Abstract
Understanding the process of evolution is one of the great challenges in biology. Cave animals are one group with immense potential to address the mechanisms of evolutionary change. Amazingly, similar morphological alterations, such as enhancement of sensory systems and the loss of eyes and pigmentation, have evolved multiple times in a diverse assemblage of cave animals. Our goal is to develop an invertebrate model to study cave evolution so that, in combination with a previously established vertebrate cave system, we can address genetic questions concerning evolutionary parallelism and convergence. We chose the isopod crustacean, Asellus aquaticus, and generated a genome-wide linkage map for this species. Our map, composed of 117 markers, of which the majority are associated with genes known to be involved in pigmentation, eye, and appendage development, was used to identify loci of large effect responsible for several pigmentation traits and eye loss. Our study provides support for the prediction that significant morphological change can be mediated through one or a few genes. Surprisingly, we found that within population variability in eye size occurs through multiple mechanisms; eye loss has a different genetic basis than reduced eye size. Similarly, again within a population, the phenotype of albinism can be achieved by two different genetic pathways—either by a recessive genotype at one locus or doubly recessive genotypes at two other loci. Our work shows the potential of Asellus for studying the extremes of parallel and convergent evolution—spanning comparisons within populations to comparisons between vertebrate and arthropod systems.
Keywords: arthropods, regressive evolution, subterranean, mapping
Cave animals have long interested biologists because of their bizarre and other-worldly appearance. Common characteristics of cave animals include the absence or great reduction of eyes, the elimination or near disappearance of pigmentation, and the elongation of limbs (1). These characteristics can be found in cave animals as diverse as salamanders, fish, spiders, shrimp, beetles, and collembolans. Most studies looking at the genetic basis of parallel or convergent evolution examine either closely related species with the same phenotypes or independently evolved populations of the same species with similar phenotypes (reviewed in ref. 2). The cave system, however, is unique in that we can examine, and eventually compare, the genetic and molecular basis for the loss of eyes and pigmentation in vastly different (i.e., both invertebrate and vertebrate) taxa.
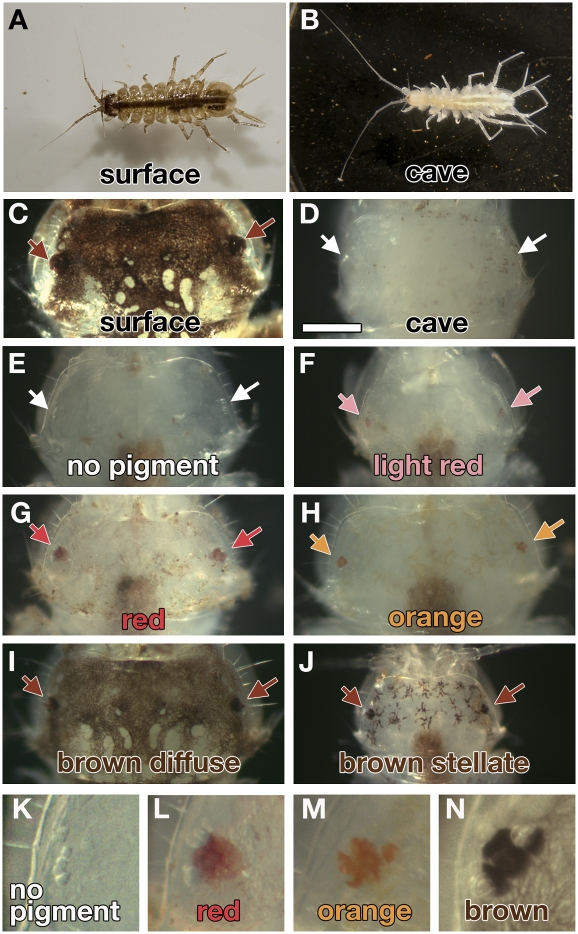
A vertebrate cave fish, Astyanax mexicanus, has been established as an emerging model species for genetic and developmental analyses (reviewed in refs. 3 and 4). Mapping, using this species, has identified regions of the genome that are responsible for cave-associated morphological changes (5, 6). We decided to develop an invertebrate cave model that will provide an independent example of cave adaptation. Of the invertebrates, crustaceans have been particularly successful in adapting to the cave environment with >2,900 known obligate cave-dwelling species (7) and are thought to be the most common aquatic subterranean organisms (8). We decided to examine Asellus aquaticus, a freshwater isopod crustacean with both surface and cave forms (Fig. 1 A and B).
Fig. 1.
Pigmentation differences in cave, surface, and backcross individuals of A. aquaticus. (A) Surface male from Planina Polje surface waters. (B) Cave male from Planina cave (Pivka channel). Heads of surface (C), cave (D), and backcross (E–J) animals. Arrows show the eye spots. (E and K) No eye or head pigmentation. Brown dots are debris on the exterior of the animal. (F) Faint red eye and no head pigmentation. (G and L) Red eye and head pigmentation. (H and M) Orange eye and head pigmentation. (I and N) Brown eye pigment and diffuse brown pigmentation. (J) Brown eye pigment and stellate brown pigmentation. (Scale bar: 0.25 mm; applies to C–J). The gut in individuals (E–H and J) is brown from eating decaying leaves.
Surface populations of A. aquaticus are found throughout most of Europe in many freshwater habitats ranging from small brooks and ephemeral ponds to rivers and lakes. Cave populations of this species can be found in Slovenia, Italy, and Romania (9). Advantages of this species include the presence of multiple cave populations allowing for studies of parallel evolution (10–12), the ability to produce hybrid offspring (13), and the many morphological differences between the cave and surface forms (14, 15). The cave forms often show an increase in body length, an increase in the length of certain appendages, an increase in the length of aesthetascs (thought to be chemoreceptors), a decrease in the degree of body and eye pigmentation, a decrease in the size of the eye, and a changed setal pattern (14, 15). The heritability of these traits was unknown. Offspring from individuals collected from mixed caves were analyzed for pigmentation and eye phenotypes in 1940 (16), but it is unclear whether those populations exist today and whether they are related to the populations we are studying. Our goal is to investigate the genetic basis of morphological evolution in A. aquaticus by combining a genetic mapping approach with a candidate gene analysis.
Results
Crosses Generated Between Cave and Surface Forms.
The hybrid cross we used to generate our backcross families was composed of 17 offspring, all female. In A. aquaticus, the method of sex determination is unclear. There is some evidence for maternal sex determination and broods of a single sex or of a skewed sex ratio have been observed (17). We believe that the female sex bias of the hybrid cross was brood-specific and not intrinsic to hybrid animals because subsequent hybrid broods contained both males and females. We mated a subset of these hybrid females to cave males to generate 194 backcross offspring (Table S1).
Markers Coalesced into a Linkage Map with Eight Linkage Groups.
To provide a genetic framework for studying A. aquaticus, we needed to identify markers in this species and generate a linkage map. Our map was generated from single-nucleotide polymorphisms (SNPs) in 88 genes and 29 SNPs from sequenced restriction site-associated DNA (RAD) tags (Table S2). For the vast majority of the markers, two genotypes were present within backcross progeny: homozygous for the cave allele and heterozygous for the cave and surface alleles. Seventeen markers were either uninformative or had three genotypes, reminiscent of the genotypes in a F2 cross, for a subset of the backcross families. The noninformative genotypes and the genotypes that showed a F2 inheritance pattern were excluded from the analysis.
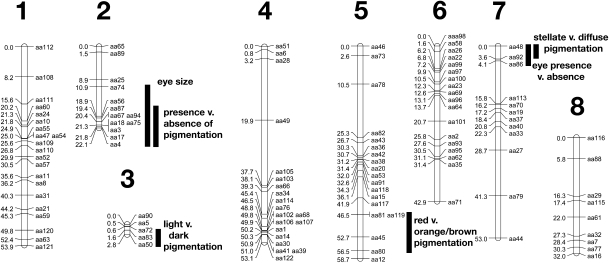
Using the program JoinMap 4 (18), all markers grouped into eight linkage groups (Fig. 2). Five markers, aa9, aa13, aa84, aa85, and aa104, were excluded from the analysis because they made it difficult to resolve the order of markers on some of the linkage groups. The number of linkage groups is equal to the number of chromosomes in this species (19). In addition, multiple clusters of markers were observed. The tightest cluster was an area on LG2 encompassing nine markers over a distance of 3.2 cM.
Fig. 2.
Linkage map of A. aquaticus. Linkage group (LG) number is listed above each linkage group diagram. Placement in centimorgans is to the left and marker name to the right. The 1.5 LOD support intervals for each trait are shown with black vertical bars.
Four Different Pigmentation Traits Map to Large-Effect Loci.
In this work, we focus exclusively on eye and pigmentation phenotypes that have evolved in the cave form of A. aquaticus. With respect to pigmentation, surface animals have dark brown pigmented chromatophores in the epithelium (Fig. 1C). The Planina cave form harbors no pigmentation, either in the body or the eyes (Fig. 1D). Our backcross animals exhibited five discrete color phenotypes. Of 194 animals, 53% were unpigmented (Fig. 1 E and K), 3% had light red eye pigmentation with no other pigmentation (Fig. 1F), 15% had red pigmentation in the eye and head (Fig. 1 G and L), 7% had orange eye and head pigmentation (Fig. 1 H and M), and 22% had brown eye and body pigmentation (Fig. 1 I, J, and N). The color phenotypes are not specific to either sex as both females and males of each color are present.
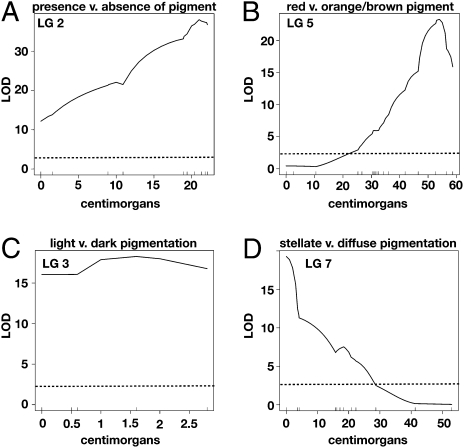
Using both binary and nonparametric interval mapping methods in R/qtl (20), we examined various pigmentation traits. First, we used the qualitative description of pigmented versus unpigmented individuals. We observed a locus on LG2 with a highly significant, by permutation test, LOD score (Fig. 3A and Fig. S1A). The area on the linkage group with the highest LOD score encompasses nine closely linked markers that include a number of pigmentation candidate genes: pale (tyrosine hydroxylase), rosy (xanthine dehydrogenase), white, and scarlet.
Fig. 3.
Loci responsible for pigmentation traits. LG is listed at the top of all graphs. (A) LOD score or measure of significance for the trait of presence vs. absence of pigmentation. (B) LOD plot for the trait of red vs. orange/brown pigment. (C) LOD plot for the trait of light vs. dark. Note that the LOD score is high across the entire linkage group because of the small size (2.5 cM) of the linkage group. (D) LOD plot for the trait of stellate vs. diffuse pigment pattern. The graphs were generated by using the binary method in R/qtl. The dotted lines in all graphs are the genome-wide significance levels (α < 0.05) by permutation test, LOD = 2.41. The vertical lines above the x axis are the placement of all markers across the linkage group.
For the next trait, we documented the color of the pigmented animals. Animals fell into two groups—those with red pigmentation (either red eye and head pigmentation or light red eyes and no head pigmentation) and those with orange and brown pigmentation. A locus responsible for a large amount of the variation in this trait mapped to LG5 with a peak LOD at the marker aa45 (cytochrome p450) (Fig. 3B and Fig. S1B).
The next phenotype analyzed was a light versus dark phenotype. There were two light phenotypes, faint red eyes with no head pigmentation, and orange eyes with orange head pigmentation. The two corresponding dark phenotypes were red eyes and red head pigmentation and brown eyes and brown head pigmentation. Examining the light versus dark phenotypes as a qualitative trait, we observed a large effect locus on LG3 at the marker aa83 (eyes absent) (Fig. 3C and Fig. S1C).
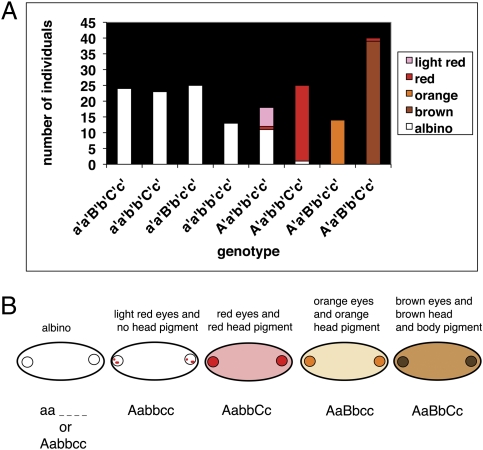
To summarize the results for pigmentation, we discovered three loci of large effect determining color in Asellus (Table S3). The association between genotype at the peak marker and phenotype for each of these traits was very close, 93%, 97%, and 98%, respectively, for presence vs. absence of pigmentation, red vs. orange/brown pigmentation, and light vs. dark pigmentation. We decided to look at the genotype distribution for these three loci, assuming that single genes cause the traits we observed. Because we have mapped these traits to a region rather than a single gene, we used the genotypes for this analysis from the marker that had the highest LOD score for each trait: aa75 rosy for presence vs. absence of pigmentation, aa45 cytochrome p450 for red vs. brown and orange pigmentation, and aa83 eyes absent for the light vs. dark trait. Comparing these genotypes and the pigmentation phenotypes (Fig. 4A), we have developed the following model (Fig. 4B). First, we have named the three genes involved in coloration A (closest linked marker, rosy), B (closest linked marker, cytochrome p450), and C (closest linked marker eyes absent). We found that gene A causes albinism when homozygous recessive and the aa genotype of gene A is epistatic to the other two genes, B and C—gene B when homozygous recessive causes red eyes and red head pigment and gene C when homozygous recessive causes orange pigmentation in Bb animals. Heterozygotes in all three genes had brown pigmentation. One surprising result was that 11 of the 12 animals (two-tailed P value = 1.24e−9) that were albino and not homozygous recessive in gene A were homozygous recessive in genes B and C. This same genotype (Aabbcc) was also present in seven additional animals, six that had faint red eyes and no red head pigment. Therefore, it appears that the genotype of Aabbcc causes two major phenotypes—albinism and faint red eyes with no head pigment. We think that unmapped loci distinguish between the two phenotypes (Discussion). To further confirm our model, we used the scantwo test in R/qtl to examine whether there were interactions between the regions of the genome in which genes A, B, and C were found (respectively, linkage groups 2, 5, and 3). We found that chromosomes 2 and 5 interact (P = 0.029) and chromosomes 2 and 3 also interact (P = 0.07) under a model that allows for the possibility of epistasis.
Fig. 4.
Pigmentation genotypes and three gene model of color. (A) Genotypes at three different markers—aa75, aa45, and aa83—and corresponding phenotypes of backcross individuals. A′ a′ are alleles for marker aa75, B′ b′ for marker aa45, and C′ c′ for marker aa83. Most genotypic classes have one predominant phenotype. (B) Model to explain the genotypic basis of the different pigmentation phenotypes. Circles represent eye spots. Five phenotypic classes are observed: albino, faint red eyes and no head pigment, red eyes and red head pigmentation, orange eyes and orange head pigmentation, and brown eyes and brown head and body pigmentation. The predicted genotype of each phenotypic class is shown at the bottom of the schematics. A and a are the surface and cave alleles, respectively, of the unknown gene responsible for presence vs. absence of pigmentation. B and b are the alleles of the unknown gene responsible for red vs. orange/brown pigmentation. C and c are the alleles of the unknown gene responsible for light vs. dark pigmentation.
The final pigmentation trait we examined regarded pattern rather than color. Individuals either had a diffuse pattern (Fig. 1I) of pigment cells or a stellate pattern of pigment cells (Fig. 1J). Pigment development in this species is a two-stage process involving the development of arm-like extensions of melanophores and then an increase in the number of melanophores (16). It is likely that the animals that have the stellate pattern are deficient in the second step of pigment development. A locus of large effect was observed on LG7 with a peak LOD score at aa48 (Fig. 3D and Fig. S1D).
Complete Eye Loss Phenotype Maps to a Major Locus.
Ommatidia are individual units of the compound eye in arthropods. The surface form of A. aquaticus has four ommatidia per eye (Fig. 5A), and the protruding part of each ommatidium is composed of up to four crystalline cone cells (21). The Planina cave contains animals with diverse eye phenotypes. Some cave individuals have no external eye structures, which we will call “eyeless,” and others have small, unfused crystalline cones of various numbers and shapes, which we will call “fragmented ommatidia.” Deficiencies of the eye structure in cave animals have been documented in the crystalline cones, photoreceptors, and the optic nerve (16). Three of our backcross families (encompassing a total of 143 individuals) exhibited some individuals with no external eye structures (Fig. 5 B and E). The other three families contained no individuals with complete eye loss although individuals with very reduced eyes were observed (Fig. 5 C and F). The distribution of eye size for families without complete eye loss showed a large amount of variation (Fig. 5F). The families with complete eye loss had a large number of individuals with no eyes and a large number of individuals with medium to large eyes (Fig. 5E).
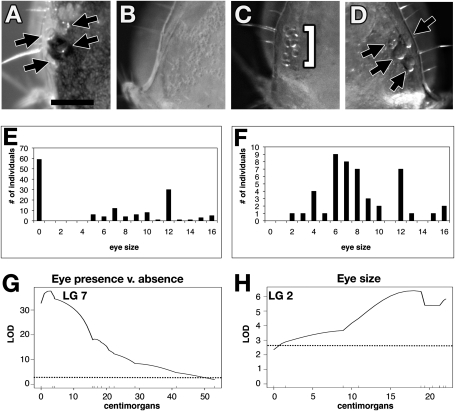
Fig. 5.
Eye size phenotypes and distribution in the backcross families. (A) A pigmented surface individual with four ommatidia. (B) A backcross individual with no external ommatidia. (C) A backcross individual with eight ommatidial fragments. (D) A backcross individual with four ommatidia. (E) Distribution of eye size for backcross families that have the eyeless phenotype. (F) Distribution of eye size for backcross families that do not have the eyeless phenotype. (G) Locus responsible for the qualitative phenotype of eye loss versus any sized eye using the binary method in R/qtl. Dotted line shows the genome-wide significance levels (α < 0.05) by permutation test, LOD = 2.41. (H) QTL for eye size in the families that have complete eye loss using Haley–Knott regression in R/qtl. Dotted line shows the genome-wide significance levels (α < 0.05) by permutation test, LOD = 2.42. Linkage group is listed at the top of the graph (G and H). (Scale bar: 0.125 mm; applies to A–D).
For eye traits, we treated the families without complete eye loss and those with eye loss separately. For families without eye loss (encompassing 51 individuals), we did not see any significant quantitative trait loci (QTL) for the eye size phenotype. However, for the families with eye loss, we saw a highly significant locus associated with the qualitative phenotype of eye loss on LG7 with the peak LOD score at aa92, lim1 (Fig. 5G and Fig. S1E). In 99% of backcross animals, the genotype at lim1 was correlated with the phenotype of eye loss. We also saw a QTL responsible for a percentage of the variance in eye size (looking only at animals with ommatidia or ommatidial fragments) with a peak LOD score of 6.3 on LG2 (Fig. 5H and Fig. S1F).
Evaluation of Bias.
Two potential biases in our analysis are the Beavis and the Noor effects. In the Beavis effect, an overestimation of effect sizes is present in analyses with small sample sizes (22). QTL of large effect are less likely to be overestimated than loci of small and medium effect (23). We calculated the odds ratio for the five binary traits (Table S3 and Table S4). Furthermore, the agreement between peak marker genotype and phenotype for all five traits was between 93% and 99%. A Beavis effect might be present for the eye size QTL but is unlikely to be present for these loci of large effect involved in the pigmentation and complete eye loss traits. Because of the small size of our analysis, we are more likely to discover QTL of large effect than QTL of small effect. In regions of low recombination, the combined effects of multiple QTL can behave like a locus of large effect, the Noor effect (24). To address this possibility, we examined the marker density around all of the loci of large effect and found that it was not significantly different from the average marker density in the map (P = 0.23).
Discussion
Genome-Wide Linkage Map in A. aquaticus.
There were several surprising aspects of our linkage map. First, with only 117 markers, we were able to generate a map with eight linkage groups, the same number as chromosomes. Second, the size of our map is 318.5 cM, which is unusually small. Third, a large number of markers are tightly linked. The seemingly complete and relatively small size of our linkage map could be explained by an extremely low recombination rate in A. aquaticus or, perhaps, there could be a low recombination rate in females (the heterozygous sex in our crosses). Another unique feature of the map was the dense clusters of markers. It is possible that the clusters are due to hotspots in recombination. The opposite extreme is also possible, that there are regions that recombine very infrequently because of genomic incompatibility between the Planina cave and surface genomes. Therefore, the markers we observe in clusters on our map may be deceptively far apart in distance on the physical map.
Genes of Large Effect Are Responsible for Eye and Pigmentation Phenotypes.
There are many examples of single genes or genes of large effect being responsible for significant morphological changes; a large number of these examples affect pigmentation (reviewed in ref. 2). In this study, we observed an extreme situation in which genes of large effect were responsible for four different pigmentation traits and the trait of eye loss.
Three loci of large effect associated with color were found (Fig. 2). One for presence vs. absence of pigmentation (LG 2, near aa75), a second for red vs. brown and orange pigmentation (LG 5, near aa45), and a third for dark vs. light pigmentation (LG 3, near aa83). Interestingly, albinism can occur through homozygosity at the first locus, or through double homozygosity at the second and third loci (Fig. 4B). A fourth locus was found that is associated with the stellate vs. diffuse pattern of pigmentation (LG 7, near aa48). A locus was also found that is associated with the presence vs. absence of eyes (LG 7, near aa92). To our knowledge, this is the first example of a locus of large effect responsible for eye loss in a naturally occurring population.
The five traits above were qualitative traits. The one quantitative trait we examined was eye size. For the families that did not show complete eye loss, we did not observe any QTL associated with eye size, which is probably due to the small number of individuals assayed. For the families that did show some individuals with complete eye loss, we saw a QTL responsible for eye size that mapped to LG 2 (near aa56). Systems with eye reduction that can be examined genetically are rare, making this an interesting trait for further study.
Candidate Genes and Linkage to Eye and Pigmentation Phenotypes.
We included many candidate genes in our analysis to improve our ability to identify the genes and mutations responsible for morphological change. We have seen close association between certain loci and candidate genes. The area of the genome with the peak LOD score for the presence vs. absence of pigmentation phenotype contained three markers with identical genotypes to each other, rosy, white, and disconnected. Between these three markers and Gene A there is only one potential recombinant. However, considering our three-gene model of pigmentation, the recombinant individual could have resulted from a recombination event between Gene C and the nearby marker, eyes absent. Rosy encodes xanthine dehydrogenase, which is involved in drosopterin synthesis (a component of Drosophila melanogaster eye pigment). Drosophila mutants in rosy have brownish eyes (defective red pigment) (25). Furthermore, there is evidence that pteridine synthesis is linked to both melanin and ommochrome synthesis in Bombyx mori (26). In Drosophila, the white gene causes a defect in ommochrome and pteridine synthesis and null mutants have white eyes (reviewed in ref. 27). Both ommochromes and melanins have been proposed to be present in the Asellus integument (13, 28). Further investigation of rosy and white should assist in determining whether either rosy or white, or a different closely linked gene, is responsible for the unpigmented phenotype.
Another candidate gene, lim1, is very closely linked to the complete eye loss phenotype. The presence of one recombinant, however, makes it likely that a gene located near lim1, but not lim1 itself, causes the eye loss phenotype.
Two Regions of the Genome Contain Closely Linked Loci Responsible for both Eye and Pigment Variation.
With the six traits we describe, there are two areas of the genome that have loci responsible for both eye and pigmentation traits. First, the locus responsible for presence versus absence of albinism and a QTL responsible for eye size are on LG2. The phenotypes of albinism and smaller eyes (and pigmented and larger eyes) are significantly correlated with a point biserial correlation coefficient of 0.58 (significant at α = 0.0001). Second, LG7 contains the locus responsible for complete loss of eyes and the locus responsible for stellate versus diffuse pigmentation. The eyeless and stellate phenotypes (and the eyed and diffuse phenotypes) are also significantly correlated (α = 0.0001) with a ϕ correlation coefficient of 0.80. It is remarkable that a close association between loci responsible for eye and pigment reduction occurs twice on independent linkage groups. All of the 10 Asellus populations known to have invaded caves independently show reduced eyes and pigmentation, whereas other cave related traits do not occur as regularly (11, 14, 15). It is possible that inheritance of the phenotypes of eye loss and pigment loss in tandem by close genetic linkage might allow for the rapid and widespread evolution of these phenotypes. Interestingly, genetic linkage of eye and pigmentation traits is not unique to Asellus; in A. mexicanus, the albino locus and a QTL responsible for eye size map to the same region (6). Therefore, the genetic architecture of eye and pigment loss might be commonly intertwined in cave animals and influence the timescale and mechanism of evolution of these characteristics.
At Least Three Genes Are Responsible for the Color Phenotypes.
Regarding pigmentation, the one genotype that appears to have multiple phenotypes from our model is heterozygous at gene A and homozygous recessive at genes B and C. In our cross, we observed that the majority of individuals with this genotype (17/18) were either completely albino or had very faint red eyes and no head or body pigmentation. In general, there was much variability in the intensity of the animals with red eyes and red head pigmentation: Some had large red eyespots and others had a small amount of red pigment in the eye region. Therefore, we believe additional gene(s) modify levels of red pigment.
One surprising result was that the ratios of animals with the different genotypes for genes A, B, and C were not as predicted by the model that assumes three single genes are responsible for color. The expected phenotypic ratios were 1/2 albino, 1/8 either albino or faint red eyes with no head pigment, 1/8 with brown pigmentation, 1/8 with red pigmentation, and 1/8 with orange pigmentation. Instead, we found fewer orange individuals (AaBbcc) and a decreased number of albino individuals of the genotype aabbcc. Certain combinations of alleles, in different regions of the genome, may be incompatible, causing this bias both in phenotype and genotype.
Comparisons with the Cavefish A. mexicanus.
One of the goals of our work was to compare the evolution of cave characteristics in vertebrate and invertebrate systems. In Asellus, we saw an unusual situation where multiple genetic mechanisms were responsible for pigment loss within a single population. Remarkably, this scenario is mirrored in the cavefish, A. mexicanus, where some populations have both a mutation in Oca2 causing albinism and a mutation in Mc1r causing reduced pigmentation (29, 30).
What is different in Asellus is that the identical phenotype of albinism can be achieved in two different genetic ways within a single cave population involving three different genes. In A. mexicanus, the same single gene, Oca2, seems to be involved in the evolution of albinism in three independently evolved cave populations (30).
In the cave population of Asellus we examined, we also saw that eye loss and eye reduction have different genetic bases. Complementation experiments between different populations of A. mexicanus result in offspring with larger eyes than either parent, indicating that eye reduction in these caves have different genetic mechanisms (31–33), again similar to the within population variation in Asellus. Furthermore, eye reduction in A. mexicanus is a polygenic trait with no single locus responsible for the majority of the variation (4, 6). We similarly predict eye reduction to be polygenic in Asellus. However, the “eyeless” phenotype in Asellus is likely governed by a single gene.
In this work, we investigated A. aquaticus, a species with populations containing individuals of very different morphologies. We have adapted this species for laboratory analyses, set up crosses, generated several hundred markers including 100 gene-specific markers, constructed a linkage map, and mapped multiple loci associated with numerous eye and pigmentation traits. We have demonstrated that eye and pigmentation traits have evolved in A. aquaticus via loci of large effect. We have also shown that within a cave population, both eye and pigment loss characteristics arise through diverse underlying mechanisms. A. aquaticus provides many of the desired characteristics of an emerging model organism including ease of rearing in the laboratory, a wealth of morphological diversity, and the ability to study both gain and loss of function characteristics. In addition, A. aquaticus belongs to a group of arthropods, crustaceans, which remain largely underrepresented and underexplored in genetic and genomic analyses. Finally, there is immense potential in studying parallel evolution in A. aquaticus, ranging from studies of population diversity to comparisons with the cavefish A. mexicanus and other vertebrate cave systems.
Materials and Methods
Crosses.
We collected “surface” animals (Fig. 1A) from Planina Polje, the surface river outside the Planina cave, and “cave” animals (Fig. 1B) from the Pivka channel of the Planina cave in Slovenia. According to a molecular clock approach, the divergence time is probably <100,000 y (11). The animals were kept in the laboratory at 10–13 °C, raised in artificial Daphnia media (34), and fed locally collected decaying leaves.
Crosses were set up between cave males and surface females because the surface females mated with a much higher frequency than cave females. One of our crosses resulted in 17 adult F1 animals, all females. Five of these F1 females were mated to 4 different males achieving a total of 194 offspring (Table S1). Some of these F1 females produced multiple broods. All backcross animals were raised in 13 °C incubators in darkness (except when removed for water changes) until the smallest individuals in the brood achieved a body length of 4 mm. For harvesting, animals were anesthetized in clove oil (10 μL in 50 mL of artificial Daphnia media) and the head, thorax, and abdomen were separated by using forceps. The head, abdomen, and all appendages were placed in 100% ethanol for further phenotyping, and the thorax minus appendages was used for DNA extraction by using the QIAamp DNA micro kit (Qiagen).
Genetic Markers.
Total RNA was extracted from surface adults by using TRIzol Reagent (Gibco BRL). cDNA was made by using SuperScript (Gibco BRL). Degenerate primers to candidate genes were designed (SI Materials and Methods and Dataset S1). A Genome Walker library (Clontech) was made by using surface DNA and EcoR1. Primer3 (http://frodo.wi.mit.edu) was used to design primers from the cloned degenerate fragment. The fragments subsequently amplified contained a portion of coding sequence as well as intronic or flanking genomic sequence. Each fragment was sequenced from surface, hybrid, and cave individuals. A genetic difference between cave and surface forms was confirmed only when a SNP was present in homozygous form in the cave and surface forms and the F1 hybrid individuals harbored the heterozygous genotype (Dataset S2). To confirm the identity of our markers, we performed a BLAST search of all of our fragments (Table S2). A subset of our SNPs were identified from sequence fragments generated by the company, Floragenex (Dataset S2). These include several of the gene associated SNPs (those missing from Dataset S2) and 29 additional SNPs. Several hundred SNPs were identified by Floragenex.
Genotyping.
The Genotypic analysis was performed by using iPlex Extend and MALDI TOF mass spectrometry (Sequenom). One hundred nine SNPs in different genes were genotyped as well as 29 additional SNPs generated by the company Floragenex. One hundred ninety-four backcross animals, the surface grandmother, cave grandfather, and a hybrid mother were genotyped for all markers. Some markers were excluded from our mapping analysis because either the Sequenom assay failed or the SNP was uninformative. One hundred nine SNPs from the Sequenom data were ultimately used for the analysis. We genotyped eight additional markers either by allele specific PCR (aa87, 88, 89, 90, 92, and 93) or size polymorphism (aa86, aa91) (Table S5). We amplified these markers from all 194 backcross animals and visualized the products by agarose gel electrophoresis.
Phenotypic Analysis.
Photographs were taken of all backcross animals by using a SPOT flex camera and a Zeiss stemi DRC dissecting microscope. We recorded eye color and head color, type of pigment pattern (stellate or diffuse), and size of the eye. Eye size was measured by counting the number of ommatidial fragments. Absence of ommatidial fragments (i.e., eyeless) was given the value of 0. Every ommatidial fragment had a value of 1. A complete ommatidium was given a value of 4 (thus an intact eye has a score of 16).
Mapping.
We used JoinMap 4 software (18) with Kosambi's mapping function, a LOD threshold of 1.0, a recombination threshold of 0.450, a jump threshold of 5.0, and a ripple after each marker was added to create a linkage map (with LOD 6.0 grouping) by using a locus file of 122 SNPs and 194 backcross progeny. We used the binary method and nonparametric interval mapping in R/qtl to analyze the qualitative traits. For the quantitative trait of eye size, we used both Haley–Knott regression and nonparametric interval mapping (20). Genome wide significance levels (α < 0.05) were determined by permutation analysis for each method with 1,000 permutations in R/qtl (20). The 1.5 LOD support intervals were computed in R/qtl (20). Scan-two tests were performed in R/qtl (20), and significance was determined by using 1,000 permutations.
Bias.
For more information, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank reviewers for their helpful comments; G. Bračko and C. Fišer for help collecting animals; L. Bauman, B. Borowsky, R. Borowsky, K. Broman, C. Chaw, D. Culver, D. Fong, J. Gross, D. Hendrix, W. Lin, P. Liu, C. Miller, S. Prevorčnik, H. Protas, D. Richter, D. Schluter, J. Štrus, and J. Yang for help and advice; H. Marques-Souza, R. Parchem, J. Serano, and M. Vargas-Vila for supplying degenerate primers; J. Dang from the Genome Analysis Core Facility, Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco for Sequenom genotyping; and J. Boone from Floragenex for RAD-tag sequencing. M.E.P. was supported by a postdoctoral fellowship from the National Institute of Health, and P.T. was supported by grants from the Slovenian Research Agency.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013850108/-/DCSupplemental.
References
- 1.Culver DC. Cave Life. Cambridge, MA: Harvard Univ Press; 1982. [Google Scholar]
- 2.Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra HE. Convergence in pigmentation at multiple levels: Mutations, genes and function. Philos Trans R Soc Lond B Biol Sci. 2010;365:2439–2450. doi: 10.1098/rstb.2010.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeffery WR. Chapter 8. Evolution and development in the cavefish Astyanax. Curr Top Dev Biol. 2009;86:191–221. doi: 10.1016/S0070-2153(09)01008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkens H. Evolution and genetics of epigean and cave Astyanax (Characidae, Pisces) Evol Biol. 1988;23:271–367. [Google Scholar]
- 5.Borowsky R, Wilkens H. Mapping a cave fish genome: polygenic systems and regressive evolution. J Hered. 2002;93:19–21. doi: 10.1093/jhered/93.1.19. [DOI] [PubMed] [Google Scholar]
- 6.Protas M, et al. Multi-trait evolution in a cave fish, Astyanax mexicanus. Evol Dev. 2008;10:196–209. doi: 10.1111/j.1525-142X.2008.00227.x. [DOI] [PubMed] [Google Scholar]
- 7.Gibert J, Deharveng L. Subterranean ecosystems: A truncated functional biodiversity. Bioscience. 2002;52:473–481. [Google Scholar]
- 8.Sket B. The nature of biodiversity in hypogean waters and how it is endangered. Biodivers Conserv. 1999;8:1319–1338. [Google Scholar]
- 9.Verovnik R, Prevorčnik S, Jugovic J. Description of a neotype for Asellus aquaticus Linné, 1758 (Crustacea: Isopoda: Asellidae), with description of a new subterranean Asellus species from Europe. Zool Anz. 2009;248:101–118. [Google Scholar]
- 10.Verovnik R, Sket B, Prevorčnik S, Trontelj P. Random amplified polymorphic DNA diversity among surface and subterranean populations of Asellus aquaticus (Crustacea: Isopoda) Genetica. 2003;119:155–165. doi: 10.1023/a:1026085716777. [DOI] [PubMed] [Google Scholar]
- 11.Verovnik R, Sket B, Trontelj P. Phylogeography of subterranean and surface populations of water lice Asellus aquaticus (Crustacea: Isopoda) Mol Ecol. 2004;13:1519–1532. doi: 10.1111/j.1365-294X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- 12.Turk-Prevorčnik & Blejec A Asellus aquaticus infernus, new subspecies (Isopoda:Asellota:Asellidae), from Romanian hypogean waters. J Crustac Biol. 1998;18:763–773. [Google Scholar]
- 13.Baldwin E, Beatty R. The pigmentation of cavernicolus animals. J Exp Biol. 1941;18:136–143. [Google Scholar]
- 14.Turk S, Sket B, Sarbu S. Comparison between some epigean and hypogean populations of Asellus aquaticus. Hydrobiologia. 1996;337:161–170. [Google Scholar]
- 15.Prevorcnik S, Blejec A, Sket B. Racial differentiation in Asellus aquaticus (L.) (Crustacea: Isopoda: Asellidae. Arch Hydrobiol. 2004;160:193–214. [Google Scholar]
- 16.Kosswig C, Kosswig L. Die Variabilität bei Asellus aquaticus, unter besonderer Berücksichtigung der Variabilität in isolierten unter- und oberirdischen Populationen. Revue de Facultie des Sciences (Istanbul) 1940;B5:1–55. [Google Scholar]
- 17.Vitagliano E, Marchetti E, Vitagliano G. Skewed sex-ratio, monogeny, and maternal sex determination in two geographical populations of Asellus aquaticus (L., 1758) (Isopoda) Crustaceana. 1996;69:455–475. [Google Scholar]
- 18.VanOoijen JW. Wageningen, the Netherlands: Kyazma, B. V.; 2006. JoinMap 4, Software for the calculation of genetic linkage maps in experimental populations. [Google Scholar]
- 19.Salema H. The chromosomes of Asellus aquaticus (L.) - A technique for isopod karyology. Crustaceana. 1979;36:316–318. [Google Scholar]
- 20.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson HL. The fine structure of the compound eyes of shallow-water asellotes, Jaera albifrons Leach and Asellus aquaticus L. (Crustacean: Isopoda) Acta Zoologica. 1978;59:69–84. [Google Scholar]
- 22.Beavis W.D. Proceedings of the Corn and Sorghum Industry Research Conference. Washington, DC: Am Seed Trade Assoc; 1994. The power and deceit of QTL experiments: Lessons from comparative QTL studies; pp. 250–266. [Google Scholar]
- 23.Albert AY, et al. The genetics of adaptive shape shift in stickleback: Pleiotropy and effect size. Evolution. 2008;62:76–85. doi: 10.1111/j.1558-5646.2007.00259.x. [DOI] [PubMed] [Google Scholar]
- 24.Noor MA, Cunningham AL, Larkin JC. Consequences of recombination rate variation on quantitative trait locus mapping studies. Simulations based on the Drosophila melanogaster genome. Genetics. 2001;159:581–588. doi: 10.1093/genetics/159.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reaume AG, Knecht DA, Chovnick A. The rosy locus in Drosophila melanogaster: Xanthine dehydrogenase and eye pigments. Genetics. 1991;129:1099–1109. doi: 10.1093/genetics/129.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato T, Sawada H, Yamamoto T, Mase K, Nakagoshi M. Pigment pattern formation in the quail mutant of the silkworm, Bombyx mori: Parallel increase of pteridine biosynthesis and pigmentation of melanin and ommochromes. Pigment Cell Res. 2006;19:337–345. doi: 10.1111/j.1600-0749.2006.00316.x. [DOI] [PubMed] [Google Scholar]
- 27.Lloyd V, Ramaswami M, Krämer H. Not just pretty eyes: Drosophila eye-colour mutations and lysosomal delivery. Trends Cell Biol. 1998;8:257–259. doi: 10.1016/s0962-8924(98)01270-7. [DOI] [PubMed] [Google Scholar]
- 28.Needham A, Brunet PC. The integumental pigment of Asellus. Cell Mol Life Sci. 1957;13:207–209. [Google Scholar]
- 29.Gross JB, Borowsky R, Tabin CJ. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet. 2009;5:e1000326. doi: 10.1371/journal.pgen.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Protas ME, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- 31.Borowsky R. Restoring sight in blind cavefish. Curr Biol. 2008;18:R23–R24. doi: 10.1016/j.cub.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Wilkens H, Strecker U. Convergent evolution of the cavefish Astyanax (Characidae, Teleostei): Genetic evidence from reduced eye-size and pigmentation. Biol J Linn Soc Lond. 2003;80:545–554. [Google Scholar]
- 33.Jeffery WR. Adaptive evolution of eye degeneration in the Mexican blind cavefish. J Hered. 2005;96:185–196. doi: 10.1093/jhered/esi028. [DOI] [PubMed] [Google Scholar]
- 34.Lynch M, Weider LJ, Lampert W. Measurement of the carbon balance in Daphnia. Limnol Oceanogr. 1986;31:17–33. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.