Abstract
DNA methylation is critical for normal development and plays important roles in genome organization and transcriptional regulation. Although DNA methyltransferases have been identified, the factors that establish and contribute to genome-wide methylation patterns remain elusive. Here, we report a high-resolution cytosine methylation map of the murine genome modulated by Lsh, a chromatin remodeling family member that has previously been shown to regulate CpG methylation at repetitive sequences. We provide evidence that Lsh also controls genome-wide cytosine methylation at nonrepeat sequences and relate those changes to alterations in H4K4me3 modification and gene expression. Deletion of Lsh alters the allocation of cytosine methylation in chromosomal regions of 50 kb to 2 Mb and, in addition, leads to changes in the methylation profile at the 5′ end of genes. Furthermore, we demonstrate that loss of Lsh promotes—as well as prevents—cytosine methylation. Our data indicate that Lsh is an epigenetic modulator that is critical for normal distribution of cytosine methylation throughout the murine genome.
Keywords: methylated DNA immunoprecipitation, chIP-sequencing, epigenetics
Cytosine methylation patterns are established during early mammalian development and play an important role in genome organization, gene expression, and cellular differentiation (1–3). Cytosine methylation is critical for suppression of retroviral elements and is involved in the process of X-inactivation and genomic imprinting. Moreover, hypermethylation of CpG islands at tumor suppressor genes is closely associated with gene silencing (4). Several studies have recently investigated genome-wide cytosine methylation patterns in eukaryotes, including Arabidopsis thaliana and human cell lines (5–14). The predominant cytosine methylation in mice and humans is present at CpG sites, but substantial cytosine methylation in a non-CpG context has been found in human embryonal stem cells (9). Although DNA methyltransferases responsible for mammalian cytosine methylation have been identified (15, 16), it still remains unclear how genome-wide cytosine methylation patterns are established and maintained.
Lsh and DDM1 encompass a distinct group of the SNF2 family of chromatin remodeling proteins, controlling cytosine methylation in the genomes of mouse and A. thaliana, respectively (17–19). In addition, both proteins can affect histone methylation patterns at certain genomic loci (20, 21). Lsh is crucial for normal development, and targeted deletion of Lsh in mice leads to early lethality (19, 22). Repeat sequences are among the major genomic sites affected by loss of Lsh and DDM1 (17, 23). A decrease in cytosine methylation leads to reactivation of endogenous retroviral elements in Lsh−/− (24) or DDM1 mutants (25), the latter also showing transposition. Thus, cytosine methylation has been proposed to act as a defense mechanism against parasitic elements in the genome. In addition to repeat sequences, some developmental genes, such as the stem cell genes Oct4 and Nanog (26), or genes targeted by polycomb proteins, such as the HoxC cluster (27, 28), show substantial reduction in CpG methylation in Lsh-depleted cells. In contrast, several genomic imprinted sites (e.g., H19, KvDMR1), that require the maintenance DNA methyltransferase Dnmt1 for correct allele-specific expression, are not affected by Lsh (29). Unlike Dnmt1, Lsh does not associate with the replication fork in early S-phase (30). Furthermore, Lsh is crucial for the acquisition, rather than the maintenance of CpG methylation at episomal target sequences (31). Thus, it is hypothesized that the functional role of Lsh is not simple maintenance of cytosine methylation. The precise role of Lsh in genome-wide methylation, however, is currently poorly defined.
To investigate how genome-wide DNA methylation patterns are established in mice and how they may specifically depend on Lsh, we generated a comprehensive genomic map of cytosine methylation for wild-type (WT) and Lsh−/− mouse embryonic fibroblasts (MEFs) using methylated DNA immunoprecipitation (MeDIP) (12) combined with whole-genome tiling microarray. In addition, we generated a histone 3 lysine 4 trimethylation (H3K4me3) chromatin map using chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) and also evaluated genome-wide gene expression using cDNA microarrays.
Results and Discussion
Our previous assessment of global cytosine methylation had been biased toward specific sequences through the analysis of restriction enzyme sites (17); therefore, we applied HPLC analysis to examine total cytosine methylation changes in dependence of Lsh. DNA derived from diverse Lsh−/− embryonic tissues from day 13.5 gestation exhibited up to 50% reduction in cytosine methylation compared with WT tissues (Fig. 1A). This finding indicated a profound effect on global cytosine methylation caused by Lsh deletion. MEF cell lines derived from day 13.5 Lsh−/− embryos or WT littermate controls were used for subsequent analyses (32). Genomic DNA from MEFs was immunoprecipitated with an antibody that specifically recognizes methyl-cytosine (12). Immunoprecipitated DNA was pooled from multiple experiments and subjected to PCR analysis to validate the enrichment of methylated DNA. As expected, diverse repeat sequences (17) and specific Hox genes (27, 28) were enriched in WT samples in contrast to Lsh−/− samples (Fig. S1). Genomic imprinted sites, such as H19, KvDMR1, and Igf2R, were enriched in WT as well as in Lsh−/− samples, suggesting that methylation enrichment by MeDIP was in agreement with our previous results (29). The MeDIP-enriched DNA and the unfractionated input genomic DNA was then co-hybridized onto a murine genomic microarray that contains 13 × 106 probes with a resolution of one 50-mer probe for every 100-bp region. The array spans over 2.4 × 109 bp of the murine genome (for chr1-19) and excludes repeat sequences.
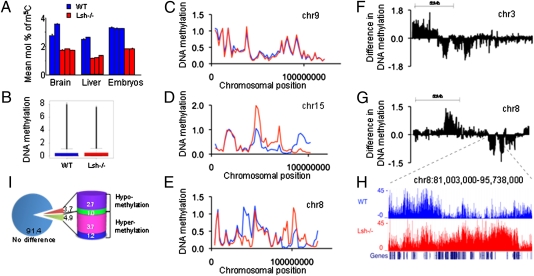
Fig. 1.
Chromosomal maps comparing cytosine methylation in WT and Lsh−/− MEFs. (A) HPLC analysis for detection of methylated cytosine comparing genomic DNA from diverse WT (blue) and Lsh−/− (red) tissues derived from day 13.5 embryos. (B) Comparison of methylated cytosine enrichment (−log10 P value) after hybridization on a murine genome tiling array comparing genomic DNA from WT and Lsh−/− MEF cell lines derived from day 13.5 embryos. WT: median 0.04, mean 0.036, third quartile 0.45. Lsh−/−: median 0.04, mean 0.37, third quartile 0.48. (C–E) Chromosomal maps at 2 Mb resolution comparing WT (blue) to Lsh−/− samples (red). (F and G) Chromosomal maps that illustrate differential methylation at 50 kb resolution. The graph shows the difference between WT mean methylation on values at 50 kb bins and Lsh−/− samples; thus, chromosomal regions above zero represent areas hypermethlated in WT and those below zero are regions hypermethylated in Lsh−/− MEFs. (H) Representation of differential methylated regions in the University of California Santa Cruz genome browser. The region indicated in the graph (G) on chromosome 8:81,003,000 to 95,738,000 is shown. Gene density is presented at the bottom of the graph. (I) Percentage of differentially methylated regions. The pie graph represents the fraction of the genome with a ratio of Lsh−/− over WT cytosine methylation larger than 10- or 5-fold (hypermethylated in Lsh−/−) or less than 10- or 5-fold (hypomethylated in Lsh−/−).
Initially, we investigated if the MeDIP array results corresponded to known CpG methylation differences between WT and Lsh−/− cells at specific genomic sites that we had previously analyzed. As expected, methylation enrichment at the Oct4 and Nanog promoters was readily detected in WT samples, but Lsh−/− cells showed reduction of cytosine methylation at the same sites (Fig. S2 A and B). In contrast, genomic imprinted sites, such as the ICR upstream of H19 or KvDMR1 revealed a similar pattern of methylation enrichment when comparing WT with Lsh−/− samples (Fig. S2 C and D). Larger regions, such as the HoxC cluster, displayed greatly decreased cytosine methylation in Lsh−/− MEFs, whereas a randomly chosen area of similar range showed a comparable methylation pattern (Fig. S2 E and F). Thus, at those specific genomic sites, the methylation enrichment generated by the MeDIP array data were in agreement with previous results (Fig. S3 A and B) (26–29).
Chromosomal Maps.
Total MeDIP enrichment of the array was of similar magnitude when comparing WT with the Lsh−/− sample, indicating that there was not a major net loss of cytosine methylation at unique sequences (Fig. 1B). Because HPLC analysis includes all repeat sequences that make up 30 to 50% of the murine genome, part of the global cytosine reduction in Lsh−/− samples may be a result of those repeat sequences (17, 19) (Fig. S1D). To investigate if Lsh deletion affects cytosine methylation at specific genomic regions, other than the previously identified loci, we generated individual chromosomal maps based on mean methylation enrichment values at 2 Mb windows. Methylation enrichment in WT MEFs was not uniformly distributed along the chromosomal axis (11, 12), and was present at gene-poor regions (Fig. S3 C–E). Chromosomal maps at 2 Mb resolution generated for Lsh−/− MEFs were similar to WT maps for most, but not all, chromosomes (Fig. 1 C–E and Fig. S4 A and B). For example, chr8 and ch15 encompass large regions that were differentially methylated (Fig. 1 D and E). Some areas were distinctively enriched for methylated cytosine in WT compared with Lsh−/− samples, but several domains were comparatively hypermethylated in the absence of Lsh. To gain better insight into the extent of DNA methylation changes, we created differential methylation maps for chromosomes at higher resolution. Mean methylation values at 50 kb windows (n = 49,295) for Lsh−/− MEFs were subtracted from mean WT values to illustrate the regions of dissimilar methylation (Fig. 1 F and G). Comparatively hyper- or hypomethylated 50-kb windows were contiguous and extended up to 2 Mb (with 5% of all contiguous regions between 1 and 2 Mb). The average size for the majority of differentially methylated regions was 50 to 200 kb (about 85% of domains). The transition points at differentially methylated regions appeared well defined (Fig. 1H), suggesting that discrete chromosomal domains share a common epigenetic pathway controlled by Lsh. To quantify the cytosine methylation changes between WT and Lsh−/− MEFs, the ratios of DNA methylation enrichment (Lsh−/− values over WT values) were computed for single values. For the whole genome, about 6.4 or 8.6% of 100-bp bins were differentially methylated (more than 10-fold changes or more than 5-fold, respectively) with fairly similar proportions being hypo- or hypermethylated in the absence of Lsh (Fig. 1I, and for single chromosomes Table S1).
Taken together, these data show that Lsh controls not only cytosine methylation at repeat sequences, but also globally regulates methylation at unique sequences. Lsh promotes as well as prevents cytosine methylation, and thus plays a crucial role in the normal distribution of cytosine methylation across the genome.
Methylation Profiles of 5′ and 3′ Ends.
To characterize cytosine methylation levels at defined gene features, we aligned genes at their 5′ ends and plotted mean methylation enrichment at 100-bp resolution covering a 5 kb region upstream and 5 kb downstream of their transcriptional start site (TSS) (Fig. 2A). The 5′ end profile in WT MEFs displayed a methylation depletion around the TSS, similar to the methylation profiles recently reported using bisulfite sequencing (5, 7, 9, 13, 14). Although each gene shows distinct methylation peaks in the promoter region or the gene body, the mean methylation levels of the TSS flanking regions, comprising promoter regions or the gene body, are comparable. Likewise, the 3′ end of genes in WT MEFs (Fig. 2B) showed a reduction in cytosine methylation, indicating that both ends of protein-coding genes are marked by distinct cytosine methylation patterns in the murine genome, as has been previously described for many eukaryotes, including human and A. thaliana (7, 9, 11, 13, 14).
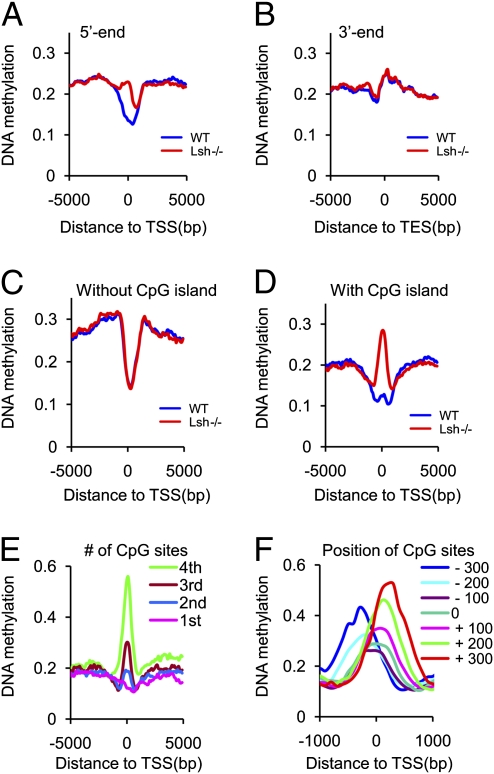
Fig. 2.
Cytosine methylation pattern at the 5′ and 3′ ends in Lsh−/− MEFs. (A) Cytosine methylation profiles comparing WT (blue) and Lsh−/− MEFs (red) at 100 bp resolution around 5′ ends (TSS) (n = 44,742) and flanking regions [5 kb upstream and 5 kb downstream (gene body)]. (B) Cytosine methylation distribution around the 3′ end of genes (n = 48,680) with flanking regions [5 kb upstream (gene body) and 5 kb downstream] comparing WT (blue) and Lsh−/− (red) MEFs at 100 bp resolution. (C and D) Cytosine methylation distribution around the 5′ ends and flanking regions comparing WT (blue) and Lsh−/− (red) at genes with CpG island (C) or without CpG island (D). (E) Cytosine methylation distribution in Lsh−/− MEFs at the 5′ end of genes classified into four groups (first to fourth) based on their density of CpG sites, with first being the lowest. (F) Cytosine methylation distribution in Lsh−/− MEFs at the 5′ end of CpG island genes that are classified into six groups based on the position of the center of the CpG islands: −300 bp (blue), −200 bp (pale blue), −100 bp (purple), 0 (green), 100 bp (pink), 200 bp (pale green), 300 bp (red).
When we examined the distribution of cytosine methylation around the TSS and flanking regions in Lsh−/− MEFs, we found it distinctly distorted compared with WT MEFs (Fig. 2A). In contrast, the 3′ end cytosine methylation was indistinguishable when comparing Lsh−/− with WT samples (Fig. 2B). The aberrant distribution pattern at 5′ ends was found throughout the genome, without an apparent preference for chromosomal position (for example chr1) (Fig. S4C). To determine which subclass of genes was primarily affected by Lsh loss, we classified genes based on the presence of CpG islands. Genes with and without CpG islands displayed a dip of cytosine methylation around the TSS in WT cells, suggesting that a paucity of methylated cytosines around the TSS is a common feature of genes. When we compared Lsh−/− MEFs with WT MEFs, we found that the 5′ end cytosine methylation distribution for non-CpG island genes was indistinguishable (Fig. 2C). In contrast, CpG island genes exhibited a hypermethylation at their 5′ ends (Fig. 2D), indicating that this subset is predominantly affected by Lsh. To evaluate the degree to which the methylation distribution at gene ends differs between WT and Lsh−/− samples, we calculated the mean correlation between WT and Lsh−/− methylation values at single 100-bp windows, as similar methylation profiles (as in Fig. 2 B and C) may mask any potential differences at single genes. The correlation profile revealed the most dramatic difference in correlation values between WT and Lsh−/− at CpG island promoter regions (Fig. S4D); the correlation was lowest at TSS sites (Pearson R = 0.39), where CpG islands are preferentially localized and increased away from the TSS (at position −5 kb, R = 0.63). In contrast, genes without CpG islands (Fig. S4D) or the 3′ end of genes (Fig. S4E) showed an overall high-correlation value between Lsh−/− and WT samples. This finding supports the notion that Lsh deletion influences DNA methylation primarily at genes with CpG islands.
To investigate further which CpG island genes are affected the most by Lsh, we sorted all genes based on CpG densities. The peak of cytosine methylation at the 5′ end of genes in Lsh−/− MEFs was most prominent at those genes with the highest CpG densities (Fig. 2E). To determine if the distribution of cytosine methylation in the absence of Lsh depends on the position of CpG islands or the TSS, we classified genes based on the position of the CpG island toward the TSS. We found that the cytosine methylation peaks in Lsh−/− MEFs shifted upstream or downstream of the TSS following the center of the CpG island (Fig. 2F). This finding suggests that the observed DNA hypermethylation was associated with the CpG island rather than the TSS.
Taken together, these data show that Lsh−/− MEFs display an aberrant distribution of cytosine methylation at the 5′ end of a subset of genes that are rich in CpG sites.
H3K4me3 Changes in Dependence of Lsh.
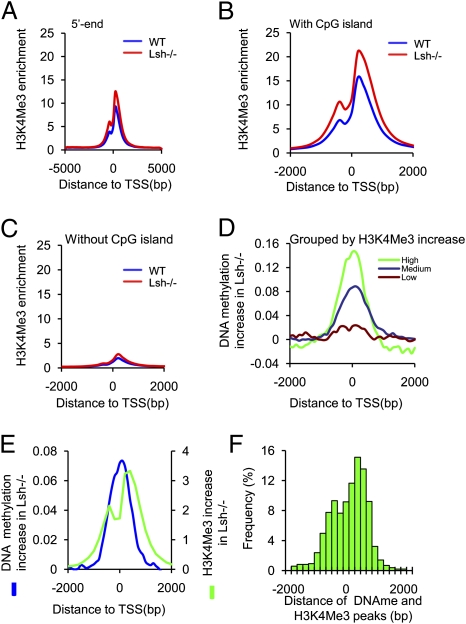
To characterize Lsh-affected genes further, and to determine the relationship of cytosine methylation to other epigenetic changes, we examined H3K4me3 modifications. Whole-genome ChIP-sequencing showed an increased frequency of H3K4me3 peaks in Lsh−/− cells compared with WT control, indicating an overall elevation of H3K4me3 modification caused by Lsh depletion (Fig. S5A). This observation is in agreement with previously reported increases of H3K4 methylation in DDM1 or Lsh mutants (20, 21). Mapping of H3K4me3 enrichment to regions flanking TSS revealed a rise in H3K4me3 enrichment in Lsh−/− MEFs compared with WT MEFs, indicating that changes in H3K4me3 occurred, at least in part, at expected sites, namely around the TSS (Fig. 3A). Increases of H3K4me3 modifications were most pronounced at CpG island genes and not detectable at non-CpG island genes (Fig. 3 B and C), suggesting that the same class of genes that exhibited aberrant cytosine methylation showed altered H3K4me3 enrichment. To investigate a link between increases in H3K4me3 and DNA methylation, we ranked genes based on their level of H3K4me3 gain in Lsh−/− cells (Fig. S5B). Those genes with the highest increases in H3K4me3 in Lsh−/− MEFs also showed the largest increase in DNA methylation at their 5′ ends (mean 0.10837 vs. 0.04592, P = 1.6 × 10−28) (Fig. 3D). Among genes that were ranked based on increase in DNA methylation in Lsh−/− cells (top 10%, n = 2,365), about two-thirds (70%) had an increase in H3K4me3 as well, but it should be noted that a subset of genes (22%) displayed a moderate loss of H3K4me3 (see below). The distribution of H3K4me3 and cytosine methylation increases at the TSS and flanking regions in Lsh−/− MEFs revealed dissimilar profiles for each epigenetic modification. Although H3K4me3 increases in Lsh−/− cells showed a slight gap around the TSS, possibly because of Pol II occupancy (33), the peak of DNA methylation gain was falling between the H3K4me3 peaks (Fig. 3E). For genes that were subgrouped according to their position of CpG islands (Fig. 2F), cytosine methylation and H3K4me3 increases shifted with the position of the CpG island and peaks were juxtaposed to each other (Fig. S5 C–G). The majority of genes had a distance of about 400 bp between peaks (Fig. 3F), suggesting that both epigenetic changes can occur at the same genes in Lsh−/− MEFs with distinct distribution patterns.
Fig. 3.
H3K4me3 changes in dependence of Lsh. (A) H3K4me3 methylation distribution at the 5′ ends (TSS) (n = 23,365) and flanking regions (5 kb upstream and 5 kb downstream) comparing WT (blue) and Lsh−/− MEFs (red) at 100 bp resolution. (B and C) H3K4me3 methylation distribution at the 5′ ends and flanking regions comparing WT (blue) and Lsh−/− MEFs (red) at genes with CpG island (B) or without CpG island (C). (D) Link between increases in DNA methylation and H3K4me3 gains at the 5′ ends. All genes (unique genes n = 23,365) were classified into three groups based on their gain of H3K4me3 in Lsh−/− cells (Fig. S5B) into high (green), medium (blue), or no gain (brown). The graph depicts the change of DNA methylation (the difference between Lsh−/− cytosine methylation and WT methylation) at the 5′ ends. (E) Distribution of H3K4me3 changes and DNA methylation changes at the 5′ ends. The gain of H3K4me3 in Lsh−/− MEFs (difference of Lsh−/− values minus WT values) and the gain of cytosine methylation in Lsh−/− MEFs (difference of Lsh−/− values minus WT values) were plotted around the 5′ end to illustrate the position of each modification. (F) Histogram illustrating the distance between H3K4me3 peaks and cytosine methylation peaks for single genes and the frequency with which they occur.
Gene Expression.
To investigate if and how the aberrant pattern of cytosine methylation in the absence of Lsh may link to transcriptional changes, we generated gene expression profiles of WT and Lsh−/− MEFs. In particular, we were interested in those genes that had acquired H3K4me3 and cytosine methylation in Lsh−/− MEFs. When we examined the three groups that had been classified based on their H3K4me3 changes in Lsh−/− MEFs (Fig. 3D and Fig. S5B), we found that the group with the most dramatic change in H3K4me3, which was associated with increases in cytosine methylation, showed significantly higher gene expression in WT MEFs (Fig. 4A). The group with low H3K4me3 changes in Lsh−/− MEFs (including moderate losses of H3K4me3) displayed a low gene-expression level in WT MEFs, suggesting that Lsh depletion resulted in epigenetic changes at those genes with extreme expression levels and not at those with intermediate expression levels. Among the subset of genes with high increases of H3K4me3 and high levels of cytosine methylation gain, we observed a mild trend of up-regulation in Lsh−/− MEFs, whereas for those genes with no H3K4me3 gain a trend to down-regulation in Lsh−/− MEFs was observed (Fig. 4A). Overall, for the majority of genes the expression level was not dramatically changed, in agreement with our previous observations in brain and liver tissues (24).
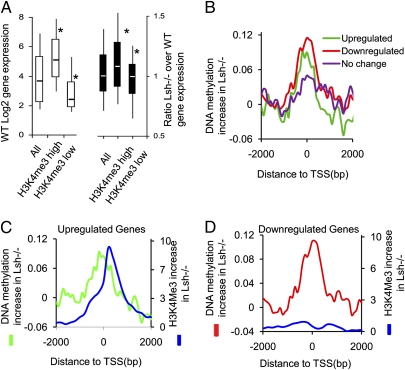
Fig. 4.
Relation of cytosine methylation changes and gene expression. (A) Box plot to demonstrate changes of gene expression changes for all genes (All) and those that were subgrouped based on changes in H3K4me3 and cytosine methylation. Genes were ranked according to their H3K4me3 changes in Lsh−/− MEFs (Fig 3D and Fig. S5B) from highest (H3K4me3 high) to lowest gain (H3K4me3 low, including those genes with H3K4me3 losses). (Left) Gene-expression level in WT MEFs according to the classification. (Right) Ratio of gene expression (Lsh−/− expression over WT expression), with values above 1 indicating up-regulation and below 1 indicating down-regulation. *P < 0.005. (B) Distribution of cytosine methylation changes in Lsh−/− MEFs at the 5′ end of genes that are twofold down-regulated in Lsh−/− MEFs (red, n = 581), twofold up-regulated in Lsh−/− MEFs (green, n = 320), or a control group without any change (purple, n = 543). (C) Distribution of cytosine methylation changes (green) and H3K4me3 changes (blue) in Lsh−/− MEFs at the 5′ end of genes that are twofold up-regulated in Lsh−/− MEFs. (D) Distribution of cytosine methylation changes (red) and H3K4me3 (blue) changes in Lsh−/− MEFs at the 5′ end of genes that are twofold down-regulated in Lsh−/− MEFs.
To gain further insight into how changes in cytosine methylation may be linked to gene expression, we analyzed the subset of genes that were differentially expressed more than twofold (5% of genes, n = 901; of total analyzed genes, n = 16,660). Gene ontology analysis of those differentially expressed genes in Lsh−/− MEFs showed an enrichment for genes involved in angiogenesis (P = 1.4 × 10−8), lung development (P = 1.010−6), skeletal system development (P = 6.6 × 10−8), embryonic morphogenesis (P = 8.6 × 10−7), and organ development (P = 4.3 × 10−7), pointing to a role of Lsh in murine development (22). This subset of genes had significant increases of cytosine methylation around the TSS in Lsh−/− MEFs compared with a control group with unaltered gene expression (mean increase is 0.073 vs. 0.037, respectively, P < 0.05). The twofold differentially regulated genes showed up- (1.9%, n = 320) as well as down-regulation (3.5%, n = 581). Unexpectedly, genes that were up-regulated and those that were down-regulated displayed an enrichment of cytosine methylation at the TSS in Lsh−/− MEFs (Fig. 4B). When we determined the H3K4me3 distribution for those genes that were differentially expressed in Lsh−/− MEFs, we found two distinct patterns. Whereas up-regulated genes had H3K4me3 increases in conjunction with DNA methylation changes (Fig. 4C), down-regulated genes displayed no significant gain of H3K4me3 in the immediate TSS region in Lsh−/− MEFs (Fig. 4D). This finding suggests that the combination of both epigenetic marks may determine the outcome of gene expression and, in addition, that DNA methylation increases in Lsh−/− MEFs can be linked to up- or down-regulation. Likewise, a recent study reported up- as well as down-regulation of Dnmt3a-targeted genes after Dnmt3a depletion, implying a dualistic role of DNA methylation in gene expression (34).
Finally, bisulfite sequencing was performed to confirm cytosine hypermethylation in Lsh−/− MEFs at several genes. We detected increases in CpG methylation corresponding to enhanced cytosine methylation assessed in the MeDIP array for six genes (Fig. S6). Those genes also showed a loss of H3K4me3 methylation, and thus a reciprocal relationship of those two epigenetic marks, as has been previously reported (10). In addition, those CpG hypermethylated genes were down-regulated in Lsh−/− MEFs, supporting a link between CpG hypermethylation and transcriptional repression. Unexpectedly, for six other genes non-CpG methylation was detected at the TSS in the absence of Lsh (Fig. S7A). Non-CpG methylation is a characteristic of embryonal stem cells and is positively correlated with gene transcription (9). The increase of non-CpG methylation was accompanied by a twofold up-regulation in gene expression for four genes (Fig. S7B). A slight enrichment of H3K4me3 was detected at those genes with non-CpG methylation, and the histone modification showed a distinct distribution around the TSS compared with the cytosine methylation profile at those genes (Fig. S8). Our findings suggest the possibility that the distinct pattern of CpG or non-CpG methylation in combination with H3K4me3 modifications may influence transcription in Lsh−/− MEFs. However, because conventional bisulfite sequencing is biased, only full genome sequencing after bisulfite conversion can ultimately determine to what extent non-CpG methylation contributes to the pattern of hypermethylation in the absence of Lsh.
It has been previously demonstrated that Lsh, similar to its A. thaliana homolog DDM1, controls cytosine methylation and transcriptional repression at repeat sequences (17–19). Our results here demonstrate that Lsh additionally controls genome-wide allocation of cytosine methylation at unique sequences, indicating that Lsh is critical for establishing normal methylation pattern during development. Changes of DNA methylation following Lsh deletion occur not only at single genes, but also at larger chromosomal domains, suggesting that regional patterns of cytosine methylation underlie a common epigenetic pathway. Differentially methylated regions of 150 kb mean length have been recently reported, comparing embryonal stem cells with somatic cells (9), indicating that those domains are regulated in the transition from pluripotency to somatic cell. It remains to be elucidated if those differentially methylated domains in the absence of Lsh are indicative of a failed differentiation process, because Lsh reduction impairs the differentiation of embryonal stem cells in vitro (26).
Lsh deletion leads to hypo- as well as hypermethylation of cytosine. This observation, together with the fact that regionally specific effects are found after Lsh deletion, indicates that Lsh is not simply involved in the maintenance of methylation patterns. The changes in cytosine methylation are in part due to increases of non-CpG methylation. A recent study examining DNA methylation at promoter regions in Lsh−/− MEFs (35) supports previous findings of hypomethylation at stem cell-specific genes (28) (Fig. S2), but did not report non-CpG methylation. This study used a methyl-CpG binding domain for enrichment of methylated DNA, a technique that is not suitable for detection of non-CpG methylation in contrast to MeDIP, as used in this study (36). Interestingly, mutants of the Lsh homolog DDM1 in A. thaliana also show hypermethylation in the context of non-CpG sites (37), revealing yet another similarity in the biological role of Lsh and DDM1. The role of Lsh in cytosine methylation is more complex than solely promoting recruitment of methyltransferases to chromatin targets, an assumption that was based on the observation that Dnmt3b association with Hox genes or stem-cell genes is reduced in the absence of Lsh (26, 28). This phenomenon of hyper- as well as hypomethylation may involve differences in the ability of Lsh to retarget Dnmt3a vs. Dnmt3b to specific chromatin sites, or may be because of Lsh affecting insulators, chromatin barriers, and heterochromatin spreading, and thus may control higher-order chromatin structure.
Finally, we show here that Lsh specifically alters cytosine methylation patterns at the 5′ end (TSS) but not the 3′ end in a subset of CpG island genes. The 5′ and 3′ ends are both marked by cytosine depletion and have previously been linked to Pol II engagement in A. thaliana (14). It remains to be shown whether Lsh alters genome-wide Pol II binding and transition to early elongation at those sites.
Cytosine methylation is evolutionarily conserved and displays species- and tissue- specific patterns. Elucidation of those factors that contribute to and facilitate the setting of these methylation patterns will enhance our understanding of the function of DNA methylation during normal development and in diseases.
Materials and Methods
Hybridization to NimbleGen Arrays.
Genomic DNA of WT (Lsh+/+) or Lsh−/− MEFs (32) was sonicated into fragments (300–1,000 bp), precipitated with anti-5-methylcytidine (Eurogentec), as previously described (12, 26), and DNA pooled for whole genome array hybridization (NimbleGen Service Laboratory). The murine whole-genomic array contains about 13 × 106 probes with a resolution of one 50-mer probe for every 100-bp region. Data were extracted from scanned images by using NimbleScan 2.3 extraction software. Cytosine methylation enrichment was evaluated using normalized log2 ratio intensity values (the ratio of the MeDIP to the input sample) and then −log10 P values from one-sided Kolmogorov-Smirnov tests performed to determine if probes displayed a significantly more positive distribution over the rest of the array. For details of the MM8 tiling array and calculation of data, please refer to www.nimblegen.com. Data calculation and detailed methods are found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Nancy Colburn, Amar Klar, and Michael Kuehn for critical comments on the manuscript. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health (Contract HHSN261200800001E) and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017000108/-/DCSupplemental.
References
- 1.Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 2.Ooi SK, O'Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J Cell Sci. 2009;122:2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rauch TA, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci USA. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 13.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 14.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 15.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 16.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 17.Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 19.Sun LQ, et al. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 2004;18:1035–1046. doi: 10.1101/gad.1176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- 21.Yan Q, Huang J, Fan T, Zhu H, Muegge K. Lsh, a modulator of CpG methylation, is crucial for normal histone methylation. EMBO J. 2003;22:5154–5162. doi: 10.1093/emboj/cdg493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiman TM, et al. Lsh, a SNF2 family member, is required for normal murine development. Biochim Biophys Acta. 2001;1526:211–220. doi: 10.1016/s0304-4165(01)00129-5. [DOI] [PubMed] [Google Scholar]
- 23.De La Fuente R, et al. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat Cell Biol. 2006;8:1448–1454. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- 24.Huang J, et al. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res. 2004;32:5019–5028. doi: 10.1093/nar/gkh821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 26.Xi S, et al. Lsh participates in DNA methylation and silencing of stem cell genes. Stem Cells. 2009;27:2691–2702. doi: 10.1002/stem.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao Y, Xi S, Briones V, Muegge K. Lsh mediated RNA polymerase II stalling at HoxC6 and HoxC8 involves DNA methylation. PLoS ONE. 2010;5:e9163. doi: 10.1371/journal.pone.0009163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xi S, et al. Lsh controls Hox gene silencing during development. Proc Natl Acad Sci USA. 2007;104:14366–14371. doi: 10.1073/pnas.0703669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan T, Hagan JP, Kozlov SV, Stewart CL, Muegge K. Lsh controls silencing of the imprinted Cdkn1c gene. Development. 2005;132:635–644. doi: 10.1242/dev.01612. [DOI] [PubMed] [Google Scholar]
- 30.Yan Q, Cho E, Lockett S, Muegge K. Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin. Mol Cell Biol. 2003;23:8416–8428. doi: 10.1128/MCB.23.23.8416-8428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu H, et al. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25:335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan T, et al. Lsh-deficient murine embryonal fibroblasts show reduced proliferation with signs of abnormal mitosis. Cancer Res. 2003;63:4677–4683. [PubMed] [Google Scholar]
- 33.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myant K, et al. LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res. 2011;21:83–94. doi: 10.1101/gr.108498.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris RA, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saze H, Kakutani T. Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 2007;26:3641–3652. doi: 10.1038/sj.emboj.7601788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.