Abstract
Dendritic cell (DC) homing to the lymphatics and positioning within the lymph node is important for adaptive immunity, and is regulated by gradients of CCL19 and CCL21, ligands for CCR7. Despite the importance of DC chemotaxis, it is not well understood how DCs interpret gradients of these chemokines in a complex 3D microenvironment. Here, we use a microfluidic device that allows rapid establishment of stable gradients in 3D matrices to show that DC chemotaxis in 3D can respond to CCR7 ligand gradients as small as 0.4%, which helps explain how DCs sense lymphatic vessels in an environment where broadcast distance for chemokine diffusion is hindered by convective flows into the vessel. Interestingly, DCs displayed similar sensitivities to both chemokines at small gradients (≤ 60 nM/mm), but migrated more efficiently towards higher gradients of CCL21, which unlike CCL19 binds strongly to matrix proteoglycans and signals without the need for internalization. Furthermore, cells preferentially migrated towards CCL21 when exposed to equal and opposite gradients of CCL21 and CCL19 simultaneously, even when matrix-binding of CCL21 was prevented. Although these ligands have similar binding affinity to CCR7, our results demonstrate that, in a 3D environment, CCL21 is a more potent directional cue for DC migration than CCL19. These findings provide new quantitative insight into DC chemotaxis in a physiological 3D environment and suggest how CCL19 and CCL21 may signal differently to fine-tune DC homing and positioning within the lymphatic system. These results also have broad relevance to other systems of cell chemotaxis, which remain poorly understood in the 3D context.
Keywords: cell migration, chemoinvasion, in vitro, microfluidics, leukocyte
Dendritic cells (DCs) are considered the most potent and professional antigen-presenting cells. DCs are positioned throughout the periphery, and when activated, migrate to lymphatic vessels and into lymph nodes, where they can direct antigen-specific T cell responses. Upon activation or maturation, DCs upregulate the C-C chemokine-receptor CCR7, which allows them to sense and home towards CCR7 ligand-secreting lymphatic vessels and lymph nodes (1). The two known ligands for CCR7 are the C-C chemokines CCL21 and CCL19 (2); both are secreted by stromal cells in the lymph node paracortex to properly position DCs with CCR7+ naïve T cells for their activation. CCL21, but not CCL19, is also expressed by the endothelium of lymphatic vessels in the periphery (1). Thus, CCR7-mediated chemotaxis is critical for DC homing to, and positioning within, lymph nodes and for T cell activation there (3).
CCL19 and CCL21 function as directional signals presumably by virtue of concentration gradients (∇C) that guide DCs towards areas of increasing concentrations. Interestingly, CCL19 and CCL21 have similar binding affinities for CCR7 (4–6) and similar chemotactic potential for DCs and T cells under 2D conditions (7, 8), but differ in their internalization (8, 9) as well as their binding affinity to extracellular matrix (ECM) proteoglycans. Specifically, the positively charged C terminus of CCL21 gives it strong binding affinity to a wide range of proteoglycans [as well as type IV collagen (10)], unlike CCL19 (11–13). Additionally, matrix-bound CCL21 can activate DC and T cell adhesion to adhesion ligands like ICAM-1 (14, 15). This study addresses the question of how DCs respond to gradients of CCR7 ligands in 3D environments—in particular, whether they differentiate between soluble vs. matrix-bound signals, and the relative importance of gradient steepness vs. average concentration.
DCs, like most migratory cells, exist and move in a 3D environment. However, our current understanding of DC chemotaxis is largely qualitative and limited to 2D (16–19). Furthermore, while it is well established that cell migration requires different mechanisms in 3D vs. 2D (20–25), nearly all chemotaxis studies to date have been performed in 2D or 2.5D conditions largely because of the lack of model systems in which well defined gradients can be created in 3D while evaluating cell invasion (especially because DC migration is relatively fast compared to the time scale of diffusion for gradient establishment). Here, we describe the chemotactic behavior of DCs in 3D to well defined gradients of CCL21 and CCL19, revealing their chemosensitivity to ligand gradient strength, insensitivity to ligand state (bound vs. soluble) and enhanced response to CCL21 vs. CCL19. To this end, we used a recently described microfluidic device in which stable and well defined gradients can be preestablished before cells and matrix are introduced, and live cell migration can be visualized (26).
Results and Discussion
Characterizing Stable, Well Defined Gradients in an Agarose-Based Microfluidic Device.
To observe DC chemotaxis in 3D environments in response to gradients of CCR7 ligands, we used a biological extracellular matrix of 1.5 mg/mL type I collagen and 10% Matrigel (MG), which had the appropriate stiffness and composition for optimal DC migration (25), while also containing heparan sulfate proteoglycans to allow natural CCL21 binding. This matrix differs from 3D matrices in vivo, which are more complex, composed of higher matrix densities, and heterogeneous in composition and architecture (27); however, physiological processes like tumor cell invasion and dendritic cell migration can be mimicked with close similarity to the in vivo situation in such matrices (21, 23, 25).
We first modified our agarose-based microfluidic gradient culture device (26) to rapidly (within 2 min) establish stable and well defined chemokine gradients within the DC-loaded 3D ECM, due to gradient-buffering capabilities of the agarose scaffold (Fig. 1). Specifically, the injected ECM contained the average concentration of the two side channels, eliminating sharp concentration gradients initially; this was particularly important for CCL21, where matrix-binding increases the time necessary for gradient development. The rapid establishment of the gradient, necessary for evaluating relatively fast-moving cells like DCs, distinguishes our gradient buffer chamber from other recently described devices that develop gradients more slowly and after cells are in place (28–30). This device proved to be robust, reproducible, and easy to use, allowing us to carry out up to 12 experiments in one day; this was critical for studies with primary DCs, whose isolation, differentiation, and maturation had to be precisely timed for each batch.
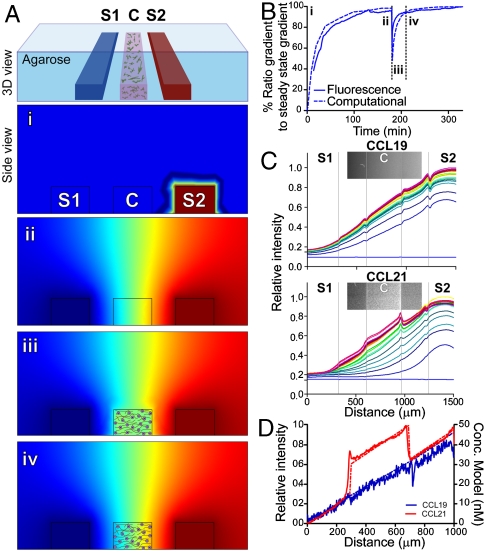
Fig. 1.
Establishment and characterization of matrix bound and soluble protein gradients in a microfluidic device. (A) Schematic view of one functional unit of the microfluidic device. The cell-matrix mixture was seeded in the center channel (C), while the chemokines/buffers flowed along the side channels (S1 and S2) to create chemical gradients across the center channel (i–iv). Computational results of CCL19 concentration gradients at various times, red and blue correspond to maximum and minimum concentrations, respectively. (i) At t = 0, chemokine solutions were pumped through the side channels (in this case, CCL19 on the right and plain medium on the left). (ii) After 180 min., a steady-state diffusion concentration gradient was reached. (iii) At this time, the cell-matrix mixture was added to the center channel, containing an average chemokine concentration of the two side channels. (iv) After several minutes, the gradient is reestablished within the center channel, before cell tracking began. (B) Temporal gradient establishment of CCL19 from experiment (solid line) and computation (dashed line). (C) Spatial gradient establishment at different time points of CCL19 (top) and CCL21 (bottom) to a collagen-MG matrix using fluorescently tagged chemokines; inserts show confocal images at 300 min. Darkest blue and red lines indicate 0 and 180 min., respectively, with 10-min. intervals shown between. (D) CCL19 (blue) and CCL21 (red) gradients at steady-state from experiment (solid lines) and computation (dashed lines).
Next, we estimated the kinetics of gradient establishment for CCL19 and CCL21 in this device using computational models (SI Text). Because CCL21 has been reported to bind various ECM proteoglycans with different affinities (11–13), we first determined the binding constant of CCL21 to our specific matrix to be 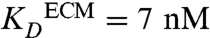 (± 8 nM) using a modified ELISA assay (SI Text, Fig. S1); this was consistent with previous measurements of 5.5 nM to heparin (12). The CCL21 was primarily binding to the proteoglycan-rich MG, because its affinity to collagen alone was below detection. No binding to the ECM could be detected for CCL19 (Fig. S1).
(± 8 nM) using a modified ELISA assay (SI Text, Fig. S1); this was consistent with previous measurements of 5.5 nM to heparin (12). The CCL21 was primarily binding to the proteoglycan-rich MG, because its affinity to collagen alone was below detection. No binding to the ECM could be detected for CCL19 (Fig. S1).
Using these binding constants, together with an assumed matrix-binding 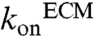 of 9.3 × 104 M-1 s-1 (31), we estimated the extracellular concentration profiles of CCL21 and CCL19 in space and time both computationally, using Comsol (Fig. 1B, dotted line) as well as experimentally, using FITC-labeled chemokines in the chamber (Fig. 1B, continuous line and Fig. 1C). A steady-state gradient was reached after ∼120 min (Fig. 1B,i), thus in all experiments we introduced the cell-ECM solution after 180 min (Fig. 1B, i). Introducing the cells destabilized the gradient (Fig. 1B, ii), but because of the gradient-buffering capacity of the agarose and addition of the average chemokine concentration to the gel initially, the gradient restabilized to within 90% of its steady-state value after only 2 min (Fig. 1B, iv). As indicated by the smooth transition from the source channel to the adjacent agarose, no significant chemokine binding to the agarose scaffold was detected (Fig. 1C, top), while CCL21 accumulated in the matrix as expected (Fig. 1C, bottom). We estimated the concentration of binding sites as 25 nM from these confocal images (Fig. 1C, insets).
of 9.3 × 104 M-1 s-1 (31), we estimated the extracellular concentration profiles of CCL21 and CCL19 in space and time both computationally, using Comsol (Fig. 1B, dotted line) as well as experimentally, using FITC-labeled chemokines in the chamber (Fig. 1B, continuous line and Fig. 1C). A steady-state gradient was reached after ∼120 min (Fig. 1B,i), thus in all experiments we introduced the cell-ECM solution after 180 min (Fig. 1B, i). Introducing the cells destabilized the gradient (Fig. 1B, ii), but because of the gradient-buffering capacity of the agarose and addition of the average chemokine concentration to the gel initially, the gradient restabilized to within 90% of its steady-state value after only 2 min (Fig. 1B, iv). As indicated by the smooth transition from the source channel to the adjacent agarose, no significant chemokine binding to the agarose scaffold was detected (Fig. 1C, top), while CCL21 accumulated in the matrix as expected (Fig. 1C, bottom). We estimated the concentration of binding sites as 25 nM from these confocal images (Fig. 1C, insets).
With this data, we could precisely predict and control the gradients of bound and soluble chemokines that formed in the center channel after chemokine solutions were imposed in the side channels.
Dendritic Cell Characterization.
Because primary cultures of murine DCs were used, which are inhomogeneous and variable, we characterized each batch for differentiation, maturity, and expression of CCR7 and CCL19 (SI Text). Bone-marrow derived cell populations typically consisted of 70% DCs (as indicated by CD11c-expression) of which 80% were CCR7+ after lipopolysaccharide treatment (Fig. S1 A–B). Compared to immature DCs, mature DCs expressed higher levels of MHCII, CD86, and CCR7, as expected (32). Finally, we found that autologous secretion of CCL19 (33), which could potentially interfere with exogenous gradient sensing, was at least an order of magnitude less than the average concentrations of added chemokines (Fig. S2F), allowing us to consider only the exogenous gradients in interpreting the results.
CCR7 Signaling Causes only Mild Chemokinesis in Dendritic Cells.
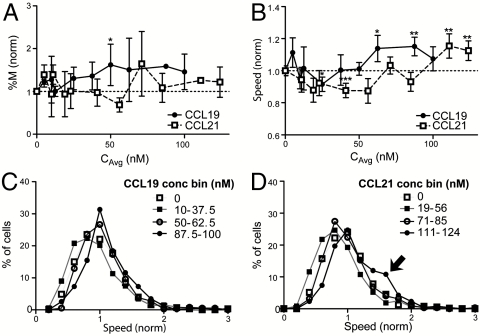
Because CCR7 signaling can increase motility in T cells (15) and in some cases DCs in 2D culture (34, 35), we next examined the influence of CCL19 and CCL21 on DC motility in a 3D environment both in terms of activating cells to be motile as well as affecting their average (random) speed. We found that in 3D, chemokinetic effects on DCs were relatively small (Fig. 2). Statistically significant increases in the percent of migrating cells and average cell speed were seen only at CCL19 concentrations above 50 nM, and only above 100 nM for CCL21. Furthermore, a biphasic trend was observed in cell speed, where lower concentrations (0–50 nM for CCL19; 0–71 nM for CCL21) decreased migration speed, and higher concentrations (> 50 nM) slightly increased speed compared to that in the absence of chemokine. This pattern was also reflected in the cell speed frequency distributions, grouped into four concentration ranges (Fig. 2 C and D). For CCL19, the cells responded in a normal distribution as seen by bell-shaped curves whose peaks shift with different concentrations (Fig. 2C). In contrast, for CCL21 only a small population (∼10%) exhibited pronounced increases in cell speed at the highest concentration range (111–124 nM). Thus, while CCL19 slightly increased the overall cell speeds at higher concentrations, CCL21 caused a small population of cells to move faster; however, these effects were minor.
Fig. 2.
Dendritic cells display mild chemokinesis to CCR7 ligands. (A) Percentage of migrating cells (%M); *P = 0.0366. (B) Average cell speed; *P < 0.05 and **P < 0.01. (A and B) All values were normalized to controls for each experiment, and comparisons were made using one-way ANOVA and Tukey posttest. (C, D) Histograms of normalized cell speeds in various concentration ranges of (C) CCL19 and (D) CCL21. (A–D) For each data point, n = 2–14, each representing the average of 71–594 cells.
Dendritic Cells Chemotact Differentially to CCL19 and CCL21 at Higher Concentration Gradients.
Chemokine gradients can be sensed by cells in a number of ways (36, 37), and CCR7-driven DC chemotaxis in 3D has not to date been quantitatively described. We sought to quantitate and compare DC chemotactic behavior in 3D in different gradients of CCL21 and CCL19, considering that the absolute concentration gradient (∇C), average concentration (CAvg), and percent gradient (∇C/CAvg) may all contribute to how DCs sense and respond to the chemokine.
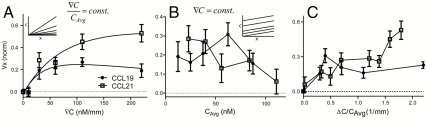
We first subjected DCs to increasing ∇C, while keeping the relative gradient (∇C/CAvg) constant. To quantify the chemotactic sensitivity, we measured the average cell velocity parallel to the imposed gradient (Vx). Interestingly, Vx became positive only at gradients ≥25 nM/mm for both chemokines, but continued to increase at higher gradients only for CCL21 (Fig. 3A). It has been hypothesized that cellular chemosensitivity is proportional to the difference of the number of ligand-receptor bound states in the front vs. back of the cell, supported by data from 2D chemotaxis studies (38, 39). At equilibrium of ligand-receptor binding, the percentage of ligand-bound receptors is C/(C + KD), where KD is the effective binding constant (koff/kon) of ligand to receptor. The difference in the fractional ligand-receptor bound state between the front and back of the cell is thus proportional to ∇(C/(C + KD)), or ∇C/(C + KD)2, where ∇ refers to the spatial gradient along the x direction in our case (see SI Text).
Fig. 3.
Dendritic cell chemosensitivity to CCL21 is greater than to CCL19. (A) Directed migration velocity (Vx) as a function of chemokine gradient (∇C). Solid lines show the fit of data to Eq. 1. (B) Vx as a function of the average concentration (CAvg). (C) Vx as a function of the relative gradient (∇C/CAvg). (A–C) For each data point, n = 2 - 14, each representing an average of 70–850 cells. Error bars indicate 95% confidence interval.
To test whether DC chemosensitivity to CCR7 ligands in 3D gels was consistent with this mechanism, we thus fit both sensitivity curves to the following equation:
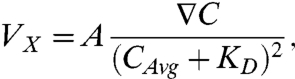 |
[1] |
where A is a constant. Both sets of data showed excellent fit to Eq. 1, indicating that ligand-receptor binding kinetics play important roles in DC chemotaxis in gradients of CCR7 ligands. At the highest ∇C tested (220 nM/mm), the chemotactic response appeared to decrease for CCL19 but not CCL21; this may reflect differences in receptor availability due to CCR7 recycling only after binding CCL21 but not CCL19 (8, 9, 40). Such receptor recycling differences would lead to a higher apparent KD of CCL21 than CCL19 for binding CCR7, which is supported by the effective KD values estimated from fitting the data in Fig. 3A to Eq. 1: namely, 100 nM (± 40 nM) for CCL19 and 250 nM (± 110 nM) for CCL21. This effective binding constant is not equal to the direct binding constant of the chemokine-receptor pair but rather reflects its apparent binding constant as reflected by the functional outcome (chemotactic behavior), taking into account receptor internalization and recycling kinetics (40, 41). These values are higher than already reported KD values for CCR7 [144 pM-4.7 nM; (4, 5)], which probably reflects the internal receptor processing as mentioned above.
We then kept ∇C constant at 30 nM/mm while increasing CAvg [in this case, the % gradient (∇C/CAvg) decreases with increasing CAvg]. We found a poor correlation between CAvg and chemosensitivity at a fixed ∇C. However, chemosensitivity generally decreased with increasing CAvg (Fig. 3B) for CCL21, but not for CCL19, suggesting that cells may be more adaptable to varying CCL19 levels than to CCL21. The higher adaptability to CCL19 vs. CCL21 levels could have implications for long-range signaling, because CCL19 concentrations strongly vary between tissues (42, 43) and its relatively non-matrix-binding characteristics imply shallower gradients over longer distances, while matrix-bound CCL21 gradients are more strongly localized to lymphatic vessels and the T cell zone of the lymph node (14).
Finally, we compared the effects of the % gradient (∇C/CAvg) on chemosensitivity (Fig. 3C). Here, as with absolute gradient (Fig. 3A), we observed a positive correlation for chemotaxis towards CCL21 at increasing % gradients, but a flat profile for chemotaxis towards CCL19 as a function of % gradient from 0.4 to 2 mm-1. For both chemokines, chemosensitivity was only nonzero when CAvg > 5 nM (Fig. 3A); this concentration threshold for gradient sensing may relate to the fact that DCs secrete small amounts of CCL19 themselves (∼1–3 nM, Fig. S2F), which the exogenous concentration must overcome. Classic experiments from ∼30 years ago estimated that neutrophils in 2D can sense a chemokine gradient of formyl-Met-Leu-Phe as small as 1–2% difference between front and back (36). Here, we show that DCs can sense a gradient as little as 0.4% for an average chemokine concentration bigger than 12.5 nM.
We also found that the DC persistence P (ratio of net distance traveled to total distance migrated) correlates well with the directed migration Vx, indicating the absence of an additional directional bias (e.g., chamber geometry; Fig. S3 A and B). Furthermore, by fitting the sensitivity curves of % gradient differences, we estimated similar KD values—namely, 84 nM (± 29 nM) for CCL19 and 255 nM (± 188) for CCL21 as those seen for ∇C.
Dendritic Cells Chemotact Preferentially Towards CCL21 vs. CCL19.
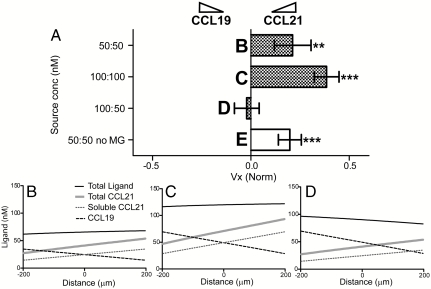
In their physiological microenvironment, DCs are often exposed to gradients of both CCL19 and CCL21 (1, 43). We thus asked whether DCs differentially respond to these chemokines when exposed to two opposing gradients simultaneously and compared DC migration in several scenarios. First, the side channels were flushed with equal concentrations of 50 or 100 nM of each chemokine, which led to equivalent and opposite gradients of soluble chemokines, while the matrix-bound CCL21 only affected total CCL21 gradients differently (Fig. 4 A–C). In the lower concentration (CAvg = 25 nM), we expect that the matrix-binding sites are not saturated and a gradient of bound CCL21 forms that, when added, to a slightly increased gradient of total CCL21 than CCL19 (60 vs. 50 nM/mm, respectively). In this scenario, DC migration was heavily skewed towards CCL21 (Fig. 4B). At the higher CAvg( ∼ 50 nM), all binding sites should be saturated and the bound fraction of CCL21 should only raise the CAvg but not affect ∇C. Interestingly, DCs still migrated towards the CCL21 gradient (Fig. 4C), displaying an even higher bias towards CCL21 than in the lower concentration (Fig. 4B). However, when the CCL19 gradient was doubled compared to that of CCL21 (111 nM/mm vs. 65 nM/mm; Fig. 4D), there was no directional bias towards either chemokine, even though the CAvg of both chemokines were in a similar range (50 nM and 40 nM respectively). Thus, the CCL19 gradient needed to be twice that of CCL21 to prevent chemotaxis towards CCL21. This result differs from that of a recent DC-chemotaxis study in 2D under flow (19), where DCs are only weakly adhesive.
Fig. 4.
Dendritic cell migration is preferentially skewed towards CCL21 vs. CCL19. (A) The average velocity Vx of cells in competing gradients of CCL19 and CCL21 as indicated. Dark columns: 1.5 mg/mL collagen ± 10% MG, white columns: collagen only. *P < 0.05, **P < 0.01, and ***P < 0.001 compared to group D with one-way ANOVA and Tukey posttest. Error bars show 95% confidence intervals. For each data point, n = 2, each representing an average of 84–123 cells. (B–E) Computationally estimated concentration gradients corresponding to the scenarios in (A). Lines indicate concentrations of CCL19 (dotted black line), soluble CCL21 (solid gray line), and total CCR7 ligand (solid black line).
Because the cells may have experienced different signaling from bound vs. soluble chemokines (14), we next asked whether the CCL21 needed to be matrix-bound to drive preferential DC chemotaxis. MG provides many more binding sites for CCL21 compared to type I collagen, we repeated the experiment in MG-free matrix and still found a strong chemotactic preference for CCL21 (Fig. 4E), even though ∇C and CAvg were matched for both chemokines (Table 1).
Table 1.
Quantification of competing gradients of CCL19 and CCL21 for the cases studied in Fig. 4.
| Case | ∇C (nM/mm) | ||||
| CCL19 | CCL21 soluble | CCL21 bound | Total Ligand | Preference | |
| i | 50 | 50 | 10 | 10 | CCL21 |
| ii | 100 | 101 | 6 | 7 | CCL21 |
| iii | 100 | 50 | 10 | −40 | Neutral |
| iv | 50 | 50 | 0 | 0 | CCL21 |
Thus the preferential chemotaxis of DCs towards gradients of CCL21 vs. CCL19 did not appear to be dependent on the bound state of CCL21. Both ligands have similar affinity to CCR7 (4, 5), but drive different fates: CCL19 signaling leads to receptor internalization and temporal desensitization while CCL21 can signal from the cell membrane (6, 9, 40, 44, 45), a well described ligand-dependent process for G-protein-coupled receptors. This differential fate of activated CCR7 could lead to increased receptor density and signaling on the cell side dominated by CCL21 (46), consistent with our observation of increased DC response to CCL21 gradients above 50 nM (Fig. 3A), where receptor desensitization would occur with CCL19. Furthermore, there are some indications that receptor desensitization could take place in a ligand specific manner (8) as described for other seven-transmembrane receptors (47). As mature DCs are secreting CCL19, CCL19-specific desensitization might be also responsible for the observed CCL21 preference (Fig. 4).
In summary, this study addressed the question of how DCs interpret such gradients in a 3D environment to drive directed invasion, as well as how they differentially respond to CCL21 vs. CCL19 in 3D, which was previously unknown. We demonstrated that DC chemoinvasion behavior towards increasing ligand gradients is consistent with general receptor binding dynamics, and with a stronger propensity for CCL21 than CCL19, particularly at higher concentrations and gradient strengths. When exposed to equal but opposite gradients of the two CCR7 ligands, chemoinvasion was always directed towards CCL21, even when matrix-binding was inhibited. We hypothesize that the preference towards CCL21 over CCL19 was likely due to differences in receptor recycling vs. degradation rather than binding affinity of the ligands to CCR7. These findings not only shed new insight into CCR7-driven DC chemoinvasion, but they also provide a general example of quantitative leukocyte chemotaxis up well defined chemokine gradients in 3D, which to date has been lacking.
Methods
DC Isolation and Culture.
Bone marrow was isolated from two to three month old C57BL/6 female mice as described (48) and cultured in RPMI medium 1640 containing 10% FBS, 1% penicillin/streptavidin, and 0.5% supernatant from mGM-CSF-secreting J558L cells (all from Gibco). On day 6, DCs were matured with 0.25 μg/mL LPS (Sigma-Aldrich) and used for migration experiments on day 7.
Agarose-Based Microfluidic Chemotaxis Device.
The design and construction of the agarose-based microfluidic device is described elsewhere (26). Medium containing chemokine (mCCL21 or mCCL19, R and D Systems) was pumped at a flow rate of 1 μL/ min; after 180 min., the gradient reached steady state and at this time the gel-cell mixture (106 cells/mL in 1.5 mg/mL type I collagen and 10% MG) was added to the center channel. The entire device was then placed in a 37 °C, 5% CO2 chamber on the microscope (Axiovert 200M, Zeiss) and after 30 min, images were collected every minute for 120 min (AxioCam MRm, Zeiss).
Migration Analysis.
Using the image sequence, cells were tracked using ImageJ (NIH), and we computed the cell speed V (i.e., average position change per min.), net velocity vector in the direction parallel to the imposed gradient Vx, and the persistence length P (i.e., net displacement divided by total distance traveled). Only cells that migrated > 10 μm/h were tracked. Actual values are shown with standard error of mean unless otherwise stated.
Chemokine Conjugation.
mCCL21 and mCCL19 were conjugated to FITC (Sigma-Aldrich) as described (49) to visualize diffusion gradients inside the matrix, and to determine the density of CCL21 binding sites in the collagen-MG matrix.
Estimation of Matrix-Binding Site Density.
The concentration of binding sites (CBS) present in the collagen-MG scaffold was estimated with an integrated Comsol-Matlab analysis in combination with a gradient image interpolation. Briefly, after introducing fluorescently labeled chemokine in one source channel, normalized voxel intensity values from a confocal image were taken and integrated into a computational matrix. Starting from an initial CBS value, randomly chosen, simulations were run using the model described above, setting CCL21 concentrations in the right and in the left source channels to 1 and 0, respectively. The resulting concentration values were compared to the imaged gradients, and iterative computations were run until differences between experiment and computational estimate reached < 5%.
Supplementary Material
Acknowledgments.
The authors thank A. Shieh, B. Dixon, M. Avermann, Y. Kalinin, S. Chen, V. Greenwood, J. Favre, and H. Madi for technical assistance and advice. Funding was provided to M.A.S. by the European Research Council (DC-LYMPH 206653-2) and the Swiss National Science Foundation (310010) and to M.W. by the National Institutes of Health (NIH) (NCI-R21CA138366). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014920108/-/DCSupplemental.
References
- 1.Randolph GJ, Angeli V, Swartz MA. Dendritic-cell trafficking to lymph nodes through lymphatic vessels. Nat Rev Immunol. 2005;5:617–628. doi: 10.1038/nri1670. [DOI] [PubMed] [Google Scholar]
- 2.Campbell JJ, et al. Chemokines and the arrest of lymphocytes rolling under flow conditions. Science. 1998;279:381–384. doi: 10.1126/science.279.5349.381. [DOI] [PubMed] [Google Scholar]
- 3.Forster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8:362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan SK, McGrath DA, Grigoriadis D, Bacon KB. Pharmacological and signaling analysis of human chemokine receptor CCR-7 stably expressed in HEK-293 cells: high-affinity binding of recombinant ligands MIP-3beta and SLC stimulates multiple signaling cascades. Biochem Biophys Res Commun. 1999;263:685–690. doi: 10.1006/bbrc.1999.1442. [DOI] [PubMed] [Google Scholar]
- 5.Ott TR, et al. The N-terminal domain of CCL21 reconstitutes high affinity binding, G protein activation, and chemotactic activity, to the C-terminal domain of CCL19. Biochem Biophys Res Commun. 2006;348:1089–1093. doi: 10.1016/j.bbrc.2006.07.165. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida R, et al. Secondary lymphoid-tissue chemokine is a functional ligand for the CC chemokine receptor CCR7. J Biol Chem. 1998;273:7118–7122. doi: 10.1074/jbc.273.12.7118. [DOI] [PubMed] [Google Scholar]
- 7.Campbell JJ, et al. 6-C-kine (SLC), a lymphocyte adhesion-triggering chemokine expressed by high endothelium, is an agonist for the MIP-3beta receptor CCR7. J Cell Biol. 1998;141:1053–1059. doi: 10.1083/jcb.141.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardi G, Lipp M, Baggiolini M, Loetscher P. The T cell chemokine receptor CCR7 is internalized on stimulation with ELC, but not with SLC. Eur J Immunol. 2001;31:3291–3297. doi: 10.1002/1521-4141(200111)31:11<3291::aid-immu3291>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 9.Byers MA, et al. Arrestin 3 mediates endocytosis of CCR7 following ligation of CCL19 but not CCL21. J Immunol. 2008;181:4723–4732. doi: 10.4049/jimmunol.181.7.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang BG, et al. Binding of lymphoid chemokines to collagen IV that accumulates in the basal lamina of high endothelial venules: its implications in lymphocyte trafficking. J Immunol. 2007;179:4376–4382. doi: 10.4049/jimmunol.179.7.4376. [DOI] [PubMed] [Google Scholar]
- 11.Patel DD, et al. Chemokines have diverse abilities to form solid phase gradients. Clin Immunol. 2001;99:43–52. doi: 10.1006/clim.2000.4997. [DOI] [PubMed] [Google Scholar]
- 12.Uchimura K, et al. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 2006;7:2. doi: 10.1186/1471-2091-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirose J, Kawashima H, Yoshie O, Tashiro K, Miyasaka M. Versican interacts with chemokines and modulates cellular responses. J Biol Chem. 2001;276:5228–5234. doi: 10.1074/jbc.M007542200. [DOI] [PubMed] [Google Scholar]
- 14.Schumann K, et al. Immobilized chemokine fields and soluble chemokine gradients cooperatively shape migration patterns of dendritic cells. Immunity. 2010;32:703–713. doi: 10.1016/j.immuni.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 15.Stachowiak AN, Wang Y, Huang YC, Irvine DJ. Homeostatic lymphoid chemokines synergize with adhesion ligands to trigger T and B lymphocyte chemokinesis. J Immunol. 2006;177:2340–2348. doi: 10.4049/jimmunol.177.4.2340. [DOI] [PubMed] [Google Scholar]
- 16.Mosadegh B, Saadi W, Wang SJ, Jeon NL. Epidermal growth factor promotes breast cancer cell chemotaxis in CXCL12 gradients. Biotechnol Bioeng. 2008;100:1205–1213. doi: 10.1002/bit.21851. [DOI] [PubMed] [Google Scholar]
- 17.Guarnieri D, et al. Covalently immobilized RGD gradient on PEG hydrogel scaffold influences cell migration parameters. Acta Biomater. 2009;6:2532–2539. doi: 10.1016/j.actbio.2009.12.050. [DOI] [PubMed] [Google Scholar]
- 18.Nasreen N, et al. Pleural mesothelial cell transformation into myofibroblasts and haptotactic migration in response to TGF-beta1 in vitro. Am J Physiol Lung Cell Mol Physiol. 2009;297:L115–124. doi: 10.1152/ajplung.90587.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricart BG, John B, Lee D, Hunter CA, Hammer DA. Dendritic cells distinguish individual chemokine signals through CCR7 and CXCR4. J Immunol. 2011;186:53–61. doi: 10.4049/jimmunol.1002358. [DOI] [PubMed] [Google Scholar]
- 20.Schmalstieg FC, Rudloff HE, Hillman GR, Anderson DC. Two-dimensional and three-dimensional movement of human polymorphonuclear leukocytes: two fundamentally different mechanisms of locomotion [corrected] J Leukocyte Biol. 1986;40:677–691. doi: 10.1002/jlb.40.6.677. [DOI] [PubMed] [Google Scholar]
- 21.Lammermann T, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 22.Behnsen J, et al. Environmental dimensionality controls the interaction of phagocytes with the pathogenic fungi Aspergillus fumigatus and Candida albicans. PLoS Pathog. 2007;3:e13. doi: 10.1371/journal.ppat.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lammermann T, et al. Cdc42-dependent leading edge coordination is essential for interstitial dendritic cell migration. Blood. 2009;113:5703–5710. doi: 10.1182/blood-2008-11-191882. [DOI] [PubMed] [Google Scholar]
- 24.Wolf K, Friedl P. Molecular mechanisms of cancer cell invasion and plasticity. Br J Dermatol. 2006;1(154 Suppl):11–15. doi: 10.1111/j.1365-2133.2006.07231.x. [DOI] [PubMed] [Google Scholar]
- 25.Wolf K, et al. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haessler U, Kalinin Y, Swartz MA, Wu M. An agarose-based microfluidic platform with a gradient buffer for 3D chemotaxis studies. Biomed Microdevices. 2009;11:827–835. doi: 10.1007/s10544-009-9299-3. [DOI] [PubMed] [Google Scholar]
- 27.Pedersen JA, Swartz MA. Mechanobiology in the third dimension. Ann Biomed Eng. 2005;33:1469–1490. doi: 10.1007/s10439-005-8159-4. [DOI] [PubMed] [Google Scholar]
- 28.Vickerman V, Blundo J, Chung S, Kamm R. Design, fabrication and implementation of a novel multi-parameter control microfluidic platform for three-dimensional cell culture and real-time imaging. Lab Chip. 2008;8:1468–1477. doi: 10.1039/b802395f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abhyankar VV, et al. A platform for assessing chemotactic migration within a spatiotemporally defined 3D microenvironment. Lab Chip. 2008;8:1507–1515. doi: 10.1039/b803533d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saadi W, et al. Generation of stable concentration gradients in 2D and 3D environments using a microfluidic ladder chamber. Biomed Microdevices. 2007;9:627–635. doi: 10.1007/s10544-007-9051-9. [DOI] [PubMed] [Google Scholar]
- 31.Shields JD, et al. Autologous chemotaxis as a mechanism of tumor cell homing to lymphatics via interstitial flow and autocrine CCR7 signaling. Cancer Cell. 2007;11:526–538. doi: 10.1016/j.ccr.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 32.Lutz MB, et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 33.Del Prete A, et al. Regulation of dendritic cell migration and adaptive immune response by leukotriene B4 receptors: a role for LTB4 in up-regulation of CCR7 expression and function. Blood. 2007;109:626–631. doi: 10.1182/blood-2006-02-003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kellermann SA, Hudak S, Oldham ER, Liu YJ, McEvoy LM. The CC chemokine receptor-7 ligands 6Ckine and macrophage inflammatory protein-3 beta are potent chemoattractants for in vitro- and in vivo-derived dendritic cells. J Immunol. 1999;162:3859–3864. [PubMed] [Google Scholar]
- 35.Riol-Blanco L, et al. The chemokine receptor CCR7 activates in dendritic cells two signaling modules that independently regulate chemotaxis and migratory speed. J Immunol. 2005;174:4070–4080. doi: 10.4049/jimmunol.174.7.4070. [DOI] [PubMed] [Google Scholar]
- 36.Zigmond SH. Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J Cell Biol. 1977;75:606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin F, et al. Neutrophil migration in opposing chemoattractant gradients using microfluidic chemotaxis devices. Ann Biomed Eng. 2005;33:475–482. doi: 10.1007/s10439-005-2503-6. [DOI] [PubMed] [Google Scholar]
- 38.Herzmark P, et al. Bound attractant at the leading vs. the trailing edge determines chemotactic prowess. Proc Natl Acad Sci USA. 2007;104:13349–13354. doi: 10.1073/pnas.0705889104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zigmond SH. Consequences of chemotactic peptide receptor modulation for leukocyte orientation. J Cell Biol. 1981;88:644–647. doi: 10.1083/jcb.88.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otero C, Eisele PS, Schaeuble K, Groettrup M, Legler DF. Distinct motifs in the chemokine receptor CCR7 regulate signal transduction, receptor trafficking and chemotaxis. J Cell Sci. 2008;121:2759–2767. doi: 10.1242/jcs.029074. [DOI] [PubMed] [Google Scholar]
- 41.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315:683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 42.Krumbholz M, et al. CCL19 is constitutively expressed in the CNS, up-regulated in neuroinflammation, active and also inactive multiple sclerosis lesions. J Neuroimmunol. 2007;190:72–79. doi: 10.1016/j.jneuroim.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 43.Luther SA, et al. Differing activities of homeostatic chemokines CCL19, CCL21, and CXCL12 in lymphocyte and dendritic cell recruitment and lymphoid neogenesis. J Immunol. 2002;169:424–433. doi: 10.4049/jimmunol.169.1.424. [DOI] [PubMed] [Google Scholar]
- 44.Kohout TA, et al. Differential desensitization, receptor phosphorylation, beta-arrestin recruitment, and ERK1/2 activation by the two endogenous ligands for the CC chemokine receptor 7. J Biol Chem. 2004;279:23214–23222. doi: 10.1074/jbc.M402125200. [DOI] [PubMed] [Google Scholar]
- 45.Otero C, Groettrup M, Legler DF. Opposite fate of endocytosed CCR7 and its ligands: recycling versus degradation. J Immunol. 2006;177:2314–2323. doi: 10.4049/jimmunol.177.4.2314. [DOI] [PubMed] [Google Scholar]
- 46.Marchese A, Paing MM, Temple BR, Trejo J. G protein-coupled receptor sorting to endosomes and lysosomes. Annu Rev Pharmacol Toxicol. 2008;48:601–629. doi: 10.1146/annurev.pharmtox.48.113006.094646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Violin JD, et al. beta2-adrenergic receptor signaling and desensitization elucidated by quantitative modeling of real time cAMP dynamics. J Biol Chem. 2008;283:2949–2961. doi: 10.1074/jbc.M707009200. [DOI] [PubMed] [Google Scholar]
- 48.Inaba K, et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hermanson GT. Bioconjugate Techniques. New York: Academic Press; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.