Abstract
Methylation on lysine 9 of histone H3 (H3K9me) and DNA methylation play important roles in the transcriptional silencing of specific genes and repetitive elements. Both marks are detected on class I and II endogenous retroviruses (ERVs) in murine embryonic stem cells (mESCs). Recently, we reported that the H3K9-specific lysine methyltransferase (KMTase) Eset/Setdb1/KMT1E is required for H3K9me3 and the maintenance of silencing of ERVs in mESCs. In contrast, G9a/Ehmt2/KMT1C is dispensable, despite the fact that this KMTase is required for H3K9 dimethylation (H3K9me2) and efficient DNA methylation of these retroelements. Transcription of the exogenous retrovirus (XRV) Moloney murine leukemia virus is rapidly extinguished after integration in mESCs, concomitant with de novo DNA methylation. However, the role that H3K9 KMTases play in this process has not been addressed. Here, we demonstrate that G9a, but not Suv39h1 or Suv39h2, is required for silencing of newly integrated Moloney murine leukemia virus–based vectors in mESCs. The silencing defect in G9a−/− cells is accompanied by a reduction of H3K9me2 at the proviral LTR, indicating that XRVs are direct targets of G9a. Furthermore, de novo DNA methylation of newly integrated proviruses is impaired in the G9a−/− line, phenocopying proviral DNA methylation and silencing defects observed in Dnmt3a-deficient mESCs. Once established, however, maintenance of silencing of XRVs, like ERVs, is dependent exclusively on the KMTase Eset. Taken together, these observations reveal that in mESCs, the H3K9 KMTase G9a is required for the establishment, but not for the maintenance, of silencing of newly integrated proviruses.
Keywords: epigenetics, covalent histone modification, long terminal repeat
Retroviruses have colonized all classes of vertebrates and are responsible for a range of pathologies in mammals, including cancer in distantly related species and acquired immunodeficiency syndrome in humans. Although productive retroviral infection by a subset of retroviruses is cytopathic, proviral elements of the acute- and slow-transforming classes induce tumorigenesis by expressing viral oncogenes and perturbing the expression of cellular genes, respectively. Given the potential deleterious effects of retroviral infection, a number of cell autonomous pathways that act at the transcriptional or post-transcriptional stages of the retroviral replicative cycle have evolved to inhibit retroviral expression (1, 2).
Retroviral vectors based on the exogenous retrovirus (XRV) Moloney murine leukemia virus (MLV), such as the MFG vector (3), have been used extensively in the laboratory and in the clinic as vehicles for gene delivery. In both settings, transcriptional silencing has proven to be a major impediment to stable/long-term proviral expression (1, 4), due in part to the presence of negative regulatory elements in the proviral LTR that are bound by transcriptional repressors (5, 6) and the binding of chromatin proteins, including histone H1 (7, 8) and macroH2A1 (9). MLV is repressed with particular efficiency in embryonic carcinoma (EC) and embryonic stem (ES) cells (10), due primarily to the presence of a potent repressor-binding site that overlaps with the primer-binding site (PBS) complementary to the 3′ end of proline tRNA (tRNAPro) in the WT MLV virus (11–13). The Kruppel-associated box (KRAB) zinc finger protein ZFP809 and the KRAB-associated protein 1 (KAP-1/Trim28/Tif1-β), a potent transcriptional corepressor, were recently identified as subunits of the stem cell-specific repressor complex (13) that binds to the repressor-binding site (14, 15). To circumvent transcriptional silencing, a number of vectors harboring mutations in these negative regulatory elements have been generated (16–18). The murine stem cell virus (MSCV) vector also includes an introduced SP1-binding site within the LTR, along with a tRNAGln PBS in place of tRNAPro (18). Although these “optimized” retroviral vectors are silenced less efficiently than WT vectors such as MFG in EC, ES, and hematopoietic stem cells, proviral silencing still occurs in these cells (7, 16, 17, 19).
In addition, transcriptional silencing of MLV-based vectors is frequently accompanied by proviral de novo DNA methylation (20, 21) and binding of the methyl DNA-binding domain protein MeCP2 (8, 22), implicating DNA methylation in the silencing process. Indeed, treatment of EC cells with the DNA methylation inhibitor 5-azacytidine leads to transcriptional reactivation of MLV (21). However, the kinetics of de novo methylation (20, 23) and the persistence of proviral silencing in murine ES cells (mESCs) deficient in the de novo DNA methyltransferases Dnmt3a and Dnmt3b (7) indicate that DNA methylation-independent silencing pathways must exist as well.
Intriguingly, genome-wide studies have shown that endogenous retroviral elements (ERVs) are marked by the repressive histone marks H3K9me2 and/or H3K9me3 in mESCs (24–26). Recently, we demonstrated that although ERVs exhibit reduced H3K9me2 (but not H3K9me3) and DNA methylation in mESCs lacking the “euchromatic” H3K9 KMTase G9a (27), these parasitic elements remain transcriptionally inert. Subsequently, we found that Eset, an H3K9 trimethylase, is required for maintaining class I and II ERVs in a silent state (28). Little is known about the role of H3K9 KMTases in the establishment of silencing of newly integrated proviruses, however. Here we show that G9a plays a vital role in the establishment of silencing of MLV-based XRVs, but is dispensable for the maintenance of silencing of these elements.
Results
G9a Is Required for the Establishment of Silencing of MLV-Based Vectors.
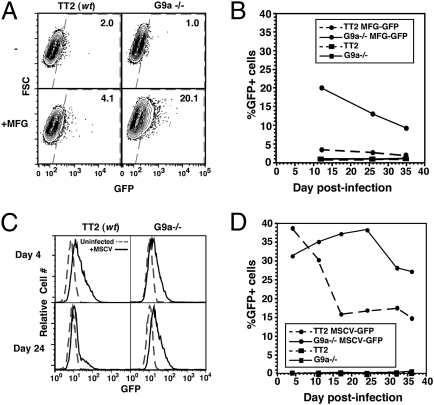
To determine whether G9a plays a role in repressing the expression of newly integrated proviral elements, WT (TT2) and G9a−/− mESCs (29) (Fig. S1) were transduced with the MLV-based retroviral vector MFG-GFP (3) (Fig. 1A), which is poorly expressed in WT pluripotent cells relative to differentiated cells, and passaged for further analysis (Fig. 1B). Intriguingly, whereas <5% of infected WT cells expressed GFP at day 12 postinfection (PI), >20% of infected G9a−/− cells expressed GFP at the same time point, as measured by flow cytometry (Fig. 2A). Furthermore, although the percentage of GFP+ cells in both lines diminished over time, the percentage of GFP+ cells in G9a−/− cells remained higher than in the WT line at all time points analyzed (Fig. 2B), indicating that G9a is required for the efficient establishment of silencing of this MLV-based provirus.
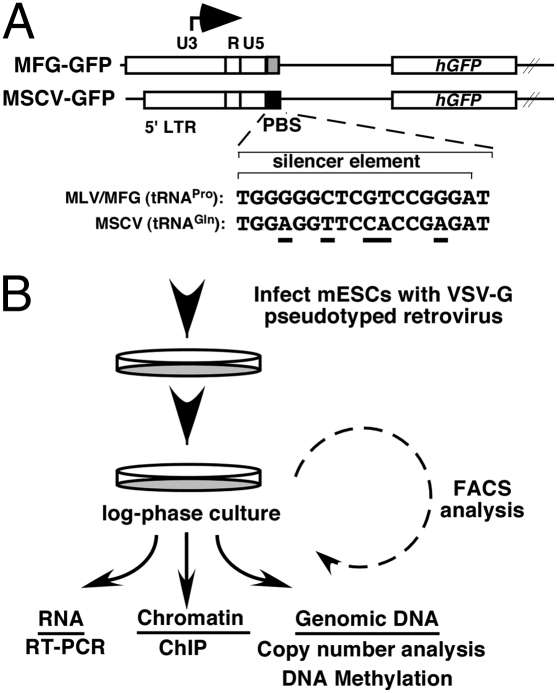
Fig. 1.
Retroviral vectors and infection schema. (A) Maps of the 5′ region of the MFG-GFP and MSCV-GFP retroviral vectors. The stem cell-specific silencer binds to the MFG/tRNAPro PBS, but not to the MSCV/tRNAGln PBS. Sequence differences are underscored. (B) mESCs were infected with MFG-GFP or MSCV-GFP and passaged for further analyses, as shown.
Fig. 2.
G9a−/− mESCs show a defect in silencing of MLV provirus. (A) TT2 (WT) and G9a−/− mESCs were infected with MFG-GFP (MFG) and analyzed by flow cytometry at day 12 PI. Uninfected cells were analyzed in parallel. Data are presented as forward scatter (FSC) versus GFP. (B) Analysis of GFP expression by flow cytometry at successive time points PI. (C and D) TT2 and G9a−/− mESCs were infected with MSCV-GFP and analyzed by flow cytometry as above.
To determine whether G9a plays a role in silencing of an alternative MLV vector that is “optimized” for expression in mESCs, we carried out similar experiments with the MLV-based vector MSCV. This retroviral vector harbors mutations in the LTR as well as a tRNAGln PBS in place of the tRNAPro PBS (Fig. 1A), which ablates binding of the stem cell-specific repressor complex (12, 18) to the overlapping silencer element. These mutations confer a significantly higher level of expression than the MFG vector (7, 30) and reduce the rate of silencing in mESCs (30). G9a−/− and WT cells were transduced with MSCV-GFP virus, and the percentage of cells expressing GFP was analyzed on a weekly basis by flow cytometry. Although the MSCV vector was expressed in ∼40% of infected WT cells early after infection, a dramatic decrease in the percentage of GFP+ cells was observed over time in culture (Fig. 2 C and D). In contrast, this vector was not efficiently silenced over time in G9a−/− cells. Consistent with the flow cytometry data, quantitative RT-PCR (qRT-PCR) analysis on day 15 PI revealed a significantly higher level of GFP expression in the G9a−/− line than the WT parent line (Fig. S2A). Relative to the WT pool, the G9a−/− pool of infected cells harbored approximately the same number of proviral copies per cell (Fig. S2B), ruling out the possibility that the observed difference in expression is the result of a difference in proviral load.
WT, but Not Catalytically Inactive G9a, Rescues the Proviral Silencing Defect in G9a−/− mESCs.
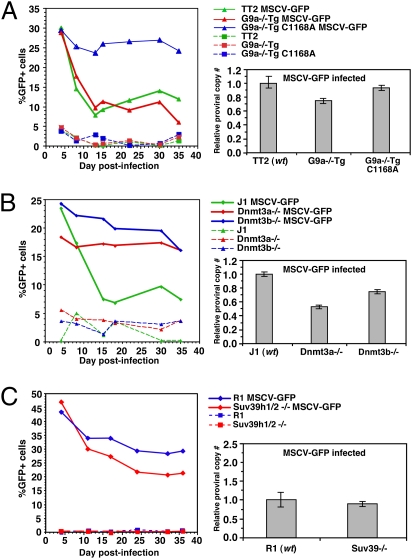
To verify that deletion of G9a per se is responsible for the observed phenotype, and to determine whether proviral silencing is dependent on G9a catalytic activity, G9a−/− cells stably expressing either a WT (G9a−/−Tg) or a catalytically inactive (G9a−/−Tg C1168A) G9a transgene (31) (Fig. S1) were infected with MSCV-GFP virus, as described above. Expression of exogenous WT G9a “rescues” the silencing defect observed in G9a−/− mESCs, but expression of the catalytic mutant does not (Fig. 3A), indicating that catalytically active G9a is critical for the silencing of newly integrated proviruses. Quantitative analysis of the relative proviral copy number reveals that the differences in proviral expression are not due to differences in proviral load.
Fig. 3.
Introduction of WT, but not of catalytically inactive G9a rescues the silencing defect, whereas Dnmt3a−/− mESCs show a defect in proviral silencing similar to that observed in G9a−/− cells. (A) TT2 (WT) and G9a−/− cells expressing a G9a transgene (G9a−/−Tg) or a catalytically inactive transgene (G9a−/−Tg C1168A) were infected in parallel with MSCV-GFP and analyzed for GFP expression by flow cytometry at successive time points, PI. Proviral load was determined by qPCR using primers specific for the GFP gene and the endogenous β-major gene as a control and the relative mean (± SD) proviral copy number/cell for each infected population normalized to the corresponding infected WT parent line. (B and C) J1 (WT), Dnmt3a−/−, and Dnmt3b−/− mESCs (B) or R1 (WT) and Suv39h1/2−/− (C) mESCs were infected in parallel and analyzed for GFP expression and proviral load as above.
The Silencing Defect Observed in G9a−/− mESCs Phenocopies That Observed in Dnmt3a−/− mESCs.
In WT mESCs, MLV-based vectors show increasing DNA methylation density with prolonged passage in culture (8), and dense DNA methylation is sufficient for proviral silencing in somatic and EC cells (8, 22, 30). To determine whether mESCs deficient in Dnmt3a and Dnmt3b have a silencing defect similar to that observed in the G9a−/− line, WT (J1) mESCs were infected with the MSCV-GFP virus in parallel with Dnmt3a and Dnmt3b double-knockout (Dnmt3a/b−/−) cells (Fig. S3). Consistent with our observations in WT TT2 mESCs, proviral expression in WT J1 mESCs decreased substantially in the first 2 wk PI. In contrast, Dnmt3a/b−/− cells demonstrated a silencing defect similar to that observed in G9a−/− mESCs. To determine which of the de novo DNMTs is required for silencing, Dnmt3a−/− (6aa) and Dnmt3b−/− (8bb) (32) mESCs were infected and analyzed by FACS at successive days PI, as above. Intriguingly, whereas Dnmt3a−/− cells demonstrated a similar silencing defect to that observed in G9a−/− cells, Dnmt3b−/− cells showed a relatively modest silencing defect (Fig. 3B). Quantitative analysis of the relative proviral copy number showed that the differences in the percentages of GFP+ cells in each line cannot be explained by differences in proviral load. Taken together with the observation that Dnmt1-deficient cells have only a modest defect in de novo DNA methylation of newly integrated proviruses (33), these results indicate that both G9a and Dnmt3a2 [the predominant isoform of Dnmt3a in mESCs (34)] are required for the establishment of proviral silencing in mESCs.
Suv39h1 and Suv39h2 Are Not Required for Silencing of Newly Integrated Proviruses.
To determine whether mESCs deficient in the “heterochromatic” H3K9 KMTases, Suv39h1 and Suv39h2, play a role in proviral silencing, WT (R1) mESCs were infected with MSCV-GFP in parallel with Suv39h1 and Suv39h2 double-deficient (Suv39h1/2−/−) mESCs, as above, and analyzed on successive days PI. In contrast to G9a−/− mESCs, and consistent with a previous report showing no decrease in DNA methylation of endogenous MLV in this line (35), the MSCV-GFP vector was effectively silenced in Suv39h1/2−/− mESCs (Fig. 3C). In fact, proviral silencing is somewhat more efficient in the Suv39h1/2−/− line than in the WT line, perhaps due to the relocalization of HP1 proteins (36) and/or Dnmt3b (35) away from the pericentromeric compartment in these cells.
H3K9me2 Is Decreased in the 5′ LTR/Promoter Region of the MSCV-GFP Provirus in G9a−/− mESCs.
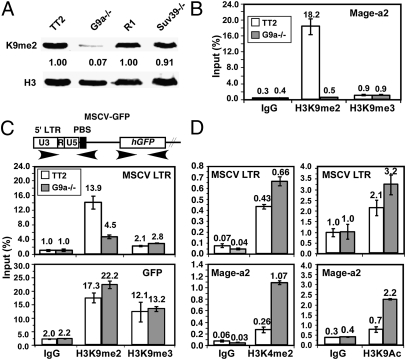
As expected, G9a−/− cells were globally depleted of H3K9me2, whereas Suv39h1/2−/− cells showed no decrease in this epigenetic mark (26) (Fig. 4A). To determine whether the silencing defect observed in G9a−/− cells is associated with reduced proviral H3K9 methylation, native ChIP experiments were conducted on chromatin isolated from WT TT2 and G9a−/− cells on day 22 PI, using antibodies raised against H3K9me2 or H3K9me3. The specificity of these antibodies for ChIP analyses was confirmed using primers specific for the Mage-a2 gene promoter and major satellite repeats, respectively (Fig. 4B and Fig. S4). Consistent with previous observations (27, 29), the Mage-a2 gene exhibited a high level of G9a-dependent H3K9me2 enrichment relative to the IgG control, but very low H3K9me3 enrichment. In contrast, major satellite repeats, previously found to be marked by H3K9me3 independent of G9a (26, 27), showed a high level of H3K9me3 in WT and G9a−/− cells (Fig. S4).
Fig. 4.
H3K9me2 is reduced at the proviral LTR in G9a−/− mESCs. (A) Depletion of H3K9me2 in G9a−/− cells, but not in Suv39h1/2−/− cells, was confirmed by Western blot analysis. ChIP was conducted on MSCV-GFP–infected TT2 and G9a−/− mESCs using H3K9me2- and H3K9me3-specific antibodies or IgG as a control. (B) qPCR was conducted using primers specific for the endogenous Mage-a2 gene. The mean level of enrichment of technical replicates [expressed as a percentage (± SD) of input] is shown. (C) Amplification with primers specific for the MSCV 5′ LTR and GFP regions revealed a decrease in H3K9me2 at the 5′ LTR in the G9a−/− line. (D) ChIP was conducted with H3K4me2- and H3K9ac-specific antibodies and analyzed by qPCR, as above.
Analysis of the MSCV LTR in the same chromatin isolates revealed a more than threefold decrease in H3K9me2 enrichment in the G9a−/− line (Fig. 4C) on day 22 PI. In contrast, the level of H3K9me3 enrichment in the same region, although greater than that observed with control IgG, did not differ significantly between the two lines, indicating that an alternative KMTase is responsible for the H3K9me3 of XRVs. Although whether distinct subsets of proviral integrants are marked by H3K9me2 or H3K9me3 is unclear, these results are consistent with our previous finding that Eset is responsible for H3K9me3 of the MSCV-GFP provirus in mESCs (28). Analysis of the GFP region revealed high levels of both H3K9me2 and H3K9me3 in both cell lines, indicating that H3K9me2 and H3K9me3 mark the transcribed region of the provirus independent of G9a (Fig. 4C). The presence of H3K9me2 and/or H3K9me3 in gene bodies has been reported previously (37, 38).
ChIP analysis using antibodies specific for H3K4me2 and H3K9ac (marks typically associated with transcriptionally active genes) revealed the opposite pattern, with higher levels of enrichment in the Mage-a2 and proviral 5′ LTR promoter regions in the G9a−/− line than in the WT line (Fig. 4D). The presence of these active marks in the proviral LTR in the WT line at levels significantly above background presumably reflects the presence of constitutively expressing proviral integrants in this pool of infected cells (Fig. 2D). Taken together, these results indicate that G9a contributes to proviral silencing in mESCs by directly marking chromatin associated with newly integrated proviruses.
Catalytically Active G9a Is Required for Efficient DNA Methylation of MLV-Based XRVs.
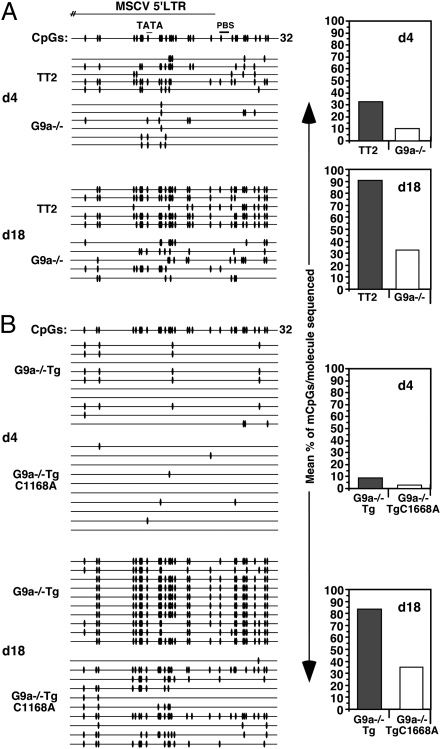
We previously showed that the 5′ LTR/PBS and GFP regions of the MFG-GFP provirus are hypomethylated in G9a−/− relative to WT cells on day 18 PI (27). An even more dramatic de novo methylation defect across the proviral LTR was observed in MFG-GFP–infected Dnmt3a/b−/− mESCs isolated on day 18 PI (27); these bisulfite data are summarized in Fig. S5A. To examine whether DNA methylation of the MSCV vector is also dependent on G9a, genomic DNA was isolated from MSCV-GFP infected TT2 and G9a−/− cells on days 4 and 18 PI and bisulfite analysis was conducted using primers specific for the MSCV 5′ LTR/PBS region. Although an increase in proviral DNA methylation was detected in both TT2 and G9a−/− cells with passage in culture, an approximately threefold lower level of DNA methylation was detected in G9a−/− cells compared with WT cells at both time points (Fig. 5A). Notably, the level of proviral DNA methylation in G9a−/− cells on day 18 PI was similar to that of the WT cells on day 4 PI, indicating that DNA methylation accumulates at a much slower rate in the absence of G9a. To determine whether efficient proviral DNA methylation is dependent on the catalytic activity of G9a, the pools of infected G9a−/−Tg and G9a−/−TgC1168A cells were analyzed by bisulfite sequencing on days 4 and 18 PI. A significantly lower level of DNA methylation was observed only in the G9a−/−TgC1168A line on day 18 PI (Fig. 5B), confirming that de novo DNA methylation of newly integrated provirus requires the enzymatic activity of G9a.
Fig. 5.
The rate of proviral de novo DNA methylation is reduced in G9a−/− and G9a−/−TgC1168A cells. TT2, G9a−/−, G9a−/−Tg, and G9a−/−TgC1168A mESCs were infected with MSCV-GFP. Genomic DNA from TT2 and G9a−/− (A) or G9a−/−Tg and G9a−/−TgC1168A (B) mESCs was isolated at d4 and d18 PI and analyzed by bisulfite sequencing using primers specific for the proviral 5′ LTR. The presence of a methylated CpG within each sequenced molecule is indicated with a black oval, and the mean percentage of methylated CpGs/molecule sequenced is shown for each data set.
In contrast, bisulfite analysis of genomic DNA isolated from WT R1 and Suv39h1/2−/− cells on day 18 PI revealed a <1.2-fold difference in DNA methylation within the MSCV LTR (Fig. S5B), consistent with a previous report showing no decrease in DNA methylation of endogenous MLV in Suv39h1/2−/− cells (35). Taken together, these results indicate that whereas catalytically active G9a is required for H3K9me2 and efficient de novo DNA methylation of MLV-based proviral elements in mESCs, Suv39h1 and Suv39h2 are dispensable for these processes.
Maintenance of Proviral Silencing in mESCs Is Not Dependent on G9a.
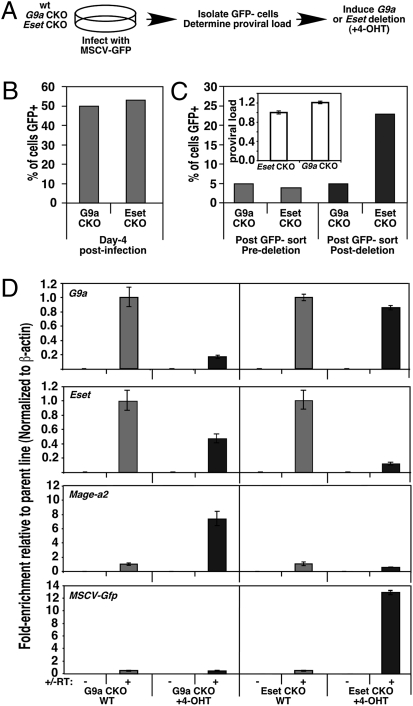
Having shown that G9a is required for the establishment of XRV silencing, we next wished to determine whether this KMTase also plays a role in the maintenance of XRV silencing in mESCs, particularly in light of our previous observation that Eset, but not G9a, is required for the maintenance of silencing of ERVs (28). G9a conditional knockout (CKO) (39) and Eset CKO (28) mESCs were infected in parallel with MSCV-GFP (Fig. 6A), and GFP− cells, representing ∼50% of each population (Fig. 6B), were isolated via flow cytometry on day 14 PI. The stability of proviral silencing in these infected pools, which harbor roughly equivalent proviral loads, was confirmed by FACS (Fig. 6C). Subsequently, these GFP− pools were treated with 4-hydroxytamoxifen (4-OHT), which induces Cre-ER recombinase-mediated deletion of G9a and Eset, respectively. Strikingly, although deletion of G9a had no effect on the percentage of GFP+ cells, deletion of Eset induced a fivefold increase in the percentage of GFP+ cells (Fig. 6C). Depletion of G9a and Eset RNA in the 4-OHT–treated CKO lines was confirmed by qRT-PCR using primers specific for the deleted exons (Fig. 6D). Furthermore, although expression of the Mage-a2 gene, previously shown to be induced in G9a−/− mESCs (29), was clearly induced after deletion of G9a, no change in proviral expression was observed in these cells (Fig. 6D), consistent with the flow cytometry results. In contrast, a 12-fold increase in proviral expression was observed in the Eset CKO line. Taken together, these results indicate that whereas Eset plays a critical role in the maintenance of proviral silencing in mESCs, G9a is dispensable for this process.
Fig. 6.
G9a is not required for the maintenance of proviral silencing in mESCs. (A) Schema for the generation of cells harboring silent XRVs. (B) Pools of G9a and Eset CKO mESCs infected with MSCV-GFP were analyzed by flow cytometry at d4 PI, and GFP− cells were isolated by FACS at day 14 PI. (C) GFP− pools were treated in parallel with 4-OHT to induce deletion of G9a and Eset, respectively and analyzed by FACS before and on day 7 post-deletion. Mean relative proviral load (± SD) was determined by qPCR using primers specific for the GFP gene, normalized to the β-major gene (Inset). (D) RNA was isolated from untreated and 4-OHT–treated CKO cells, and the mean (± SD) fold-enrichment in expression of G9a, Eset, Mage-a2, and MSCV-GFP (normalized to β-actin) relative to the untreated WT parent lines was determined by qRT-PCR.
Maintenance of Proviral DNA Methylation in MEFs Is Not Dependent on G9a.
To determine whether G9a influences proviral DNA methylation in a somatic cell type, we analyzed murine embryonic fibroblasts (MEFs), which express Dnmt1, Dnmt3a1, and Dnmt3b (40). Primary MEF cultures were established from embryos that carry conditional alleles of G9a, and immortalized clones showing conditional depletion of G9a and a concomitant loss of H3K9me2 were identified (Fig. S6A). Although the MSCV-GFP provirus is not efficiently de novo methylated in MEFs (41) and is not silenced in these cells (Fig. S6B), ERVs are methylated in MEFs in a Dnmt3b-dependent manner (40). Thus, we compared the DNA methylation status of ERVs in G9a-deficient MEFs and mESCs. Although Southern blot analysis revealed decreased DNA methylation of MLV and IAP elements in G9a−/− mESCs (Fig. S6C), as reported previously (27), no decrease in DNA methylation of either ERV was detected in G9a−/− MEFs (Fig. S6D). Taken together, these results indicate that G9a plays a critical role in de novo methylation of proviral elements in mESCs, but is not required for maintenance DNA methylation of proviral elements in mESCs or MEFs.
Discussion
H3K9 methylation acts upstream of DNA methylation in plants and filamentous fungi and plays a critical role in silencing of transposable elements in these eukaryotes (42). In mESCs, ERVs are marked by H3K9me2 and H3K9me3 (24–26), and Eset is required for H3K9me3 and silencing of these elements (28), indicating that a similar pathway exists in mammals. Curiously, however, although G9a is required for DNA methylation of ERVs in mESCs independent of its catalytic activity (27), it is dispensable for the maintenance of silencing of these elements. In contrast, catalytically active G9a is required for the establishment of silencing and DNA methylation of newly integrated XRVs. To reconcile these observations, we propose that G9a plays two separable yet critical roles: the establishment of transcriptional silencing, which initially requires its catalytic activity, and de novo DNA methylation, which does not. According to this model, the catalytic activity of G9a is required for DNA methylation of XRVs but not of ERVs, because only the former are initially transcriptionally active.
MLV-based retroviral vectors similar to those used in this study have been used extensively for gene transfer studies and to deliver genes for therapeutic purposes (43). Despite the fact that MLV preferentially integrates within or near the promoter regions of genes (44), DNA methylation and transcriptional silencing of such vectors are frequently observed (1), explaining in part why they have shown limited efficacy. The results presented here indicate that G9a plays an important role in the silencing of such vectors in mESCs and raises the question of whether G9a plays a similar role in other cell types.
We previously showed that G9a is required for recruitment of Dnmt3a to ERVs (27). Although G9a has been reported to interact directly with each DNMT (45, 46), our results are most consistent with a role for G9a in regulating Dnmt3a-mediated de novo methylation in mESCs. Indeed, recent experiments revealed that in mESCs, the establishment of silencing of an MLV vector closely related to MSCV requires Dnmt3l (47), a key regulator of Dnmt3a2-mediated de novo DNA methylation in vivo (48). Thus, G9a may act in concert with Dnmt3l to promote de novo methylation by Dnmt3a2. Notably, Dnmt3a2 and Dnmt3l generally are not expressed in somatic cells, perhaps explaining why depletion of G9a in MEFs has no effect on DNA methylation of ERVs.
Recently, ZFP809 (15) and the corepressor KAP1 were identified as components of the stem cell-specific repressor complex that binds to the MLV PBS (13, 14). The efficiency of MLV vector silencing is reduced in KAP1-deficient EC cells, indicating that this repressor plays an important role in proviral silencing. However, in the MSCV vector, the WT PBS was replaced with a sequence complementary to tRNAGln (Fig. 1A), which ablates stem cell-specific repressor binding (13, 14). Given that the MSCV vector is nevertheless silenced over time in WT cells, a G9a-dependent silencing pathway also must operate in mESCs independent of the PBS silencer element.
Although G9a is clearly required for the establishment of DNA methylation and silencing of XRVs, this KMTase is dispensable for the maintenance of proviral silencing in mESCs. Similarly, in MEFs, where DNA methylation of IAP elements is dependent on Dnmt1 and Dnmt3b, but not on Dnmt3a (40, 49), G9a is not required to maintain these elements in a densely methylated state. Furthermore, in contrast to the silencing defect observed for newly integrated MLV-based vectors, G9a−/− mESCs show no increase in expression of endogenous MLV (27). To explain why integrated XRVs demonstrate a silencing defect in G9a−/− cells, whereas ERVs and XRVs introduced before deletion of G9a remain in a repressed state after depletion of G9a, we propose that G9a functions primarily in the establishment of proviral silencing, by depositing the repressive dimethyl mark on H3K9 and promoting recruitment of Dnmt3a2, perhaps in a Dnmt3l-dependent manner, to the newly integrated proviruses.
Interestingly, the chromatin remodeler LSH interacts with G9a to establish the silencing of many genes in mESCs (50). Because LSH is also required for the silencing of retrotransposons, G9a may establish proviral silencing in a pathway that also involves chromatin remodeling. Once established, the silencing of XRVs is sustained by DNA methylation and/or H3K9me3, which are maintained in a G9a-independent manner by Dnmt1 and Eset, respectively. Taken together, our data reveal a previously unappreciated interplay between H3K9 KMTases and de novo DNA methylation in the establishment and maintenance of proviral silencing.
Materials and Methods
Viral Infections and Flow Cytometry.
MFG-GFP and MSCV-GFP retroviral constructs were introduced into target cells via the Phoenix A system and analyzed as described previously (28). Proviral copy number was determined by qPCR using genomic DNA isolated from infected cells and primers specific for the GFP gene. All primer sequences are listed in Table S1.
Establishment of G9a CKO MEFs and mESCs.
Derivation of G9a CKO mESCs was done as described previously (39). The protocol for establishment of G9a CKO MEFs is described in SI Materials and Methods.
DNA methylation analyses.
Genomic DNA were isolated and subjected to sodium bisulfite conversion using the EZ DNA Methylation-Gold Kit (Zymo Research) according to the manufacturer's instructions, followed by seminested PCR with primers specific for the 5′ LTRs of the MFG and MSCV vectors. The detailed protocol is described in SI Materials and Methods. Southern blot analysis was conducted as described previously (27).
Western Blot Analysis, Native ChIP, RNA Extraction, and RT-PCR
Detailed protocols are presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the University of British Columbia Flow Cytometry and Nucleic Acid Protein Service facilities and Drs. En Li and Thomas Jenuwein for the mESC lines. This work was supported by Canadian Institutes of Health Research Grants 77805 (to M.C.L.) and 92090 (to M.C.L. and D.L.M.), and by a grant-in-aid from the Ministry of Education, Science, Technology and Culture of Japan (to M.T. and Y.S.). M.C.L. is a Michael Smith Foundation for Health Research Scholar and a Canadian Institutes of Health Research New Investigator.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014660108/-/DCSupplemental.
References
- 1.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 2.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 3.Dranoff G, et al. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci USA. 1993;90:3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bestor TH. Gene silencing as a threat to the success of gene therapy. J Clin Invest. 2000;105:409–411. doi: 10.1172/JCI9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flanagan JR, Krieg AM, Max EE, Khan AS. Negative control region at the 5′ end of murine leukemia virus long terminal repeats. Mol Cell Biol. 1989;9:739–746. doi: 10.1128/mcb.9.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukiyama T, Niwa O, Yokoro K. Mechanism of suppression of the long terminal repeat of Moloney leukemia virus in mouse embryonal carcinoma cells. Mol Cell Biol. 1989;9:4670–4676. doi: 10.1128/mcb.9.11.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pannell D, et al. Retrovirus vector silencing is de novo methylase- independent and marked by a repressive histone code. EMBO J. 2000;19:5884–5894. doi: 10.1093/emboj/19.21.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao S, et al. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol Ther. 2004;10:27–36. doi: 10.1016/j.ymthe.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Changolkar LN, Singh G, Pehrson JR. macroH2A1-dependent silencing of endogenous murine leukemia viruses. Mol Cell Biol. 2008;28:2059–2065. doi: 10.1128/MCB.01362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jähner D, et al. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- 11.Colicelli J, Goff SP. Isolation of a recombinant murine leukemia virus utilizing a new primer tRNA. J Virol. 1986;57:37–45. doi: 10.1128/jvi.57.1.37-45.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barklis E, Mulligan RC, Jaenisch R. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell. 1986;47:391–399. doi: 10.1016/0092-8674(86)90596-9. [DOI] [PubMed] [Google Scholar]
- 13.Petersen R, Kempler G, Barklis E. A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol Cell Biol. 1991;11:1214–1221. doi: 10.1128/mcb.11.3.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 15.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grez M, Akgün E, Hilberg F, Ostertag W. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc Natl Acad Sci USA. 1990;87:9202–9206. doi: 10.1073/pnas.87.23.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Challita PM, et al. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J Virol. 1995;69:748–755. doi: 10.1128/jvi.69.2.748-755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawley RG, Lieu FH, Fong AZ, Hawley TS. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 19.Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niwa O, Yokota Y, Ishida H, Sugahara T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983;32:1105–1113. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- 21.Stewart CL, Stuhlmann H, Jähner D, Jaenisch R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc Natl Acad Sci USA. 1982;79:4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorincz MC, Schübeler D, Groudine M. Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol Cell Biol. 2001;21:7913–7922. doi: 10.1128/MCB.21.23.7913-7922.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautsch JW, Wilson MC. Delayed de novo methylation in teratocarcinoma suggests additional tissue-specific mechanisms for controlling gene expression. Nature. 1983;301:32–37. doi: 10.1038/301032a0. [DOI] [PubMed] [Google Scholar]
- 24.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martens JH, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 27.Dong KB, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–2701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsui T, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 29.Tachibana M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swindle CS, Kim HG, Klug CA. Mutation of CpGs in the murine stem cell virus retroviral vector long terminal repeat represses silencing in embryonic stem cells. J Biol Chem. 2004;279:34–41. doi: 10.1074/jbc.M309128200. [DOI] [PubMed] [Google Scholar]
- 31.Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 33.Lei H, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 34.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 35.Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 36.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 37.Appanah R, Dickerson DR, Goyal P, Groudine M, Lorincz MC. An unmethylated 3′ promoter-proximal region is required for efficient transcription initiation. PLoS Genet. 2007;3:e27. doi: 10.1371/journal.pgen.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 39.Tachibana M, Nozaki M, Takeda N, Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 2007;26:3346–3359. doi: 10.1038/sj.emboj.7601767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dodge JE, et al. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem. 2005;280:17986–17991. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- 41.Zhu H, et al. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25:335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freitag M, Selker EU. Controlling DNA methylation: Many roads to one modification. Curr Opin Genet Dev. 2005;15:191–199. doi: 10.1016/j.gde.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 44.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 45.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 46.Estève PO, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ooi SK, et al. Dynamic instability of genomic methylation patterns in pluripotent stem cells. Epigenetics Chromatin. 2010;3:17. doi: 10.1186/1756-8935-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nimura K, et al. Dnmt3a2 targets endogenous Dnmt3L to ES cell chromatin and induces regional DNA methylation. Genes Cells. 2006;11:1225–1237. doi: 10.1111/j.1365-2443.2006.01012.x. [DOI] [PubMed] [Google Scholar]
- 49.Jackson-Grusby L, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 50.Myant K, et al. LSH and G9a/GLP complex are required for developmentally programmed DNA methylation. Genome Res. 2011;21:83–94. doi: 10.1101/gr.108498.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.