Abstract
CD8 T cells play a key role in mediating protective immunity against selected pathogens after vaccination. Understanding the mechanism of this protection is dependent upon definition of the heterogeneity and complexity of cellular immune responses generated by different vaccines. Here, we identify previously unrecognized subsets of CD8 T cells based upon analysis of gene-expression patterns within single cells and show that they are differentially induced by different vaccines. Three prime-boost vector combinations encoding HIV Env stimulated antigen-specific CD8 T-cell populations of similar magnitude, phenotype, and functionality. Remarkably, however, analysis of single-cell gene-expression profiles enabled discrimination of a majority of central memory (CM) and effector memory (EM) CD8 T cells elicited by the three vaccines. Subsets of T cells could be defined based on their expression of Eomes, Cxcr3, and Ccr7, or Klrk1, Klrg1, and Ccr5 in CM and EM cells, respectively. Of CM cells elicited by DNA prime-recombinant adenoviral (rAd) boost vectors, 67% were Eomes− Ccr7+ Cxcr3−, in contrast to only 7% and 2% stimulated by rAd5-rAd5 or rAd-LCMV, respectively. Of EM cells elicited by DNA-rAd, 74% were Klrk1− Klrg1−Ccr5− compared with only 26% and 20% for rAd5-rAd5 or rAd5-LCMV. Definition by single-cell gene profiling of specific CM and EM CD8 T-cell subsets that are differentially induced by different gene-based vaccines will facilitate the design and evaluation of vaccines, as well as enable our understanding of mechanisms of protective immunity.
Keywords: lymphocyte subsets, microarray, immune differentiation
A major aim of vaccines designed to elicit immunity against chronic viral pathogens is the generation of pathogen-specific cytotoxic T cells of sufficient magnitude and quality to mediate protection against disease (1, 2). Cytotoxic T cells can control viral load in various models of simian immunodeficiency virus (SIV) infection in rhesus macaques (3–7). However, the quantitative and qualitative features of vaccine-elicited T-cell responses that mediate protection are yet to be defined (8, 9). Recently, analysis of T cells by intracellular cytokine staining has allowed greater precision in measuring antigen-specific responses, facilitating the functional profiling of antigen-specific cells in vitro. Simultaneous measurement of IFN-γ, TNF-α, and IL-2 in vaccine-elicited CD4 T cells has shown that the proportion of cells secreting multiple cytokines, polyfunctional T cells, correlated with protection in a mouse model of leishmania infection (10). However, T-cell polyfunctionality was not a major predictor of outcome in a number of SIV protection studies in rhesus macaques (6, 11) and in other vaccine-induced protection models (12).
The generation of synthetic class I MHC:peptide tetramers has allowed the identification of vaccine elicited antigen-specific T cells responsive to vaccine epitopes (13). Analysis of surface proteins, such as the interleukin-7 receptor (IL7R) and L-selectin (CD62L), in mice has allowed the definition of three differentiation states, referred to as effector (IL7R low, CD62L low), effector memory (IL7R+, CD62L−), and central memory (IL7R+, CD62L+) states characterized by a gradient of proliferative and cytotoxic potential (14). Similar T-cell subsets have also been defined in humans (15), indicating that these subsets are evolutionarily conserved. However, the presence of such cell types does not predict vaccine efficacy with consistency. Some studies have implicated a protective role of central memory (CM) T cells, although others have suggested a protective role for effector memory (EM) cells (6, 16). These studies highlight the need to understand and discriminate among differences in CD8 T-cell responses generated by different vaccines; we have therefore compared vaccine-elicited cellular immune responses elicited by three different gene-based vaccine regimens through transcriptional profiling and the application of a technique that has allowed us to measure 96 gene-expression signals from single immune cells. We have identified qualitative differences in CD8 T cells elicited by the three vaccine regimens that cannot be detected by conventional techniques, and that allow individual CD8 T cells elicited by the three immunizations to be readily distinguished. Furthermore, we have identified unique subsets of CD8 T cells based upon analysis of gene expression of Eomes, Cxcr3, and Ccr7, or Klrk1, Klrg1, and Ccr5 in CM and EM cells, respectively, that were differentially induced by the three vaccines.
Results
Three Different Prime-Boost Vaccine Combinations Stimulate CD8 T-Cell Responses of Similar Magnitude and Cytokine Functionality.
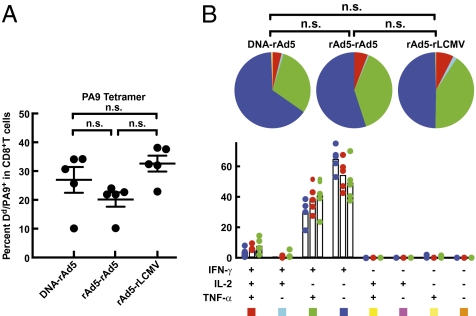
Plasmid DNA, recombinant replication-defective adenovirus and lymphocytic choriomeningitis virus (rAd5 and rLCMV, respectively), encompassing the same insert derived from a recombinant HIV-1 Env gene (17), were used to immunize BALB/c mice in different prime-boost combinations. The three different prime-boost immunization regimens (DNA-rAd5, rAd5-rAd5, and rAd5-rLCMV) elicited T-cell responses with mean frequencies ranging from 20.1to 32.6% (Fig. 1A), as measured by binding to an antigen-specific tetramer H2-Dd/PA9 (18). To confirm the specificity of tetramer binding and therefore the accuracy of subsequent tetramer-sorting experiments, we compared the frequency of tetramer-binding CD8+ T cells in rAd5-rAd5 immunized animals with that in unimmunized animals (Fig. S1 A and B). The mean percentage of CD8 T cells binding the tetramer was only 0.027% [95% CI (confidence interval) 0.007–0.047] in naive mice versus 38.3% (95% CI 29.29–47.31) in vaccinated mice, predicting a tetramer-sort purity of 99.9% (Fig. S1C). We also assessed the proportion of CM and EM CD8 T cells within the tetramer-binding populations elicited by the three vaccines, and found this to be similar for all three vaccines (P > 0.05) (Fig. S2 A and B). In vaccine studies, the quality of T-cell memory is often assessed by measuring the ability of cells to produce one or more cytokines in response to antigen (1). To assess the cytokine functionality of CD8+ T cells elicited by these vectors, splenocytes were isolated from mice 3 wk after the final boost. Cells were pulsed with PA9 peptide, and intracellularly stained for the cytokines IFN-γ, TNF-α, and IL-2. The cytokine functionality of CD8+ T cells was similar for all three vaccines (Fig. 1B) (P > 0.05), highlighting an inability to detect qualitative differences between antigen-specific T cells elicited by different vaccines using standard cytokine detection assays.
Fig. 1.
Gene-based vaccine regimens and assessment of resultant CD8+ T-cell responses. Animals were immunized intramuscularly with DNA-rAd, rAd-rAd, or rAd-LCMV encoding HIV-1 Env, titrated to elicit similar frequencies of CD8+ T cells specific for the PA9 epitope in HIV-1 Env 3 wk following boost immunization. (A) pMHC tetramer staining. H2-Dd/PA9 tetramer staining of splenocytes isolated at 3 wk postimmunization. (B) Intracellular cytokine analysis following PA9 peptide stimulation. Splenocytes were pulsed with PA9 peptide and stained intracellularly for IFN-γ, TNF-α, and IL2 after 5 h. Similar results were obtained in three independent experiments. Data show the mean of five mice per group of one representative experiment.
Distinct Sets of Genes Are Induced in CD8+ T Cells by Alternative Immunizations.
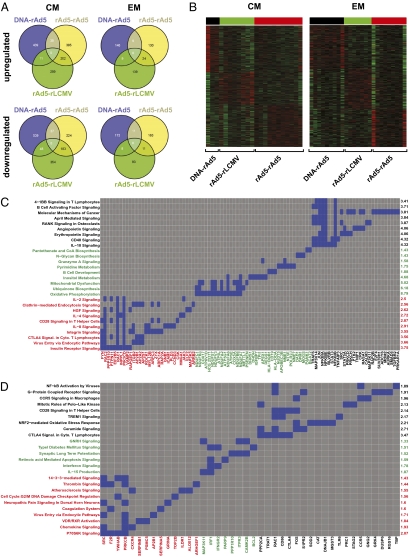
We asked whether the patterns of gene expression in antigen-specific CD8 T cells differed depending on the vaccine vectors used. Global gene-expression patterns were determined using cDNA microarray analysis of either CM or EM H2-Dd/PA9 tetramer-binding CD8+ T cells 3 wk after the final immunization. Each vaccine regimen induced a unique set of genes in either CM or EM subsets (Fig. 2 A and B and Table S1). In addition, we found that common genes were induced in CM and EM subsets when any two vectors were compared (Fig. 2 and Table S1). To gain insight into the nature of the genes that were uniquely induced by our vector regimens, we performed an enrichment analysis of the data using Ingenuity Pathway Analysis (Ingenuity Systems), which is similar to Geneset Enrichment Analysis (19). Fig. 2 C and D depict the most significantly regulated pathways induced in EM and CM cells with representative genes from these pathways. Immunization with rAd5-rAd5 selectively induced pathways involved in IL-2, IL-4, IL-8, and CTLA-4 signaling (Fig. 2 C and D, pathways in red), whereas rAd5-rLCMV activated pathways involved in IL-15, Granzyme A and IFN signaling (Fig. 2 C and D, pathways in green) and DNA-rAd5 induced pathways involved in IL-10, 41BB, CD40 (Fig. 2 C and D, pathways in black). We also observed the differential induction of pathways involved in metabolic processes (insulin receptor signaling, oxidative phosphorylation, mitochondrial dysfunction) in T-cells elicited by the different vaccines. In addition, some pathways involved in carcinogenesis and neuronal differentiation were also induced, suggesting the presence of some parallel genetic features between these phenomena and T-cell differentiation. These results demonstrate that different vaccine vectors elicit CD8 T cells in which distinct gene networks that are associated with multiple immune functions have been induced. However, whether these transcriptional differences were uniformly induced or caused by the differential induction of distinct subsets within these compartments remained unclear.
Fig. 2.
Microarray analysis of global gene expression in vaccine-elicited CD8+ T cells. (A) The three vaccine regimens uniquely induce distinct sets of genes. H2-Dd/PA9-binding CD8+ T cells from the spleens of immunized mice were isolated and gene transcription was analyzed by cDNA microarray hybridization. Venn diagrams indicate the number of uniquely up- or down-regulated genes induced by the three immunization regimens (P < 0.05) (see Table S1 for the complete list). (B) Gene expression heatmaps of the uniquely up-regulated genes in each immunization group (red indicates high and green indicates low relative expression). (C and D) Enrichment analysis of the uniquely up-regulated genes within EM and CM compartments for each immunization (black: DNA-rAd5; green: rAd5-rLCMV; red: rAd5-rAd5) using the canonical pathway gene sets from Ingenuity Pathway Analysis Software. The gray and blue colors of the figures show the representative pathway membership for each gene (blue: present in the pathway; gray: absent). The identifiers of the most significant (P < 0.05) pathways are displayed on the left side and -Log10(p) on the right side of each figure. The overrepresentation test was performed using Fisher's exact test. Microarray comparisons are based on data obtained in two independent experiments.
Multiparameter Analysis of Gene Expression in Individual Antigen-Specific CD8+ T Cells.
Our findings suggested that distinct gene sets were induced by alternative modes of immunization. However, these analyses were performed using mRNA isolated from pooled populations of vaccine-induced cells; we therefore sought to inquire whether the gene-expression differences observed were the result of differences that were uniformly present in all T cells induced by the immunizations, or whether they were the result of differential induction of subsets of T cells bearing distinct transcriptional patterns. We measured mRNA expression of 91 different transcripts, selected for their relevance to T-cell phenotype, regulation, survival, and function (Table S2) within individual resting CD8+ T cells within CM or EM subsets, and identified by H2-Dd/PA9 tetramer-binding 3 wk following boost immunization.
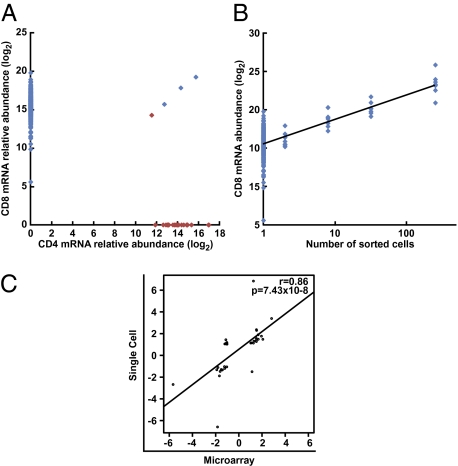
To confirm clonality of sorted cells and the specificity of the qPCR signals, we sorted individual surface-stained CD4 or CD8 single-positive lymphocytes and analyzed the expression of Cd4 and Cd8a mRNA within these cells, in addition to the other transcripts in the assay. Only 0.59% of CD8+ CD4− cells were Cd4 mRNA-positive, and only 4.34% of CD4+ CD8− sorted cells were CD8a mRNA-positive (Fig. 3A). To evaluate the sensitivity of the assay at levels of expression found within single cells, we titrated the number of sorted CD8 surface-positive cells from 128 cells to a single cell per well and measured the abundance of Cd8a mRNA. This analysis showed a strongly linear association between number of cells and the abundance of CD8a mRNA (Fig. 3B). We also validated the single-cell gene-expression data against microarray data of pooled populations of similarly sorted cells. We enumerated the proportion of single cells positive for each gene transcript among CM and EM subsets separately, and then compared these ratios with mRNA measurements from corresponding pooled populations of cells assessed by conventional cDNA microarrays. The ratio of positive cells for a given gene correlated (r = 0.86; P < 0.0001) with its relative expression by cDNA microarray hybridization when comparing CM and EM cells (Fig. 3C).
Fig. 3.
Validation of single-cell gene-expression analysis. (A) Evaluation of false-positive signals. Individual CD4 or CD8 single-positive lymphocytes were sorted by FACS and analyzed for the expression of CD4 and CD8a mRNA in addition to all other transcripts evaluated. Red dots show CD4 single-positive cells and blue dots show CD8 single-positive cells. (B) Evaluation of signal linearity. A titration of CD8+ cells through a range of cell counts from 128 cells down to single cells was sorted by FACS and analyzed for the cd8a mRNA abundance in addition to all other transcripts evaluated. (C) Validation of single-cell gene expression measurements. Single-cell gene-expression data were validated by comparison with microarray analysis of pooled populations of cells. Single H2-Dd/PA9 binding CM and EM cells were either individually sorted and the proportion of positive cells for each transcript evaluated, or cells were pooled and analyzed by conventional cDNA hybridization microarrays. The proportion of positive cells for each transcript was then compared, where possible, with microarray fold-change values. Each point reflects one gene transcript evaluated. Data analysis of single-sorted T cells is based on two independent experiments.
Efficient Discrimination of Individual CD8+ T Cells Elicited by Different Vector Combinations.
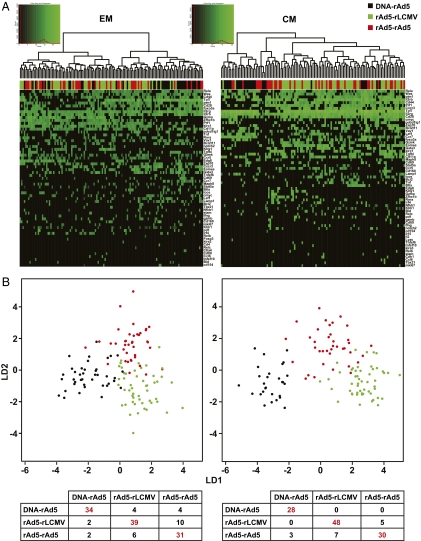
To explore the single-cell gene-expression profiles of CM and EM cells elicited by the three vaccines, we performed a two-way hierarchical cluster analysis (Fig. 4A). In both CM and EM, most of the cells elicited by DNA-rAd were found to cluster separately from the other groups, confirming results obtained by conventional gene arrays (Fig. 2A). To test whether single cells elicited by different immunizations could be distinguished based upon their single-cell gene-expression profiles, we performed a Fisher's linear discriminant analysis. Using this supervised learning algorithm, we were able to readily distinguish CD8 T cells elicited by the three immunization protocols based upon their gene-expression profiles despite separate analysis of CM and EM cells, with a low rate of misclassification (Fig. 4B) (12.4% of CM cells and 21.2% of EM cells). These analyses therefore demonstrate that the majority of T cells elicited by different gene-based vaccines are qualitatively distinct from one another, and can be readily discriminated by analysis of single-cell gene-expression profiles.
Fig. 4.
Analysis of single-cell gene expression in vaccine-elicited CD8+ T cells. Resting H2-Dd/PA9 tetramer binding cells were isolated from the spleens of immunized mice. Expression of the indicated mRNA transcripts was assessed by single cell quantitative RT-PCR. and individually sorted into 96-well plates. Following reverse transcription of cellular mRNA using specific primers, each single-cell cDNA mixture was subjected to 18 cycles of PCR preamplification before microfluidic separation into a further 96 separate reactions for the specific quantification of single-gene transcripts by quantitative RT-PCR. (A) Gene-expression heatmaps of single-cell gene-expression profiles obtained after two-way hierarchical clustering using Euclidean distance and Ward agglomeration methods. The coexpression of gene transcripts within single CD8+ T cells elicited by the three immunizations was analyzed. (B) Linear discriminant analysis. To assess whether single-cell gene-expression profiles could be used to discriminate individual EM (Left) or CM (Right) CD8+ T cells elicited by the three different immunization regimens, we performed a Fisher Linear Discriminant Analysis on the dataset. Each dot represents a single cell (black: DNA-rAd, green: rAd-rLCMV, red: rAd-rAd). The truth tables below each heatmap represent a crossvalidation of the linear discriminant analysis and compare predicted classifications (columns) with the actual immunization regimen used (rows). Both EM and CM cells elicited by the three immunizations could be readily discriminated, as indicated by a low rate of misclassification (12.4% for CM T cells and 21.2% for EM T cells). Data analysis of single cell sorted T cells is based on two independent experiments.
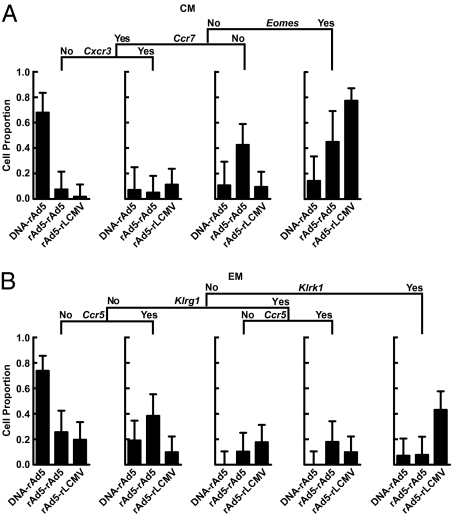
To identify the smallest set of genes that could allow classification of T cells elicited by the three different immunization regimens, we performed a supervised decision-tree analysis using binary expression values of a smaller number of genes preselected using Fisher's exact test (Tables S3 and S4). We found that a majority of surface IL7R+ CD62L+ CM cells elicited by the three immunization groups could be distinguished based upon the differential expression of a small number of genes, in particular, Eomes, Cxcr3, and Ccr7 (Fig. 5A). Notably, 77% of rAd-LCMV–elicited CM CD8 T cells were Eomes+, while only 14% of DNA-rAd–elicited CD8 T cells and 45% of rAd-rAd–elicited T cells were Eomes+ (Fig. 5A, Right). Additionally, we found that 67% of plasmid-primed (DNA-rAd–elicited) CM T-cells were Eomes− Ccr7+ Cxcr3−, but only 7% and 2% of vector-primed cells (elicited by rAd5-rAd5 and rAd-LCMV, respectively) possessed a similar transcriptional pattern (Fig. 5A, Left), indicating the presence of a subset of CM cells, the frequency of which was dependent upon the vector used to prime the immune response. Similarly, this analysis allowed subsets of EM cells to be defined based upon the expression of Klrk1, Klrg1, and Ccr5, the proportion of which varied with the immunization regimen used (Fig. 5B). We found that 73.8% of plasmid-primed (DNA-rAd5–elicited) EM cells were Klrk1− Klrg1−Ccr5−, whereas only 25.6% and 19.6% of vector-primed cells (rAd5-rAd5 and rAd5-LCMV, respectively) exhibited this transcriptional signature. Thus, analysis of single-cell gene-expression profiles allowed the majority of cells elicited by the three different vaccines to be distinguished, despite analysis within EM and CM compartments independently.
Fig. 5.
Decision-tree analysis of single-cell gene expression profiles. Gene-expression data were analyzed to determine whether assessment of single-cell coexpression in a smaller subset of genes could allow the classification of cells elicited by the three immunization groups. Bar graphs showing leaf statistics represent the proportion of cells elicited by each immunization group which satisfy the given transcriptional pattern as indicated in the regression tree. (A) CM, (B) EM CD8 T cells. The genes used for decision tree analysis were selected following Fisher's exact test to identify the top seven and top four most significantly discriminating genes for CM (Table S3) and EM (Table S4) respectively (Bonferroni threshold 0.0008).
Discussion
In this study, we analyzed gene expression in pooled antigen-specific cells and within single cells comprising these populations to assess differences in cellular immunity resulting from immunization with three different prime-boost vaccine regimens. Although the immune responses were indistinguishable by assays conventionally used in vaccine studies, we found that the majority of CD8 T cells elicited by the different vaccines could be readily distinguished based upon analysis of their single-cell gene-expression profiles, despite the fact that we analyzed cells within phenotypically defined subsets. These results support the view that quantitative parameters, such as response magnitude, or qualitative paramaters, such as cytokine functionality and phenotype, may not identify correlates of protective T-cell immunity in vaccine studies, because they are insensitive to the extent of heterogeneity that can be measured in CD8 T-cell responses.
In our analysis, cells elicited by DNA-rAd5 and rAd5-rAd5 could be readily distinguished from each other, indicating the substantial role of the priming immunization in shaping the quality of the T-cell response. Beneficial protective outcomes have been observed since the description of heterologous prime boosting using gene-based viral vectors (20). Recently, Liu et al. found that rhesus macaques immunized with rAd26/rAd5 encoding SIV Gag in prime-boost combination exhibited greater reductions of peak and setpoint viraemia, as well as decreased AIDS-related mortality compared with macaques that received rAd5/rAd5 or rAd35/rAd5 following challenge with SIV(MAC251) (4). Although some modest differences in the magnitude and cytokine profile of elicited T cells were reported, no qualitative parameter could be defined that substantially predicted outcome. Our results provide a strategy for the evaluation of the independent roles of prime and boost immunization in determining the quality of the subsequent T-cell response and for identifying correlates of immune protection in such studies and others (3–7).
Based on single-cell gene-expression measurements, we were able to define unique subsets of CD8 T cells that were differentially induced by the three vaccines. We observed that Klrk1− Klrg1−Ccr5− EM cells were induced with greater frequency as a result of priming with plasmid DNA rather than a viral vector. Indeed, each of these genes was seen to be progressively up-regulated in a recently reported experiment where the global gene-expression profiles of CD8 T cells repetitively restimulated in vivo was assessed (21). Additionally, KLRG1 has been previously characterized as a marker of terminal differentiation in CD8 T cells, the expression of which is induced with increased antigen-receptor signaling (22). These data suggest that plasmid-primed EM responses may exist in an earlier maturational state than viral vector-primed responses as a result of weaker stimulation, consistent with the weaker T-cell immunogenicity of plasmid DNA compared with viral vectors (23, 24). However, although Eomes− Ccr7+ Cxcr3− CM cells were also induced with greater frequency as a result of priming with plasmid DNA rather than a viral vector, all of these genes were progressively downregulated with increasing stimulation, indicating the presence of other factors, in addition to antigen strength, that may also determine the quality of T-cell memory.
Both microarray analysis of pooled cell populations and single-cell gene-expression analyses were able to resolve differences in T-cell responses generated by the three immunization regimens. Although microarrays allowed an unbiased evaluation of transcriptional differences between cellular immune responses elicited by the three vaccines, single-cell measurements allowed the analysis of coordinate gene expression, and thus the discrimination of subsets of cells that would not have been discriminated based upon a unidimensional analysis of gene transcription (Fig. 4). Furthermore, multidimensional analysis of single-cell gene-expression profiles allowed the resolution of vast heterogeneity at a transcriptional level within both CM and EM compartments. Just as the breadth of T-cell epitope specificity is evaluated in vaccine-induced responses (25, 26), measurement of the diversity of the T-cell memory response induced by vaccination is possible through analysis of single-cell gene-expression profiles, a parameter that cannot be measured by microarray analysis of pooled cell populations, and this may be of substantial utility in identifying correlates of protective outcomes in future studies.
Our attempts to design and evaluate CD8 T-cell vaccines are confounded by our inability to resolve major qualitative differences between the T cells elicited by different vaccines, even when divergent protective outcomes are observed upon infectious challenge (3–7, 11, 16). Being able to resolve differences between T cells elicited by different vaccines is therefore critically important if effective T-cell vaccines are to be developed against such infectious pathogens as HIV, tuberculosis, and malaria. In this study, the analysis of gene transcription within single immune cells has allowed the qualitative discrimination of CD8+ T-cell responses from three vaccines that could not be resolved using conventional assays. Such analyses of vaccine-elicited T cells in clinical efficacy trials and nonhuman primate studies will likely allow the identification of correlates of protective immunity in these studies.
Materials and Methods
Animals and Immunization Protocols.
Mice at 6 to 10 wk of age were immunized with DNA-rAd5, rAd5-rAd5, or rAd5-rLCMV prime-boost vaccine regimens, and splenocytes were analyzed by flow cytometry at 3 wk following the final immunization by surface or intracellular staining, or by microarray or nanofluidic single cell quantitative RT-PCR following FACS as described below and in SI Materials and Methods.
Flow Cytometry and Intracellular Staining.
The phenotype and function of antigen-specific immune responses were measured by H2Dd/PA9 tetramer staining and intracellular cytokine staining following PA9 peptide stimulation, as described in SI Materials and Methods.
Measurement of Gene Expression Within Single Cells and Microarray Analyses.
Microarray analyses were performed on pooled populations of antigen-specific cells, as described in SI Materials and Methods.
Measurement of Gene Expression Within Single Cells.
Single tetramer-binding CD8+ T cells were stained and individually sorted using a BD FACS Aria into 96-well plates. Following cellular lysis and specific generation of single cell cDNA libraries (27, 28), each sample was split nanofluidically into 96 separate chambers for specific qRT-PCR for the individual transcripts indicated in Table S2, as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Steve Perfetto, Richard Nguyen, David Ambrozak, Pratip Chattopadhyay, Enrico Lugli, Yolanda Mahnke, and Carl Hogerkorp for help and advice with cell sorting, microarray, and real time PCR; Jeff Skinner for his advice regarding hierarchical clustering of gene expression data; Derek Macallan and George Griffin, Martin Pieprzyk, Tomer Kalisky, Daniel Danila, and Nicholas Restifo for their help and advice; and Brenda Hartman for help with the figure preparation and Ati Tislerics for support in manuscript editing. This research was supported in part by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health. L.F. was supported by the Schwyzer Stiftung.
Footnotes
*This Direct Submission article had a prearranged editor.
Conflict of interest statement: M.L. is a research technician for Fluidigm Corporation.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013084108/-/DCSupplemental.
References
- 1.Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: Implications for vaccine design. Nat Rev Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 2.Sekaly RP. The failed HIV Merck vaccine study: A step back or a launching point for future vaccine development? J Exp Med. 2008;205(1):7–12. doi: 10.1084/jem.20072681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson NA, et al. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J Virol. 2009;83:6508–6521. doi: 10.1128/JVI.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, et al. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature. 2009;457(7225):87–91. doi: 10.1038/nature07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reynolds MR, et al. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J Exp Med. 2008;205:2537–2550. doi: 10.1084/jem.20081524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen SG, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15:293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukamoto T, et al. Impact of cytotoxic-T-lymphocyte memory induction without virus-specific CD4+ T-Cell help on control of a simian immunodeficiency virus challenge in rhesus macaques. J Virol. 2009;83:9339–9346. doi: 10.1128/JVI.01120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letvin NL. Correlates of immune protection and the development of a human immunodeficiency virus vaccine. Immunity. 2007;27:366–369. doi: 10.1016/j.immuni.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Friedrich TC, Watkins DI. Wanted: Correlates of vaccine-induced protection against simian immunodeficiency virus. Curr Opin HIV AIDS. 2008;3:393–398. doi: 10.1097/COH.0b013e3282faa461. [DOI] [PubMed] [Google Scholar]
- 10.Darrah PA, et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13:843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 11.Engram JC, et al. Vaccine-induced, simian immunodeficiency virus-specific CD8+ T cells reduce virus replication but do not protect from simian immunodeficiency virus disease progression. J Immunol. 2009;183:706–717. doi: 10.4049/jimmunol.0803746. [DOI] [PubMed] [Google Scholar]
- 12.Reyes-Sandoval A, et al. Prime-boost immunization with adenoviral and modified vaccinia virus Ankara vectors enhances the durability and polyfunctionality of protective malaria CD8+ T-cell responses. Infect Immun. 2010;78(1):145–153. doi: 10.1128/IAI.00740-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274(5284):94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 14.Kaech SM, et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 15.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 16.Acierno PM, et al. Preservation of functional virus-specific memory CD8+ T lymphocytes in vaccinated, simian human immunodeficiency virus-infected rhesus monkeys. J Immunol. 2006;176:5338–5345. doi: 10.4049/jimmunol.176.9.5338. [DOI] [PubMed] [Google Scholar]
- 17.Chakrabarti BK, et al. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J Virol. 2002;76:5357–5368. doi: 10.1128/JVI.76.11.5357-5368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honda M, et al. Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity. J Immunol. 2009;183:2425–2434. doi: 10.4049/jimmunol.0900581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irvine KR, et al. Enhancing efficacy of recombinant anticancer vaccines with prime/boost regimens that use two different vectors. J Natl Cancer Inst. 1997;89:1595–1601. doi: 10.1093/jnci/89.21.1595. [DOI] [PubMed] [Google Scholar]
- 21.Wirth TC, et al. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8(+) T cell differentiation. Immunity. 2010;33(1):128–140. doi: 10.1016/j.immuni.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarkar S, et al. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao SS, et al. Comparative evaluation of three different intramuscular delivery methods for DNA immunization in a nonhuman primate animal model. Vaccine. 2006;24:367–373. doi: 10.1016/j.vaccine.2005.07.072. [DOI] [PubMed] [Google Scholar]
- 24.Kong WP, et al. Immunogenicity of multiple gene and clade human immunodeficiency virus type 1 DNA vaccines. J Virol. 2003;77:12764–12772. doi: 10.1128/JVI.77.23.12764-12772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seaman MS, et al. Multiclade human immunodeficiency virus type 1 envelope immunogens elicit broad cellular and humoral immunity in rhesus monkeys. J Virol. 2005;79:2956–2963. doi: 10.1128/JVI.79.5.2956-2963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, Kong WP, Nabel GJ. Enhanced breadth of CD4 T-cell immunity by DNA prime and adenovirus boost immunization to human immunodeficiency virus Env and Gag immunogens. J Virol. 2005;79:8024–8031. doi: 10.1128/JVI.79.13.8024-8031.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foygel K, et al. A novel and critical role for Oct4 as a regulator of the maternal-embryonic transition. PLoS ONE. 2008;3:e4109. doi: 10.1371/journal.pone.0004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diehn M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.