Abstract
Anthracyclines are frequently used for the treatment of breast cancer and topoisomerase II alpha (TOP2A) is considered to be the molecular target. Numerous studies have evaluated the predictive value of TOP2A using different methodological approaches and inconsistent results have been reported. Indeed, the correlation between techniques for the assessment of TOP2A status has not been well evaluated. In this study, we determined TOP2A status in 61 breast tumor samples by real-time PCR, DNA microarrays, immunohistochemistry (IHC), and fluorescence in situ hybridization (FISH), and then evaluated these results with clinical-pathological features and breast cancer intrinsic subtypes. First, we observed a statistical significant correlation of TOP2A gene expression between real-time PCR and microarrays (Pearson coefficient, 0.816; P < 0.001), and both predicted TOP2A IHC results fairly well (area under the curve > 0.74). In contrast, poor agreement between FISH and IHC data was observed (k: 0.134). Secondly, TOP2A expression was found significantly associated with cell proliferation, and with the highly proliferative Luminal B, Her2-enriched and Basal-like intrinsic subtypes. In conclusion, TOP2A expression in breast cancer was associated with high proliferation and aggressive tumor subtypes and appears to be independent of its amplification status. All of these features should be taken into consideration when assessing the predictive value of TOP2A for anthracycline-based chemotherapy.
Anthracyclines are some of the most powerful agents available for the treatment of breast cancer and are frequently used in the adjuvant setting. However, anthracyclines are associated with severe adverse effects, such as cardiac toxicity1 and bone marrow dysfunction, including acute leukemia and myelodysplasia.2 Therefore, there is a need to identify the subset of patients who might benefit from these drugs.
The mechanism of action of these compounds is related to the inhibition of the nuclear enzyme DNA topoisomerase II (Topo II),3 although other antitumor mechanisms have also been described.4 Topoisomerase II is an important enzyme for cell division because it releases torsional stress in double-stranded DNA by inducing transient breaks that are then subsequently resealed.5 The catalytic activity of Topo II in mammalian cells is mediated by two isoforms [ie, Topo II α (TOP2A) and Topo II β].6 In vitro studies7–10 have suggested that sensitivity to Topo II inhibitors is dependent on the expression level of TOP2A in target cancer cells. Cells with a low concentration of TOP2A protein are less sensitive to Topo II–inhibiting drugs versus cells containing a high concentration of TOP2A. Interestingly, the TOP2A gene itself is frequently found coamplified with v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) (ERBB2) in human breast cancer cells,11 and several studies12–16 have shown that anthracycline-containing regimens improve survival in women with ERBB2-positive breast cancer.
Numerous retrospective studies have investigated the predictive value of TOP2A using different methods. Some of these approaches have focused on determining TOP2A expression,17,18 whereas others have evaluated TOP2A copy number.19,20 In any case, the results of these studies are contradictory and it is still unclear whether investigating TOP2A provides useful clinical information concerning anthracycline benefit. Current techniques for measuring TOP2A are not standardized, which may account, at least partly, for these apparently contradictory results. In addition, the agreement among techniques and the correlation between TOP2A expression values measured by different methods is also not well established. On the other hand, TOP2A is considered a surrogate marker of cell proliferation and its expression has been associated with high tumor grade21,22 and the absence of hormone receptors,23 suggesting that overexpression of TOP2A could be characteristic of a specific type of breast tumor. Investigation of the predictive significance of TOP2A for anthracycline-containing regimens requires knowing the correlation between different methods and the characterization of TOP2A-expressing tumors to control potential confounding factors.
The aim of this study was to evaluate the status of TOP2A in breast cancer tumors by four different methods [ie, fluorescence in situ hybridization (FISH), gene expression via DNA microarrays, gene expression via real-time PCR (Q-PCR), and immunohistochemistry (IHC)]. We also sought to assess the correlation and agreement among these techniques when classifying the tumors as TOP2A positive or negative. In addition, TOP2A status was correlated to clinical features, such as cell proliferation, hormone receptor status, and breast cancer intrinsic subtypes, that identified many significant correlations.
Material and Methods
Study Population
Tumor biopsy specimens were obtained from a set of 61 pretreated patients diagnosed as having locally advanced breast cancer; these patients participated in a neoadjuvant clinical trial (registered at the following Web site: http://www.clinicaltrials.gov; identifier NCT00123929). The clinical trial was approved by the Hospital Clínico San Carlos Ethics Committee, Madrid, Spain. Briefly, eligibility criteria included the following: women aged between 18 and 78 years; clinical stage IIB, IIIA, or IIIB breast cancer; and palpable breast tumors not amenable to breast-preserving surgery. Before the start of the trial, an informed consent was obtained from every patient. A total of 226 registered patients were enrolled in the trial; 204 underwent a complete evaluation. No significant differences were observed between the subset of 61 patients and the whole cohort of 226 patients. Clinical and pathological characteristics of each of the 61 patients are available in Supplemental Table S1 (at http://ajp.amjpathol.org).
RNA Isolation and Microarray Expression Profiling
Total RNA was extracted from tumor biopsy specimens using a kit (Qiagen RNeasy Mini Kit; Qiagen Inc., Valencia, CA), following the instructions of the manufacturer. The biopsy specimens were obtained before neoadjuvant chemotherapy. To check cellularity, an H&E image was obtained from all tumors; only samples with more than 80% tumor cells were used. The amount of RNA was assessed with a device (Nanodrop ND-1000 UV Spectrophotometer; Thermo Fisher Scientific, Wilmington, DE). RNA integrity was assessed using a kit (RNA 6000 Nano Chip kit), followed by analysis (Bioanalyzer 2100; Agilent Technologies, Santa Clara, CA). Whole human genome oligo 4 × 44 microarrays (Agilent Technologies) were hybridized after low RNA input fluorescence amplification of tumor total RNA, according to the manufacturer's protocol. Arrays were scanned (GenePix 4000B scanner; Molecular Devices Corporation, Sunnyvale, CA), analyzed using software (GenePix 5.1), and uploaded into a University of North Carolina microarray database, in which a Lowess normalization of the log2 ratio (Cyanine 5/Cyanine 3) intensity values is automatically performed. For all analyses, genes were filtered by requiring the Lowess-normalized intensity values in both channels to be greater than 10; only genes that reported values in 70% or more of the samples were included. The genes were median centered across all samples. Breast cancer subtypes were assigned as described by Parker et al.24 In addition, the recently identified subtype named Claudin-low was assigned using a separate centroid-based predictor25; therefore, tumors were categorized into Luminal A, Luminal B, Basal-like, Her2-enriched, Claudin-low, and Normal-like. The primary microarray data presented in this study are available in the Gene Expression Omnibus database (accession No. GSE21997).
Q-PCR Analysis
Total RNA, 200 ng, was used as a template to obtain first-strand cDNA using a system (SuperScript First-Strand Synthesis System for RT-PCR; Invitrogen, Parsley, UK), following the manufacturer's instructions. To determine that cDNA was synthesized, the first-strand cDNA was further amplified by conventional PCR using primers for β-actin, as previously described.26 Real-time PCR was performed using gene expression assays (TaqMan; Applied Biosystems, Foster City, CA) and a PCR system (ABI PRISM 7900HT Fast Real Time PCR System; Applied Biosystems). Briefly, 4 μL of cDNA template was mixed with 1 μL of ×20 TOP2A primers, probes labeled with 5,6 carboxifluoresceina reporter dye (assay Hs00172214_m1), 1 μL of ×20 human β-actin primers, and probes labeled with VIC-reported dye (TaqMan endogenous control, part 4326315E; Applied Biosystems), 10 μL of mix (TaqMan Universal PCR Mastermix), and 4 μL of RNase-free water. Human β-actin served as an endogenous control to normalize the TOP2A mRNA levels in the subsequent quantitative analysis. The PCR conditions were those recommended by the manufacturer. A pool of RNA from 11 breast tissue samples of healthy individuals who underwent cosmetic or plastic surgery was used as a control. Each sample was measured in triplicate. The data were analyzed by the comparative Ct method, in which the amount of TOP2A, normalized to β-actin and relative to control, is given as follows: 2−(ΔCt sample − ΔCt control). Tumors that yielded values of two or greater were considered as TOP2A positive.
IHC and Tumor Grading
Paraffin-embedded tumor samples from core biopsy specimens were evaluated by IHC analysis for TOP2A (TOP2A-mouse monoclonal antibody Novocastra Leica-TopoIIA, clone 3F6, 1:40; Leica Microsystems, Wetzlar, Germany); estrogen receptor (ER) (clone 1D5, 1:35; Dako Cytomation, Glostrup, Denmark), progesterone receptor (clone PgR 636, 1:50; Dako Cytomation), and Ki-67 (clone MIB-1, 1:75; Dako Cytomation). After incubation with the primary antibodies, immunohistochemical studies were performed using a Bond-Max immunostainer (Vision BioSystems, Hingham, MA) with polymer-defined peroxidase detection.
Because no cutoff for positivity has been validated to define TOP2A overexpression, we analyzed this variable based on a cutoff of 20% or greater (mean value in this study) and median values (10% of stained cells). The Ki-67 positivity was defined as 20% or greater of stained cells because the staining in our normal control breast tissue (from the tissue bank of Hospital Clínico San Carlos) was always lower than this value. The cut points for ER and progesterone receptor positivity were established at 10% or greater of stained cells. All tumors were graded by the study pathologist (JALGA) by using Elston-Ellis histological grading.27
Measurement of TOP2A and ERBB2 Amplification
The amplification of TOP2A and ERBB2 was measured by FISH. The probes used were as follows: locus-specific identifier TOP2A, labeled in orange; centromere enumeration probe 17, labeled in green; and locus-specific identifier ERBB2 probe, labeled in orange (Vysis-Abbott, Downers Grove, IL). Slides were prepared according to the manufacturer's instructions for paraffin sections. A positive result was defined as an ERBB2 and TOP2A genes/chromosome 17 ratio of 2.2 or greater (for ERBB2) or greater than 2 (for TOP2A). The cutoff for the deletion was established as a TOP2A gene/chromosome 17 ratio of 0.5 or lower. Images were visualized on a fluorescence microscope and captured on a workstation (MetaSystems, Altlussheim, Germany). A minimum of 100 nuclei were counted per case. All cut points were predefined before the correlations were performed; in all of the cases, the pathologist (JALGA) was blinded from the patient's identity and outcome.
Statistical Analysis
Qualitative variables were summarized by their frequency distribution, and quantitative variables were summarized by their mean ± SD. The continuous values of Q-PCR were log transformed. For qualitative variables, comparison was evaluated by the χ2 or Fisher exact test in cases in which more than 25% of the expected values were less than five. The association between the expression of TOP2A, measured by gene expression microarrays or Q-PCR, and other qualitative variables was assessed by t-test. To compare between more than two groups, a one-way analysis of variance for quantitative variables was used. The correlation among the quantitative values of TOP2A expression, measured by Q-PCR and gene expression microarrays, was evaluated with simple linear regression analysis. The comparison between TOP2A expression data, assessed by IHC, and gene expression microarrays or Q-PCR was performed using receiver operating characteristic curve analysis. The area under the curve, sensitivity, and specificity were also determined. The adjusted false-positive, false-negative, and error rates of the determined cutoffs were estimated using a bootstrapping method. Simple Cohen's κ coefficients and percentage agreement with its 95% confidence interval were used to assess the agreement when classifying the tumors as TOP2A positive or TOP2A negative. The strength of agreement is considered to be slight when κ values are between 0.00 and 0.20; fair, 0.21 and 0.40; moderate, 0.41 and 0.60; good, 0.61 and 0.80; and almost perfect, 0.81 and 1.00. All statistical tests were two sided, and P < 0.05 was considered significant. The statistical analysis was performed using software (SPSS 17.0 and R 2.10.1).
Results
Clinicopathological Characteristics of the Study Population
Table 1 summarizes the pathological characteristics of the 61 tumor samples/patients and the distribution of characteristics by TOP2A status. As shown, TOP2A copy number alterations were significantly associated with tumor size, stage, and ERBB2 amplification status. Interestingly, TOP2A expression, as assessed by Q-PCR or microarray, was not significantly associated with any of these characteristics. On the other hand, TOP2A status, by Q-PCR and gene expression microarrays, was significantly associated with Ki-67 positivity (P = 0.008 and P = 0.005, respectively). Similarly, according to IHC data, TOP2A protein overexpression was more frequent in Ki-67–positive tumors, although Ki-67 positivity was not significantly associated with TOP2A IHC positivity (P = 0.08).
Table 1.
Characteristics of Study Participants and Distributions of Characteristics by TOP2A Status
| Characteristics | FISH data |
Q-PCR data |
Microarray data |
IHC data |
|||||
|---|---|---|---|---|---|---|---|---|---|
| No. of participants (n = 61) | No. of tumors with TOP2A CNAs (n = 15) | P value | TOP2A expression⁎ | P value | TOP2A expression⁎ | P value | No. of TOP2A-positive tumors (n = 25) | P value | |
| Tumor size (cm) | |||||||||
| ≤5 | 24 | 2 | 0.03† | 0.92 (1.8) | 0.44 | 0.08 (1.0) | 0.45 | 9 | 0.79 |
| >5 | 37 | 13 | 0.57 (1.7) | −0.09 (0.7) | 16 | ||||
| Histological feature | |||||||||
| Ductal | 48 | 13 | 0.87 | 0.93 (1.8) | 0.12 | 0.051 (0.9) | 0.38 | 21 | 0.78 |
| Lobular | 10 | 2 | −0.35 (1.5) | −0.37 (0.8) | 3 | ||||
| Other | 3 | 0 | 0.72 (1.5) | −0.11 (1.0) | 1 | ||||
| Stage at diagnosis | |||||||||
| II | 21 | 2 | 0.03† | 0.58 (2.0) | 0.91 | 0.07 (1.0) | 0.84 | 8 | 0.28 |
| IIIA | 20 | 4 | 0.73 (1.6) | −0.06 (0.8) | 6 | ||||
| IIIB | 20 | 9 | 0.83 (1.8) | −0.08 (0.8) | 11 | ||||
| Histological grade | |||||||||
| II | 40 | 9 | 0.76 | 0.56 (1.8) | 0.38 | −0.11 (0.8) | 0.26 | 10 | <0.01† |
| III | 21 | 6 | 0.99 (1.8) | 0.15 (0.9) | 15 | ||||
| Ki-67 | |||||||||
| Low | 18 | 3 | 0.52 | −0.21 (1.4) | <0.01† | −0.48 (0.7) | <0.01† | 4 | 0.086 |
| High | 43 | 12 | 1.10 (1.8) | 0.17 (0.8) | 21 | ||||
| Estrogen receptor | |||||||||
| Positive | 36 | 12 | 0.07 | 0.55 (1.8) | 0.39 | −0.07 (0.9) | 0.59 | 13 | 0.43 |
| Negative | 25 | 3 | 0.95 (1.7) | 0.05 (0.8) | 12 | ||||
| Progesterone receptor | |||||||||
| Positive | 37 | 10 | 0.76 | 0.67 (1.9) | 0.86 | 0.01 (0.9) | 0.76 | 12 | 0.11 |
| Negative | 24 | 5 | 0.76 (1.6) | −0.06 (0.7) | 13 | ||||
| ERBB2 status | |||||||||
| Positive | 19 | 8 | 0.05† | 0.58 (1.9) | 0.71 | −0.11 (0.9) | 0.60 | 8 | 1.0 |
| Negative | 42 | 7 | 0.76 (1.7) | 0.02 (0.9) | 17 | ||||
CNA, copy number alteration.
Data are given as the mean (SD).
Significant difference.
In addition, we found that TOP2A mRNA expression, as assessed by Q-PCR or gene expression microarrays, varied significantly with intrinsic subtype (P < 0.001 in both cases). High-proliferative subtypes, such as Basal-like, Luminal B, and Her2-enriched, expressed higher levels of TOP2A than Luminal A, Claudin-low, or Normal-like tumors (Figure 1, A and B). In the same way, according to IHC data, the proportion of TOP2A-positive tumors was statistically different depending on tumor subtype (P = 0.002). As shown in Figure 1C, Basal-like, Her2-enriched, and Luminal B tumors had the most TOP2A-positive tumors, whereas Luminal A tumors were mostly TOP2A negative and all Normal-like tumors were TOP2A negative. Therefore, TOP2A expression can be considered a proliferative marker that is highly characteristic of the rapidly growing tumor subtypes. Finally, we observed that TOP2A copy number alterations occurred in Luminal A, Luminal B, Her2-enriched, and Claudin-low subtypes; no alterations were found in Basal-like and Normal-like tumors (Figure 1D).
Figure 1.
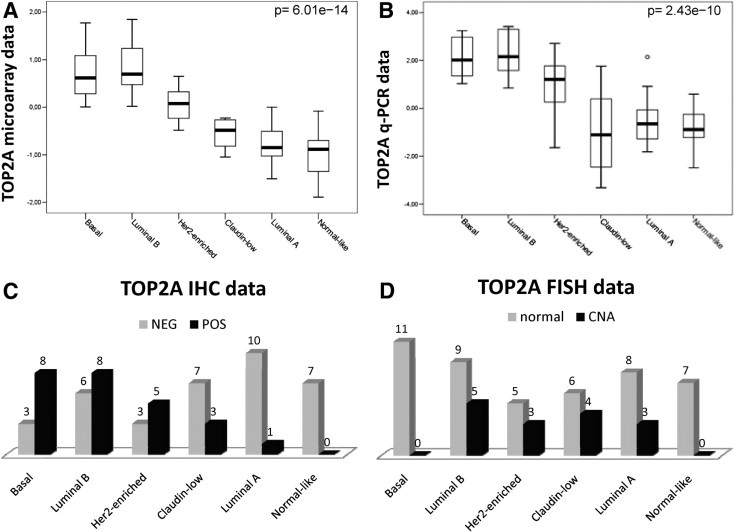
A: Box plot. The TOP2A mRNA expression values, measured by gene expression microarrays, according to genomic subtype. B: Box plot. The TOP2A mRNA expression values, measured by Q-PCR, according to genomic subtype. C: Bar chart. The number of TOP2A-positive (POS) and TOP2A-negative (NEG) tumors, according to IHC, for each subtype. D: Bar chart. The number of TOP2A-positive and TOP2A-negative tumors, according to FISH, for each subtype. CNA indicates copy number alteration.
Correlation Between TOP2A Status Assessed by Q-PCR, Gene Expression Array, and IHC
The TOP2A expression values measured by Q-PCR and gene expression microarrays significantly correlated with each other (Pearson correlation coefficient, 0.816; 95% confidence interval, 0.71 to 0.89; P = 1.2 × 10−15) (Figure 2). In addition, tumors with positive TOP2A expression by IHC had relatively higher mean levels of TOP2A expression, as measured by Q-PCR (P = 0.002) or gene expression microarrays (P < 0.001).
Figure 2.
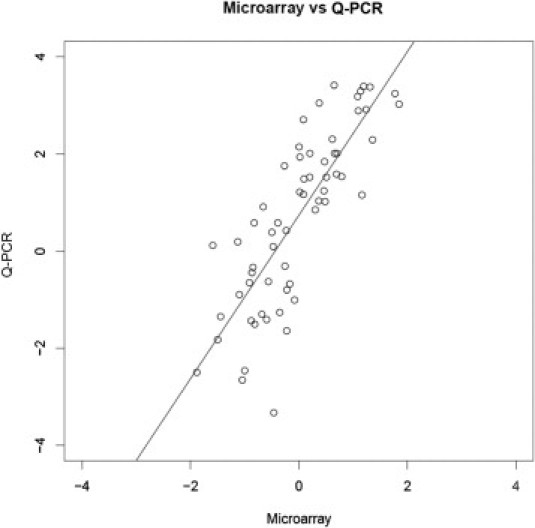
Correlation between TOP2A expression values, assessed by gene expression microarrays and Q-PCR.
To demonstrate the concordance of TOP2A expression status, determined by these techniques, we performed receiver operating characteristic curve analysis to assess how TOP2A gene expression by Q-PCR or microarrays can predict TOP2A-positive status by IHC. For Q-PCR, the area under the curve was 0.741 (sensitivity, 80%; specificity, 72%; best cutoff value, 1.04; adjusted error rate, 27%; adjusted false-positive rate, 23%; adjusted false-negative rate, 30%); for gene expression microarrays, the area under the curve was 0.791 (sensitivity, 100%; specificity, 53%; best cutoff value, −0.48; adjusted error rate, 30%; adjusted false-positive rate, 4%; adjusted false-negative rate, 49%) (Figure 3).
Figure 3.
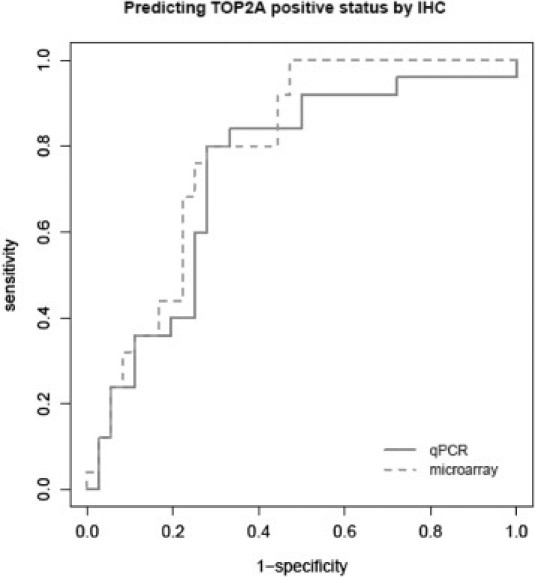
Receiver operating characteristic curves. Prediction of TOP2A IHC positivity according to gene expression microarrays and Q-PCR data.
TOP2A Copy Number Alteration and TOP2A Expression
The TOP2A was amplified in 13 cases (21.3%) and deleted in 2 cases (3.3%). Unlike RNA and protein levels, no significant association was observed between TOP2A copy number alterations and the expression of the gene measured by Q-PCR or gene expression microarrays. In addition, we did not find agreement between IHC and FISH when assessing TOP2A status in tumor samples (κ = 0.134, P = 0.26) (Table 2).
Table 2.
The TOP2A-Positive and TOP2A-Negative Tumors According to FISH and IHC Results
| FISH results | IHC results |
κ (P) value⁎ | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 8 (13) | 7 (11) | 0.134 (0.26) |
| Negative | 17 (28) | 29 (48) | |
Data are given as number (percentage) of 61 tumors unless otherwise indicated.
Cohen's κ coefficient for agreement between FISH and IHC results.
Discussion
In this study, TOP2A gene expression, as assessed by DNA microarray or Q-PCR, was significantly associated with Ki-67 IHC positivity. When TOP2A was assessed by IHC, a similar tendency was observed. Taken together, these data suggest that TOP2A expression across all breast cancers is likely a proliferation marker, which is concordant with previous reports about breast cancer28–30 and tumor types.31,32 Consistent with these data, we found higher TOP2A gene expression in highly proliferative subtypes, such as Basal-like, Luminal B, and Her2-enriched tumors, when compared with Luminal A, Normal-like, and Claudin-low tumors. Therefore, tumor proliferation may have a confounding effect on the predictive value of TOP2A and, therefore, TOP2A expression may possibly need to be adjusted for Ki-67 or other proliferative markers.
There is growing evidence supporting that breast cancer is a heterogeneous disease, rather than a single disease; and breast cancer subtypes have specific clinicopathological characteristics with different prognoses.24,25,33,34 The relative benefit of anthracyclines has not been tested for each specific breast cancer molecular subtype, although therapeutic outcome might be affected by molecular subtype.35 Different results among researchers who have assessed the predictive value of TOP2A expression might be, in part, because of different proportions of breast cancer subtypes between study populations; we demonstrated that TOP2A expression is substantially different across the known subtypes.
In our study, TOP2A amplification was associated with tumor size, stage, and ERBB2 positivity. Interestingly, none of these characteristics was associated with TOP2A expression. This might suggest that TOP2A protein–overexpressing tumors and tumors with TOP2A copy number alterations might be biologically different. Indeed, we found that the distribution of TOP2A-amplified tumors among subtypes is different and that the protein expression is also uniquely different; thus, it is not surprising to find that the FISH and IHC data are not that highly correlated. The expression of TOP2A has been extensively studied in human breast cancers by different methods. However, the correlation between techniques has been scarcely reported. As expected, our results showed a good correlation among Q-PCR and gene expression microarrays (Pearson correlation coefficient, 0.816).
A threshold value for TOP2A expression that could define subgroups associated with treatment outcome remains to be established. Some researchers17,36–38 have used an arbitrary IHC cutoff of greater than 10%, whereas others39 have used a 25% cutoff. We used a cutoff of 20% because staining in our control healthy breast tissue was always lower than this value; however, similar results were obtained when considering the cutoff of 10% (data not shown). We found that both microarrays and Q-PCR predicted fairly well the TOP2A expression values measured by IHC. In addition, our results showed a significant association between TOP2A mRNA and protein levels. However, IHC is potentially a more subjective technique and may have a lower precision when assessing the status of TOP2A relative to Q-PCR. Interestingly, no association between TOP2A copy number alterations and the expression of the gene was observed, which is in agreement with previous reports.17,40
Because TOP2A is considered the molecular target of anthracyclines,3 the predictive value of this gene in patients with breast cancer has been widely studied. Several retrospective studies12–16 have shown that ERBB2-positive tumors are rather sensitive to anthracyclines. In addition, amplification of ERBB2 is variable in size. Some flanking genes in the 17q12-q21 region, such as TOP2A, are frequently either coamplified or deleted in breast cancers with ERBB2 amplification.41–43 Because of this coincidence, many researchers have hypothesized that TOP2A amplification might become the gold standard predictive factor for anthracyclines, rather than ERBB2; and many retrospective studies have been conducted to assess its predictive value. However, although several studies19,39,42,44 have shown that TOP2A alterations are associated with an increased responsiveness to anthracycline-containing regimens, others20,45 have been inconclusive.
Our data suggest that TOP2A amplification and TOP2A overexpression are indicative of different biological processes because these two events are not associated and are characteristic of different tumor subtypes. For example, Basal-like tumors typically overexpressed TOP2A protein and/or mRNA, but no Basal-like tumor had TOP2A DNA copy number changes. This might suggest that the predictive value of TOP2A copy number may not be because of a TOP2A protein function. Rather, it may be indicative of some other biological process or genomic aberration that could be characteristic of ERBB2-amplified tumors because TOP2A amplification occurs almost exclusively within ERBB2-amplified tumors. In support of this hypothesis, a previous study,39 performed in a cohort of 245 patients, found that TOP2A amplification predicted benefit from adjuvant anthracyclines only in the subset of ERBB2-positive breast cancer but failed to predict response when the entire population of the study was considered. Similarly, the TOP trial showed that TOP2A amplification, which was always associated with ERBB2 amplification, predicted benefit from regimens containing anthracyclines in ER-negative tumors.46 In both studies, TOP2A expression did not correlate with treatment outcome. Contrary to these results, we recently observed that TOP2A expression is predictive of response to a single agent, doxorubicin, but not TOP2A amplification.47 Because the target of anthracyclines is the TOP2A protein, not the gene, it seems that the study of the expression (RNA and protein) might predict more accurately the treatment outcome with anthracyclines than TOP2A copy number alterations (amplification or deletion); further studies on this point are clearly warranted.
In summary, TOP2A copy number alterations do not correlate with gene expression in breast cancers, and the expression of the gene/protein varies significantly depending on the tumor subtype and proliferation status. These facts could explain, in part, the previously reported discrepancies regarding the predictive value of TOP2A for anthracycline-based chemotherapy regimens.
Footnotes
Supported by grants FIS07/00316 and RTICC 06/0020/0021, Instituto de Salud Carlos III, Spanish Ministry of Science and Innovation, and Fondo Europeo de Desarrollo Regional; NCI Breast SPORE program (P50-CA58223-09A1 to UNC-CH); and a Terry Fox Foundation postdoctoral research fellowship (M.C.U.C.).
A.R. and M.M. contributed equally to this work.
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi:10.1016/j.ajpath.2010.12.042.
Supplementary Data
References
- 1.Ryberg M., Nielsen D., Cortese G., Nielsen G., Skovsgaard T., Andersen P. New insight into epirubicin cardiac toxicity: competing risks analysis of 1097 breast cancer patients. J Natl Cancer Inst. 2008;100:1058–1067. doi: 10.1093/jnci/djn206. [DOI] [PubMed] [Google Scholar]
- 2.Diamandidou E., Buzdar A.U., Smith T.L., Frye D., Witjaksono M., Hortobagyi G.N. Treatment-related leukemia in breast cancer patients treated with fluorouracil-doxorubicin-cyclophosphamide combination adjuvant chemotherapy: the University of Texas M.D.: Anderson Cancer Center experience. J Clin Oncol. 1996;14:2722–2723. doi: 10.1200/JCO.1996.14.10.2722. [DOI] [PubMed] [Google Scholar]
- 3.Tewey K.M., Rowe T.C., Yang L., Halligan B.D., Liu L.F. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984;226:466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- 4.Minotti G., Menna P., Salvatorelli E., Cairo G., Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 5.Osheroff N. Biochemical basis for the interaction of type I and type II topoisomerases with DNA. Pharmacol Ther. 1989;41:233–241. doi: 10.1016/0163-7258(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 6.Wang J.C. DNA topoisomerase. Annu Rev Biochem. 1996;58:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 7.Davies S.M., Robson C.N., Davies S.L., Hickson I.D. Nuclear topoisomerase II levels correlate with the sensitivity of mammalian cells to intercalating agents and epipodophyllotoxins. J Biol Chem. 1988;263:17724–17729. [PubMed] [Google Scholar]
- 8.Asano T., An T., Mayes J., Zwelling L.A., Kleinerman E.S. Transfection of human topoisomerase IIa into etoposide-resistant cells: transient increase in sensitivity followed by down-regulation of the endogenous gene. Biochem J. 1996;319:307–313. doi: 10.1042/bj3190307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Withoff S., Keith W.N., Knol A.J., Coutts J.C., Hoare S.F., Mulder N.H., de Vries E.G. Selection of a subpopulation with fewer DNA topoisomerase II alpha gene copies in a doxorubicin-resistant cell line panel. Br J Cancer. 1996;74:502–507. doi: 10.1038/bjc.1996.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Withoff S., de Vries E.G.E., Keith W.N., Nienhuis E.F., van der Graaf W.T., Uges D.R., Mulder N.H. Differential expression of DNA topoisomerase II alpha and -beta in P-gp and MRP-negative VM26, mAMSA and mitoxantrone-resistant sublines of the human SCLC cell line GLC4. Br J Cancer. 1996;74:1869–1876. doi: 10.1038/bjc.1996.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Järvinen T.A., Tanner M., Rantanen V., Bärlund M., Borg A., Grénman S., Isola J. Amplification and deletion of topoisomerase II associate with HER-2 amplification and affect sensitivity to topoisomerase II inhibitor doxorubicin in breast cancer. Am J Pathol. 2000;156:839–847. doi: 10.1016/s0002-9440(10)64952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thor A.D., Berry D.A., Budman D.R., Muss H.B., Kute T., Henderson I.C., Barcos M., Cirrincione C., Edgerton S., Allred C., Norton L., Liu E.T. ErbB-2, p53, and efficacy of adjuvant therapy in lymph node-positive breast cancer. J Natl Cancer Inst. 1998;90:1346–1360. doi: 10.1093/jnci/90.18.1346. [DOI] [PubMed] [Google Scholar]
- 13.Paik S., Bryant J., Park C., Fisher B., Tan-Chiu E., Hyams D., Fisher E.R., Lippman M.E., Wickerham D.L., Wolmark N. erbB-2 and response to doxorubicin in patients with axillary lymph node-positive, hormone receptor-negative breast cancer. J Natl Cancer Inst. 1998;90:1361–1370. doi: 10.1093/jnci/90.18.1361. [DOI] [PubMed] [Google Scholar]
- 14.Paik S., Bryant J., Tan-Chiu E., Yothers G., Park C., Wickerham D.L., Wolmark N. ERBB2 and choice of adjuvant chemotherapy for invasive breast cancer: National Surgical Adjuvant Breast and Bowel Project Protocol B-15. J Natl Cancer Inst. 2000;92:1991–1998. doi: 10.1093/jnci/92.24.1991. [DOI] [PubMed] [Google Scholar]
- 15.Pritchard K.I., Shepherd L.E., O'Malley F.P., Andrulis I.L., Tu D., Bramwell V.H., Levine M.N. ERBB2 and responsiveness of breast cancer to adjuvant chemotherapy. N Engl J Med. 2006;354:2103–2111. doi: 10.1056/NEJMoa054504. [DOI] [PubMed] [Google Scholar]
- 16.Gennari A., Sormani M.P., Pronzato P., Puntoni M., Colozza M., Pfeffer U., Bruzzi P. ERBB2 status and efficacy of adjuvant anthracyclines in early breast cancer: a pooled analysis of randomized trials. J Natl Cancer Inst. 2008;100:14–20. doi: 10.1093/jnci/djm252. [DOI] [PubMed] [Google Scholar]
- 17.Petit T., Wilt M., Velten M., Millond R., Rodier J.F., Borel C., Mors R., Haegelé P., Eber M., Ghnassia J.P. Comparative value of tumour grade, hormonal receptors, Ki-67, HER-2 and topoisomerase II alpha status as predictive markers in breast cancer patients treated with neoadjuvant anthracycline-based chemotherapy. Eur J Cancer. 2004;40:205–211. doi: 10.1016/s0959-8049(03)00675-0. [DOI] [PubMed] [Google Scholar]
- 18.Sparano J.A., Goldstein L., Childs B., Shak S., Brassard D., Badve S., Baehner F.L., Bugarini R., Rowley S., Perez E., Shulman L.N., Martino S., Davidson N.E., Sledge G.W., Gray R. Relationship between topoisomerase 2A RNA expression and recurrence after adjuvant chemotherapy for breast cancer. Clin Cancer Res. 2009;15:7693–7700. doi: 10.1158/1078-0432.CCR-09-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Malley F.P., Chia S., Tu D., Shepherd L.E., Levine M.N., Bramwell V.H., Andrulis I.L., Pritchard K.I. Topoisomerase II alpha and responsiveness of breast cancer to adjuvant chemotherapy. J Natl Cancer Inst. 2009;101:644–650. doi: 10.1093/jnci/djp067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris L.N., Broadwater G., Abu-Khalaf M., Cowan D., Thor A.D., Budman D., Cirrincione C.T., Berry D.A., Winer E.P., Hudis C.A., Hayes D.F., Friedman P., Ellis M., Dressler L. Topoisomerase II{alpha} amplification does not predict benefit from dose-intense cyclophosphamide, doxorubicin, and fluorouracil therapy in ERBB2-amplified early breast cancer: results of CALGB 8541/150013. J Clin Oncol. 2009;27:3430–3436. doi: 10.1200/JCO.2008.18.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuccari G., Rizzo A., Giuffrè G., Barresi G. Immunocytochemical detection of DNA topoisomerase type II in primary breast carcinomas: correlation with clinico-pathological features. Virchows Arch A Pathol Anat Histopathol. 1993;423:51–55. doi: 10.1007/BF01606432. [DOI] [PubMed] [Google Scholar]
- 22.Lynch B.J., Guinee D.G., Jr, Holden J.A. Human DNA topoisomerase II-alpha: a new marker of cell proliferation in invasive breast cancer. Hum Pathol. 1997;28:1180–1188. doi: 10.1016/s0046-8177(97)90256-2. [DOI] [PubMed] [Google Scholar]
- 23.Nakopoulou L., Lazaris A.C., Kavantzas N. DNA topoisomerase II alfa inmunoreactivity as a marker of aggressiveness in invasive breast cancer. Pathobiology. 2000;68:137–143. doi: 10.1159/000055914. [DOI] [PubMed] [Google Scholar]
- 24.Parker J.S., Mullins M., Cheang M.C., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z., Quackenbush J.F., Stijleman I.J., Palazzo J., Marron J.S., Nobel A.B., Mardis E., Nielsen T.O., Ellis M.J., Perou C.M., Bernard P.S. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prat A., Parker J.S., Karginova O., Fan C., Livasy C., Herschkowitz J.I., He X., Perou C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreuzer K.A., Lass U., Landt O., Nitsche A., Laser J., Ellerbrok H., Pauli G., Huhn D., Schmidt C.A. Highly sensitive and specific fluorescence reverse transcription-PCR assay for pseudogene-free detection of beta-actin transcripts as quantitative reference. Clin Chem. 1999;45:297–300. [PubMed] [Google Scholar]
- 27.Elston C.W., Ellis I.O. Pathological prognostic factors in breast cancer, I: the value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 28.Hellemans P., van Dan P.A., Geysken M., Oosterom A.T., Buytaert P.H., van Marck E. Immunohistochemical study of topoisomerase II α expression in primary ductal carcinoma of the breast. J Clin Pathol. 1995;48:147–150. doi: 10.1136/jcp.48.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Järvinen T.A., Kononen J., Pelto-Huikko M., Isola J. Expression of topoisomerase II alpha is associated with rapid cell proliferation, aneuploidy, and c-ERBB2 overexpression in breast cancer. Am J Pathol. 1996;148:2073–2082. [PMC free article] [PubMed] [Google Scholar]
- 30.Boege F., Andersen A., Jensen S., Zeidler R., Kreipe H. Proliferation-associated nuclear antigen ki-64 is identical with topoisomerase IIα: delineation of a carboxyl-terminal epitope with peptide antibodies. Am J Pathol. 1996;146:1302–1308. [PMC free article] [PubMed] [Google Scholar]
- 31.Shvero J., Koren R., Shvili I., Yaniv E., Sadov R., Hadar T. Expression of human DNA topoisomerase II-alpha in squamous cell carcinoma of the larynx and its correlation with clinicopathologic variables. Am J Clin Pathol. 2008;130:934–939. doi: 10.1309/AJCPROG61USKCBEI. [DOI] [PubMed] [Google Scholar]
- 32.Wong N., Yeo W., Wong W.L., Wong N.L., Chan K.Y., Mo F.K., Koh J., Chan S.L., Chan A.T., Lai P.B., Ching A.K., Tong J.H., Ng H.K., Johnson P.J., To K.F. TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer. 2009;124:644–652. doi: 10.1002/ijc.23968. [DOI] [PubMed] [Google Scholar]
- 33.Perou C.M., Sørlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., Fluge O., Pergamenschikov A., Williams C., Zhu S.X., Lønning P.E., Børresen-Dale A.L., Brown P.O., Botstein D. Molecular portraits of human breast tumors. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 34.Carey L.A., Perou C.M., Livasy C.A., Dressler L.G., Cowan D., Conway K., Karaca G., Troester M.A., Tse C.K., Edmiston S., Deming S.L., Geradts J., Cheang M.C., Nielsen T.O., Moorman P.G., Earp H.S., Millikan R.C. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 35.Rouzier R., Perou C.M., Symmans W.F., Ibrahim N., Cristofanilli M., Anderson K., Hess K.R., Stec J., Ayers M., Wagner P., Morandi P., Fan C., Rabiul I., Ross J.S., Hortobagyi G.N., Pusztai L. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–5685. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 36.Durbecq V., Paesmans M., Cardoso F., Desmedt C., Di Leo A., Chan S., Friedrichs K., Pinter T., Van Belle S., Murray E., Bodrogi I., Walpole E., Lesperance B., Korec S., Crown J., Simmonds P., Perren J.T., Leroy J., Rouas G., Sotiriou C., Piccart M., Larsimon D. Topoisomerase-II alpha expression as a predictive marker in a population of advanced breast cancer patients randomly treated either with single-agent doxorubicin or single-agent docetaxel. Mol Cancer Ther. 2004;3:1207–1214. [PubMed] [Google Scholar]
- 37.Di Leo A., Larsimont D., Gancberg D., Jarvinen T., Beauduin M., Vindevoghel A., Michel J., Focan C.H., Ries F., Gobert P.H., Closon-Dejardin M.T., Dolci S., Rouas G., Paesmans M., Lobelle J.P., Isola J., Piccart M.J. HER-2 and topo-isomerase IIα as predictive markers in a population of node-positive breast cancer patients randomly treated with adjuvant CMF or epirubicin plus cyclophosphamide. Ann Oncol. 2001;12:1081–1089. doi: 10.1023/a:1011669223035. [DOI] [PubMed] [Google Scholar]
- 38.Cardoso F., Di Leo A., Larsimont D., Gancberg D., Rouas G., Dolci S., Ferreira F., Paesmans M., Piccart M. Evaluation of HER-2, p53, bcl-2, topoisomerase II-α, heat shock proteins 27 and 70 in primary breast cancer and metastatic ipsilateral axillary lymph nodes. Ann Oncol. 2001;12:615–620. doi: 10.1023/a:1011182524684. [DOI] [PubMed] [Google Scholar]
- 39.Arriola E., Rodriguez-Pinilla S.M., Lambros M.B., Jones R.L., James M., Savage K., Smith I.E., Dowsett M., Reis-Filho J.S. Topoisomerase II alpha amplification may predict benefit from adjuvant anthracyclines in HER2 positive early breast cancer. Breast Cancer Res Treat. 2007;106:181–189. doi: 10.1007/s10549-006-9492-5. [DOI] [PubMed] [Google Scholar]
- 40.Muller R.E., Parkers R.K., Andrulis I., O'Malley F.P. Amplification of the TOP2A gene does not predict high levels of topoisomerase II alpha protein in human breast tumor samples. Genes Chromosomes Cancer. 2004;39:288–297. doi: 10.1002/gcc.20008. [DOI] [PubMed] [Google Scholar]
- 41.Olsen K.E., Knudsen H., Rasmussen B.B., Balslev E., Knoop A., Ejlertsen B., Nielsen K.V., Schönau A., Overgaard J; Danish Breast Cancer Co-operative Group. Amplification of ERBB2 and TOP2A and deletion of TOP2A genes in breast cancer investigated by new FISH probes. Acta Oncol. 2004;43:35–42. doi: 10.1080/02841860310019007. [DOI] [PubMed] [Google Scholar]
- 42.Knoop A.S., Knudsen H., Balslev E., Rasmussen B.B., Overgaard J., Nielsen K.V., Schonau A., Gunnarsdóttir K., Olsen K.E., Mouridsen H., Ejlertsen B., Danish Breast Cancer Cooperative Group Retrospective analysis of topoisomerase IIa amplifications and deletions as predictive markers in primary breast cancer patients randomly assigned to cyclophosphamide, methotrexate, and fluorouracil or cyclophosphamide, epirubicin, and fluorouracil: Danish Breast Cancer Cooperative Group. J Clin Oncol. 2005;23:7483–7490. doi: 10.1200/JCO.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 43.Park K., Han S., Gwak G.H., Kim H.J., Kim J., Kim K.M. Topoisomerase II-alpha gene deletion is not frequent as its amplification in breast cancer. Breast Cancer Res Treat. 2006;98:337–342. doi: 10.1007/s10549-006-9170-7. [DOI] [PubMed] [Google Scholar]
- 44.Scandinavian Breast Group Trial 9401. Tanner M., Isola J., Wiklund T., Erikstein B., Kellokumpu-Lehtinen P., Malmström P., Wilking N., Nilsson J., Bergh J. Topoisomerase IIalpha gene amplification predicts favorable treatment response to tailored and dose-escalated anthracycline-based adjuvant chemotherapy in HER-2/neu-amplified breast cancer: Scandinavian Breast Group Trial 9401. J Clin Oncol. 2006;24:2428–2436. doi: 10.1200/JCO.2005.02.9264. [DOI] [PubMed] [Google Scholar]
- 45.Hannemann J., Kristel P., van Tinteren H., Bontenbal M., van Hoesel Q.G., Smit W.M., Nooij M.A., Voest E.E., van der Wall E., Hupperets P., de Vries E.G., Rodenhuis S., van de Vijver M.J. Molecular subtypes of breast cancer and amplification of topoisomerase IIa: predictive role in dose intensive adjuvant chemotherapy. Br J Cancer. 2006;95:1334–1341. doi: 10.1038/sj.bjc.6603449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desmedt C, Azambuja E, Larsimont D, Haibe-Kains FL, Rouas G, Selleslags J, Delaloge S, Duhem C, Kains J-P, Carly B, Maerevoet M, Vindevoghel A, Cardoso F, Durbecq V, Nogaret J-M, Veys I, Scobbens J-C, Noterman D, Salgado R, Di Leo, A, D'Hondt V, Piccart M, Sotiriou C: Predicting the efficacy of anthracyclines in breast cancer (BC): The results of the TOP trial and their validation in the BIG 00-01 trial. Abstract 1893. Presented at the AACR Annual Meeting, April 18–22, 2009, Denver, CO
- 47.Chia SK, Ung K, Bramwekk VH, Tu D, Perou CM, Ellis MJ, Pernard PS, Vickery T, Shepherd LE, Nielsen TO: Prognostic and Predictive impact of biologic classification by qRT-PCR with a 50-gene subtype predictor (PAM50) for adjuvant tamoxifen in premenopausal breast cancer: Results from the NCIC CTG MA.12 randomized trial. Abstract 508. Presented at the ASCO Annual Meeting, June 4–8, 2010, Chicago, IL
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


