Abstract
Previously, we identified the calcium-activated nucleotidase 1 (CANT1) transcript as up-regulated in prostate cancer. Now, we studied CANT1 protein expression in a large cohort of nearly 1000 prostatic tissue samples including normal tissue, prostatic intraepithelial neoplasia (PIN), primary carcinomas, metastases, and castrate-resistant carcinomas, and further investigated its functional relevance. CANT1 displayed predominantly a Golgi-type immunoreactivity with additional and variable cytoplasmic staining. In comparison to normal tissues, the staining intensity was significantly increased in PIN lesions and cancer. In cancer, high CANT1 levels were associated with a better prognosis, and castrate-resistant carcinomas commonly showed lower CANT1 levels than primary carcinomas. The functional role of CANT1 was investigated using RNA interference in two prostate cancer cell lines with abundant endogenous CANT1 protein. On CANT1 knockdown, a significantly diminished cell number and DNA synthesis rate, a cell cycle arrest in G1 phase, and a strong decrease of cell transmigration rate and wound healing capacity of CANT1 knockdown cells was found. However, on forced CANT1 overexpression, cell proliferation and migration remained unchanged. In summary, CANT1 is commonly overexpressed in the vast majority of primary prostate carcinomas and in the precursor lesion PIN and may represent a novel prognostic biomarker. Moreover, this is the first study to demonstrate a functional involvement of CANT1 in tumor biology.
Prostate cancer is the most common malignancy in men in western countries,1–2 and its carcinogenesis is still incompletely understood. For the development of more efficient therapies, elucidating the molecular processes of prostate cancer progression is of primordial importance. Furthermore, biomarkers that facilitate the diagnosis of prostate cancer at an early stage and allow the differentiation between insignificant and potentially aggressive carcinomas are urgently needed. Previously, we have conducted an array-based transcript analysis of matched normal tissue and prostate cancer to identify differentially expressed genes as candidates for further research. Among the top up-regulated genes in prostate cancer was calcium-activated nucleotidase 1 (CANT1), which has hitherto not been characterized further in human neoplasias. In a multitissue screen, Smith et al3 described expression of CANT1 mRNA in various organs being strongest in testis, placenta, small intestine, and prostate. The CANT1 protein acts as apyrase and hydrolyzes di- and triphosphates in a calcium-dependent manner, preferably UDP, GDP, and UTP.3–4 Since CANT1 is androgen-regulated,5 its analysis in a primarily androgen-dependent tumor is of particular interest and, to our knowledge, has not been conducted so far.
The objective of this study was to clarify the diagnostic and prognostic properties and the functional role of CANT1 in prostate cancer. CANT1 protein expression was analyzed in two independent cohorts of clinically characterized human prostate cancer cases, together representing nearly 1000 patients. A recurrent overexpression of CANT1 protein in human prostate cancer tissues and already in prostatic intraepithelial neoplasia (PIN) lesions is demonstrated, indicating that CANT1 up-regulation might be an early event during prostate carcinoma development. A subtle analysis of CANT1 overexpression further demonstrated that very high expression rates in carcinomas correlates with better patient prognosis. Cell culture studies did not reveal an increase of proliferative or migratory capacity on CANT1 overexpression. However, we show that CANT1 knockdown leads to a reduced cell proliferation and migration rate of prostate cancer cell lines and thus constitute for the first time a functional relevance of CANT1 in prostate carcinomas.
Materials and Methods
Patients and Tissue Microarray Description
Two tissue microarrays (TMAs) were used to follow CANT1 expression during prostate cancer progression and to evaluate the diagnostic and prognostic potential of CANT1 immunohistochemistry.
TMA #1 consists of formalin-fixed paraffin-embedded prostate tissues from 529 patients, who were diagnosed at the Institute of Surgical Pathology, University Hospital Zurich, between 1993 and 2006.6 Clinical follow-up data were available for 201 patients after radical prostatectomy (RPE). Seventy-nine patients (39%) experienced a prostate-specific antigen (PSA) relapse, defined as a rising PSA level exceeding 0.1 ng/mL, having reached a nadir after surgery. The median follow-up time (of all patients) was 67 months. The TMA was constructed as described;6 briefly, each case was represented by a single 0.6-mm core of tissue. A total of 349 of 529 cores were evaluable for CANT1 staining. These cases represent 30 benign hyperplasias of the prostate, 269 primary prostate carcinomas (for demographics see Table 1), and 50 advanced cases that are either derived from lymph node or organ metastases (29) or from castrate-resistant carcinomas (21). This study was approved by the Cantonal Ethics Committee of Zurich (approval number StV 25-2007).
Table 1.
Relationship between CANT1 Expression and Clinicopathological Data of Primary Prostate Carcinoma Specimens (RPE) in Cohort #1
| n = 238 Median (range)/cases (percentage) | Golgi staining |
Cytoplasmic staining |
|||||
|---|---|---|---|---|---|---|---|
| Parameter | CANT1 low | CANT1 high | P value | CANT1 low | CANT1 high | P value | |
| Age | 65 (50–77) | 0.894 | 0.782 | ||||
| ≤64 years | 68 | 46 | 78 | 36 | |||
| >64 years | 76 | 48 | 82 | 42 | |||
| Pre-OP PSA | 12.3 (0.2–209) | 0.669 | 0.026⁎ | ||||
| ≤10 ng/mL | 55 | 35 | 51 | 39 | |||
| >10 ng/mL | 67 | 49 | 84 | 32 | |||
| Gleason score | 0.764 | 0.823 | |||||
| 5–6 | 37 (15.5%) | 21 | 16 | 24 | 13 | ||
| 7 | 138 (58%) | 86 | 52 | 95 | 43 | ||
| 8–10 | 63 (26.5%) | 37 | 26 | 41 | 22 | ||
| pT status | 0.893 | 0.069 | |||||
| pT2 | 142 (59.7%) | 85 | 57 | 102 | 40 | ||
| pT3/4 | 96 (40.3%) | 59 | 37 | 58 | 38 | ||
| Margin status | 0.583 | 0.563 | |||||
| R0 | 150 (63%) | 92 | 58 | 100 | 50 | ||
| R1 | 85 (35.7%) | 49 | 36 | 60 | 25 | ||
Pre-OP PSA, preoperative prostate-specific antigen; RPE, radical prostatectomy.
Statistically significant.
Formalin-fixed, paraffin-embedded prostatectomy specimens from 640 patients who underwent radical prostatectomy between 1999 and 2005 at the Department of Urology, Charité Universitätsmedizin Berlin, were included in TMA #2 (for demographics, see Table 2), which was constructed as described7–9 and approved by the Charité University Ethics Committee (approval number EA1/06/2004) on 20 September 2004. Eighty-five patients (14.6%) experienced a PSA relapse. The median follow-up time of all cases was 47.5 months. Each case is represented by five cores of 2 mm in diameter each, encompassing benign prostatic hyperplasia (BPH) of the transitional zone, normal tissue from the peripheral zone, PIN, if present (otherwise another core from the peripheral zone), and two cores of invasive carcinoma, ideally of primary and secondary Gleason scores. CANT1 staining of normal and cancerous cores was evaluable for 618 patients; the PIN core was evaluable for 467 patients.
Table 2.
Relationship between CANT1 Expression and Clinicopathological Data of Primary Prostate Carcinoma Specimens (RPE) in Cohort #2
| n = 640 Median (range)/cases (percentage) | Golgi staining |
|||
|---|---|---|---|---|
| Parameter | CANT1 low | CANT1 high | P value | |
| Age | 62 (43–74) | 0.273 | ||
| ≤62 years | 196 | 116 | ||
| >62 years | 205 | 100 | ||
| Pre-OP PSA | 7.2 (0.8–39) | 0.701 | ||
| ≤10 ng/mL | 293 | 153 | ||
| >10 ng/mL | 104 | 59 | ||
| Gleason score | 0.054 | |||
| 5–6 | 234 (36.6%) | 136 | 84 | |
| 7 | 293 (45.8%) | 185 | 103 | |
| 8–10 | 112 (17.5%) | 80 | 29 | |
| pT status | 0.014⁎ | |||
| pT2 | 442 (69.1%) | 262 | 162 | |
| pT3/4 | 197 (30.8%) | 139 | 54 | |
| Margin status | 0.569 | |||
| R0 | 463 (72.3%) | 287 | 159 | |
| R1 | 173 (27%) | 113 | 55 | |
pre-OP PSA, preoperative prostate-specific antigen; RPE, radical prostatectomy.
Statistically significant.
Immunohistochemistry and Immunofluorescence
Freshly cut sections (3 μm) from the TMA blocks were analyzed by immunohistochemistry using the Ventana Benchmark automated staining system (Ventana Medical Systems, Tucson, AZ) and Ventana reagents according to the manufacturer's recommendations. Following antigen retrieval using the “CC1 protocol,” the CANT1 antibody (clone 2D3; 1:300; Abnova, Taipei, Taiwan) was detected by the UltraVIEW DAB detection kit. Subsequently, the slides were counterstained with hematoxylin, dehydrated, and then mounted.
Evaluation of CANT1 Immunohistochemistry
On TMA #1, CANT1 staining intensity was evaluated by two clinical pathologists (G.K., F.R.F.) using a scoring system that differentiates between negative (0), weak (1), moderate (2), and strong (3) staining. Since TMA #2 with its multiple cores and the larger diameter per core allowed a better estimation of intratumoral heterogeneity, a more differentiated evaluation scheme was applied in addition to the simpler 0 to 3 rating. A histochemical score (H-score) was set up to include the percentages of weak, moderate, and strong expression and to sum these up in a weighted manner [H-score = (1 × percentage weak) + (2 × percentage moderate) + (3 × percentage strong)]; this evaluation was performed by the pathologists C.S. and G.K.
To prove antibody specificity, two consecutive slides were stained as described above (antibody dilution, 1:1000). The antibody solution for one slide was preincubated with CANT1 recombinant protein (Q1; Abnova) 100 times in excess overnight at 4°C.
Immunofluorescence double staining was performed as described previously7 using the same CANT1 antibody as for immunohistochemistry (1:300) and a GOLPH2 polyclonal rabbit antibody (ab22209; 1:100; Abcam, Cambridge, MA).
Ultracentrifugation and Precipitation of Supernatants
After pelleting the cell debris, the supernatant was transferred to Ultra-Clear centrifuge tubes (Beckmann, Palo Alto, CA), and vesicles were separated by ultracentrifugation in a SW32.1 Ti rotor for 2.5 hours at 24,000 rpm and 4°C. Supernatants were then precipitated by adding trichloroacetic acid to a final concentration of 10% and washing the pellets with ice-cold acetone.
Serological Tests
Serum samples (n = 38) were collected from 23 prostate cancer patients (who gave informed consent) and 15 healthy individuals and submitted to an enzyme-linked immunosorbent assay (ELISA) for detection of serum CANT1. The ELISA was performed as a sandwich ELISA as follows: Maxisorb plates (NUNC, Langenselbold, Germany) were coated overnight at 4°C with 1 μg/mL purified monoclonal mouse immunoglobulin against CANT1 (clone 2D3; Abnova). The following day, plates were washed with PBS and blocked using 10% BSA and 0.5% Tween 20 (Sigma-Aldrich, St. Louis, MO) for 1 hour at room temperature. Samples were diluted 1:10 and applied in triplicate to the plate for 1.5 hours at room temperature. Recombinant CANT1 protein (P01; Abnova) was plated in increasing concentrations (0.05 μg/mL, 0.1 μg/mL, 0.2 μg/mL, 0.5 μg/mL, 1 μg/mL, 2 μg/mL, 10 μg/mL, and 20 μg/mL). Plates were then incubated for 1 hour at room temperature with polyclonal rabbit serum against CANT1 (1:500; Sigma-Aldrich) that was detected by the horseradish peroxidase–linked secondary anti-rabbit antibody (1:4000, for 45 minutes; Sigma-Aldrich). In between each incubation step, plates were extensively washed. Wells were replenished with 100 μL of staining solution (TMBsolution; Pierce Biotechnology, Rockford, IL) per well and left at room temperature in the dark for 20 minutes. The color reaction was stopped by adding 50 μL of 2N H2SO4 to each well. The absorbance at 450 nm was measured using an Emax microplate reader (Molecular Devices, Sunnyvale, CA) and analyzed with Softmax Pro V3.0 software.
Cell Culture
PC-3, DU-145 (both ATCC, Manassas, VA), and LNCaP cells (DSMZ, Braunschweig, Germany) were cultivated in Ham's F-12 medium/Kaighn's Modification, Eagle's Minimal Essential medium, and RPMI-1640 medium, respectively, each supplemented with 10% FBS. RWPE-1 cells (ATCC) were grown in keratinocyte serum-free medium mixed with bovine pituitary extract and human recombinant epidermal growth factor (all cell culture media and supplements from Invitrogen, Carlsbad, CA). All cell lines were cultured at 37°C, 5% CO2, and 100% humidity.
RNA Isolation and Quantitative Real-Time RT-PCR
RNA was isolated using the RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions, followed by cDNA synthesis applying the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Quantitative real-time PCR (QRT-PCR) (primer CANT1: 5′-CTGGGTGTCCAACTACAACG-3′, 5′-ACTCCAGCAGGCAGACTCAT-3′; probe #42) was performed on the 7900HT Fast Real-Time PCR System using TaqMan Mastermix (both Applied Biosystems) and the Universal Probe Library (Roche Applied Science, Mannheim, Germany) according to the recommendations of the manufacturer. Gene expression was normalized to ALAS1 (primer: 5′-TAATGACTACCTAGGAATGAGTCG-3′, 5′-CCATGTTGTTTCAAAGTGTCCA-3′, all primers were from Microsynth, Balgach, Switzerland; probe #43), which is stably expressed in prostate cancer cells.10
Cell Lysis and Western Blot Analysis
Cells were lysed in 60 mmol/L n-Octyl-β-d-glucopyranoside (Sigma-Aldrich) in the presence of protease inhibitors (complete, Mini, EDTA-free, Protease Inhibitor cocktail tablets; Roche Applied Science). Twenty micrograms of cleared lysates were separated by SDS-PAGE and transferred onto a polyvinylidene fluoride membrane (Bio-Rad, Hercules, CA). After blocking with 1% BSA in PBS-Tween, membranes were probed with primary antibodies (CANT1, clone 2D3; Abnova; Actin, MAB1501; Millipore, Billerica, MA; tGFP; OriGene, Rockville, MD) followed by horseradish peroxidase–conjugated anti-mouse secondary antibody (Pierce Biotechnology) incubation and detection via SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnology).
Transient Gene Knockdown and Overexpression
For transient gene knockdown, LNCaP cells were transfected immediately after seeding using HiPerFect transfection reagent (Qiagen, Hilden, Germany) following the FastForward Protocol provided by the manufacturer. PC-3 cells were transfected using Lipofectamine 2000 (Invitrogen) as transfection reagent. Cells were transfected at the day of seeding with a final siRNA concentration of 10 nmol/L (target sequences: unspecific 5'-AATTCTCCGAACGTGTCACGT-3', CANT1 siRNA #1 5'-CCAGATCATTGTGGCCCTCAA-3', CANT1 siRNA #2 5-'ACCCGGAATGGAATGAGTCTA-3'; Qiagen). One day after transfection, the medium of PC-3 cells was changed. Knockdown efficiency was confirmed on the RNA and protein levels using QRT-PCR and Western blot analysis, respectively.
For transient overexpression, RWPE-1 cells were transfected at 80% density with FuGENE 6 transfection reagent (Roche Applied Science) according to the manufacturer's protocol. For 6-cm dishes, 20 μL of transfection reagent and 8 μg of pCMV6-AN-GFP (named GFP) or pCMV6-XL6-CANT1 (named CANT1, both from OriGene) were used.
DNA Synthesis Rate
The DNA synthesis rate was measured based on bromodeoxyuridine (BrdU) incorporation into newly synthesized DNA strands applying the Cell Proliferation ELISA, BrdU kit (Roche Applied Science) according to the manufacturer's instructions. Briefly, cells were seeded in 96-well plates; after 6 hours, BrdU labeling reagent was added to the medium, and cells were incubated at 37°C overnight. Subsequently, cells were fixed and stained with an anti-BrdU antibody, which was detected with substrate solution. The color reaction was stopped by adding sulfuric acid, and the optical density was measured at 450 nm and at 595 nm as reference wavelength using the infinite F200 microplate reader (Tecan, Männedorf, Switzerland).
Cell Cycle Analysis
Cells were detached, washed twice with sample buffer (1% glucose in PBS), and then fixed with 70% ethanol overnight at 4°C. The next day, cells were incubated with 50 μg/mL propidium iodide (Sigma-Aldrich), 0.3 mg/mL RNaseA (Qiagen), and 0.05% Triton X-100 (Sigma-Aldrich) in sample buffer and analyzed by flow cytometry. Proportion of cells in G1, S, and G2/M phase was determined by applying the Dean/Jett/Fox model provided by the FlowJo 6.3 software.
Transmigration Assay
Haptotactic cell migration was analyzed in a modified Boyden chamber assay. Transwell chambers (Corning, Corning, NY) were coated on the bottom with 10 μg/mL fibronectin (Roche Applied Science) for 2 hours at room temperature. The lower chamber was filled with serum-free RPMI-1640 medium supplemented with 0.5% BSA (Sigma-Aldrich) for transmigration of LNCaP and PC-3 cells; keratinocyte serum-free medium without supplements with 0.5% BSA was used in case of RWPE-1 cells. Cells were seeded in the upper chamber in the corresponding medium. After 24 hours incubation at 37°C, nonmigrated cells were removed with a cotton swab, and the remaining cells were fixed with methanol/acidic acid and stained with DAPI (Sigma-Aldrich). Pictures were taken at nine defined places of the membrane and analyzed with ImageJ 1.4 software. The migration rate was quantified by determining the area of the membrane that was covered with stained nuclei.
Scratch Wound Assay
Cells were seeded in 6-cm cell culture dishes and grown to 95% density, before they were transfected with siRNAs. A wound was introduced into the cell monolayer using a pipette tip, and pictures were taken every 24 hours. The area of the wound was measured using ImageJ 1.4 software.
Statistical Analysis
Mean CANT1 staining intensities were compared between different groups of tumor stage using the Mann-Whitney test in cohort #1 and the Wilcoxon matched-pairs test in cohort #2. For analysis of the association of staining intensity with clinicopathological parameters, cross tables were calculated (χ2 test, two-tailed Fisher's exact test). Univariate survival analyses were conducted according to Kaplan-Meier (log-rank test). These statistics were calculated with PASW18 (SPSS, Chicago, IL). Mean serum ELISA levels were compared with help of the two-tailed unpaired t-test.
The cell culture results are displayed as mean ± SD. Each experiment was repeated at least three times if not otherwise indicated, and significance was tested in a two-tailed paired or unpaired t-test, if necessary, with Welch's correction, depending on the dataset.
Results
CANT1 Protein Is Overexpressed in Human Prostate Cancers
Previously, we have reported an overexpression of CANT1 mRNA in human prostate carcinoma tissue compared to adjacent normal tissue.11 To confirm the results on protein level and to evaluate the diagnostic and prognostic potential of CANT1 expression, two TMAs representing 989 patients were stained for CANT1. Before, the specificity of the CANT1 monoclonal antibody was verified by preincubation with CANT1 recombinant protein, which led to the complete blocking of the immunohistochemical staining (see Supplemental Figure S1 at http://ajp.amjpathol.org).
In general, CANT1 immunoreactivity was detected in the cytoplasm apically, close to the nucleus, and resembling a Golgi apparatus staining pattern (Figure 1A). Additionally, a diffuse cytoplasmic staining was observed in some cases (Figure 1, C and D), which was recorded separately on TMA #1. In normal tissue, CANT1 staining was only detected in secretory epithelia, whereas basal cells were generally negative for CANT1 (Figure 1A).
Figure 1.
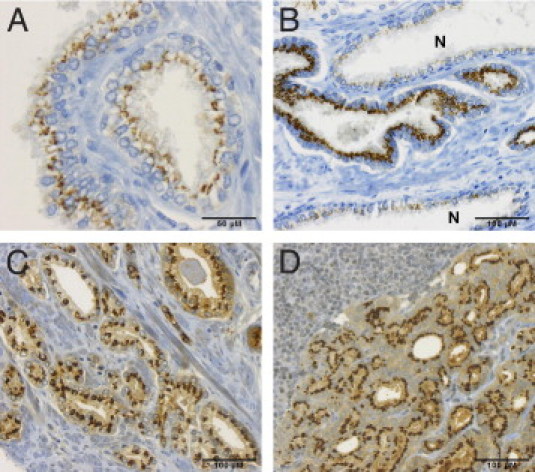
CANT1 immunohistochemistry of human prostatic tissues. Chromogenic immunohistochemistry of human prostatic tissues, magnification ×200 if not otherwise indicated. A: Normal prostate glands, magnification ×400. B: Prostate adenocarcinoma infiltrating inbetween normal glands (N). C: Prostate adenocarcinoma; note the strong diffuse cytoplasmic staining. D: Prostate cancer metastasis in the lung; note the lymphocytes in the upper left corner.
On TMA #1, which contains samples from 30 normal cases, 269 primary carcinomas, 29 metastases, and 21 castrate-resistant prostate carcinomas, CANT1 staining intensity was monitored throughout carcinoma progression. The mean intensity of Golgi and cytoplasmic staining was 1.4 and 0.4 in normal tissue, 2.3 and 1.2 in primary carcinomas, 2.3 and 1.3 in metastases (lymph node metastasis in Figure 1D), and 1.9 and 0.7 in castrate-resistant carcinomas, respectively (Figure 2A). The difference in the mean staining intensity of CANT1 between normal tissues and primary carcinomas, metastases, and all cancerous tissues was highly significant for Golgi staining as well as for cytoplasmic staining. However, only the Golgi staining intensity was significantly different between normal tissues and castrate-resistant carcinomas (Table 3).
Figure 2.
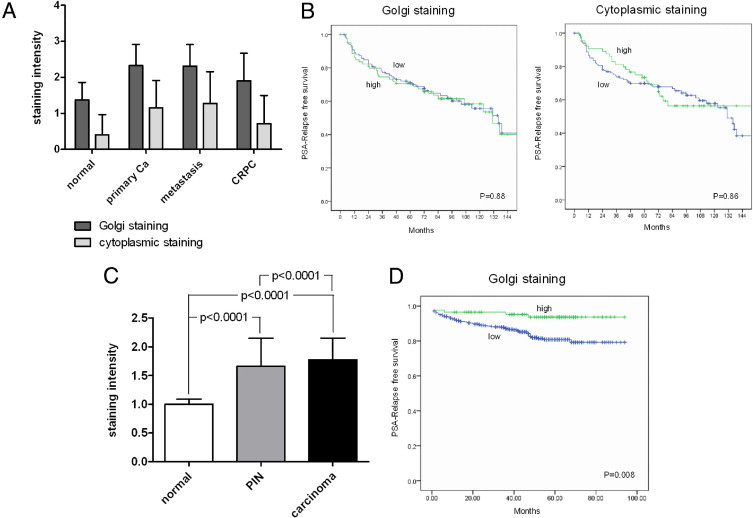
CANT1 staining intensity in human prostatic tissue samples and univariate survival analysis. A: Mean CANT1 staining intensity on TMA #1 during neoplasia progression, P values are put together in Table 3. B: Kaplan-Meier curves of CANT1 Golgi and cytoplasmic expression in cohort #1, dichotomized by the median in low (blue) and high (green) levels. C: Mean CANT1 staining intensity on TMA #2 in normal, prostate intraepithelial neoplastic, and cancer tissue. D: Kaplan-Meier curve of CANT1 Golgi expression in cohort #2, green: upper quartile, blue: lower three quartiles of H-score in cancer tissue. Staining scores: 0 = negative, 1 = weak, 2 = moderate, 3 = strong. Ca = carcinoma; CRPC = castrate-resistant prostate carcinoma; PIN = prostatic intraepithelial neoplasia.
Table 3.
Statistics of Staining Intensities on TMA #1 (P Values)
| Normal | Primary Ca | Metastasis | CRPC | Tumor | |
|---|---|---|---|---|---|
| Golgi staining | |||||
| Normal | — | <0.0001⁎ | <0.0001⁎ | 0.009⁎ | <0.0001⁎ |
| Primary Ca | — | 0.889 | 0.008⁎ | — | |
| Metastasis | — | 0.053 | — | ||
| Cytoplasmic staining | |||||
| Normal | — | <0.0001⁎ | 0.001⁎ | 0.152 | <0.0001⁎ |
| Primary Ca | — | 0.489 | 0.012⁎ | — | |
| Metastasis | — | 0.028⁎ | — |
The category “tumor” includes primary carcinomas, metastases and castrate resistant carcinomas.
Ca, carcinoma; CRPC, castrate-resistant prostate carcinoma.
Statistically significant.
On TMA #2, the mean intensity of CANT1 Golgi staining was 1.0 in the normal cores, 1.7 in the PIN cores, and 1.8 in the carcinoma cores (Figure 2C). In addition, the composition of TMA #2 allowed the direct comparison of CANT1 staining intensity between cancerous and adjacent normal tissue from each patient. To also detect subtle differences between carcinoma and normal cores that would in some cases be assigned the same staining score, it was also reported whether the cancer area stained stronger than adjacent normal glands. This was found in 97.2% of the cases (example in Figure 1B).
Altogether, these data confirm the overexpression of CANT1 in human prostate carcinomas compared to normal prostate epithelium on protein level, which was reported on the mRNA level before. Additionally, it was shown that this up-regulation occurs already in PIN lesions. Further, a slight reduction of CANT1 expression was noted in castrate-resistant carcinomas.
CANT1 Expression, Clinicopathological Associations, and Survival Analysis
For statistical analyses of cohort #1 (cross tables and survival analyses), both staining qualities were dichotomized by the median into low and high groups (Golgi pattern: 0, 1, 2 vs. 3; cytoplasmic pattern: 0, 1 vs. 2, 3). No correlations of Golgi and cytoplasmic CANT1 staining with patient age, Gleason score, pT stage, and margin status were found. However, cytoplasmic CANT1 staining correlated negatively with serum PSA levels (P = 0.026) (Table 1). In cohort #2, CANT1 staining intensity (dichotomized by H-score, 1.9) did not correlate with patient age, serum PSA-levels, and margin status. A trend between high Gleason grade and low H-score was noted, as well as a significant correlation between high pT stage and low H-score (Table 2).
On univariate analysis of PSA relapse-free survival times, no association of CANT1 tissue levels with disease relapse was found on TMA #1, neither for the Golgi nor the cytoplasmic staining (Figure 2B). However, due to the larger cores of TMA #2, it was possible to also consider heterogeneity of CANT1 staining and to derive a more differentiated H-score. A careful analysis of these data revealed a good prognosis of patients with the highest CANT1 expression (the upper quartile), whereas the majority of the patients experienced a PSA relapse much earlier (Figure 2D). However, this survival difference was not detectable when only the simpler overall intensity score (0 to 3) was analyzed (data not shown).
Cellular Localization of CANT1
In transfected CHO cells, the full-length rat homologue of CANT1 localizes to the membranes of the endoplasmic reticulum (ER) and pre-Golgi intermediates,12 whereas a truncated form of human CANT1 nucleotidase was detected in the supernatant of transfected COS-1 cells, indicating cleavage and secretion.3 To analyze the localization of CANT1 in human tissues, double immunofluorescence staining of normal and cancerous prostate tissue with GOLPH2, a known Golgi protein,13 was performed. An extensive overlap of both staining patterns was seen, illustrating that CANT1 is predominantly localized in the Golgi apparatus. An additional diffuse CANT1 staining was observed in the cytoplasm in some cases (Figure 3).
Figure 3.
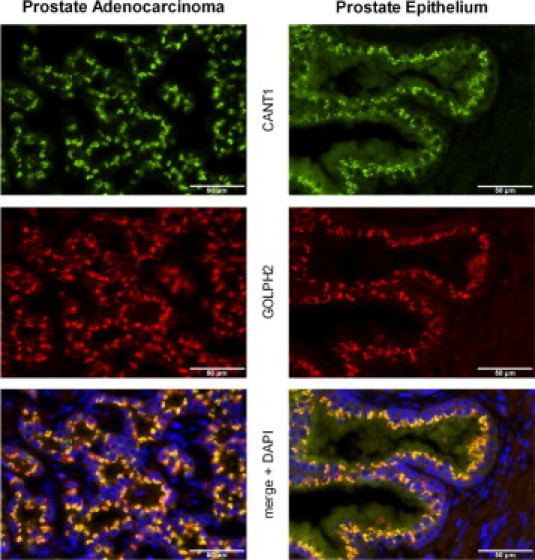
Cellular localization of CANT1. Double immunofluorescence of CANT1 and GOLPH2 in human prostatic tissues: left, prostate adenocarcinoma, right, normal prostate epithelium, magnification ×400.
We next examined whether CANT1 is also secreted by cells that express the protein endogenously. Thus, serum-free supernatants from LNCaP and PC-3 cells were collected and precipitated. The Western blot analysis showed that a shortened form of CANT1, which was approximately 2 kDa smaller compared to the band from the cell lysate (compare lane 2 and 3 to lane 1 in Figure 4A), was detectable in the supernatants of both cell lines before as well as after ultracentrifugation.
Figure 4.
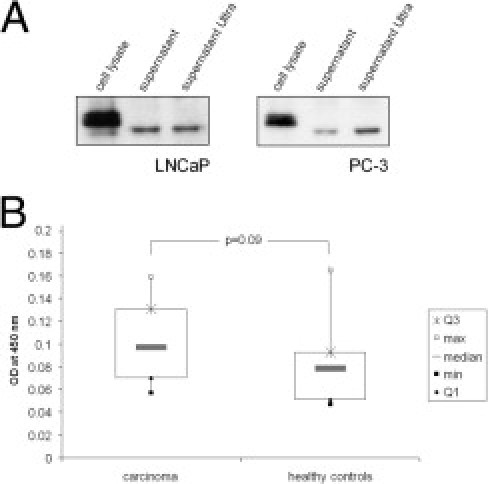
Secretion of CANT1 and ELISA of human sera. A: A total of 600,000 LNCaP and 400,000 PC-3 cells were seeded in 6-cm dishes, at 70% density; complete medium was washed away and replaced by serum-free medium. Twenty-four hours later, supernatants were cleared from cell debris, and afterward, vesicles were pelleted by ultracentrifugation. Soluble CANT1 was precipitated and analyzed by Western blotting together with lysates from the same cells. B: CANT1 was detected by a sandwich ELISA in 38 serum samples (15 healthy controls and 23 prostate cancer patients). Displayed are box plots of the different groups.
CANT1 Is Detectable in an ELISA Assay
The confirmation of the secretion of CANT1 prompted us to establish a sandwich ELISA assay to clarify whether CANT1 is detectable in human serum. When testing commercially available antibodies, we found the combination of mouse monoclonal CANT1 antibody (Abnova) for capturing the antigen and rabbit polyclonal CANT1 antibody (Sigma-Aldrich) for detection to be the most reproducible design. Using this ELISA, serum CANT1 levels appeared to be too low for quantification in this system, with a detection range from 0.1 to 20 μg/mL. Therefore, results are shown as measurement of absorbance at 450 nm.
Serum CANT1 levels in 23 tested patients with prostate cancer were slightly higher than in 15 healthy controls; however, this difference failed to reach statistical significance (median patients: 0.096, median controls: 0.061; Figure 4B). No correlation was noted between serum PSA and serum CANT1 levels (data not shown).
Characterization of CANT1 Expression in Prostatic Cell Lines
We next sought to test whether CANT1 overexpression has a functional relevance in prostate cancer. Therefore, cell lines were evaluated for endogenous CANT1 expression levels and selected for subsequent functional studies. The expression on the gene and protein level was determined in an immortalized prostatic epithelial cell-line, RWPE-1, and in three metastasis-derived prostate cancer cell lines, LNCaP, DU-145, and PC-3. As observed by Western blot analysis, CANT1 protein was strongly expressed in LNCaP and PC-3 cells, whereas it was hardly detectable in DU-145 cells. A clear, but weak, band was detected in the lysate of the epithelium-derived cell line RWPE-1. Similarly, mRNA expression was strongest in LNCaP and PC-3 cells. High levels of CANT1 mRNA were also detected in DU-145 and RWPE-1 cells (Figure 5A). On the basis of the strong CANT1 expression on the mRNA and protein levels, LNCaP and PC-3 cells were chosen for knockdown studies. RWPE-1 cells were chosen for overexpression studies because of the low CANT1 protein level and the noncancerous background.
Figure 5.
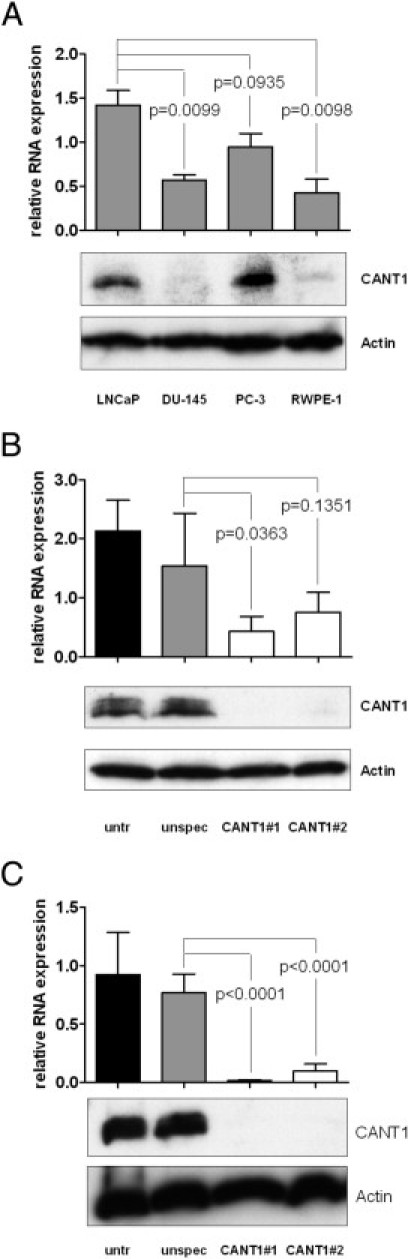
Characterization of CANT1 expression in prostatic cell lines. Cells were cultivated and treated as described below, and CANT1 mRNA and protein expression was determined by QRT-PCR (top) and Western blot analysis (bottom), respectively. A: LNCaP, DU-145, PC-3, and RWPE-1 cells were cultivated in T75 cell culture flasks until a density of 70% to 80%. A total of 300,000 LNCaP (B) and 200,000 PC-3 cells (C) were seeded into 6-well plates, transfected with siRNAs as indicated, and incubated for 72 hours.
A knockdown protocol based on RNA interference was established for LNCaP and PC-3 cells. QRT-PCR and Western blot analysis revealed a potent CANT1 knockdown on the mRNA and protein levels, respectively, 72 hours after siRNA transfection (Figure 5, B and C), the knockdown efficiency being higher with CANT1 siRNA #1 than #2.
The strong reduction of the Western blot band on transfection with both CANT1-specific siRNAs further substantiated the specificity of the CANT1 antibody, which was used as well for immunohistochemistry and in the ELISA assay, and showed that there is no cross-reactivity with other molecules.
CANT1 Knockdown Reduces Cell Proliferation
One of the main characteristics of cancer cells is uncontrolled proliferation.14 To determine whether CANT1 expression has an influence on prostate cancer cell proliferation, LNCaP and PC-3 cells were transfected with CANT1-specific siRNAs, and cells were counted. Compared to unspecific siRNA transfected cells, the cell number of CANT1 siRNA #1 and #2 transfected LNCaP cells was reduced significantly by 17% and 21%, respectively. Although siRNA treatment itself strongly affected the propagation of PC-3 cells, the number of CANT1 siRNA #1 and #2 transfected cells was further decreased by 38% and 26% compared to control cells (Figure 6, A and B). To test whether the reduction of cell number on CANT1 knockdown was caused by a reduction of cell proliferation rate, DNA synthesis rate was determined by measuring BrdU incorporation. As shown in Figure 6C, 32% and 36% less BrdU was detected after transfection of LNCaP cells with CANT1 siRNA #1 and #2, respectively. The DNA synthesis rate of PC-3 cells was decreased highly significantly by 67% and 30% in CANT1 siRNA #1 and #2 transfected cells compared to unspecific siRNA transfected cells (Figure 6D). Taken together, these results show a potent reduction of proliferation of LNCaP and PC-3 cells after CANT1 knockdown.
Figure 6.
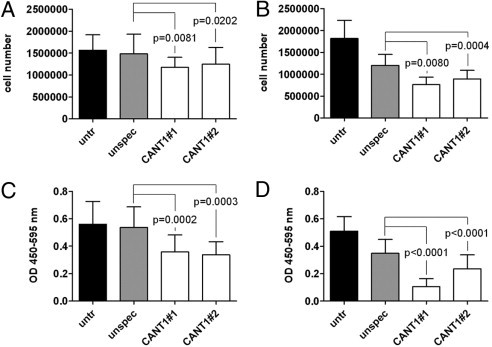
Proliferation of prostate cancer cell lines on CANT1 knockdown. A total of 300,000 LNCaP (A) and 200,000 PC-3 cells (B) were seeded in 6-well plates, transfected with siRNAs as indicated, and counted after 72 hours. A total of 600,000 LNCaP (C) and 400,000 PC-3 cells (D) were seeded in 6-cm dishes and transfected with siRNAs as indicated. After 72 hours, 20,000 LNCaP and 10,000 PC-3 cells were reseeded in quadruplicates into 96-well plates, and DNA synthesis rate was measured.
Reduced Cell Proliferation Is Associated with G1 Arrest
The observed reduction of cell proliferation on CANT1 knockdown involved the question of whether this was reflected in a change of cell cycle distribution. Hence, the proportion of cells in G1, S, and G2/M phase was determined by flow cytometry of propidium iodide–stained cells. The proportion of LNCaP cells in G1 phase increased from 68% in control cells to 76% in CANT1 knockdown cells, whereas the proportion of cells in S phase declined from 20% to 13% in the respective groups; the proportion of cells in G2/M phase remained unchanged (Figure 7, A, C, and E, see Supplemental Figure S2A at http://ajp.amjpathol.org). The differences were even more dramatic in PC-3 cells. Sixty-three percent of the cells treated with unspecific siRNA were in G1 phase in contrast to 81% and 76% of the cells treated with CANT1-specific siRNA #1 and #2, respectively. Different from the LNCaP cells, the proportion of cells in S as well as G2/M phase was altered in PC-3 knockdown cells. S phase cells decreased from 19% in the control cells to 6% and 12% in the knockdown cells; in parallel, G2/M phase cells decreased from 17% to 11% in the corresponding groups (Figure 7, B, D, and F, see Supplemental Figure S2B at http://ajp.amjpathol.org).
Figure 7.
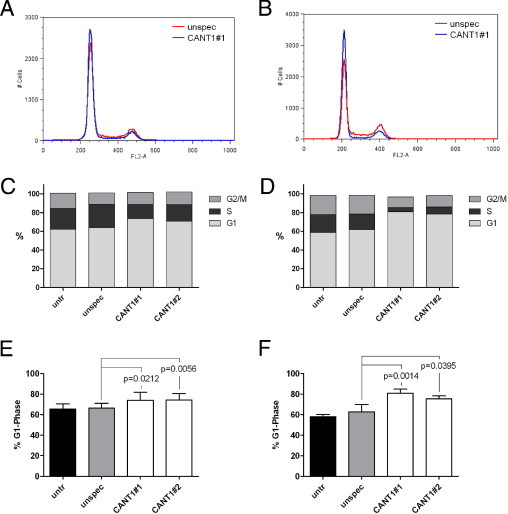
Cell cycle distribution in CANT1 knockdown cells. A total of 500,000 LNCaP cells (A, C, and E) and 200,000 PC-3 cells (B, D, and F) that were left untreated (due to higher proliferation rates, compare Figure 6, A and B) or 600,000 LNCaP cells (A, C, and E) and 400,000 PC-3 (B, D, and F) cells that were transfected with siRNAs as indicated were seeded in 6-cm dishes. After 72 hours, cell cycle analysis was performed. A and B: Representative histograms of cells that were treated either with unspecific siRNA (red) or CANT1 siRNA #1 (blue) are shown; note the higher G1 phase peak and lower S phase plateau and G2 phase peak in the CANT1 siRNA histogram compared to the unspecific siRNA histogram. C and D: Representative distribution of G1, S, and G2/M phase. E and F: Proportion of cells in G1 phase from all replicates.
Summarized, these data clearly indicate a G1 arrest on CANT1 knockdown in LNCaP as well as PC-3 cells.
Cell Migration Is Inhibited on CANT1 Knockdown
An elevated migration rate is another important feature of malignant cells.14 The relevance of CANT1 expression during cell migration was elucidated in a transmigration assay. In particular, migration toward fibronectin was investigated. Down-regulation of CANT1 reduced highly significantly migration of LNCaP cells through the porous membrane by 55% and 20% using siRNA #1 and #2 compared to control cells. The reduction of PC-3 cell migration accounts for 61% and 13% with the respective siRNAs (Figure 8, A and B). Migration of PC-3 cells on a plain surface was assessed in a scratch wound assay. The area, which was not re-covered with migrated cells within 24 hours after introducing the wound, was 1.4 and 1.6 times larger following CANT1 down-regulation with specific siRNA #1 and #2, respectively, compared to unspecific siRNA treatment (Figure 8C), indicating a diminished cell motility. Altogether, these data show that the reduction of CANT1 expression induces strong inhibition of LNCaP and PC-3 cell migration.
Figure 8.
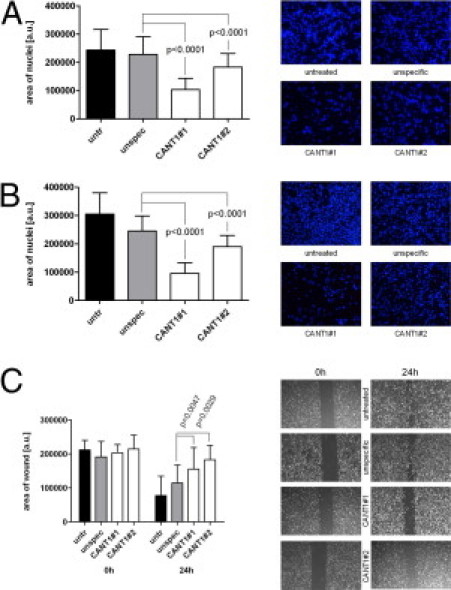
Migration of prostate cancer cell lines on CANT1 knockdown. A total of 600,000 LNCaP (A) and 400,000 PC-3 cells (B) were seeded in 6-cm dishes and transfected with siRNAs as indicated. After 72 hours, the transmigration assay was performed using 50,000 cells. On the right, representative pictures of the DAPI-stained membranes are displayed (magnification ×100). C: A total of 400,000 PC-3 cells were seeded in 6-cm dishes, and the scratch wound assay was conducted. On the right, representative pictures of the wounds are shown (magnification ×25).
CANT1 Overexpression Influences Neither Cell Proliferation nor Cell Migration
To check whether enforced expression of CANT1 in contrast to CANT1 knockdown increases the proliferative and migratory capacity of prostatic cells, CANT1 was transiently transfected into the prostate epithelium-derived, benign cell line RWPE-1, and DNA synthesis rate and transmigration were measured. An increasing expression level of CANT1 was reached by transfecting the cells with a GFP vector as negative control, with a mixture of GFP and CANT1 vector, which consisted of 33% CANT1 vector and 67% GFP vector to keep the DNA content constant, and with pure CANT1 vector. However, neither the DNA synthesis rate nor the transmigration rate of RWPE-1 cells changed on CANT1 overexpression (Figure 9). Similar results were obtained with the prostate cancer cell line DU-145, which expresses very low levels of CANT1 as well as with PC-3 cells, where a further increase of CANT1 expression was achieved by transient transfection (data not shown). These results show that increased expression of CANT1 does not increase the tumorigenic potential of prostatic cell lines.
Figure 9.
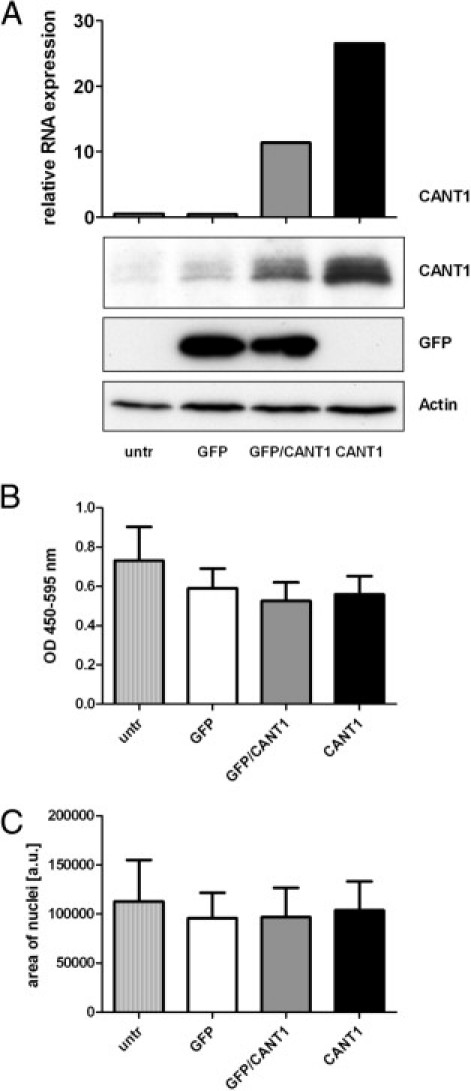
Proliferation and migration of a prostate epithelium cell line on CANT1 overexpression. A total of 1.2 × 106 RWPE-1 cells were seeded in 6-cm dishes and cultured for 3 days before transfection with GFP or CANT1 vector as indicated; the GFP/CANT1 mixture contained 5.33 μg of GFP vector and 2.67 μg of CANT1 vector. A: The day after transfection CANT1 mRNA expression was measured by QRT-PCR (top, one measurement), and protein expression of CANT1 and GFP was determined by Western blot analysis (bottom, representative blot from three independent experiments), respectively. B: The day after transfection 20,000 cells were reseeded in quadruplicates into 96-well plates and DNA synthesis rate was measured. C: The day after transfection, the transmigration assay was performed using 100,000 cells.
Discussion
Although most tumors of the prostate are usually slowly growing, and the majority of patients can be cured by either surgery or radiotherapy,15 a significant fraction of patients experiences disease progression. For these, treatment options are limited, and the survival rates for late advanced, castrate-resistant prostate carcinomas are low.16–19 To address these challenges, intensive research on reliable disease markers and elucidation of disease development on the molecular level is necessary. Previously, we identified CANT1 as a highly up-regulated gene in human prostate cancer compared to adjacent normal tissue.11 Further, CANT1 gene expression has been shown to be significantly higher in tumors of the prostate compared to 28 other tumor entities.20 However, the clinical significance of CANT1 expression in prostate cancer tissues and the functional significance of CANT1 expression in cancer cells have not been investigated yet. Here, we report the first comprehensive analysis, to our knowledge, of CANT1 protein expression in prostatic tissues and cell lines. Main findings include a consistent overexpression of CANT1 protein in human prostate cancer that is already detectable in PIN lesions, a good prognosis for patients with very high rates of CANT1 in prostate cancer following prostatectomy and a reduction of cell proliferation and migration on CANT1 down-regulation in vitro, clearly showing a tumorbiological relevance of CANT1.
After development of a valid immunohistochemistry protocol to allow specific detection of CANT1, nearly 1000 prostate tissue samples were analyzed in a TMA format as described.6–9 CANT1 immunoreactivity was detected in the Golgi apparatus region as well as in the cytoplasm. Evaluation of TMA #1 showed ubiquitous CANT1 expression in normal prostate epithelial tissue; however, the staining intensities of the Golgi apparatus and the cytoplasm were generally stronger in carcinomas, thus suggesting CANT1 as a candidate diagnostic marker for prostate carcinomas. Slightly lower levels of CANT1 were noted in castrate-resistant prostate carcinomas. The unique design of TMA #2 with matching normal and cancer tissues allowed the individual evaluation of the diagnostic value of CANT1. Here, the up-regulation of CANT1 in carcinomas could be convincingly confirmed. Moreover, the direct case comparison of cancerous and adjacent normal glands revealed a gain of CANT1 expression in 97.2% of the cases, clearly substantiating the applicability of CANT1 as potentially helpful ancillary marker to ascertain a cancer diagnosis in a suspicious lesion. Nevertheless, the diagnostic applicability of CANT1 as a prostate cancer marker is hampered by its basal expression in normal glands and its inability to discriminate PIN glands from invasive carcinoma glands.
The prognostic value of CANT1 appears to be limited, since the initial survival analysis of CANT1 expression categorized into negative, weak, moderate, or strong expression did not show significant differences in Kaplan-Meier analyses. However, a significant impact toward better prognosis in cases with strong CANT1 overexpression was noted in the larger study cohort (TMA #2) using a sophisticated semiquantitative evaluation scheme, which included area and intensity to compensate for expression heterogeneity. This more differentiated analysis allowed the separation of the upper quartile in the range of expression values, which was not feasible with the simpler three-tier scoring system applied to cohort #1. This finding was further substantiated by the tendential and significant correlation between lower H-scores and higher Gleason scores and pT stages, respectively. These results nicely fit the observation that CANT1 expression is lower in the aggressive group of castrate-resistant carcinomas compared to primary carcinomas and suggests that progression to castrate resistance is either accompanied or caused by a loss of CANT1.
In accordance with the overexpression of CANT1 in prostate carcinomas, we detected higher CANT1 levels in patient sera compared to age-matched healthy controls using a newly constructed sandwich ELISA assay, although statistical significance was not reached. Unfortunately, serum levels of CANT1 were in the range of the lower detection threshold of our assay (detection range from 0.1 to 20 μg/mL), which precluded exact quantification. Thus, we conclude that ELISA is not the ideal technique for serum measurements of CANT1 and that this protein may not be an ideal biomarker for routine laboratory tests.
In conclusion, we demonstrated a ubiquitous gain of CANT1 expression in prostate carcinomas compared to normal prostate epithelium in two independent cohorts. Yet, on account of the virtually equally intensive up-regulation in PIN, CANT1 is not recommended as a diagnostic tool for the surgical pathologist. However, a detailed analysis of CANT1 staining under consideration of its heterogeneity may help to differentiate between insignificant and potentially aggressive carcinomas at the time of prostatectomy.
The recapitulation of the course of CANT1 expression during prostate cancer progression, being low in normal prostate epithelium and strong already in PIN lesions and remaining more or less constant in primary and advanced carcinomas, suggests that CANT1 up-regulation is an early event. Therefore, it was encouraging to investigate whether CANT1 has a tumor-promoting function. For this, we set up an in vitro system to test cell proliferation and migration behavior in CANT1 high and low cells. To analyze whether CANT1 has any transforming capacities, the prostate epithelial cell line RWPE-1, which expresses low CANT1 protein levels, was transfected with a CANT1 vector. The analysis of cell proliferation rate via DNA synthesis rate measurement as well as the analysis of cell migration rate in a modified Boyden chamber assay revealed no changes on CANT1 overexpression. We next tested whether up-regulation of CANT1 leads to enhanced in vitro tumorigenicity of prostate cancer cells. This was not the case for DU-145 cells that are characterized by very low CANT1 protein levels. Similarly, proliferation and migration rates of PC-3 cells that endogenously express abundant CANT1 protein remained constant on further enhancement of CANT1 expression (data not shown). We further investigated whether down-regulation of CANT1 influences the tumorigenic potential of prostate cancer cells. Interestingly, CANT1 knockdown heavily impaired the function of both PC-3 cells and LNCaP cells, the latter being also characterized by high endogenous CANT1 protein levels. We showed that suppression of CANT1 expression slowed down propagation of LNCaP and PC-3 cells, which was caused by a reduced cell proliferation rate as demonstrated by DNA synthesis rate measurement. Cell cycle distribution analysis revealed that this reduction in turn was caused by an arrest in G1 phase. Consistent with the measurement of DNA synthesis rate, a similar reduction of the S phase cell population was observed in the cell cycle study (compare Figure 7, C and D, and Supplemental Figure S2 at http://ajp.amjpathol.org). Additionally, a decreased motility of both cell lines on CANT1 down-regulation was observed in the transmigration assay, which could be verified for PC-3 cells in the scratch wound assay. The latter assay is less reliably applicable to LNCaP cells, as these do not build a dense monolayer. In most of the assays, the effect of CANT1 siRNA #1 was stronger than the effect of siRNA #2, which is consistent with the stronger reduction of CANT1 expression on transfection with siRNA #1. This dose dependency underlines the specificity of the effects, being indeed related to changes in CANT1 expression. These cell culture studies indicate that CANT1 overexpression alone is not sufficient to induce or enhance in vitro tumorigenicity of prostate epithelial or prostate cancer cells, respectively. However, CANT1 expression is essential to sustain cancer cell proliferation and migration. Thus, CANT1 appears to be relevant for the maintenance of prostate carcinomas.
The questions arise, what leads to CANT1 up-regulation during carcinogenesis and what is the reason for CANT1 decline in castrate-resistant carcinomas? Which mechanisms underlie the diminished tumorigenic potential of prostate cancer cell lines on CANT1 knockdown? Why does the overexpression of CANT1 not induce an increase of in vitro tumorigenicity of prostatic cell lines? To answer these questions, knowledge about CANT1's cellular function would be helpful. However, so far, very little is known about the biology of CANT1. In a study about MAPK and NFκB signaling,21 CANT1 was identified as a novel candidate gene involved in these important cancer-related pathways. This genome-wide study based on a luciferase reporter assay using constructs with promoters of MAPK target genes or promoters that contain NFκB binding sites revealed that CANT1 activates both pathways,21 which are frequently deregulated in tumorigenesis, including prostate cancer.22–26
The main function of CANT1 discussed currently is not related to carcinogenesis in particular, but regards the essential general cellular process of protein glycosylation. In its function as nucleoside-diphosphate (NDP) hydrolase, CANT1 is suggested to be required for a continuous import of NDP-sugars into the ER or Golgi apparatus. The proposed mechanism is that CANT1 hydrolyzes NDP, which is a cleavage by-product after transfer of the sugar to the glycosylated protein. The resulting nucleoside-monophosphate (NMP) is then exported into the cytosol in exchange for a further NDP-sugar molecule.12,27 Depending on its intracellular localization, CANT1 might thus influence three important cellular processes that are related to glycosylation: protein folding in the ER, proteoglycan synthesis in the Golgi apparatus and protein glycosylation, which involves both compartments. The double immunofluorescence stainings clearly indicate a Golgi localization of CANT1; moreover, the diffuse cytoplasmic CANT1 staining, which was observed in these immunofluorescence stainings as well as in the immunohistochemical stainings, might indicate additional ER localization. Various studies dealing with Caenorhabditis elegans, CHO cells, or patients that suffer from a rare type of Desbuquois chondrodysplasia mainly point to an involvement of CANT1 in protein folding and proteoglycan synthesis. APY-1, the C. elegans homologue of CANT1, is induced on ER stress, and knockdown of APY-1 leads to a constitutive unfolded protein response (UPR).27 Additionally, the rat homologue of CANT1 colocalizes with an UPR sensor in the pre-Golgi intermediates when transfected into CHO cells.12 In patients suffering from Desbuquois dysplasia, who frequently display mutations in the CANT1 gene,28 inclusion bodies were detected in distended rough ER,29 which might be a hint for impaired protein folding and subsequent accumulation of misfolded proteins. Hence, by interfering with protein folding, CANT1 knockdown possibly causes the disruption of various cellular processes, including, among others, cell proliferation and migration. It is tempting to speculate that this explains the significantly reduced rate of proliferation and migration, which we observed in prostate cancer cell lines on knockdown of CANT1. A participation of CANT1 in proteoglycan synthesis is substantiated by the finding that bone specimens of Desbuquois dysplasia patients are characterized by a decreased amount of proteoglycans.30 Furthermore, C. elegans APY-1 loss-of-function mutants display pharyngeal alterations resembling mutants defective in proteoglycan synthesis.27 An impaired proteoglycan synthesis on CANT1 down-regulation could imply a miscomposed extracellular matrix and thus affect cell motility. Finally, the influence of CANT1 expression on glycosylation patterns of proteins has not been investigated yet and ought to be clarified in future studies.
In confirmation of Smith et al,3 we demonstrated that a soluble form of CANT1 is secreted, since it was detectable in the supernatant after separation of vesicles by ultracentrifugation. The function of this truncated form of the protein is currently unknown.
To what extent the possible involvement of CANT1 in NFκB and MAPK signaling, in protein folding, proteoglycan synthesis and protein glycosylation refers to the herein described effects remains to be elucidated in future studies. The fact that CANT1 overexpression, in contrast to knockdown, does not influence cell proliferation or migration indicates that CANT1 function is saturable and thus favors a role of CANT1 in glycosylation rather than in NFκB or MAPK signaling.
Another open question is how CANT1 itself is regulated in prostate cancer. Probably, this involves androgen receptor signaling, as CANT1 was shown to be induced on androgen treatment.5 Within this study, the expression of two CANT1 transcripts coding for the same protein was described in the prostate, one of them being prostate specific. Whether CANT1 regulation is transcript specific or whether other mechanisms play a role, will also be a matter of future research. Further, a mutation analysis of CANT1 in human prostate cancers could contribute to explain CANT1 regulation and function.
In summary, this study depicted detailed CANT1 expression patterns in different stages of prostate carcinogenesis. The protein level is low in normal prostate epithelium, increases at the early stage of PIN, remains constant in primary prostate carcinomas, and metastases and decreases to an intermediate level in castrate-resistant carcinomas. Under consideration of heterogeneity, a corresponding correlation between very high CANT1 levels in prostatectomy specimens and good prognosis was revealed. More important, we show that CANT1 expression is involved in prostate cancer cell proliferation and migration and thus demonstrate a functional relevance of CANT1 in a neoplasia. These results contribute to a better understanding of prostate cancer molecular biology and recommend further studies to clarify the mechanisms of CANT1 function and regulation in prostate carcinogenesis.
Acknowledgments
We thank Martina Storz (tissue microarray core facility) for excellent technical assistance and Peter Schraml (tissue biobank) for providing the patient material of cohort #1.
Footnotes
Supported in part by Oncosuisse and Krebsliga Zürich.
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2010.12.046.
Supplementary data
Blocking of immunohistochemistry with recombinant protein. A and B: Consecutive slides of Prostate adenocarcinoma stained with CANT1 antibody, magnification ×25. B. Antibody pre-incubated with CANT1 recombinant protein.
Cell cycle distribution in CANT1 knockdown cells (SPhase). 500,000 LNCaP cells (A) and 200,000 PC-3 cells (B) that were left untreated (due to higher proliferation rates; compare to Figure 6, A and B), or 600,000 LNCaP cells and 400,000 PC-3 cells that were transfected with siRNAs as indicated were seeded in 6-cm dishes. After 72 hours cell cycle analysis was performed as described above. Proportion of cells in S-phase from all replicates is shown.
References
- 1.Boyle P., Levin B., editors. WHO World Cancer Report. IARC; Lyon, France: 2008. http://www.iarc.fr/en/publications/pdfs-online/wcr/ last accessed February 18, 2011. [Google Scholar]
- 2.Jemal A., Siegel R., Ward E., Hao Y., Xu J., Thun M.J. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Smith T.M., Hicks-Berger C.A., Kim S., Kirley T.L. Cloning, expression, and characterization of a soluble calcium-activated nucleotidase, a human enzyme belonging to a new family of extracellular nucleotidases. Arch Biochem Biophys. 2002;406:105–115. doi: 10.1016/s0003-9861(02)00420-4. [DOI] [PubMed] [Google Scholar]
- 4.Murphy D.M., Ivanenkov V.V., Kirley T.L. Bacterial expression and characterization of a novel, soluble, calcium-binding, and calcium-activated human nucleotidase. Biochemistry. 2003;42:2412–2421. doi: 10.1021/bi026763b. [DOI] [PubMed] [Google Scholar]
- 5.Hermans K.G., Bressers A.A., van der Korput H.A., Dits N.F., Jenster G., Trapman J. Two unique novel prostate-specific and androgen-regulated fusion partners of ETV4 in prostate cancer. Cancer Res. 2008;68:3094–3098. doi: 10.1158/0008-5472.CAN-08-0198. [DOI] [PubMed] [Google Scholar]
- 6.Tischler V., Fritzsche F.R., Wild P.J., Stephan C., Seifert H.H., Riener M.O., Hermanns T., Mortezavi A., Gerhardt J., Schraml P., Jung K., Moch H., Soltermann A., Kristiansen G. Periostin is up-regulated in high grade and high stage prostate cancer. BMC Cancer. 2010;10:273. doi: 10.1186/1471-2407-10-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kristiansen G., Fritzsche F.R., Wassermann K., Jager C., Tolle A., Lein M., Stephan C., Jung K., Pilarsky C., Dietel M., Moch H. GOLPH2 protein expression as a novel tissue biomarker for prostate cancer: implications for tissue-based diagnostics. Br J Cancer. 2008;99:939–948. doi: 10.1038/sj.bjc.6604614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritzsche F.R., Stephan C., Gerhardt J., Lein M., Hofmann I., Jung K., Dietel M., Kristiansen G. Diagnostic and prognostic value of T-cell receptor gamma alternative reading frame protein (TARP) expression in prostate cancer. Histol Histopathol. 2010;25:733–739. doi: 10.14670/HH-25.733. [DOI] [PubMed] [Google Scholar]
- 9.Tischler V., Fritzsche F.R., Gerhardt J., Jager C., Stephan C., Jung K., Dietel M., Moch H., Kristiansen G. Comparison of the diagnostic value of fatty acid synthase (FASN) with alpha-methylacyl-CoA racemase (AMACR) as prostatic cancer tissue marker. Histopathology. 2010;56:811–815. doi: 10.1111/j.1365-2559.2010.03535.x. [DOI] [PubMed] [Google Scholar]
- 10.Ohl F., Jung M., Xu C., Stephan C., Rabien A., Burkhardt M., Nitsche A., Kristiansen G., Loening S.A., Radonic A., Jung K. Gene expression studies in prostate cancer tissue: which reference gene should be selected for normalization? J Mol Med. 2005;83:1014–1024. doi: 10.1007/s00109-005-0703-z. [DOI] [PubMed] [Google Scholar]
- 11.Kristiansen G., Pilarsky C., Wissmann C., Kaiser S., Bruemmendorf T., Roepcke S., Dahl E., Hinzmann B., Specht T., Pervan J., Stephan C., Loening S., Dietel M., Rosenthal A. Expression profiling of microdissected matched prostate cancer samples reveals CD166/MEMD and CD24 as new prognostic markers for patient survival. J Pathol. 2005;205:359–376. doi: 10.1002/path.1676. [DOI] [PubMed] [Google Scholar]
- 12.Failer B.U., Braun N., Zimmermann H. Cloning, expression, and functional characterization of a Ca(2+)-dependent endoplasmic reticulum nucleoside diphosphatase. J Biol Chem. 2002;277:36978–36986. doi: 10.1074/jbc.M201656200. [DOI] [PubMed] [Google Scholar]
- 13.Kladney R.D., Bulla G.A., Guo L., Mason A.L., Tollefson A.E., Simon D.J., Koutoubi Z., Fimmel C.J. GP73, a novel Golgi-localized protein upregulated by viral infection. Gene. 2000;249:53–65. doi: 10.1016/S0378-1119(00)00136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 15.Schulz W.A., Burchardt M., Cronauer M.V. Molecular biology of prostate cancer. Mol Hum Reprod. 2003;9:437–448. doi: 10.1093/molehr/gag064. [DOI] [PubMed] [Google Scholar]
- 16.Di Lorenzo G., Autorino R., Figg W.D., De Placido S. Hormone-refractory prostate cancer: where are we going? Drugs. 2007;67:1109–1124. doi: 10.2165/00003495-200767080-00002. [DOI] [PubMed] [Google Scholar]
- 17.Uzzo R.G., Haas N.B., Crispen P.L., Kolenko V.M. Mechanisms of apoptosis resistance and treatment strategies to overcome them in hormone-refractory prostate cancer. Cancer. 2008;112:1660–1671. doi: 10.1002/cncr.23318. [DOI] [PubMed] [Google Scholar]
- 18.Petrylak D.P., Tangen C.M., Hussain M.H., Lara P.N., Jr, Jones J.A., Taplin M.E., Burch P.A., Berry D., Moinpour C., Kohli M., Benson M.C., Small E.J., Raghavan D., Crawford E.D. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351:1513–1520. doi: 10.1056/NEJMoa041318. [DOI] [PubMed] [Google Scholar]
- 19.Tannock I.F., de Wit R., Berry W.R., Horti J., Pluzanska A., Chi K.N., Oudard S., Theodore C., James N.D., Turesson I., Rosenthal M.A., Eisenberger M.A. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 20.Han B., Mehra R., Dhanasekaran S.M., Yu J., Menon A., Lonigro R.J., Wang X., Gong Y., Wang L., Shankar S., Laxman B., Shah R.B., Varambally S., Palanisamy N., Tomlins S.A., Kumar-Sinha C., Chinnaiyan A.M. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer Res. 2008;68:7629–7637. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuda A., Suzuki Y., Honda G., Muramatsu S., Matsuzaki O., Nagano Y., Doi T., Shimotohno K., Harada T., Nishida E., Hayashi H., Sugano S. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene. 2003;22:3307–3318. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- 22.Domingo-Domenech J., Mellado B., Ferrer B., Truan D., Codony-Servat J., Sauleda S., Alcover J., Campo E., Gascon P., Rovira A., Ross J.S., Fernandez P.L., Albanell J. Activation of nuclear factor-kappaB in human prostate carcinogenesis and association to biochemical relapse. Br J Cancer. 2005;93:1285–1294. doi: 10.1038/sj.bjc.6602851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boutros T., Chevet E., Metrakos P. Mitogen-activated protein (MAP) kinase/MAP kinase phosphatase regulation: roles in cell growth, death, and cancer. Pharmacol Rev. 2008;60:261–310. doi: 10.1124/pr.107.00106. [DOI] [PubMed] [Google Scholar]
- 24.Andela V.B., Gordon A.H., Zotalis G., Rosier R.N., Goater J.J., Lewis G.D., Schwarz E.M., Puzas J.E., O'Keefe R.J. NFkappaB: a pivotal transcription factor in prostate cancer metastasis to bone. Clin Orthop Relat Res. 2003:S75–S85. doi: 10.1097/01.blo.0000093048.96273.aa. [DOI] [PubMed] [Google Scholar]
- 25.Uchino R., Madhyastha R., Madhyastha H., Dhungana S., Nakajima Y., Omura S., Maruyama M. NFkappaB-dependent regulation of urokinase plasminogen activator by proanthocyanidin-rich grape seed extract: effect on invasion by prostate cancer cells. Blood Coagul Fibrinolysis. 2010;21:528–533. doi: 10.1097/MBC.0b013e32833a9b61. [DOI] [PubMed] [Google Scholar]
- 26.Shen H.M., Tergaonkar V. NFkappaB signaling in carcinogenesis and as a potential molecular target for cancer therapy. Apoptosis. 2009;14:348–363. doi: 10.1007/s10495-009-0315-0. [DOI] [PubMed] [Google Scholar]
- 27.Uccelletti D., Pascoli A., Farina F., Alberti A., Mancini P., Hirschberg C.B., Palleschi C. APY-1, a novel Caenorhabditis elegans apyrase involved in unfolded protein response signalling and stress responses. Mol Biol Cell. 2008;19:1337–1345. doi: 10.1091/mbc.E07-06-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faden M., Al-Zahrani F., Arafah D., Alkuraya F.S. Mutation of CANT1 causes Desbuquois dysplasia. Am J Med Genet A. 2010;152A:1157–1160. doi: 10.1002/ajmg.a.33404. [DOI] [PubMed] [Google Scholar]
- 29.Huber C., Oules B., Bertoli M., Chami M., Fradin M., Alanay Y., Al-Gazali L.I., Ausems M.G., Bitoun P., Cavalcanti D.P., Krebs A., Le Merrer M., Mortier G., Shafeghati Y., Superti-Furga A., Robertson S.P., Le Goff C., Muda A.O., Paterlini-Brechot P., Munnich A., Cormier-Daire V. Identification of CANT1 mutations in Desbuquois dysplasia. Am J Hum Genet. 2009;85:706–710. doi: 10.1016/j.ajhg.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faivre L., Le Merrer M., Al-Gazali L.I., Ausems M.G., Bitoun P., Bacq D., Maroteaux P., Munnich A., Cormier-Daire V. Homozygosity mapping of a Desbuquois dysplasia locus to chromosome 17q25.3. J Med Genet. 2003;40:282–284. doi: 10.1136/jmg.40.4.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Blocking of immunohistochemistry with recombinant protein. A and B: Consecutive slides of Prostate adenocarcinoma stained with CANT1 antibody, magnification ×25. B. Antibody pre-incubated with CANT1 recombinant protein.
Cell cycle distribution in CANT1 knockdown cells (SPhase). 500,000 LNCaP cells (A) and 200,000 PC-3 cells (B) that were left untreated (due to higher proliferation rates; compare to Figure 6, A and B), or 600,000 LNCaP cells and 400,000 PC-3 cells that were transfected with siRNAs as indicated were seeded in 6-cm dishes. After 72 hours cell cycle analysis was performed as described above. Proportion of cells in S-phase from all replicates is shown.


