Abstract
Ants are some of the most abundant and familiar animals on Earth, and they play vital roles in most terrestrial ecosystems. Although all ants are eusocial, and display a variety of complex and fascinating behaviors, few genomic resources exist for them. Here, we report the draft genome sequence of a particularly widespread and well-studied species, the invasive Argentine ant (Linepithema humile), which was accomplished using a combination of 454 (Roche) and Illumina sequencing and community-based funding rather than federal grant support. Manual annotation of >1,000 genes from a variety of different gene families and functional classes reveals unique features of the Argentine ant's biology, as well as similarities to Apis mellifera and Nasonia vitripennis. Distinctive features of the Argentine ant genome include remarkable expansions of gustatory (116 genes) and odorant receptors (367 genes), an abundance of cytochrome P450 genes (>110), lineage-specific expansions of yellow/major royal jelly proteins and desaturases, and complete CpG DNA methylation and RNAi toolkits. The Argentine ant genome contains fewer immune genes than Drosophila and Tribolium, which may reflect the prominent role played by behavioral and chemical suppression of pathogens. Analysis of the ratio of observed to expected CpG nucleotides for genes in the reproductive development and apoptosis pathways suggests higher levels of methylation than in the genome overall. The resources provided by this genome sequence will offer an abundance of tools for researchers seeking to illuminate the fascinating biology of this emerging model organism.
Keywords: Hymenoptera, invasive species, transcriptome, chemoreception, sociality
Ants are pivotal players in the Earth's terrestrial ecosystems. They include >14,000 described species, comprise about half of all insect biomass in the tropics, are the most important predators of insects and other arthropods, and turn and aerate more soil than earthworms (1). The diversity of lifestyles displayed by ants is equally impressive, including minute, acorn-dwelling species (and the ants that parasitize them), fungus-growing leafcutters with complex division of labor, trap-jaw ants that exhibit the fastest animal movements (2), slave-making ants that kidnap and enslave the young of other ants, and the countless teeming multitudes of army ants that prowl the leaf litter and subterranean habitats (1).
Some ant species have been introduced to new geographic ranges by human activities, and a few have emerged as damaging and destructive invasive species (3). The Argentine ant (Linepithema humile) is one of the most widely distributed of these invaders and is established in nearly every Mediterranean-type climate in the world (4). Introduced Argentine ants form enormous “supercolonies,” often across hundreds or thousand of kilometers (5), and workers from the largest supercolonies on different continents even accept each other as colonymates (6). At a local scale, the absence of aggression among workers within supercolonies allows them to direct resources toward colony growth (7) and attain high population densities. These introduced populations are then able to outcompete and eliminate native ants, which imperils plants and animals that normally interact with the native ants (5). In contrast, Argentine ants in their native South American range display high levels of aggression and genetic structure among colonies across substantially smaller spatial scales (8, 9). Argentine ants are also significant pests and thus are targets of heavy insecticidal control, leading to environmental contamination and harm to nontarget organisms (10).
Despite their importance and prominence, there are essentially no genomic resources for Argentine ants. Here we report the transcriptome (GenBank accession no. 46575) and de novo genome sequence of the Argentine ant (GenBank accession no. 45799), generated using a combination of Roche 454 and Illumina sequencing. Project data are archived at the Hymenoptera Genome Database (http://hymenopteragenome.org/linepithema/genome_consortium). This project (GenBank accession no. 45805) represents an organizational advance in emerging model organism genomics (11), as it was not supported by a federal grant or genome sequencing center but, rather, was accomplished using small sums of discretionary funds provided by members of the insect genomics and Argentine ant research communities. This genome sequence provides a powerful resource for future analysis of gene families and phenotypes and candidate SNP markers for future population genetic and association mapping studies.
Results and Discussion
Genomic Features.
We sequenced the Argentine ant genome (2n = 16) to ∼23× coverage using source material from a single nest of the large California supercolony (Table 1 and SI Appendix S1, Table S1). The combined assembly of Roche 454 (∼9× coverage) and Illumina (∼14×) sequencing resulted in 215.6 megabases (Mb) of scaffolded sequence (86% of the 250.8-Mb genome; ref. 12). We also recovered 12,516 bp of the mitochondrial genome in three scaffolds (SI Appendix S1, Fig. S1). We assembled the combined 454 and Illumina data using the Roche gsAssembler (454 Life Sciences) and Celera CABOG assemblers (13) (Table 1, SI Appendix S1, Table S1). Early 454-only assemblies contained homopolymer errors, but addition of Illumina sequence data produced marked improvements (Table 1) with an overall error rate estimated at 0.012% (SI Appendix). Overall, the Celera assembler yielded the best assembly, and addition of the Illumina data dramatically increased contig and scaffold length (Table 1). We also generated a 454 transcriptome sequence from ants of mixed age, caste, and geographic location and used it to train hidden Markov models for a custom MAKER (14) annotation pipeline (SI Appendix S1, Table S2).
Table 1.
Genome and sequencing statistics for three successive assemblies
| Roche gsAssembler | Celera V0.3 | Celera V0.4 | |
| Total scaffolded sequence, bp* | 199,810,258 (80%) | 218,892,451 (87%) | 215,552,578 (86%) |
| No. of scaffolds | 3,093 | 3,180 | 3,030 |
| No. of contigs | 48,934 | 22,086 | 18,227 |
| N50 contig size, bp | 18,503† | 28,104 | 35,858 |
| N50 scaffold size, bp | 453,083 | 1,427,074 | 1,386,360 |
| Total contig coverage | 25× | 14.6× | 23× |
| Total reads used in scaffolds | 38,902,428 | 21,970,969 | 46,779,980 |
| 454 | |||
| 3-kb paired-end | 1,613,759 | 1,276,042 | 1,255,603 |
| 8-kb paired-end | 1,789,614 | 298,804 | 292,277 |
| Unpaired | 5,040,847 | 4,440,267 | 4,346,871 |
| Illumina | |||
| 3-kb paired-end | NA | 12,088,916 | 1,2265,551 |
| 8-kb paired-end | NA | 3,866,940 | 4,047,251 |
| Unpaired | 31,222,237 | 0 | 24,572,427 |
NA, not applicable.
*Values in parentheses indicate percent of the total genome size (12).
†Contigs > 500 bp; genome size, 250.8 Mb (0.26 pg).
The assembled genome appears to be relatively complete. First, our genome assembly captured 99% (246 of 248) of the core CEGMA genes (15), and 96% of them (239 of 248) were complete. Second, annotation of the cytoplasmic ribosomal protein genes revealed 83 genes, including the full set of 79 cytoplasmic ribosomal proteins (16, 17), and 4 duplicated genes (RpS16, RpS23, RpS28, and RpS30) (SI Appendix S1, Table S2). Third, annotation of the 67 nuclear-encoded oxidative phosphorylation genes shows that the L. humile genome assembly is only missing cox7a (SI Appendix S1, Table S2). This gene is also absent from the Apis mellifera genome, suggesting that it has been lost from the L. humile genome.
We generated a de novo repeat library of 523 elements and performed a whole-genome repeat annotation (Table 2). We found that 41 predictions were classified as retroid transposable elements (TEs) and 42 as DNA transposons, and 424 could not be classified. Although it has been hypothesized that the paucity of TEs in the A. mellifera genome is a product of eusociality, their abundance in the L. humile genome suggests otherwise. The L. humile genome also possesses remnants of viruses and viroids from a variety of families (SI Appendix S1, Table S4). The total repeat content was 28.7 Mb (13%), but without de novo repeat libraries, only 3.78% (8.3 Mb) of the L. humile assembly was identified as repetitive, with only 1.4% (3 Mb) identified as TEs. The L. humile genome was 62% AT, which is intermediate between A. mellifera (67%) and Nasonia vitripennis (58%). When combined with the 37.7 Mb of missing and ambiguous nucleotides, the total repetitive fraction (∼66.4 Mb; 30.8% of the genome) was significantly higher than A. mellifera (10%), but lower than N. vitripennis (40%).
Table 2.
Genes, repeats, and annotated features
| No. of features | Length occupied, bp | % of genome | |
| Genes | 16,331 | 20,828,920 | 8.30 |
| OGS 1.1 Genes | 16,123 | ND | ND |
| OGS 1.1 Transcripts | 16,177 | 20,754,334 | 8.28 |
| miRNA genes | 71 | ND | ND |
| Pseudogenes | 58* | ND | ND |
| snRNA | 84 | 45,847 | 0.02 |
| rRNA | 53 | 28,739 | 0.01 |
| Assembled transcriptome contigs† | 20,070 | 7,634,808 | 3.04 |
| Assembled transcriptome isotigs‡ | 5,352 | 4,845,691 | 1.93 |
| Repeats | 352,005 | 58,953,684 | 23.5 |
| Non-interspersed repeats | 88,703 | 5,125,662 | 2.04 |
| Simple repeats | 33,764 | 1,704,425 | 0.68 |
| Low complexity | 54,932 | 3,420,628 | 1.36 |
| Satellite | 7 | 609 | 0 |
| Microsatellites§ | 30 | 58,679 | 0.02 |
| Interspersed repeats | 85,324 | 23,613,393 | 9.27 |
| LTR | 4,021 | 2,324,263 | 0.93 |
| LINE | 3,068 | 1,432,386 | 0.57 |
| DNA | 13,307 | 3,947,143 | 1.57 |
| SINE | 739 | 152,262 | 0.06 |
| Helitron | 182 | 117,105 | 0.05 |
| Novel | 67,928 | 17,057,139 | 6.8 |
| Predictions | 523¶ | ND | ND |
| RECON | 348¶ | ND | ND |
| RepeatScout | 132¶ | ND | ND |
| PILER-DF | 43¶ | ND | ND |
ND, not done.
*Derived from manually curated gustatory, odorant, and ionotropic receptor, ribosomal, and oxidative phosphorylation genes.
†Assembled with Roche gsAssembler Version 2.0.
‡Assembled with Roche gsAssembler Version 2.3.
§Obtained from GenBank.
¶Not including predictions screened out as redundant or false positive.
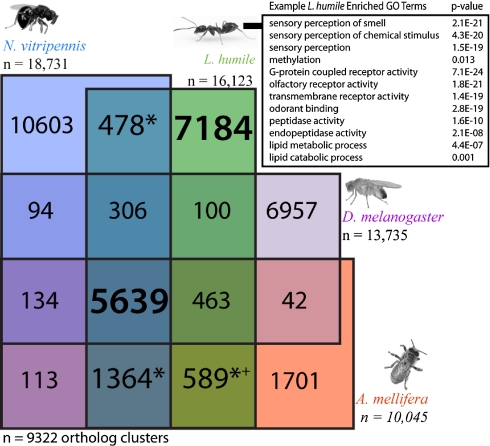
We described 16,123 genes (16,177 transcripts) in the L. humile official gene set (OGS1.1); 8,303 (51%) were supported by EST evidence, which confirmed 70% and 76% of the splice sites and exons for these genes, respectively. Of these genes, 1,364 were shared with A. mellifera and N. vitripennis, but not Drosophila melanogaster (Fig. 1), and 589 were shared by the two aculeate Hymenoptera, L. humile and A. mellifera, but not with Nasonia (the outgroup genome for the Aculeata) or Drosophila. Although L. humile and A. mellifera are both social, Aculeata also contains solitary species. A total of 7,184 genes (45%) were unique to L. humile relative to these three other species.
Fig. 1.
Venn diagram of predicted orthologs. L. humile (green), A. mellifera (red), N. vitripennis (blue), and D. melanogaster (purple) protein sets are shown. (Inset) A subset of the 99 GO terms enriched specifically in L. humile. Bold numbers represent genes common to all species studies or present in L. humile only (green). Predicted orthologs only present in the social hymenoptera tested (+) or in two or three of the hymenoptera tested (*) are indicated.
We used Interproscan (18) and KEGG (19) (SI Appendix S1, Table S5 and Fig. S2) to identify putative functional domains and compared Gene Ontology (GO; SI Appendix S1, Fig. S3) terms for all L. humile genes relative to D. melanogaster, A. mellifera, and N. vitripennis (SI Appendix S1, Tables S6 and S7) and for the L. humile-specific genes. Of 16,123 genes in the OGS1.1, a total of 7,514 (44%) were annotated with at least one GO term (average = 3). Of the 7,184 (16%) genes unique to L. humile, 1,174 (Fig. 1 and SI Appendix S1, Fig. S3) had ontology terms. In the subset of genes found only in L. humile, 99 terms were enriched (P < 0.05) relative to all L. humile genes (SI Appendix S1, Fig. S3). These included odorant receptors, peptidases that may play a role in venom production, genes associated with lipid activities that could be involved in cuticular hydrocarbon (CHC) synthesis or catabolism, and DNA methylation genes, which may play a role in caste development.
Relative to other species, six cellular location terms were enriched, with most associated with the synapse (P < 1.96E−04) or postsynaptic membrane (P < 4.74E−04; SI Appendix S1, Table S7). Of the 17 genes associated with enriched molecular functions, 6 (all P < 1.87E−08 for GO categories) included cation binding and may be involved in neurological or other signal transduction (P < 1.36E−04) processes associated with odorant binding and olfaction, learning, memory, and behavior. Lipid catabolism (P < 1.59E−05), lipase (P < 8.88E−03), and phospholipid function (3; all P < 0.04) enrichments may include genes for the synthesis of CHCs involved in social communication. Electron transport, heme, and cation binding functions characteristic of cytochrome P450s were also enriched, consistent with the observed expansion of these genes.
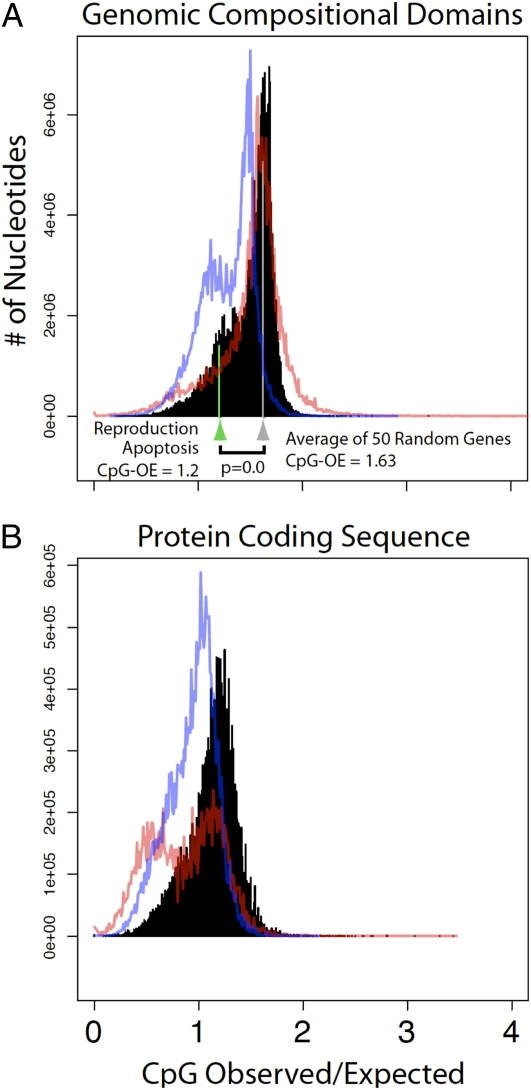
The mean GC content of the L. humile genome was 37.7%, and the ratio of observed to expected CpG nucleotides [CpG(O/E)] was 1.55 (Fig. 2); both values are within the ranges reported for other Hymenoptera (A. mellifera and N. vitripennis; refs. 20 and 21). A comparison of GC compositional-domain lengths among L. humile, A. mellifera, N. vitripennis, Tribolium castaneum, Anopheles gambiae, and D. melanogaster shows that the hymenopterans have the smallest proportion (0.1–0.5%) of long compositional domains (>100 kb) and the widest range in GC content (SI Appendix S1, Fig. S7 and Table S9). Similar to the other sequenced Hymenoptera, genes in L. humile occur in more GC-poor regions (SI Appendix S1, Fig. S8). Although the mean CpG(O/E) values of hymenopteran genomes are among the highest known, the distribution of CpG(O/E) within genomes is not consistent among the three hymenopterans (SI Appendix S1, Figs. S9 and S10).
Fig. 2.
Distribution of CpG(OE) predictions in genomic compositional domains (A) and protein coding regions (B) for L. humile (black), A. mellifera (red line), and N. vitripennis (blue line). The combined average CpG(OE) for all genes involved in either reproductive (mean = 1.21) or apoptosis processes (mean = 1.22) (green arrow and line) is significantly lower than the average CpG(OE) for 50 random genes resampled 1,000 times as a control (gray arrow and line; P << 0.05, statistical randomization test).
Sociality.
The sophisticated social structures of ant colonies are regulated by a complex interplay of chemical signaling, perception of those signals, and behavioral responses. Through these interactions, ants coordinate the activities of myriad colony members, allowing division of labor among behavioral and morphological castes. To clarify the genetic and genomic contributions to these aspects of ant sociality, we analyzed genes for the production of chemical signals (desaturases), the reception of chemical signals, gene networks that underlie differential wing and reproductive development between castes, and the yellow/major royal jelly protein gene family (which plays a key role in the development of honey bee queens vs. workers).
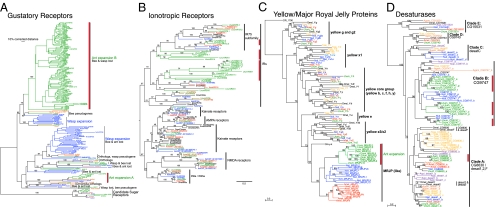
As in many social insects, CHCs are colony recognition cues for L. humile (22). The number and location of double bonds in unsaturated CHCs are regulated by desaturase genes. We found 16 complete ∆9 desaturase genes in the L. humile genome, including 7 with EST support (Fig. 3D and SI Appendix S1, Fig. S15). We also identified 17 partial or fragmentary ∆9 desaturase genes, including 6 supported by ESTs. Compared with N. vitripennis and A. mellifera, which possess 16 and 7 ∆9 desaturase genes, respectively, L. humile has a large expansion in ∆9 desaturase genes. The details of how and when these genes are expressed and the consequences for ant CHC profiles will provide insights into how these chemicals mediate social interactions within and among colonies.
Fig. 3.
Gene expansions in the Argentine ant genome. (A) Gustatory receptors. (B) Ionotropic receptors. (C) Yellow/major royal jelly proteins. (D) Desaturases. Vertical red bars indicate L. humile gene duplications and expansions. The L. humile genome also possesses an enormous expansion of odorant receptors (Ors; SI Appendix S2, Fig. S12). Green, L. humile; red, A. mellifera; blue, N. vitripennis; black, D. melanogaster.
The detection of chemical signals, such as CHCs, is mediated by a battery of gustatory receptors (Grs), olfactory binding proteins (OBPs), odorant receptors (Ors), and ionotropic receptors (IRs). Our annotation revealed a remarkable proliferation of these chemosensory genes in the Argentine ant. We found and manually annotated 116 L. humile Grs (including 20 pseudogenes; SI Appendix S1, Table S10), compared with the 11 AmGrs reported in Apis (23) and the 58 NvGrs in Nasonia (24). Major subfamily gene expansions have occurred in L. humile subfamily A, related to the Am/NvGr8/9 genes, and subfamily B, which is related to the NvGr48-50 genes and a subfamily of fragmented pseudogenes in Apis (Fig. 3A). These two expanded subfamilies may include bitter taste receptors for defensive plant compounds, which may be functionally lost from A. mellifera (subfamily B) or unexpanded in A. mellifera and Nasonia (subfamily A).
The Or family of 7-transmembrane proteins mediates most of insect olfaction, and ants are expected to have a large Or repertoire. We manually annotated 367 Or genes (337 genes and 30 pseudogenes), revealing several ant-specific gene subfamily expansions, as well as losses from ant, bee, and wasp (SI Appendix S1, Table S11 and SI Appendix S2, Fig. S12). A large 9-exon gene subfamily is highly expanded in L. humile and may encode the receptors for the CHCs that are used for colonymate recognition (SI Appendix).
The ionotropic glutamate receptor-related chemosensory receptors (IRs) are also likely involved in Argentine ant chemosensory behaviors. In the L. humile genome, we found 32 IR genes (Fig. 3B, SI Appendix S1, Table S12), whereas A. mellifera and N. vitripennis each possess only 10 (25). Five L. humile IRs appear to be orthologous to conserved IRs in other insect genomes that are expressed in olfactory organs (Fig. 3B; IR25a, IR8a, IR93a, IR76b, and IR68a; ref. 26). Other IRs appear to be ant-specific, with low homology to other insect receptors, and may be used for species-specific recognition behaviors (25).
Caste differentiation and division of labor within colonies are key innovations underlying the proliferation of ants, both numerically and taxonomically. Although the processes underlying caste differentiation are not fully understood, CpG DNA methylation is an important process for transcriptional regulation in many animals (27–29) and plays a role in the development of honey bee queens vs. workers (30). L. humile possesses a fully intact methylation toolkit with all three Dnmt genes (SI Appendix S1, Table S2 and Fig. S4). The only other sequenced insect genomes with the de novo methyltransferase Dnmt3 are the pea aphid, A. mellifera, and N. vitripennis (20, 31). Interestingly, all three genes occur as single copies in L. humile, whereas A. mellifera and N. vitripennis have two and three copies of Dnmt1, respectively. Although inherited and de novo methylation have not yet been tested in L. humile, active methylation occurs in at least two other ant subfamilies (32).
To test for a genomic signature of CG methylation, we performed two dinucleotide analyses (SI Appendix S1, Figs. S5–S10 and Table S8) that revealed overall CG bias similar to A. mellifera, but no indication of exon or intron-specific methylation as seen in A. mellifera or N. vitripennis, respectively. We also found that dinucleotide transition SNPs associated with DNA methylation (i.e., CG↔TG) were 10-fold more prevalent in the L. humile SNP data compared with either of the transversions at this site (i.e., CG↔AG, CG↔GG). We observed 21,623 SNPs at the C position of the CG/TG sites, which should yield an expected 17,428 CG↔TG transition SNPs (80.6%) based on the overall observed genome rate of 4.15 transition mutations per transversion. However, we measured 18,611 CG↔TG transitions (86%), which indicated significant bias of CG↔TG SNPs relative to all other mutations at this site (χ2 = 80.3, P < <0.001). This bias in dinucleotide CG↔TG mutation relative to all other dinucleotide mutations is consistent with the variable distribution seen for the CG dinucleotide in the genome and suggestive of in vivo methylation (SI Appendix S1, Figs. S5, S6, and S10). Genes ranked with the largest numbers of CG↔TG SNPs included major facilitator superfamily transporters, male sterility proteins, several classes of zinc finger transcription factors, and several Ig superfamily cell adhesion proteins implicated in neuronal development (SI Appendix S1, Table S8).
Wing polyphenism and reproductive division of labor between queens and workers are two important features associated with eusociality in ants. The gene networks that underlie wing and reproductive development have evolved to be differentially expressed between winged reproductive castes and wingless sterile worker castes in response to environmental factors (33, 34). We found that the mean CpG(O/E) (35) for genes involved in reproductive system development (n = 38; mean = 1.21; P << 0.05) and apoptosis (n = 18; mean = 1.22; P << 0.05) were significantly lower than the mean CpG(O/E) (mean = 1.63) for 50 random genes resampled 10,000 times. (Fig. 2 and SI Appendix). Genes in the wing polyphenism network were not significantly different from the average gene CpG(O/E) (n = 37; mean = 1.50; P = 0.4865). The mean CpG(O/E) of the Drosophila orthologs (coding regions) that underlie wing development (mean = 0.95; P = 0.86), reproduction (mean = 0.98; P = 0.98), and apoptosis (mean = 1.00; P = 0.99) were not significantly different (SI Appendix S1, Fig. S11) than average gene CpG(O/E) in Drosophila. These results indicate that developmental genes in the reproduction and apoptosis networks have distinct germ-line methylation signatures relative to the rest of the genome, whereas genes in the wing polyphenism network do not.
Yellow genes occur in insects as well as some bacteria and fungi, but they are curiously absent in all noninsect metazoans (36, 37). The initially described yellow-y gene (Y-y) functions in cuticle pigmentation in D. melanogaster (38), but these genes have also been implicated in processes such as male courtship behavior (39), follicle cell function, and egg development (40). Proteins from a gene expansion of the ancestral yellow-e3 gene in A. mellifera [the major royal jelly (mrjp) subfamily] are involved in a regulating reproductive division of labor, but also have age-, sex-, caste-, and brain-specific expression (37, 41).
In the L. humile assembly we detected 10 yellow genes and 10 major royal jelly protein-like genes (Fig. 3C and SI Appendix S1, Fig. S14). The 8 yellow genes (LhumY-y,-b,-c,-e,-e3,-g,-g2, and -h) are similar to those of D. melanogaster and are likely orthologs. The remaining 2 are putative orthologs of the yellow-x1 and -x2 genes, which have been found only in the Hymenoptera. Interestingly, the L. humile genome contains an independent radiation of major royal jelly protein-like (mrjpl) genes similar to those in A. mellifera and N. vitripennis. The mrjp and mrjpl gene sets of all three focal taxa each form their own strongly supported clade within the monophyletic mrjp subfamily (Fig. 3C and SI Appendix S1, Fig. S14). These independent radiations in different hymenopteran lineages may indicate that the ancestral gene had a tendency to proliferate, allowing mrjpls to take on new functions and to respond quickly to new forms of selection (42).
Invasion Biology.
Although Argentine ants are a widespread and familiar invasive species, many details of their invasion biology remain shrouded in mystery. To clarify some of the biological processes that may be related to their invasive success, we annotated and analyzed genes that are likely to be involved in Argentine ant immune processes, insecticide resistance, and dietary detoxification. We also identified a large number of candidate SNP markers that will be valuable tools for identifying source populations in the Argentine ant's native range, reconstructing the history of introductions around the world, and identifying current routes of transport.
The Argentine ant's unicolonial social structure allows introduced populations to attain enormous population densities in its introduced range. However, it is not clear how Argentine ants control the proliferation of pathogens and parasites in these high-density populations, particularly in light of the extremely low levels of genetic diversity that typify introduced populations (5, 8). We annotated immune genes primarily from the Toll, Imd, JNK, and JAK/STAT signaling pathways. In total, 90 of 202 immune genes had reciprocal best matches (SI Appendix S1, Tables S2 and S16). Of these 202 genes, 152 are present in the Drosophila genome and 78 in Apis. Thus, L. humile appears similar to Apis and Nasonia in having markedly fewer immune genes than Drosophila. Moreover, the many hygienic behaviors and chemical secretions described for the social Hymenoptera may play key roles in controlling pathogens.
Introduced populations of Argentine ants are also frequently targets of heavy insecticidal control, suggesting that genes conferring insecticide resistance may be under strong positive selection. Like many invasive ants, Argentine ants are also extreme dietary generalists, and this plasticity may require an underlying ability to detoxify many different components of their food sources. Cytochrome P450s are a family of heme-thiolate enzymes that catalyze a diverse array of chemical reactions in nearly all organisms (43), and they have been implicated in both insecticide resistance and dietary detoxification.
The L. humile genome encodes 111 cytochrome P450s (SI Appendix S1, Table S2 and SI Appendix S2, Fig. S13), substantially more than A. mellifera (46 genes) (44) and N. vitripennis (92 genes) (45). It has been hypothesized that the paucity of P450 genes in the genome of A. mellifera is a product of its eusocial life history (44), but their abundance in L. humile suggests that eusociality alone cannot explain their scarcity in Apis. The L. humile genome encodes 69 CYP3 clan P450s (SI Appendix S2, Fig. S13), the most in any sequenced insect genome. CYP3 P450s are associated with oxidative detoxification of xenobiotics (46), and their abundance in L. humile may be an adaptation to a variety of toxins encountered in the diet of this generalist ant, compared with the rather specialized diet of the honey bee.
To facilitate the development of population genetic markers for future analyses of the origin, history, and movement of Argentine ants, we scanned the genome for SNPs. Because the source material contained a single queen pupa and ∼100 workers from a single nest (SI Appendix S1, Table S1), we were able to identify predicted polymorphic sites. We discovered 231,248 SNPs (0.9 SNP/kb) that occurred in at least three reads and at least 10% of overlapping reads. A total of 5,734 genes (36%) had at least 1 SNP, with 14,136 SNPs (6%, 0.7 SNP/kb) in exons, 26,720 (12%, 0.9 SNP/kb) in introns, 66,830 (29%, 2.8 SNP/kb) in annotated repeats, and 123,492 (53%, 1.1 SNP/kb) in nonrepeat intergenic regions. Although 77% of exonic SNPs would be nonsynonymous (23% synonymous) if all substitutions were equally likely, we observed that only 54% of SNPs were nonsynonymous and 46% were synonymous (P < 0.001). When each type of SNP was normalized to the total number of possible sites for that type, we observed 4.9 × 10−4 SNPs per nonsynonymous site and 1.9 × 10−3 SNPs per synonymous site. As expected, we saw a 3.87-fold excess of synonymous SNPs in the OGS1.1 gene set.
We also manually investigated the genes with the highest number of SNPs per kb of exon sequence (SI Appendix S1, Table S3). Interestingly, six L. humile genes with Interproscan-predicted functions similar to male sterility and gametogenesis genes in Drosophila showed a high degree of polymorphism in L. humile (24–38 SNPs/kb). These polymorphic genes may be under sexual selection or diversifying selection and could be useful for studying different patriline lineages in native vs. invasive ants. Cytochrome P450s, lipid metabolism genes, and transcription factors were also ranked highly, with >75 SNPs in the exon and intronic regions.
Conclusion.
With the sequencing and annotation of the Argentine ant genome and the development of associated genomic resources, this species is well positioned to become a model organism in which powerful genetic approaches can be coupled with a wealth of natural history and behavioral and ecological knowledge. Given the immense financial and ecological costs associated with introductions of Argentine ants, these tools will likely find widespread application and produce tangible benefits for agriculture, societies, and ecosystems. Finally, the evolutionary forces driving some of the unusual and remarkable genomic patterns reported here remain unknown and will be productive avenues for future research that explores the basis of eusociality and the causes and consequences of biological invasions.
Materials and Methods
Source Material.
We collected an Argentine ant colony fragment from a residential orchard in Santa Clara County, CA. We confirmed that these ants belong to the large supercolony that dominates the introduced range in California using behavioral assays, microsatellite genotyping, and analysis of CHC profiles (SI Appendix S1, Table S14 and Fig. S17).
Library Preparation and Sequencing.
Transcriptome.
cDNA was generated from mixed source material and sequenced using Roche 454 Genome Sequencer LR70 FLX technology (Table 1 and SI Appendix S1, Table S1). This process yielded ∼128 Mb of DNA sequence, which was assembled into 20,070 contigs.
Genomic.
Genomic DNA was extracted and purified from a single queen pupa (Saratoga) and sequenced using seven runs of 454 FLX Titanium sequencing. Additional worker-derived genomic libraries were constructed and sequenced on the 454 and Illumina platforms.
Assembly.
We created several assemblies using Newbler and CABOG assembler (13), as described in SI Appendix.
Annotation.
We first used the automatic annotation pipeline MAKER (14) to annotate the genome of L. humile. We also manually annotated ∼1,000 genes (SI Appendix S1, Table S2), including a number that are related to fundamental biological processes (oxidative phosphorylation, ribosomal), complex behaviors (learning, memory and aggression), sensory biology (vision, chemoreception), insecticide resistance, immunity, and developmental networks (wings, reproduction).
Supplementary Material
Acknowledgments
We thank G. Anderson for providing the ants used in this project; A. Smith for assistance with transcriptome sequencing; L. Tonkin and the V. Coates Genomic Sequencing Facility and the Center for High Performance Computing for assistance and use of facilities; G. Robinson for support; M. Wong for script support; M. Goodisman for valuable discussion; B. Hunt and S. Yi for generously sharing useful data; and B. Moore for assistance with SNP analysis. Infrastructure for this work was supported in part by National Human Genome Research Institute National Institutes of Health (NIH) Grant 1R01HG004694 (to M.D.Y.), National Institute of Mental Health NIH Grant 5SC2MH086071 (to C.D.S.), and the University of Illinois at Urbana–Champaign Romano Professorial Scholarship (H.M.R.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. 46575 (transcriptome), 45799 (genome), and 45805 (genome project)].
See Commentary on page 5477.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008617108/-/DCSupplemental.
References
- 1.Hölldobler B, Wilson EO. The Ants. Cambridge, MA: Belknap Press; 1990. p. 733. [Google Scholar]
- 2.Patek SN, Baio JE, Fisher BL, Suarez AV. Multifunctionality and mechanical origins: Ballistic jaw propulsion in trap-jaw ants. Proc Natl Acad Sci USA. 2006;103:12787–12792. doi: 10.1073/pnas.0604290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holway DA, Lach L, Suarez AV, Tsutsui ND, Case TJ. The causes and consequences of ant invasions. Annu Rev Ecol Syst. 2002;33:181–233. [Google Scholar]
- 4.Suarez AV, Holway DA, Case TJ. Patterns of spread in biological invasions dominated by long-distance jump dispersal: Insights from Argentine ants. Proc Natl Acad Sci USA. 2001;98:1095–1100. doi: 10.1073/pnas.98.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suarez AV, Holway DA, Tsutsui ND. Genetics and behavior of a colonizing species: The invasive Argentine ant. Am Nat. 2008;172(Suppl 1):S72–S84. doi: 10.1086/588638. [DOI] [PubMed] [Google Scholar]
- 6.van Wilgenburg E, Torres CW, Tsutsui ND. The global expansion of a single ant supercolony. Evolutionary Applications. 2010;3:136–143. doi: 10.1111/j.1752-4571.2009.00114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holway DA, Suarez AV, Case TJ. Loss of intraspecific aggression in the success of a widespread invasive social insect. Science. 1998;282:949–952. doi: 10.1126/science.282.5390.949. [DOI] [PubMed] [Google Scholar]
- 8.Tsutsui ND, Suarez AV, Holway DA, Case TJ. Reduced genetic variation and the success of an invasive species. Proc Natl Acad Sci USA. 2000;97:5948–5953. doi: 10.1073/pnas.100110397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsutsui ND, Suarez AV, Holway DA, Case TJ. Relationships among native and introduced populations of the Argentine ant (Linepithema humile) and the source of introduced populations. Mol Ecol. 2001;10:2151–2161. doi: 10.1046/j.0962-1083.2001.01363.x. [DOI] [PubMed] [Google Scholar]
- 10.Weston DP, Holmes RW, You J, Lydy MJ. Aquatic toxicity due to residential use of pyrethroid insecticides. Environ Sci Technol. 2005;39:9778–9784. doi: 10.1021/es0506354. [DOI] [PubMed] [Google Scholar]
- 11.Smith CR, Dolezal A, Eliyahu D, Holbrook CT, Gadau J. Cold Spring Harb Protoc. 2009. Ants (Formicidae): Models for social complexity. 7:pdb emo125. [DOI] [PubMed] [Google Scholar]
- 12.Tsutsui ND, Suarez AV, Spagna JC, Johnston S. The evolution of genome size in ants. BMC Evol Biol. 2008;8:64. doi: 10.1186/1471-2148-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller JR, et al. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24:2818–2824. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cantarel BL, et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 16.Uechi T, Tanaka T, Kenmochi N. A complete map of the human ribosomal protein genes: Assignment of 80 genes to the cytogenetic map and implications for human disorders. Genomics. 2001;72:223–230. doi: 10.1006/geno.2000.6470. [DOI] [PubMed] [Google Scholar]
- 17.Marygold SJ, et al. The ribosomal protein genes and minute loci of Drosophila melanogaster. Genome Biol. 2007;8:R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quevillon E, et al. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005;33(Web Server issue):W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35(Web Server issue):W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werren JH, et al. Nasonia Genome Working Group. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinstock GM, et al. Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres CW, Brandt M, Tsutsui ND. The role of cuticular hydrocarbons as chemical cues for nestmate recognition in the invasive Argentine ant (Linepithema humile) Insectes Soc. 2007;54:363–373. [Google Scholar]
- 23.Robertson HM, Wanner KW. The chemoreceptor superfamily in the honey bee, Apis mellifera: Expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006;16:1395–1403. doi: 10.1101/gr.5057506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson HM, Gadau J, Wanner KW. The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis. Insect Mol Biol. 2010;19(Suppl 1):121–136. doi: 10.1111/j.1365-2583.2009.00979.x. [DOI] [PubMed] [Google Scholar]
- 25.Croset V, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Field LM, Lyko F, Mandrioli M, Prantera G. DNA methylation in insects. Insect Mol Biol. 2004;13:109–115. doi: 10.1111/j.0962-1075.2004.00470.x. [DOI] [PubMed] [Google Scholar]
- 28.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 29.Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 30.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, et al. An annotated cDNA library and microarray for large-scale gene-expression studies in the ant Solenopsis invicta. Genome Biol. 2007;8:R9. doi: 10.1186/gb-2007-8-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kronforst MR, Gilley DC, Strassmann JE, Queller DC. DNA methylation is widespread across social Hymenoptera. Curr Biol. 2008;18:R287–R288. doi: 10.1016/j.cub.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 33.Abouheif E, Wray GA. Evolution of the gene network underlying wing polyphenism in ants. Science. 2002;297:249–252. doi: 10.1126/science.1071468. [DOI] [PubMed] [Google Scholar]
- 34.Khila A, Abouheif E. Evaluating the role of reproductive constraints in ant social evolution. Philos Trans R Soc Lond B Biol Sci. 2010;365:617–630. doi: 10.1098/rstb.2009.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elango N, Hunt BG, Goodisman MAD, Yi SV. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci USA. 2009;106:11206–11211. doi: 10.1073/pnas.0900301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arakane Y, et al. Identification, mRNA expression and functional analysis of several yellow family genes in Tribolium castaneum. Insect Biochem Mol Biol. 2010;40:259–266. doi: 10.1016/j.ibmb.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Drapeau MD, Albert S, Kucharski R, Prusko C, Maleszka R. Evolution of the Yellow/Major Royal Jelly Protein family and the emergence of social behavior in honey bees. Genome Res. 2006;16:1385–1394. doi: 10.1101/gr.5012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wittkopp PJ, True JR, Carroll SB. Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development. 2002;129:1849–1858. doi: 10.1242/dev.129.8.1849. [DOI] [PubMed] [Google Scholar]
- 39.Drapeau MD, Radovic A, Wittkopp PJ, Long AD. A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain. J Neurobiol. 2003;55:53–72. doi: 10.1002/neu.10196. [DOI] [PubMed] [Google Scholar]
- 40.Claycomb JM, Benasutti M, Bosco G, Fenger DD, Orr-Weaver TL. Gene amplification as a developmental strategy: Isolation of two developmental amplicons in Drosophila. Dev Cell. 2004;6:145–155. doi: 10.1016/s1534-5807(03)00398-8. [DOI] [PubMed] [Google Scholar]
- 41.Schmitzová J, et al. A family of major royal jelly proteins of the honeybee Apis mellifera L. Cell Mol Life Sci. 1998;54:1020–1030. doi: 10.1007/s000180050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson BR, Linksvayer TA. Deconstructing the superorganism: Social physiology, groundplans, and sociogenomics. Q Rev Biol. 2010;85:57–79. doi: 10.1086/650290. [DOI] [PubMed] [Google Scholar]
- 43.Isin EM, Guengerich FP. Complex reactions catalyzed by cytochrome P450 enzymes. Biochim Biophys Acta. 2007;1770:314–329. doi: 10.1016/j.bbagen.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Claudianos C, et al. A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol Biol. 2006;15:615–636. doi: 10.1111/j.1365-2583.2006.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oakeshott JG, et al. Metabolic enzymes associated with xenobiotic and chemosensory responses in Nasonia vitripennis. Insect Mol Biol. 2010;19(Suppl 1):147–163. doi: 10.1111/j.1365-2583.2009.00961.x. [DOI] [PubMed] [Google Scholar]
- 46.Li XC, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.