developmental pathway has not been explored. We have found that CG31357, CG7611, and CG3295, which are all involved in Notch signaling (44), bind directly to M6, raising the possibility that this is a functional complex. Although some of the interactions we found may be specific to insects, other associations are likely conserved, such as those with the trafficking proteins Kermit and CG3529. Drosophila M6 presumably shares functions with its mammalian counterparts, and the identification of interactions allows for the investigation and direct testing of those shared functions in mammalian systems. Characterization of other binding partners with a variety of assays, such as what we have demonstrated with Cornetto, should reveal additional roles for M6 and help explain some of its described functions. Notably, we did not recover Disabled (Dab), the Drosophila orthologue of the myosin VI binding protein Dab2, on our column. The myosin VI binding region of human Dab2 is outside of the conserved phosphotyrosine-binding domain, and the residues critical for binding (46) do not appear to be in the Dab sequence. In addition, Drosophila M6 contains LWY in the position of the WWY motif required for Dab2 binding (47); therefore, an interaction between the Drosophila proteins is not necessarily expected.
Cooperation of Cornetto and Myosin VI in Hedgehog Secretion.
The involvement of M6 and Cornetto in export of lipid-modified Hedgehog raises intriguing questions about the function of this complex. The phenotypes we observe in embryos are of low penetrance, which suggests that the complex may facilitate, but is not absolutely required for, Hh trafficking in vivo. Although it is possible that this low penetrance is due to variations in expression of the transgenic constructs, we suspect that is not the case and favor the possibility that the M6–Cornetto complex plays an accessory role in this system. How might an actin-based motor and a presumed microtubule-binding protein cooperate in the Hh transport pathway? Perhaps they work together at or near the Golgi, a known hub of actin and microtubule networks (48). If this is the case, does this complex function in any other types of secretion? The cholesterol-deficient Hh construct does not appear to require either M6 or Cornetto for export, so the complex may be specialized for the transport of lipid-modified proteins in the context of membranous carriers.
Alternatively, M6 and Cornetto could be involved in the export or recycling of extracellular matrix materials, such as the heparan sulfate proteoglycans (HSPGs) that are known to influence the spread of lipid-modified Hh (reviewed in ref. 49). Such an activity could also explain our observation that at higher temperatures, far fewer embryos produce cuticle, a secreted extracellular structure (50), when expressing cornetto or jaguar (M6) RNAi than in controls (Fig. S4). Although these data could also indicate a failure to reach the appropriate stage, they are consistent with previous observations (16). It is not clear whether HSPGs would be required for the release of Hh in cultured cells, but the Hh trafficking pathway is an area of active research. Determining the mechanistic role of the M6–Cornetto complex in this secretion process will be the subject of future work.
Discovery of Binding Partners as a Means to Identify Functions for Motor Proteins.
We have presented data indicating that affinity chromatography yields valuable information about motor protein cargoes. Though other screening methods can be used to obtain such information, there tends to be little overlap between two-hybrid and affinity methods, for example (22), and our screen may have captured a distinct set of cargoes. From our work on Cornetto, we have found that at least one constitutes a relatively weak (micromolar range) affinity protein–protein interaction (51), which, although expected for a dynamically associating motor-cargo complex, could be easily missed with other techniques, though the affinity between native M6 and Cornetto proteins may be higher. Nonetheless, the combined sensitivity of affinity chromatography and mass spectrometry enabled us to identify a biologically interesting binding interaction.
The discovery of binding partners is an important step toward understanding how motors function. Because each protein has specific domains, expression patterns, documented interactions with other proteins, and predicted or known functions, we can use their identities to form hypotheses about how they interact with myosins in cells. These data are especially useful when genetic predictions of protein function are not available, as is often the case with myosins because of functional redundancies that lead to weak or confusing phenotypes. Affinity chromatography and mass spectrometry, in addition to direct binding assays, have provided us with many candidates to explain the role of myosin VI in Drosophila development, and we expect that this method will be useful for other motors in the future.
Methods
Detailed methods are described in SI Methods.
Protein Expression and Purification.
GST-tagged M6 (amino acids 833–1,256), M5 (amino acids 1,392–1,792), and Corn 5 (amino acids 770–978) were purified with glutathione agarose. For column construction, M6 and M5 were further purified by ion exchange chromatography. For binding assays, M6 and Corn 5 were cleaved from GST on resin using GST-HRV 3C protease and then subjected to ion exchange chromatography.
Preparation of Embryonic Extract.
Embryos were lysed in the presences of protease inhibitors, ATP, and sodium orthovanadate, followed by a low-speed spin, dilution, and ultracentrifugation.
Column Construction and Affinity Chromatography.
BSA and GST (approximately 100 mg) were each coupled to 10 mL of Affi-gel resin; GST-M6 and GST-M5 (approximately 10 mg) were each coupled to 1 mL Affi-gel resin. Extracts were cleared by two passes over each of the BSA and GST resins, split into two fractions, passed over myosin columns, recombined, split, and passed again over the myosin columns. Columns were subjected to washes with a buffer containing 100 mM KCl followed by stepwise elutions with 250 mM, 500 mM, and 1 M KCl. Fractions were trichloroacetic acid (TCA)-precipitated, pelleted, and then frozen or prepared for SDS-PAGE.
Mass Spectrometry.
Mass spectrometry sample handling and protein identification were performed as a paid service by NextGen, Inc. on TCA-precipitated pellets from 1 M elutions with an LTQ Orbitrap XL mass spectrometer and Mascot search engine; data were provided as a searchable Scaffold file.
Direct Binding Assays.
Myc-tagged candidates were transcribed and translated in vitro and applied to GST-M6 or GST-M5 on glutathione agarose in a 150 mM NaCl buffer. Binding was allowed to proceed for 1 h, followed by three washes in a 300 mM NaCl buffer. Retention by either resin was assessed by anti-myc Western blot.
Antibody Production and Purification.
Antibodies were raised against M6 and M5 in rabbits by Cocalico Biologicals, Inc., and against Corn 5 in chickens by Josman Labs, according to standard techniques. Each was affinity purified over columns made from purified antigen.
Coimmunoprecipitations.
S2R+ cells were lysed and centrifuged, and the supernatants were applied to IgG or IgY on Sepharose resin for 1.5 h. Resins were washed three times in a 300 mM NaCl buffer and then prepared for SDS-PAGE followed by anti-M6 Western blot.
Hedgehog Secretion Assay.
S2R+ cells were seeded onto 12-well plates. After 1 d, cells were transfected with dsRNAs targeting YFP, dispatched, jaguar (M6), or cornetto. The next day, cells were transfected with Hh or HhN DNA. On the fifth day, conditioned media was collected and subjected to SDS-PAGE followed by anti-Hedgehog Western blot.
Cuticle Preparations.
Crosses between engrailed-GAL4 females were crossed to y1w67c23, UAS-jaguar tail-CBD, or UAS-cornetto RNAi males at 28–29 °C. Fixations and cuticle preparations were performed according to standard techniques.
Supplementary Material
Acknowledgments.
We thank Drs. Aaron Straight and Phil Beachy (Stanford University) for plasmids, antibodies, and helpful suggestions. We thank Drs. Yuh-Nung Jan (University of California, San Francisco) and Roel Nusse (Stanford University) for fly stocks and use of a darkfield microscope, Dr. Mark Krasnow (Stanford University) for use of a confocal microscope, and Mark McElwain for help with Drosophila genetics. We thank members of the Spudich and Straight labs for helpful discussions and Drs. Alex Dunn, I. Robert Lehman, and Rajat Rohatgi for critical reading of the manuscript. We thank the Transgenic RNAi Project at Harvard Medical School (National Institutes of Health/National Institute of General Medical Sciences Grant R01-GM084947) for providing a plasmid vector used in this study. This work was supported by National Institutes of Health Grant GM33289 and a Human Frontiers Science Program grant to J.A.S., National Institutes of Health Cell and Molecular Biology Training Grant T32–GM007276 supporting M.A.H., and Stanford Graduate Fellowships awarded to D.F. and M.A.H.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101415108/-/DCSupplemental.
References
- 1.Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci USA. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol. 2004;16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 3.O’Connell CB, Tyska MJ, Mooseker MS. Myosin at work: Motor adaptations for a variety of cellular functions. Biochim Biophys Acta. 2007;1773:615–630. doi: 10.1016/j.bbamcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Krendel M, Mooseker MS. Myosins: Tails (and heads) of functional diversity. Physiology. 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- 5.Nambiar R, McConnell RE, Tyska MJ. Myosin motor function: The ins and outs of actin-based membrane protrusions. Cell Mol Life Sci. 2010;67:1239–1254. doi: 10.1007/s00018-009-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown ME, Bridgman PC. Myosin function in nervous and sensory systems. J Neurobiol. 2004;58:118–130. doi: 10.1002/neu.10285. [DOI] [PubMed] [Google Scholar]
- 7.Akhmanova A, Hammer JA., III Linking molecular motors to membrane cargo. Curr Opin Cell Biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karcher RL, Deacon SW, Gelfand VI. Motor-cargo interactions: The key to transport specificity. Trends Cell Biol. 2002;12:21–27. doi: 10.1016/s0962-8924(01)02184-5. [DOI] [PubMed] [Google Scholar]
- 9.Sweeney HL, Houdusse A. What can myosin VI do in cells? Curr Opin Cell Biol. 2007;19:57–66. doi: 10.1016/j.ceb.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Buss F, Spudich G, Kendrick-Jones J. Myosin VI: Cellular functions and motor properties. Annu Rev Cell Dev Biol. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- 11.Roberts R, et al. Myosin VI: Cellular functions and motor properties. Philos Trans R Soc Lon B. 2004;359:1931–1944. doi: 10.1098/rstb.2004.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez OC, Cheney RE. A new direction for myosin. Trends Cell Biol. 2000;10:307–311. doi: 10.1016/s0962-8924(00)01797-9. [DOI] [PubMed] [Google Scholar]
- 13.Mermall V, Miller KG. The 95F unconventional myosin is required for proper organization of the Drosophila syncytial blastoderm. J Cell Biol. 1995;129:1575–1588. doi: 10.1083/jcb.129.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millo H, Leaper K, Lazou V, Bownes M. Myosin VI plays a role in cell-cell adhesion during epithelial morphogenesis. Mech Dev. 2004;121:1335–1351. doi: 10.1016/j.mod.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Hicks JL, Deng WM, Rogat AD, Miller KG, Bownes M. Class VI unconventional myosin is required for spermatogenesis in Drosophila. Mol Biol Cell. 1999;10:4341–4353. doi: 10.1091/mbc.10.12.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng W, Leaper K, Bownes M. A targeted gene silencing technique shows that Drosophila myosin VI is required for egg chamber and imaginal disc morphogenesis. J Cell Sci. 1999;112:3677–3690. doi: 10.1242/jcs.112.21.3677. [DOI] [PubMed] [Google Scholar]
- 17.Petritsch C, Tavosanis G, Turck CW, Jan LY, Jan YN. The Drosophila myosin VI Jaguar is required for basal protein targeting and correct spindle orientation in mitotic neuroblasts. Dev Cell. 2003;4:273–281. doi: 10.1016/s1534-5807(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 18.Lin HP, et al. Cell adhesion molecule Echinoid associates with unconventional myosin VI/Jaguar motor to regulate cell morphology during dorsal closure in Drosophila. Dev Biol. 2007;311:423–433. doi: 10.1016/j.ydbio.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 19.Lantz VA, Miller KG. A class VI unconventional myosin is associated with a homologue of a microtubule-binding protein, cytoplasmic linker protein-170, in neurons and at the posterior pole of Drosophila embryos. J Cell Biol. 1998;140:897–910. doi: 10.1083/jcb.140.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millo H, Bownes M. The expression pattern and cellular localisationof myosin VI during the Drosophila melanogaster life cycle. Gene Expr Patterns. 2007;7:501–510. doi: 10.1016/j.modgep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Miller KG, Field CM, Alberts BM, Kellogg DR. Use of actin filament and microtubule affinity chromatography to identify proteins that bind to the cytoskeleton. Methods Enzymol. 1991;196:303–319. doi: 10.1016/0076-6879(91)96028-p. [DOI] [PubMed] [Google Scholar]
- 22.von Mering C, et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 23.Mermall V, et al. Drosophila myosin V is required for larval development and spermatid individualization. Dev Biol. 2005;286:238–255. doi: 10.1016/j.ydbio.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 24.Bulgheresi S, Kleiner E, Knoblich JA. Inscuteable-dependent apical localization of the microtubule-binding protein Cornetto suggests a role in asymmetric cell division. J Cell Sci. 2001;114:3655–3662. doi: 10.1242/jcs.114.20.3655. [DOI] [PubMed] [Google Scholar]
- 25.Papoulas O, Hays TS, Sisson JC. The golgin Lava lamp mediates dynein-based Golgi movements during Drosophila cellularization. Nat Cell Biol. 2005;7:612–618. doi: 10.1038/ncb1264. [DOI] [PubMed] [Google Scholar]
- 26.Sisson JC, Field C, Ventura R, Royou A, Sullivan W. Lava lamp, a novel peripheral golgi protein, is required for Drosophila melanogaster cellularization. J Cell Biol. 2000;151:905–918. doi: 10.1083/jcb.151.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisman RC, Stewart N, Miller D, Kaufman TC. Centrosomin's beautiful sister (cbs) encodes a GRIP-domain protein that marks Golgi inheritance and functions in the centrosome cycle in Drosophila. J Cell Sci. 2006;119:3399–3412. doi: 10.1242/jcs.03088. [DOI] [PubMed] [Google Scholar]
- 28.Djiane A, Mlodzik M. The Drosophila GIPC homologue can modulate myosin based processes and planar cell polarity but is not essential for development. PLoS One. 2010;5:e11228. doi: 10.1371/journal.pone.0011228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reed BC, et al. GLUT1CBP(TIP2/GIPC1) interactions with GLUT1 and myosin VI: Evidence supporting an adapter function for GLUT1CBP. Mol Biol Cell. 2005;16:4183–4201. doi: 10.1091/mbc.E04-11-0978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh Y, Imakagura H, Futatsumori M, Nakayama K. Recruitment of clathrin onto endosomes by the Tom1-Tollip complex. Biochem Biophys Res Commun. 2006;341:143–149. doi: 10.1016/j.bbrc.2005.12.156. [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Pacifico S, Liu G, Finley RL., Jr DroID: The Drosophila Interactions Database, a comprehensive resource for annotated gene and protein interactions. BMC Genomics. 2008;9:461. doi: 10.1186/1471-2164-9-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 33.Schnorrer F, et al. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature. 2010;464:287–291. doi: 10.1038/nature08799. [DOI] [PubMed] [Google Scholar]
- 34.Kellerman KA, Miller KG. An unconventional myosin heavy chain gene from Drosophila melanogaster. J Cell Biol. 1992;119:823–834. doi: 10.1083/jcb.119.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mermall V, McNally JG, Miller KG. Transport of cytoplasmic particles catalysed by an unconventional myosin in living Drosophila embryos. Nature. 1994;369:560–562. doi: 10.1038/369560a0. [DOI] [PubMed] [Google Scholar]
- 36.Geething NC, Spudich JA. Identification of a minimal myosin Va binding site within an intrinsically unstructured domain of melanophilin. J Biol Chem. 2007;282:21518–21528. doi: 10.1074/jbc.M701932200. [DOI] [PubMed] [Google Scholar]
- 37.Konecna A, et al. Calsyntenin-1 docks vesicular cargo to kinesin-1. Mol Biol Cell. 2006;17:3651–3663. doi: 10.1091/mbc.E06-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puri C, et al. Overexpression of myosin VI in prostate cancer cells enhances PSA and VEGF secretion, but has no effect on endocytosis. Oncogene. 2010;29:188–200. doi: 10.1038/onc.2009.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warner CL, et al. Loss of myosin VI reduces secretion and the size of the Golgi in fibroblasts from Snell's waltzer mice. EMBO J. 2003;22:569–579. doi: 10.1093/emboj/cdg055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mann RK, Beachy PA. Novel lipid modifications of secreted protein signals. Annu Rev Biochem. 2004;73:891–923. doi: 10.1146/annurev.biochem.73.011303.073933. [DOI] [PubMed] [Google Scholar]
- 41.Burke R, et al. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99:803–815. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- 42.Sanson B. Generating patterns from fields of cells. Examples from Drosophila segmentation. EMBO Rep. 2001;2:1083–1088. doi: 10.1093/embo-reports/kve255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JJ, von Kessler DP, Parks S, Beachy PA. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- 44.Mummery-Widmer JL, et al. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shalaby NA, et al. A screen for modifiers of notch signaling uncovers Amun, a protein with a critical role in sensory organ development. Genetics. 2009;182:1061–1076. doi: 10.1534/genetics.108.099986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris SM, et al. Myosin VI binds to and localises with Dab2, potentially linking receptor-mediated endocytosis and the actin cytoskeleton. Traffic. 2002;3:331–341. doi: 10.1034/j.1600-0854.2002.30503.x. [DOI] [PubMed] [Google Scholar]
- 47.Spudich G, et al. Myosin VI targeting to clathrin-coated structures and dimerization is mediated by binding to Disabled-2 and PtdIns(4,5)P2. Nat Cell Biol. 2007;9:176–183. doi: 10.1038/ncb1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brownhill K, Wood L, Allan V. Molecular motors and the Golgi complex: Staying put and moving through. Semin Cell Dev Biol. 2009;20:784–792. doi: 10.1016/j.semcdb.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 49.Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- 50.Payre F. Genetic control of epidermis differentiation in Drosophila. Int J Dev Biol. 2004;48:207–215. [PubMed] [Google Scholar]
- 51.Phizicky EM, Fields S. Protein-protein interactions: Methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.