Abstract
Large conformational transitions play an essential role in the function of many proteins, but experiments do not provide the atomic details of the path followed in going from one end structure to the other. For the hemoglobin tetramer, the transition path between the unliganded (T) and tetraoxygenated (R) structures is not known, which limits our understanding of the cooperative mechanism in this classic allosteric system, where both tertiary and quaternary changes are involved. The conjugate peak refinement algorithm is used to compute an unbiased minimum energy path at atomic detail between the two end states. Although the results confirm some of the proposals of Perutz [Perutz MF (1970) Stereochemistry of cooperative effects in haemoglobin. Nature 228:726–734], the subunit motions do not follow the textbook description of a simple rotation of one αβ-dimer relative to the other. Instead, the path consists of two sequential quaternary rotations, each involving different subdomains and axes. The quaternary transitions are preceded and followed by phases of tertiary structural changes. The results explain the recent photodissociation measurements, which suggest that the quaternary transition has a fast (2 μs) as well as a slow (20 μs) component and provide a testable model for single molecule FRET experiments.
Keywords: conformational change, cooperativity, domain motion, protein hinges
Many proteins undergo large conformational transitions that are essential for their functions (1–3). The transitions can occur in monomers such as the myosin molecular motor (4–6), in multisubunit complexes such as the chaperone GroEL (7), and in systems such as the flagellar motor of bacteria, which is composed of several hundred proteins (8). Probably the most studied example is the T (tense) to R (relaxed) transition in the vertebrate hemoglobin tetramer (Hb) (9–11), for which the conformational transition increases the efficiency of oxygen transport. Hemoglobin, which is composed of two identical α- and two identical β-subunits, is the paradigm for the development of models of cooperativity and allosteric regulation in proteins (9–13). Phenomenologically, the positive cooperativity involves the increase in the affinity for oxygen of unliganded subunits upon the successive binding of oxygen to other subunits. This is achieved by structural changes within subunits (tertiary changes) and between subunits (quaternary changes). Based on the superposition of the crystallographic structures for the deoxy unliganded T state and the fully liganded R state of the protein, the quaternary T → R transition has been described as involving primarily an approximately 15° rotation of one “dimer” (α1β1) relative to the other (α2β2) around a virtual axis (Fig. 1A) (9, 14). Together with a small relative translation of the αβ-dimers, this reduces the central cavity between the dimers (a channel along the C2 axis of symmetry), where the heterotropic effector 2,3-bisphosphoglycerate is bound in the T state. An important aspect of these structural changes is that they are accompanied by alterations in the packing of residues at the interfaces between the subunits. There is a “switch” in the dovetailing between residues of the β1- and the α2-subunits (and correspondingly at the α1/β2-interface) (14); another contact between the dimers has been referred to as the “shift” region (14). In addition, the breaking of several salt bridges that favor the T state has been proposed as playing an important role in the energetics of the transition (9, 13, 15). Binding of oxygen to the heme groups results in destabilization of these salt bridges by a steric mechanism involving undoming of the hemes, as Perutz suggested in his model for cooperative oxygen binding (15) (for a recent review, see ref. 9) and was confirmed by atomic-level energy calculations, which introduced the concept of an allosteric core (16).
Fig. 1.
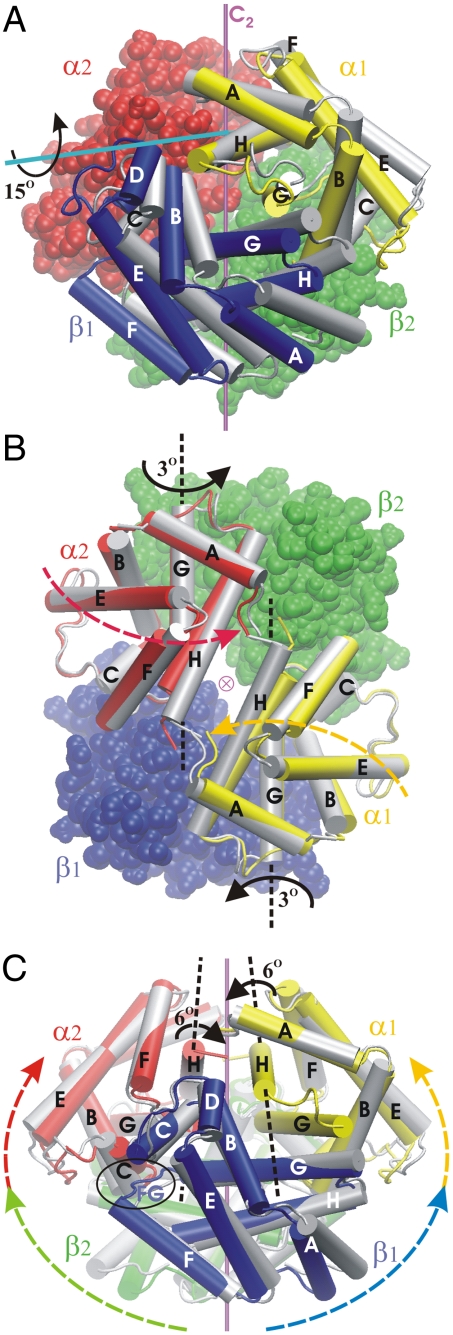
The T → R transition of hemoglobin. (A) Comparing the T (i.e., deoxy, shown in color) and R (tetraoxy, in gray) conformers (superposed by fitting their α2β2-subunits). The α1β1-dimer as a whole appears rotated by approximately 15° [the screw axis identified by DynDom (28) is shown in cyan]. The C2-symmetry axis relating the α1β1- and α2β2-dimers is in magenta. (B) First quaternary event (Q1): relative to its position in the T state (in color), each α-subunit rotates by 3° around its G helix (the two α-subunits pivoting toward each other), resulting in the intermediate halfway along the path (in gray). The view is down the central channel, along the C2-symmetry axis (⊗). (C) Second quaternary event (Q2): relative to its position halfway along the path (in color), each αβ-dimer rotates by 6° (R state shown in gray) around its αH helix. One of the two switch regions (α2C/β1FG loop) is circled. Movies S1 and S2 displaying the two quaternary transitions (Q1 and Q2) are provided in SI Text.
The comparison of two end structures does not provide direct information on the actual pathway(s) of the conformational change. Consequently, a number of calculations to simulate the quaternary transition of Hb have been made. They include a rigid body model (17), an implementation of targeted molecular dynamics (18), and a normal mode analysis (19). In this paper, we have used the conjugate peak refinement (CPR) method (20) to determine the minimum energy pathway (MEP) involved in the transition. The CPR method makes no assumptions concerning the reaction coordinate and finds a continuous path between the end states that follows the valleys and climbs over the connecting passes (saddle points) of the energy surface. Every atom is allowed to move independently and no constraints are introduced to drive the transition. Fig. S1 shows a simple illustration of the method. Examples of applications include a bacterial pore (21) and myosin (5), for which the transition mechanism found by CPR was subsequently verified by experiment (22).
The CPR method leads to the unexpected result that the pathway from the T to the R structure is more complex than the standard model based on the relative rotation/translation of the two αβ-dimers (14). It consists of two quaternary transitions (Q1 and Q2), each involving a different set of subunits and different rotation axes. Tertiary structural changes precede and accompany these quaternary transitions. The overall C2 symmetry of the αβ-dimers is preserved, but the α- and β-subunits behave very differently. These results provide an unsuspected pathway of the allosteric transition of hemoglobin and serve to explain some recent experiments on the R to T transition after photodissociation (23–26). Although the calculations were essentially completed and most of the manuscript was written by 2000, a crucial experiment (23) validating the results was published only in 2010. In answer to a referee’s comment, we note that one of the authors (M.K.) inquired of the editors of several journals about publication and was told that the manuscript would not be considered unless there was experimental evidence to support the “iconoclastic” results.
The findings are also important because they show that caution is required in evaluating the results of some widely used interpolation methods for analyzing complex conformational transitions in proteins; see, for example, ref. 27.
Results
Behavior of the Subunits.
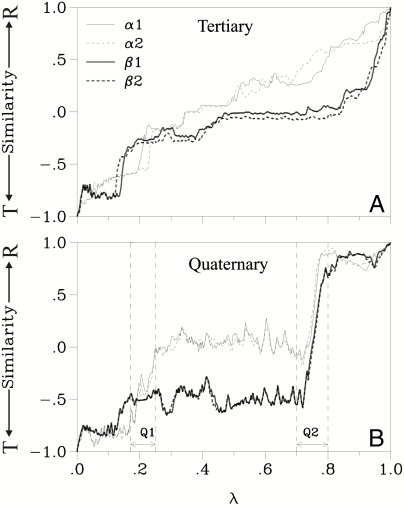
Fig. 2 shows the progress of structural change in each of the four subunits along the MEP at the tertiary and the quaternary levels (in terms of deviations relative to the T and R conformations, see Methods). The α1- and α2-subunits change nearly synchronously, particularly at the quaternary level, even though C2 symmetry was not imposed as a constraint during the calculations. The same synchrony is observed between the β-subunits. Local motion, such as side-chain rearrangements, occurs in one pair of αβ-subunits only slightly before or after the corresponding change in the other pair, resulting in small differences in progress at the tertiary level between corresponding subunits (for example, α1 relative to α2; see Fig. 2A). Thus, the global tertiary and quaternary C2 symmetry of hemoglobin is essentially maintained during the transition.
Fig. 2.
Progress along the path. The similarity to either end state (T or R) is plotted for structures taken along the minimum energy path. The reaction coordinate λ is the normalized integral (along the path) of all atomic coordinate changes (measured as a rms difference). (A) The tertiary (i.e., intrasubunit) and (B) the quaternary (i.e., intersubunit) similarity are separated for each of the four subunits, as described in Methods. The first (Q1) and second (Q2) major quaternary events are indicated. See Methods for a description of the similarity measure.
In contrast to this synchrony between the two α-subunits and the two β-subunits, the structural progress of the α-subunits is not synchronous with that of the β-subunits. This uncoupling between α- and β-subunits occurs at both the tertiary level, as indicated by the difference in the curves in Fig. 2A, and the quaternary level (Fig. 2B). Over most of the path, the quaternary progress of the α-subunits toward the R state precedes that of the β-subunits.
The Quaternary Events.
There are two distinct quaternary events, called here Q1 and Q2 (see Fig. 2B) at reaction coordinate values λ = 0.18–0.25 and λ = 0.7–0.8. Each of the major quaternary events involves a different portion of the protein. During event Q1, the protein behaves as if it was composed of three semirigid bodies: the α1-subunit, the α2-subunit, and the β1β2-dimer. The motion is a simultaneous rotation by 3° of each α-subunit relative to the “stationary” β1β2-dimer (as shown in Fig. 1B), bringing the two α-subunits closer to each other. The rotation axis of each α-subunit is close to the axis of its αG helix, which remains essentially stationary and thus functions like a hinge. Two regions that play an essential role in Q2 are modified as a result of Q1: (i) The axis of each αH helix moves significantly (see Fig. 1B) and (ii) the dovetailing at the switch interface is loosened (as described below). In the second major quaternary event, Q2, the protein behaves as if composed of two bodies: the α1β1-dimer and the α2β2-dimer. Each dimer rotates by 6° in the opposite sense of the other dimer, around a rotation axis that runs along the respective αH helix (Fig. 1C).
Summarizing, the conformational difference between the end states (Fig. 1A) is the result of two successive sets of semirigid rotations, Q1 and Q2, each involving different regions of the protein. Four different rotation axes are involved, two in each quaternary event (Fig. 1 B and C). These axes coincide approximately with elements of secondary structure, namely the αG and αH helices. It is noteworthy that the helices that move most during event Q1 (αH) serve as axes during Q2. The overall C2 symmetry is conserved because, during both Q1 and Q2, each quaternary motion in the α1β1-dimer is mirrored in the α2β2-dimer. The small overall translation of α1β1 toward α2β2 noted by Baldwin and Chothia in their comparison of the T and R structures (14) takes place in two steps here: First, α1 moves closer to α2 during Q1; then β1 moves closer to β2 during Q2.
Subunit Interfaces.
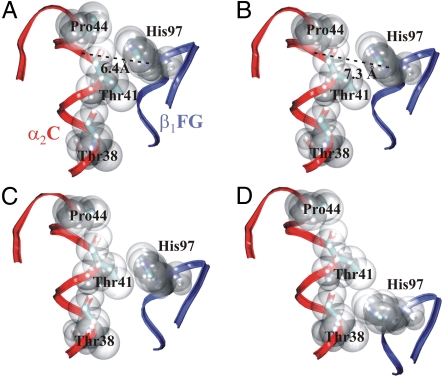
An important feature distinguishing the T and R crystal structures is the switch region, in which the dovetailing between the β1FG loop and the α2C helix (visible in Fig. 1C) at the α2/β1-interface (and respectively at the α1/β2-interface) is changed (9, 14, 15). This is an essential contact point between the α1β1- and α2β2-dimers. In the T state, the side chain of β1His97 is between the side chains of α2Pro44 and α2Thr41 (Fig. 3A); in the R state, it is between α2Thr41 and α2Thr38 (Fig. 3D). Baldwin and Chothia suggested that this change might occur as an abrupt switch between quaternary states (14), β1His97 acting as a prong that “clicks” from one groove of the α2C helix into the next. This is indeed what happens along the present minimum energy path, where the switch occurs simultaneously in both interfaces during event Q2. The residues of the switch region are closely packed in the end states (Fig. 3 A and D). Before and mostly during the quaternary event Q1, this packing loosens, and β1His97 moves out of the α2C-helix groove by nearly 1 Å (Fig. 3B). This allows β1His97 to glide past α2Thr41 during Q2 essentially without rearrangements or distortions (Fig. 3C). Thus, the motion of the α-subunits during event Q1 prepares for Q2 by loosening the α1/β2-(and α2/β1-) interface to facilitate the switch.
Fig. 3.
The switch interface. (A) T state, with β1His97 packing into the groove between α2Pro44 and α2Thr41 of helix α2C. Path intermediates: (B) Just before event Q2 (at λ = 0.72), the packing has loosened (i.e., β1His97 moved away from the helix). The distance between His97 (Cβ atom) and the helix groove (Cα of Tyr42) is indicated. (C) Intermediate halfway along the Q2 event (at λ = 0.75). (D) R state, with β1His97 in the groove between α2Thr41 and α2Thr38. The backbone of helix α2C is shown in red and of the β1FG loop in blue.
The other contact region between the dimers is between the β1C helix and the α2FG loop, which has been referred to as the shift or the “flexible joint” region. Unlike the distinct motion of the C helix relative to the FG loop that is observed in the switch region, their counterparts in the shift region move little relative to each other and without significant rearrangement of their side chains. This is because the shift region is located radially close to both the Q1 and the Q2 rotation axes, so that the two quaternary rotations produce little conformational change in the shift region.
Intermediate Structure.
Halfway along the transition between Q1 and Q2 (between λ 0.3 and 0.7 in Fig. 2), the overall structure is not simply an interpolation between the T and R conformers. At the quaternary level, the α-subunits are in a position between that of the T and R states, whereas the β-subunits are still in a more T-like state (Fig. 2B). Compared to the T state (Fig. 4A), this structure has the entrance to the central cavity reduced between the α-subunits (Fig. 4B) by nearly as much as it will be in the R state. This shrinking is mostly due to the rotation of the α-subunits toward each other during event Q1, which brings the α1 and α2 H helices closer together (as seen in Fig. 1B). The EF loops and the N-terminal segments of the α-subunits also contribute to the shrinking (Fig. 1B). In contrast, the central cavity of the intermediate structure is still fully open between the β-subunits (Fig. 4B); it closes during event Q2. Thus, between events Q1 and Q2, the protein has an intermediate quaternary conformation, which is maintained approximately during the phases of tertiary changes described in the next section.
Fig. 4.
Shrinking of the central cavity. (A) In the T state (shown as molecular surface), the central channel along the C2-symmetry axis is fully open (same view and coloring as Fig. 1B). (B) In the intermediate structure (λ = 0.5, viewed from the opposite end of the channel), the channel is nearly closed between the α-subunits (also shown as licorice in A) but still open between the β-subunit. When reaching the R state (shown as licorice in B), the channel closes also between the β-subunits.
Tertiary Phases.
The concerted motions of entire domains during events Q1 and Q2 are preceded and followed by series of mostly localized rearrangements of side chains and loops. Thus, Q1 and Q2 divide the transition into three phases of tertiary change. The amplitude of the displacements in each phase is shown in Fig. 5. The dominant motions are summarized here in the order of occurrence.
Fig. 5.
Displacements during successive phases. A serves to identify the main secondary elements: In this orientation (same as in Fig. 1A), the axes of event Q1 (αG helices, in blue) and the axes of event Q2 (αH helices, in brown) can be recognized. The βG helices are shown in yellow, βH in red. The βCD loops are shown in purple, βE and βF in orange, and βNA, βA, and βEF in gray. B–F use the same orientation (but subunits are colored as in Fig. 1). The backbone thickness is drawn proportional to the amplitude of motion during each transition phase: (B) first tertiary phase. (C) Event Q1. D) second tertiary phase. (E) Event Q2. (F) third tertiary phase.
During the first tertiary phase (λ = 0.0–0.18 in Fig. 2), the CD loops and the C termini of the β-subunits are moving the most (Fig. 5B). Perutz had identified a set of salt bridges as being important for stabilizing the T state (15), although there are additional contributions to stabilizing the T state versus the R state (10, 16). Six of the salt bridges are at the interfaces between subunits and two are internal to the β-subunits. All of these salt bridges are broken before the second quaternary phase Q2. The breaking of these salt bridges increases the motional freedom at the subunit interfaces that is required for the quaternary movement of the subunits. In Q1 the β-subunits are essentially stationary (as can be seen from the small amplitude of their motion indicated by the thin backbone trace in Fig. 5C), whereas there are large displacements in the α-subunits, each of which rotates 3° around its αG helix as described above (Fig. 1B).
During the second tertiary phase (λ = 0.25–0.7, between events Q1 and Q2), there are small fluctuations at the quaternary level (see Fig. 2B), which indicate that the tetramer is somewhat labile in this intermediate structure. These fluctuations are induced by many small tertiary rearrangements, the largest of which occur in the N-terminal segments and the EF loops of the β-subunits, and in the CD loops and the C termini of the α-chains (Fig. 5D). The distal histidine in each β-subunit (but not in the α-subunits) moves out of the heme pocket to form a hydrogen bond to the mainchain of β-Lys59 and remains in this position until after event Q2, when it returns to the heme pocket. The significance of this movement is not clear, but it could facilitate oxygen access to the β-hemes. This is followed by quaternary event Q2, in which the amplitude of motion is much larger in the β-subunits than in the α-subunits (Fig. 5E). This amplitude increases radially with distance from the αH helices, which are essentially stationary (as indicated by the thin backbone traces in Fig. 5E) and serve as rotation axes for each αβ-dimer as described above (see Fig. 1C). An effect of this rotation is to shrink the central cavity between the β-subunits (see Fig. 4B).
In the third tertiary phase (λ = 0.8–1.0), the changes occur mostly in the β-subunits and are greatest at the entrance of the central channel (Fig. 5F), including the N-terminal segment, the EF loop, and the F and H helices. This completes the shrinking of the β-entrance to the central cavity (Fig. 4B).
Discussion
The conjugate peak refinement algorithm (20) has been used to determine the tertiary and quaternary structural changes along the allosteric pathway of tetrameric hemoglobin. Using the two known end structures (that of the unligand T and the liganded R state), it has been shown that the CPR minimum energy path for the quaternary transition is fundamentally different from the textbook description. As originally noted by Baldwin and Chothia (14), when the end states are superposed by fitting their α2β2-dimers, the α1β1-dimer is related in the two end states by a 15° rotation. Although this superposition approach served to highlight the differences between the X-ray structures, the 15° rotation is shown in the present analysis to be the product of two quaternary rotations (Q1 and Q2) that occur in succession. Such complex mechanics are beyond the scope of even sophisticated multihinge finding programs [e.g., DynDom (28)] when one relies on comparing only the transition end states (see also SI Text).
In spite of the quasisymmetry relating the α-subunits to the β-subunits in the crystallographic end states, the results show that the α- and β-subunits behave very differently along the path. Along the new pathway, it is still valid (9, 14) to consider hemoglobin as a dimer of αβ-dimers in which the C2 symmetry between the two αβ-dimers is essentially maintained. This behavior is consistent with a recent NMR study showing that carbon monoxy-hemoglobin, while undergoing its quaternary transition between the R and R2 states (R2 is a low-salt form), retains C2 symmetry in solution (29). It also is consistent with the Monod–Wyman–Changeux model of cooperativity (12), in which the protein retains its symmetry at the quaternary level. Conservation of C2 symmetry means here that both quaternary events involve two equivalent axes of rotation, one in each αβ-dimer; i.e., each α-subunit rotates around its own G helix during event Q1 (Fig. 1B), and each αβ-dimer rotates around its αH helix during event Q2 (Fig. 1C). Having two successive quaternary rotation events, each with two axes related by C2 symmetry, results in a total of four rotation axes. Quaternary event Q1 brings the two αH helices into a position where they can serve as rotation axes for quaternary event Q2. If the αβ-dimers were to rotate around their αH helix before Q1, this would result in a different transformation. The loosening of the α1β2-switch interface takes place before and during event Q1 and prepares for the switch, which occurs concurrently with event Q2. This in turn defines the order of the three tertiary phases, the first two of which facilitate the quaternary rotations and the third follows the second quaternary rotation as a function of the CPR parameter λ. Given that tertiary events are on the nanosecond time scale, whereas quaternary events are in the microsecond range (see also below), the last tertiary event is expected to be essentially simultaneous with Q2.
The present results that the transition of hemoglobin from the T to the R state is composed of a minor (Q1) and a major (Q2) quaternary transition, which are preceded or accompanied by tertiary transitions, can be compared with several recent experiments. All of them study what happens to R-state hemoglobin after photodissociation of CO; i.e., the data obtained follow the R to T transition. The results can be directly compared with the reverse of the direction described in the CPR path reported here, because the minimum energy path is independent of the direction. Experimentally, it is known that the complete transition occurs on a time scale of 20 to 40 μs (30, 31). A recently published paper (23) used time-resolved wide angle X-ray scattering (TR-WAXS) to determine the time scale of major structural changes in the R to T transition after photolysis. Interestingly, the observed time constant is 2 μs, much faster than the 20 μs of the overall transition. The authors propose that the slow portion of the quaternary transition thus corresponds to subsequent smaller changes in the R to T direction that are not detectable within the accuracy of their experiment. The WAXS study complements earlier TR UV magnetic circular dichroism measurements (24) associated with Trp residues and UV resonance Raman (UVRR) measurements (25) associated with both Trp and Tyr residues. They indicate that the Trp β37-Asp α94 hydrogen bond in the shift (hinge) region of the α1β2-interface is formed in 2 μs after the photodissociation, whereas the Tyr α42-Asp β99 hydrogen bond in the switch region of the α1β2-interface is formed in 20 μs. Both of these hydrogen bonds are present only in the T structure, so the authors conclude that the quaternary transition has both a fast (2 μs) and a slow (20 μs) phase.
Although the present calculations do not provide information on the time constants of the transitions, the recent experiments (for an overview, see ref. 26) provide strong support for the two-step quaternary transition predicted by the calculations (see Fig. 2); in the R to T direction, the early large quaternary change Q2 would correspond to the 2-μs step, whereas the smaller late quaternary change Q1 and some tertiary changes would correspond to the 20-μs step.
As to the H-bond data cited above, the comparison is complicated by the fact that the energy-minimized X-ray structures used in the CPR calculations change slightly upon minimization and that the UV data (24, 25) do not provide direct information on the transition. Specifically, the Tyr α42-Asp β99 hydrogen bond in the switch region was concluded to occur as part of the slow step in the UVRR measurement (25). However, the H bond is lost in the minimized T structure, and the actual switch occurs in Q2, though the contacts in the switch region loosen during Q1 (see above). This appears to disagree with the conclusion of ref. 25 unless Q2 is associated with the slow step. The other H bond studied spectroscopically, Trp β37-Asp α94, was determined to be formed during the early fast (2-μs time constant) transition phase in the UV measurements (24, 25). Although this H bond changes very little in the transition to the R structure in the calculations and in the available X-ray structures (32–34), the main calculated change occurs during Q2. This result, together with the UV measurements, provides another indication that Q2 happens during the fast phase.
The use of silica gels to trap unstable intermediates of hemoglobin (35) also supports the present pathway. These experiments have shown that liganded hemoglobin blocked by the gel in a quaternary T state can have R-state kinetic properties, suggesting an incomplete coupling between the tertiary and quaternary changes. This uncoupling is exactly what is observed in the present CPR pathway: In the intermediate state between Q1 and Q2, the subunits have a more R-like tertiary structure (Fig. 2A), whereas the quaternary structure is clearly more T-like (Fig. 2B). Likewise, in the tertiary phase between λ = 0.8 and 1.0, large changes occur at the tertiary level (particularly in the β-subunits), whereas the quaternary structure remains R-like (see Fig. 2). The separation of tertiary and quaternary changes suggested based on the silica gel trapping experiments and found in the present study was presaged in earlier phenomenological models of the allosteric transition; see, for example, refs. 36 and 37. Both the T and R state have two tertiary structures in equilibrium with one having a low affinity for O2 and the other having a high affinity for O2.
Thus, the major unsuspected features of the calculated results (i.e., that the quaternary transition involves two separate steps rather than the single process of the accepted Baldwin–Chothia model, and that tertiary changes are not strictly coupled to the quaternary changes) are supported by recent experimental work: When the CPR path is analyzed in the R to T direction (to simplify the comparison with the photodissociation experiments), it is clear that (i) the quaternary transition Q2 comes first and (ii) is much larger than the subsequent and smaller transition Q1 (both of which are in accord with the WAXS findings), and (iii) that the tertiary and quaternary changes can be uncoupled (consistent with the gel experiments).
Because hemoglobin continues to serve as “the” system for discussions of quaternary control of biological activity in proteins, the result that the transition follows an unsuspected pathway appears to us to be of general interest. Although the experimental data given above support the proposed transition mechanism, it would be of interest to have more data concerning the transition. One possibility is based on trapping the Q1 state with a covalently linked heterotropic effector (see Fig. S2) and another on single molecule FRET measurements (38–40); see Figs. S3 and S4.
Methods
The CHARMM program (41, 42) polar hydrogen force field (parameter set 19) (42) was used (except for the aromatic side chains and the heme, represented with all hydrogens). Solvent screening of the electrostatic interactions was approximated by using distance-dependent dielectric screening and by scaling the charges of the ionic groups so as to reproduce intersubunit interactions obtained from solving the Poisson–Boltzman equation, as described previously (43). Because the calculations require a one-to-one correspondence of atoms in the reactant and product states, the oxygen molecules were removed from the oxy structure. However, even though the oxygens have been removed, the tertiary structure of the subunit changes from that in the unliganded T state to the liganded R state during the transition.
The MEP obtained by the CPR algorithm (20), which is implemented in the trajectory refinement and kinematics module of the program CHARMM (41, 42), consists of a series of intermediate structures of the protein, including the saddle point for every energy barrier crossed. CPR monitors the energy along every path segment (built by linear interpolation between adjacent intermediates) and adds enough intermediates until no energy barriers are hidden within segments (Fig. S1). Starting from the initial path built by linear interpolation in Cartesian coordinates between the energy-minimized crystal structures of T (44) and R (32) (which have rms deviations of 0.87 Å and 1.39 Å from the respective crystal structures), the final MEP is composed of about 1,700 structures and crosses 80 saddle points. Most of these saddle points are expected to be smoothed out along the free energy path due to thermal averaging (5, 45).
Methods such as the finite temperature string method (46) could, in principle, be used to determine the free energy, but are too costly to use at the present time for a system as large as hemoglobin. A recent application of the string method to a conformational transition in myosin V indicates that the minimum energy path and the minimum free energy path are very similar, though the main barrier is lowered significantly by the entropic contributions along the latter (47).
To assess the structural progress along the path in each monomeric subunit, a “similarity” measure was used in Fig. 2. It estimates whether the protein atomic coordinates xλ taken at a point λ along the path (see the caption of Figure 2 for the definition of λ) are more similar to the T state (xT) or the R state (xR): [rms(xλ,xT)rms(xλ,xR)]/rms(xT,xR), where rms(x,y) denotes the root mean square difference between structures x and y over the atoms of a given subunit. This similarity amounts to a hyperbolic projection onto the vector between the T and R coordinates. The similarity value varies between -1 and +1. For example, a value close to -1 (respectively +1) indicates that the structure is very similar to the T state (respectively, the R state); a value of zero indicates that the structure is equidistant from the two end states. Before calculating the rmsd, the two structures x and y are superimposed by using a least-squares fit. This separates the tertiary from the quaternary changes: For the tertiary change the least-squares fit is done with respect to the atoms of each subunit, whereas for quaternary change the least-squares fit involves all atoms of the protein. Contributions from intrasubunit change were removed from the quaternary similarity by substituting the internal coordinates of each subunit with those of a constant structure (chosen here as the average of T and R). Note that while the similarity described here is useful to measure the progress along a preexisting path, it is not appropriate as an order parameter (i.e., as a reaction coordinate) for free energy evaluations, as discussed previously (5).
Supplementary Material
Acknowledgments.
We thank Thomas Spiro for showing us experimental results before publication, William Eaton for helpful discussions, and Sidonia Zafiu for assistance with DynDom (28). The work was supported in part by a grant from the National Institutes of Health (to M.K.) and by a grant from the Deutsche Forschungs Gemeinschaft (to S.F.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011995108/-/DCSupplemental.
References
- 1.Alberts B. The cell as a collection of protein machines: Preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Gerstein M, Lesk AM, Chothia C. Structural mechanisms for domain movements in proteins. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 3.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc Natl Acad Sci USA. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geeves MA, Holmes KC. Structural mechanism of muscle contraction. Annu Rev Biochem. 1999;68:687–728. doi: 10.1146/annurev.biochem.68.1.687. [DOI] [PubMed] [Google Scholar]
- 5.Koppole S, Smith JC, Fischer S. The structural coupling between ATPase activation and recovery-stroke in the Myosin II motor. Structure. 2007;15:825–837. doi: 10.1016/j.str.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Cecchini M, Houdusse A, Karplus M. Allosteric communication in myosin V: From small conformational changes to large directed movements. PLoS Comput Biol. 2008;4:e1000129. doi: 10.1371/journal.pcbi.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma J, Sigler PB, Xu Z, Karplus M. A dynamic model for the allosteric mechanism of GroEL. J Mol Biol. 2000;302:303–313. doi: 10.1006/jmbi.2000.4014. [DOI] [PubMed] [Google Scholar]
- 8.Berg HC. The rotary motor of bacteria flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 9.Perutz MF, Wilkinson AJ, Paoli M, Dodson GG. The stereochemical mechanism of the cooperative effects in hemoglobin revisited. Annu Rev Biophy Biomol Struct. 1998;27:1–34. doi: 10.1146/annurev.biophys.27.1.1. [DOI] [PubMed] [Google Scholar]
- 10.Eaton WA, Henry ER, Hofrichter J, Mozzarelli A. Is cooperative oxygen binding by hemoglobin really understood? Nat Struct Biol. 1999;6:351–358. doi: 10.1038/7586. [DOI] [PubMed] [Google Scholar]
- 11.Cui Q, Karplus M. Allostery and cooperativity revisited. Protein Sci. 2008;17:1295–1307. doi: 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 13.Szabo A, Karplus M. A mathematical model for structure-function relations in hemoglobin. J Mol Biol. 1972;72:163–197. doi: 10.1016/0022-2836(72)90077-0. [DOI] [PubMed] [Google Scholar]
- 14.Baldwin J, Chothia C. Haemoglobin: Structural changes related to ligand binding and its allosteric mechanism. J Mol Biol. 1979;129:175–220. doi: 10.1016/0022-2836(79)90277-8. [DOI] [PubMed] [Google Scholar]
- 15.Perutz MF. Stereochemistry of cooperative effects in haemoglobin. Nature. 1970;228:726–734. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 16.Gelin BR, Lee AWM, Karplus M. Hemoglobin tertiary structural change on ligand binding: Its role in the co-operative mechanism. J Mol Biol. 1983;171:489–559. doi: 10.1016/0022-2836(83)90042-6. [DOI] [PubMed] [Google Scholar]
- 17.Janin J, Wodak SJ. Reaction pathway for the quaternary structure change in hemoglobin. Biopolymers. 1985;24:509–526. doi: 10.1002/bip.360240307. [DOI] [PubMed] [Google Scholar]
- 18.Mouawad L, Perahia D, Robert CH, Guilbert C. New insights into the allosteric mechanism of human hemoglobin from molecular dynamics simulations. Biophys J. 2002;82:3224–3245. doi: 10.1016/S0006-3495(02)75665-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mouawad L, Perahia D. Motions in hemoglobin studied by normal mode analysis and energy minimization: Evidence for the existence of tertiary T-like, quaternary R-like intermediate structures. J Mol Biol. 1996;258:393–410. doi: 10.1006/jmbi.1996.0257. [DOI] [PubMed] [Google Scholar]
- 20.Fischer S, Karplus M. Conjugate peak refinement: An algorithm for finding reaction paths and accurate transition-states in systems with many degrees of freedom. Chem Phys Lett. 1992;194:252–261. [Google Scholar]
- 21.Dutzler R, Schirmer T, Karplus M, Fischer S. Translocation mechanism of long sugar chains across the maltoporin membrane channel. Structure. 2002;10:1273–1284. doi: 10.1016/s0969-2126(02)00811-0. [DOI] [PubMed] [Google Scholar]
- 22.Kintses B, Yang Z, Málnási-Csizmadia C. Experimental investigation of the seesaw mechanism of the relay region that moves the myosin lever arm. J Biol Chem. 2008;283:34121–34128. doi: 10.1074/jbc.M805848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cammarata M, Levantino M, Wulff M, Cupane A. Unveiling the timescale of the R-T transition in human hemoglobin. J Mol Biol. 2010;400:951–962. doi: 10.1016/j.jmb.2010.05.057. [DOI] [PubMed] [Google Scholar]
- 24.Goldbeck RA, Esquerra RM, Kliger DS. Hydrogen bonding to Trp beta 37 is the first step in a compound pathway for hemoglobin allostery. J Am Chem Soc. 2002;124:7646–7647. doi: 10.1021/ja025855l. [DOI] [PubMed] [Google Scholar]
- 25.Balakrishnan G, et al. Time-resolved absorption and UV resonance Raman spectra reveal stepwise formation of T quaternary contacts in the allosteric pathway of hemoglobin. J Mol Biol. 2004;340:843–856. doi: 10.1016/j.jmb.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Spiro TG, Balakrishnan G. Quaternary speeding in hemoglobin. J Mol Biol. 2010;400:949–950. doi: 10.1016/j.jmb.2010.05.056. [DOI] [PubMed] [Google Scholar]
- 27.Krebs WG, Gerstein M. The morph server: A standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 2000;28:1665–1675. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayward S, Berendsen HJC. Systematic analysis of domain motions in proteins from conformational change: New results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 29.Lukin JA, et al. Quaternary structure of hemoglobin in solution. Proc Natl Acad Sci USA. 2003;100:517–520. doi: 10.1073/pnas.232715799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawicki CA, Gibson QH. Quaternary conformational changes in human hemoglobin studied by laser photolysis of carboxyhemoglobin. J Biol Chem. 1976;251:1533–1542. [PubMed] [Google Scholar]
- 31.Jones CM, et al. Speed of intersubunit communication in proteins. Biochemistry. 1992;31:6692–6702. doi: 10.1021/bi00144a008. [DOI] [PubMed] [Google Scholar]
- 32.Shaanan B. Structure of human oxyhaemoglobin at 2.1 Å resolution. J Mol Biol. 1983;171:31–59. doi: 10.1016/s0022-2836(83)80313-1. [DOI] [PubMed] [Google Scholar]
- 33.Tame JRH, Vallone B. The structures of deoxy human haemoglobin and the mutant Hb Tyr[alpha]42His at 120 K. Acta Crystallogr D. 2000;56:805–811. doi: 10.1107/s0907444900006387. [DOI] [PubMed] [Google Scholar]
- 34.Park WY, Yokoyama T, Shibayama N, Shiro Y, Tame JRH. 1.25 Å resolution crystal structures of human haemoglobin in the oxy, deoxy and carbonmonoxy forms. J Mol Biol. 2006;360:690–701. doi: 10.1016/j.jmb.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 35.Viappiani C, et al. New insights into allosteric mechanism from trapping unstable protein conformations in silica gels. Proc Natl Acad Sci USA. 2004;101:14414–14419. doi: 10.1073/pnas.0405987101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee AW, Karplus M. Structure-specific model of hemoglobin cooperativity. Proc Natl Acad Sci USA. 1983;80:7055–7059. doi: 10.1073/pnas.80.23.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henry ER, Bettati S, Hofrichter J, Eaton WA. A tertiary two-state allosteric model for hemoglobin. Biophys Chem. 2002;98:149–164. doi: 10.1016/s0301-4622(02)00091-1. [DOI] [PubMed] [Google Scholar]
- 38.Abbondanzieri EA, et al. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarsa PB, et al. Detecting force-induced molecular transitions with fluorescence resonant energy transfer. Angew Chem Int Edit. 2007;46:1999–2001. doi: 10.1002/anie.200604546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merchant KA, Best RB, Louis JM, Gopich IV, Eaton WA. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulation. Proc Natl Acad Sci USA. 2007;104:1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brooks BR, et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 42.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer S, Michnick S, Karplus M. A mechanism for rotamase catalysis by the FK506 binding protein (FKBP) Biochemistry. 1993;32:13830–13837. doi: 10.1021/bi00213a011. [DOI] [PubMed] [Google Scholar]
- 44.Fermi G, Perutz MF, Shaanan B, Fourme R. The crystal structure of human deoxyhemoglobin at 1.74 Å resolution. J Mol Biol. 1984;175:159–174. doi: 10.1016/0022-2836(84)90472-8. [DOI] [PubMed] [Google Scholar]
- 45.Neria E, Fischer S, Karplus M. Simulation of activation free energies in molecular systems. J Chem Phys. 1996;105:1902–1921. [Google Scholar]
- 46.Vanden-Eijnden E, Venturoli M. Revisiting the finite temperature string method for the calculation of reaction tubes and free energies. J Chem Phys. 2009;130:194103. doi: 10.1063/1.3130083. [DOI] [PubMed] [Google Scholar]
- 47.Ovchinnikov V, Karplus M, Vanden-Eijnden E. Free energy of conformational transition paths in biomolecules: The string method and its application to Myosin VI. J Chem Phys. 2011 doi: 10.1063/1.3544209. in-press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.