Abstract
DNA methylation is a key mechanism for repression of gene expression, including that of α-smooth muscle actin (α-SMA) gene expression in fibroblasts. However, the trans-acting factors that interact with the methylated α-SMA gene to regulate its expression have not been identified. Using gel shift and chromatin immunoprecipitation (ChIP) assays, methyl CpG binding protein 2 (MeCP2) was shown to bind to the α-SMA gene. Suppression of MeCP2 gene expression by siRNA or its deficiency in lung fibroblasts isolated from MeCP2 knockout mice caused significant reduction of α-SMA gene expression. In contrast, transient transfection of MeCP2 expression plasmid into fibroblasts enhanced α-SMA gene expression. Moreover, in vivo studies revealed that compared to their wild type littermates, MeCP2-deficient mice exhibited significantly decreased alveolar wall thickness, inflammatory cell infiltration, interstitial collagen deposition, and myofibroblast differentiation in response to endotracheal injection of bleomycin. Thus, MeCP2 is essential for myofibroblast differentiation and pulmonary fibrosis.
Induction of myofibroblast differentiation is a key feature of wound healing, tissue repair, and remodeling or fibrosis.1–6 The myofibroblast arises de novo at these sites of tissue repair, and is a characteristic cellular component of active tissue remodeling, such as that in the fibroblastic foci of affected lungs from patients with idiopathic pulmonary fibrosis.1–6 The myofibroblast is a major source of the extracellular matrix that is found in these areas of remodeling, as well as cytokines, such as transforming growth factor β (TGFβ).1–6 The myofibroblast's characteristic expression of α-SMA is widely used as a marker for its identification as well as differentiation from precursor cells, such as fibroblasts.1 In the lung the myofibroblast is found to be essential for alveolar development,7 but it is also implicated in pathogenesis of chronic fibrotic lung diseases.6 The myofibroblast is thought to promote fibrosis by engaging in cross talk with adjacent alveolar epithelial cells resulting in heightened production of fibrogenic cytokines and extracellular matrix components, with consequent distortion of normal lung architecture and mechanical properties.1–6 Given these significant developmental and pathogenic roles of the myofibroblast, obtaining insight into the mechanism of its differentiation from precursor cells would be of considerable interest. Recent studies have focused on signaling and complex/combinatorial transcriptional, regulatory mechanisms of the marker gene, α-SMA expression,8–17 but remain incompletely understood especially at the epigenetic level.
Epigenetic regulation of gene expression commonly occurs at two primary levels, namely DNA methylation and modified histone interaction with DNA.18 There is evidence that both modes of epigenetic regulation affect myofibroblast differentiation.19–22 Although inhibition of histone deacetylase is known to suppress myofibroblast differentiation, the molecular mechanism is unclear, especially vis-à-vis regulation of α-SMA gene expression.20–22 With respect to DNA methylation, there is recent evidence of indirect mechanisms affecting expression of the α-SMA gene.23,24 These studies implicate a role for the methyl CpG binding protein 2 (MeCP2), a key member of the methyl-DNA binding protein family of proteins.25–29 While MeCP2 can bind to unmethylated DNA,29 preferentially it binds methylated DNA at the 5′-CpG residues.30–31 MeCP2 is originally considered to be a transcriptional repressor in conjunction with Sin3A and histone deacetylase, but was found later to also have a significant role as a transcriptional activator, as well as in the regulation of chromatin architecture and RNA splicing.32–34 Thus, there is considerable complexity in the possible mechanisms by which MeCP2 can regulate gene expression.
The presence of three CpG islands in the α-SMA gene is recently reported and their methylation is associated with suppression of α-SMA gene expression and thus, the undifferentiated state of the precursor fibroblasts.19 In cells that do not express α-SMA, such as the alveolar epithelial type II cell, intronic regions are also highly methylated.19 This is in contrast to the situation in fibroblasts with the potential to express α-SMA, wherein only the promoter regions were significantly methylated.19 Moreover in fibroblasts, inhibition of DNA methyltransferase (Dnmt) activity or induced deficiency of Dnmt 1, 3a, and/or 3b is sufficient to induce α-SMA expression.19 TGFβ-induced myofibroblast differentiation is associated with reduction in Dnmt1 and Dnmt3a expression, while induced over-expression of all three Dnmts suppress α-SMA expression without complete suppression of TGFβ inducibility.19 Thus, DNA methylation appears to be a key mechanism for maintenance of the undifferentiated state, but the regulatory mechanism associated with gene repression is not clear.
Given that MeCP2 is implicated in mechanisms regulating gene sequences with methylated DNA and the previous identification of methylated DNA in the α-SMA gene, the objective of this study was to investigate the potential role of MeCP2 in the regulation of α-SMA gene expression in lung fibroblasts. The results indicated that MeCP2 preferentially bound the methylated α-SMA gene promoter and enhanced α-SMA gene expression. Suppression of MeCP2 gene expression in lung fibroblasts by siRNA or its deficiency in cells from MeCP2 knock out mice resulted in reduced α-SMA gene expression. Further studies indicated that MeCP2 deficient mice, compared to their wild-type littermates, exhibited significantly decreased alveolar wall thickness, inflammatory cell infiltration, interstitial collagen deposition, and myofibroblast differentiation on endotracheal injection of bleomycin. Thus, an essential role of MeCP2 in myofibroblast differentiation and pulmonary fibrosis was suggested.
Material and Methods
Animals and Cell Culture
All animal care was in accordance with the National Institutes of Health ethics, procedures, and regulations. Pathogen-free female Fisher 344 rats (7 to 8 weeks old) were purchased from Charles River Breeding Laboratories, Inc. (Wilmington, MA). The MeCP2 deficient mice and their wild-type littermates were purchased from the Jackson Laboratory (Bar Harbor, ME).32 Fibroblasts were isolated from mouse and rat lungs by enzymatic digestion and then maintained in Dulbecco's modified Eagle's medium, supplemented with 10% plasma-derived serum (Cocalico Biologicals, Inc., Reamstown, PA), antibiotics; 1% insulin, transferrin, and selenium (Sigma Chemicals, St. Louis, MO); 5 ng/mL platelet-derived growth factor (R&D Systems, Minneapolis, MN); and 10 ng/mL EGF (R&D Systems) as before.8 The adherent cells were then trypsinized and passaged for at least three times before use.
Bleomycin-induced pulmonary fibrosis was induced as previously described.11 The control group received the same volume of sterile phosphate-buffered saline only (saline treated). At 7 or 21 days after bleomycin injection, the mice were sacrificed and the lungs of some of the mice were removed and extracted for total mRNA and total protein, while the remainder of lungs was formalin-fixed and stained for routine histopathology.
Plasmid, Constructs, and Enzymes
The human MeCP2 expression cDNA construct was purchased from B-Bridge International, Inc. (Mt. View, CA). The lentivirus-based siRNA construct specific for rat MeCP2 and the corresponding negative control siRNA construct were purchased from Thermo Scientific (Huntsville, AL). The −2880 to +2803 (numbered from transcription start site, including promoter and first intron) rat α-SMA gene promoter was previously amplified by PCR and cloned into promoterless pGL3-basic vector to form α-SMApro-intron-Luc construct,19 wherein the luciferase reporter gene expression was driven by this promoter. Escherichia coli CpG methyltransferase (M. SssI) was purchased from New England Biolabs, Inc. (Ipswich, MA).
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay was done as previously described.8 Briefly, methylated and unmethylated double-stranded oligonucleotide DNA probe corresponding to the CpG islands in the indicated α-SMA promoter and intronic regions were synthesized and labeled with 32P by T4 polynucleotide kinase. These probes corresponded to the following regions: i) −735 to −646 (numbered from the transcriptional start site) with methylated sequence 5′-AAAAGAmCGGTCCTTAAGCATGATATCAAGGGTCAGmCGATAAACCAACAACATGCAmCGTGGACTGTACCTAAGGGTTAAmCGCAGTTACAGT-3′, ii) site 1 in the first intronic region from +191 to +280 of the α-SMA promoter with methylated sequence 5′-ATGATTTmCGTATCTAAAmCGGGACTAAAAATGAATmCGTGGTTTACTGGCAAAGGAGATGGAGAGGAAATTAAAGTTTGTTCATGmCGTGGCA-3′, and iii) site 2 in the first intronic region from +444 to +533 of the α-SMA promoter with methylated sequence 5′-ACmCGAGTACAGCmCGGGTTAACTGGAAGTGGATGTCAGGAGTGAACTGGmCGmCGGTTGCCTGmCGCTCTGGTTTTGGCTGAGTGGACTGmCGTT-3′. They were individually incubated with MeCP2 (Abnova, Walnut, CA) at the indicated concentrations, 100 ng of Poly dI-dC and 0.1 μg poly-L-lysine in a final volume of 25 μl in Dignam's Buffer C (20 mmol/L HEPES, pH 7.9, 0.42 M NaCl, 1 mmol/L EDTA, 0.1 mmol/L EGTA, 1.0 mmol/L DTT, 100 μmol/L sodium orthovanadate and protease inhibitors). Samples were then analyzed by electrophoresis on 4% nondenaturing polyacrylamide gels at 150 V in 1 × Tris-borate-EDTA buffer (90 mM Tris, 90 mM boric acid, 2 mM EDTA, pH. 8.0) for about 2 hours. Following electrophoresis, the gels were dried and exposed to X-ray film for 24 to 48 hours.
ChIP Assay
Chromatin immunoprecipitation (ChIP) assay was performed using a kit from Millipore Co. (Billerica, MA) following the manufacturer's protocol as previously described.9 Lung fibroblasts were treated with TGFβ or buffer only for 24 hours, fixed with 1% formaldehyde in Dulbecco's modified Eagle's medium, washed twice with PBS, and then lysed in SDS lysis buffer. After sonication to shear the DNA to an average of 1000 bp, the lysate was centrifuged and the supernatant was collected. After preclearing with protein A agarose, each sample was aliquoted (20 μl) separately for use as “input DNA” in PCR analysis. The remainder of each sample was then divided equally into three aliquots for incubation with: i) anti-MeCP2 antibody (anti-MeCP2 bound DNA fraction), ii) nonimmune rabbit IgG (nonspecific antibody background DNA fraction), or iii) PBS-buffer (no antibody background fraction). Any immune complexes formed were affinity-adsorbed with protein A agarose and collected by centrifugation. The bound DNA was washed extensively and eluted from the protein A agarose with freshly made elution buffer (1% SDS, 0.1 M NaHCO3). The eluates and the original “input DNA” sample were incubated at 65°C for 4 hours to reverse the crosslink and then used as templates for PCR analysis along with the oligonucleotide primer pairs A (5′-CGTTGACTGTCCATTGAAGC-3′) and B (5′-TGTAGTCTGGAGTCTGTGTG-3′), C (5′-CAGTCGCCATCAGGGTAAGT-3′) and D (5′-CAACACCTAAGTAGAAACAA-3′), and E (5′-TTGTTTCTACTTAGGTGTTG-3′) and F (5′-CATAGGTTTGAATCGTAAGG-3′). These primers were designed to amplify the CpG islands in the α-SMA promoter region from −917 to −531, site 1 in first intronic region from +17 to +390 and site 2 in first intronic region from +371 to +600 of the rat α-SMA gene, respectively. The PCR products were then analyzed by gel electrophoresis in 1.3% agarose along with 100 bp DNA ladder from New England Biolabs, Inc.
Transfection and Reporter Gene Assay
All transient transfections were performed using the FuGENE6 reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer's instructions as before.8 Supercoiled DNA was isolated with an endotoxin-free Qiagen column kit (Qiagen Inc., Valencia, CA). Unless otherwise indicated, cells were seeded in six-well plates at a density of 105 per well in Dulbecco's modified Eagle's medium containing 10% plasma-derived serum, and incubated at 37°C overnight. In all, 2 μg DNA of interest were transfected per culture in Dulbecco's modified Eagle's medium containing 0.5% plasma-derived serum with or without 4 ng/mL TGFβ for the indicated time. To test the effect of MeCP2 on α-SMA promoter activity, 2 μg pLenti6-MeCP2 or the control empty vector pLenti6-v5 were co-transfected with the rat α-SMA promoter luciferase construct, α-SMA pro-intron-Luc, or plasmid pRL-TK control vector (used for normalization), respectively. After 48 hours, the cells were harvested and the activity of firefly or Renilla luciferase was measured using the dual luciferase assay system from Promega Co. (Madison, WI). The relative luciferase activity was calculated by normalizing firefly luciferase activity to that of Renilla luciferase to correct for transfection efficiency. Experiments with each construct were repeated 2 to 4 times, and the relative light units were expressed as mean ± SE.
RNA Analysis by Quantitative Real-Time PCR
This was undertaken to assess gene expression using a GeneAmp 7500 Sequence Detection System (Applied Biosystems, Foster City, CA) as before.8–11 All required primers and probes were purchased from Applied Biosystems and the results were expressed as 2−ΔΔCT using 18S rRNA as the reference. The specific calibrator for each assay was indicated in the figure legends.
Western Blot Analysis
The anti-α-SMA antibody was purchased from Sigma-Aldrich (St. Louis, MO) and all of the other antibodies were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Equal amounts of cell protein extracts were loaded onto 12% SDS polyacrylamide gels and transferred onto Hybond-P membranes for Western blotting as previously described.8 After each specific blot, the membranes were stripped and re-blotted with anti-GAPDH antibody as a loading control.
Histopathology
Morphological evaluation was performed on formalin-fixed, paraffin-embedded lung tissue sections and stained with H&E, as previously described.11
Hydroxyproline Assay
As a measure of fibrosis, total lung hydroxyproline content was determined by colorimetric assay after acid hydrolysis, as previously described.11
Statistical Analysis
This was undertaken as before using analysis of variance, and followed, where appropriate, by post hoc testing using Scheffé's test. A value of P < 0.05 was used as a criterion for statistical significance in comparisons between any two groups.8–11
Results
MeCP2 Binding to α-SMA Gene
MeCP2 is known to bind preferentially to methylated DNA regions serving as either a transcriptional repressor or activator, depending on the context.27 Since methylated DNA residues are present in the α-SMA gene19 the possibility of MeCP2 binding to these α-SMA gene sequences was initially investigated by incubation of recombinant MeCP2 with methylated and unmethylated DNA probes containing the sequences corresponding to the 3 CpG islands present in the α-SMA gene. They were then analyzed on nondenaturing polyacrylamide gels in an electrophoretic mobility shift assay. The resulting X-ray showed that at least two shifted bands could be discerned, representing MeCP2 bound complexes in both methylated and unmethylated probe samples (Figure 1A). The intensity of the complexes was higher in methylated than in unmethylated probe samples, suggesting increased binding when the CpG islands were methylated. There appeared to be more binding to the promoter and intron 1 site 2 probes relative to the intron1 site 1 probe. The binding specificity was confirmed by the virtual loss of retarded bands on incubation with 100 × cold unlabeled probe. Thus MeCP2 could bind the corresponding CpG island containing sequences in the α-SMA gene.
Figure 1.
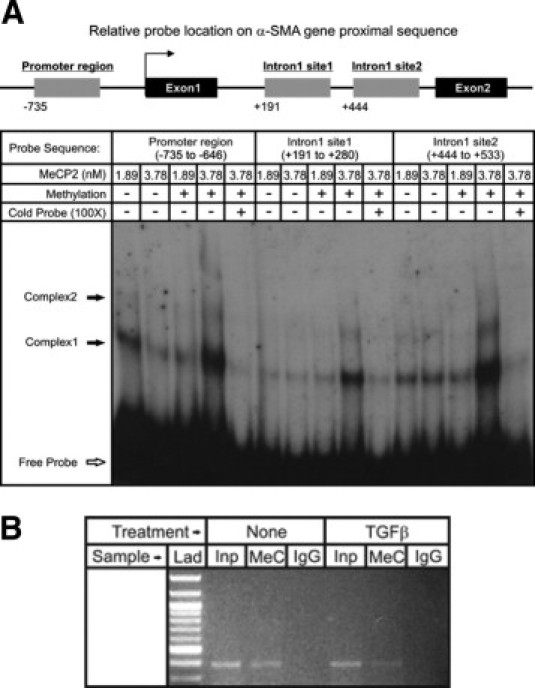
Binding of MeCP2 to α-SMA gene. A: Methylated and unmethylated oligo DNA primers spanning the CpG islands of the promoter region, intron 1 site 1 and intron 1 site 2 (corresponding locations in the α-SMA gene shown in upper panel cartoon, with numbering from the transcription start site) of the α-SMA gene were synthesized and labeled with 32P for use in an electrophoretic mobility shift assay. After incubation with the indicated doses of MeCP2 the samples were then analyzed by gel electrophoresis. Where indicated, 100-fold excess cold probe was added to the reaction mixture. The specific DNA-protein complexes were indicated by solid arrows. B: Cells were treated as indicated and then analyzed for binding of MeCP2 to the α-SMA gene promoter using a ChIP assay in conjunction with anti-MeCP2 antibodies. After incubation with the antibodies (“MeC”) or control IgG (“IgG”), the precipitated DNA, as well as unfractionated DNA (input or “Inp”), were analyzed by PCR using primers spanning the α-SMA gene promoter region. The PCR products were then separated in a 1.3% agarose gel.
To confirm binding of MeCP2 to the α-SMA gene in intact cells, anti-MeCP2 antibody was used in a ChIP assay of samples from untreated or TGFβ-treated cells. As shown in Figure 1B, a DNA fragment (∼380 bp) was amplified by PCR using anti-MeCP2 antibody precipitated DNA as a template together with primers spanning the α-SMA gene promoter region. In this region more than 70% of CpG islands were methylated in rat lung fibroblasts.19 Samples from cells treated with TGFβ showed the presence of a similar band, although with lower intensity. No band was detected when control IgG was used indicating specific precipitation by the anti-MeCP2 antibody. Moreover, no band was detected when primers spanning the intronic region were used for PCR amplification (data not shown). This intronic region was previously shown to be less than 8% methylated in lung fibroblasts.19 Thus, consistent with the gel shift data, MeCP2 was bound to the α-SMA promoter in intact cells, while the binding to intronic regions revealed by gel shift assays was undetectable in intact cells.
MeCP2 Activation of α-SMA Gene Expression
Given the observation of binding interactions between MeCP2 and the α-SMA gene promoter, the effect of the former on α-SMA expression was examined. This was undertaken by examining the effects of induced over- and under-expression of MeCP2 on α-SMA expression. First, the effect of over-expression using MeCP2 expression plasmid and corresponding negative control was analyzed by Western blotting and real-time PCR analysis for α-SMA protein and mRNA, respectively. The results showed an incremental increase in MeCP2 protein expression by the plasmid above the pre-existing levels, which were already quite significant (Figure 2A, upper panel). However, greater effect on suppression of MeCP2 protein expression was noted with the MeCP2 siRNA (Figure 2B, upper panel). The over-expression of MeCP2 caused modest but significant stimulation of α-SMA protein expression in both untreated, as well as TGFβ-treated cells (Figure 2A). TGFβ treatment alone caused the expected increase in α-SMA expression, which was further enhanced by transfection with the MeCP2 plasmid. In contrast, when the cells were transfected with MeCP2 siRNA to suppress endogenous MeCP2 expression, α-SMA expression was inhibited in both untreated and TGFβ-treated cells, although the inhibition was less and not statistically significant in the latter (Figure 2B). However, the ability of TGFβ to induce α-SMA expression appeared to be unimpaired. Similar findings were noted when effects on α-SMA mRNA levels were analyzed, although the effects of MeCP2 over-expression (Figure 2C) or under-expression (Figure 2D) were noticeably greater at the mRNA level compared to those at the protein level. Moreover, MeCP2 siRNA was able to significantly suppress α-SMA mRNA levels, even in TGFβ-treated cells (Figure 2D). These findings suggested an activator role for MeCP2 in regulation of α-SMA gene expression in fibroblasts, which might enhance or be an additive to activation by TGFβ. In addition, TGFβ induced α-SMA expression appeared to be unaffected by MeCP2 deficiency.
Figure 2.
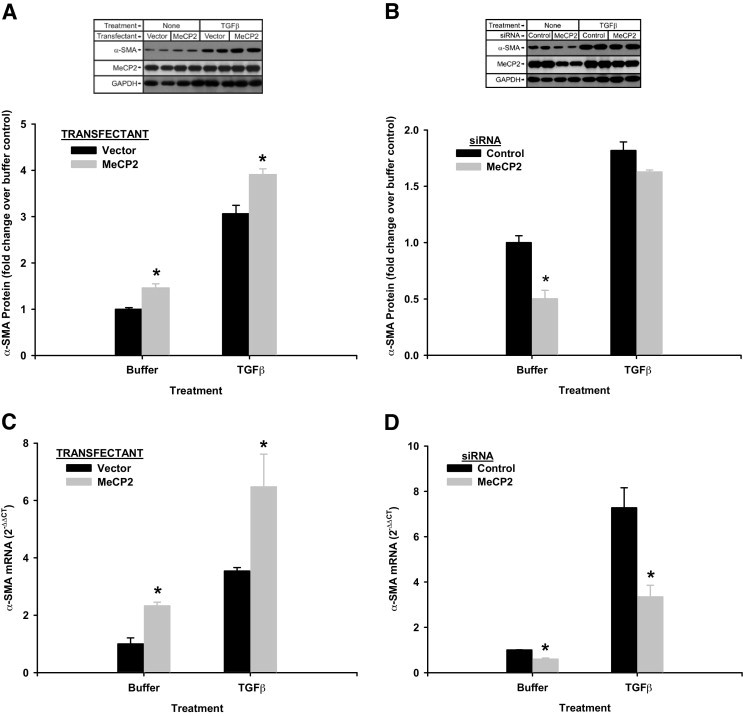
Effect of MeCP2 on α-SMA expression. Lung fibroblasts were transfected with MeCP2 expression (“MeCP2”) or control empty (“Vector”) vector (A and C), or MeCP2 (“MeCP2”) or random control (“Control”) siRNA (B and D). The cells were then treated with buffer only (“Buffer”) or with TGFβ (4 ng/mL). They were then analyzed for α-SMA protein by western blotting with GAPDH used as loading control (A and B). Representative blots are shown (upper panels in A and B), and the blots from three separate experiments were then scanned, quantified digitally, and expressed as fold change after normalization to the respective GAPDH band (lower panels, A and B). C and D: Total RNA was extracted from mouse lung fibroblasts treated and transfected as in A and B, respectively, and then analyzed for α-SMA mRNA levels by real time PCR with 18S rRNA as reference and the buffer treated control as calibrator for calculation of 2−ΔΔCT. All bar graphs showed means ± SE, with N = 3. Statistically significant difference of *P < 0.05 in comparison to the corresponding vector or siRNA control values. The effect of TGFβ treatment was statistically significant in all of the shown experiments (ie, TGFβ-treated versus respective buffer controls).
Methylation of α-SMA Promoter Inhibited the Stimulatory Effect of MeCP2
Since MeCP2 preferentially binds to methylated DNA, the effect of DNA methylation on the MeCP2 activation of α-SMA gene expression was analyzed using an α-SMA gene promoter construct. This construct contained the promoter and the first intronic regions (from −2880 to +2803), which was previously cloned and fused with a luciferase reporter gene, wherein the expression of luciferase was driven by α-SMA gene promoter.19 A sample of this construct was also methylated in vitro by CpG methyltransferase (M. SssI), which methylates all cytosine residue (C5) within the double stranded dinucleotide with recognition sequence 5′-CG-3′. The methylated and unmethylated α-SMA promoter constructs were then co-transfected with constitutively expressed MeCP2 construct or the expression vector only into rat lung fibroblasts. After 48 hours, the α-SMA gene promoter activity was determined in terms of luciferase activity. The results confirmed the activator role of MeCP2 on α-SMA gene expression, and further revealed that this activation occurred at the transcriptional level (Figure 3). However, when the promoter construct was methylated, promoter activity was significantly and markedly suppressed. This suppression could not be reversed by induced over-expression of MeCP2 using the expression plasmid.
Figure 3.
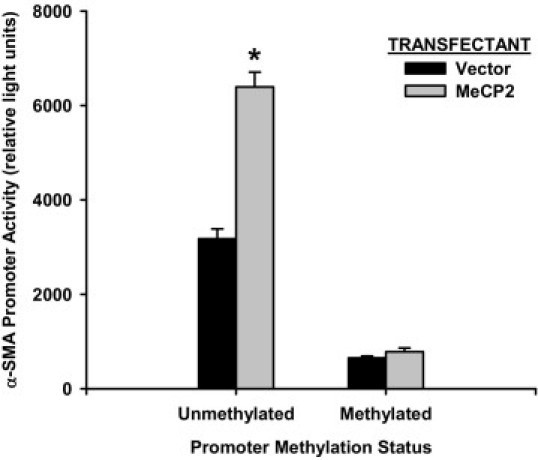
Effects of DNA methylation and MeCP2 on α-SMA promoter activity. Methylated or unmethylated α-SMA gene promoter construct α-SMApro-intron-Luc was co-transfected with MeCP2 expression plasmid (“MeCP2”) or control vector (“Vector”) into rat lung fibroblasts and then analyzed for luciferase activity 48 hours after transfection. The luciferase activity was normalized to its respective Renilla luciferase control activity and the results were expressed as relative light units shown as means ± SE of triplicates. The MeCP2 vector caused significant stimulation (*P < 0.05) stimulation of promoter activity over control vector, only in the unmethylated promoter.
MeCP2-Deficient Mice Showed Reduced Fibrotic Response on Bleomycin Treatment
Bleomycin is an anti-tumor antibiotic35 that has been used to induce an animal model of lung injury and fibrosis characterized by de novo induction of myofibroblast differentiation.11 Since myofibroblasts play a key role in the pathogenesis of pulmonary fibrosis,1–6 and MeCP2 could activate myofibroblast differentiation (Figures 2 and 3), the importance of MeCP2 on the development of pulmonary fibrosis was investigated. Mice deficient in MeCP2 and their wild-type littermates were endotracheally injected with bleomycin and analyzed for development of fibrosis.
At day 21 after treatment, morphologically there was no significant abnormality in lungs of control saline-treated MeCP2 null mice as shown by H&E staining, with no discernable difference when compared to wild-type control, lung tissue sections (Figure 4, panels A and C). Bleomycin-treated, wild-type murine lung sections showed the expected extensive dense lung fibrosis characterized by increased interstitial wall thickness, inflammatory cell infiltration, hypercellularity, and interstitial collagen deposition (Figure 4, panel B). In contrast, although scattered, more focal fibrotic lesions were observed, the fibrotic changes were markedly decreased in extent and severity in lung tissue sections from similarly treated MeCP2 null mice (Figure 4, panel D). The fibrotic lesions were less widespread and more narrowly confined to the peribronchial and perivascular areas. These results suggested that MeCP2 deficiency was associated with a reduction in bleomycin-induced pulmonary fibrosis.
Figure 4.
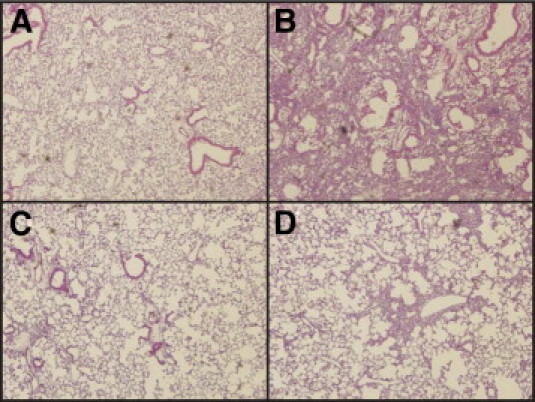
Effects of MeCP2 deficiency on lung histopathology of bleomycin-treated mice. Representative H&E stained lung tissue sections from saline or bleomycin-treated, wild-type (A and B, respectively) and MeCP2 null (C and D, respectively) mice were shown. Five mice of each group were evaluated and representative photographs were shown. All photographs represent ×20 magnification.
To confirm that MeCP2 deficiency impaired bleomycin-induced fibrosis, whole lung hydroxyproline content was determined by analyzing the hydroxyproline content of lungs from these wild-type and knockout mice at day 21 after saline or bleomycin injection. The results showed that while bleomycin treatment caused a 65% increase (over respective saline-treated controls) in the lung hydroxyproline content of wild-type mice, this bleomycin-induced increase was significantly less (33% increase greater than respective saline-treated controls) in MeCP2-deficient mice (Figure 5A). This reduction in lung hydroxyproline was also reflected in the analysis of lung type I collagen expression by Western blotting or real time PCR. Thus Western blot analysis of day 21 lung homogenate protein extracts revealed reduction in lung type I collagen in MeCP2 null mice relative to their wild-type littermates (Figure 5B). Similar reduction in type I collagen mRNA was also discernable in the lungs of MeCP2 knockout mice, with only a minor response to bleomycin treatment (Figure 5C). Thus, a role for MeCP2 in the development of fibrosis in this animal model was identified.
Figure 5.
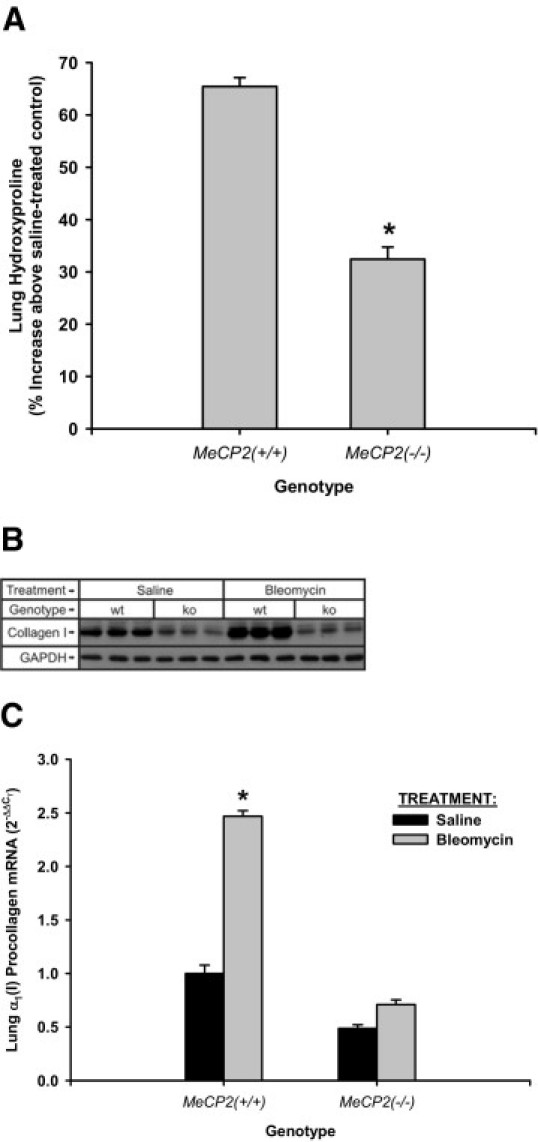
Effects of MeCP2 deficiency on pulmonary fibrosis indices. The indicated murine strains were treated with saline or bleomycin. The lungs were harvested and analyzed for hydroxyproline content (A), type I collagen protein by Western blot (B) and mRNA by real time PCR (C). A: The results were expressed as a percentage of the respective saline control values and shown as mean ± SE (N = 5). The reduction in lung hydroxyproline in the MeCP2-deficient strain was statistically significant (asterisk indicated P < 0.05). B: Each lane represented sample from a single animal. GAPDH was used as a loading control after strippng the membrane. C: α1(I) procollagen mRNA was detected by real-time PCR and results expressed as 2−ΔΔCT, with 18S rRNA used as the reference and the level in saline-treated, wild-type mice used as calibrator. Data were shown as mean ± SE from triplicate samples. Statistically significant difference compared to respective saline control (*P < 0.05).
To determine whether the reduced fibrotic response in MeCP2-deficient mice was related to suppression of myofibroblast differentiation, lung α-SMA expression was analyzed by Western blot and real time PCR analyses. As shown in Figure 6A, the reduced lung fibrosis in MeCP2 deficient mice correlated with a reduction in lung α-SMA protein expression, which was only weakly stimulated by bleomycin treatment in contrast to the robust response in wild type mice. This difference in lung α-SMA protein was also reflected at the mRNA level, with virtually no detectable induction by bleomycin treatment in knockout mice in contrast to the expected significant induction in wild-type mice (Figure 6B). Similar differences were noted in both α-SMA protein and mRNA in lung fibroblasts isolated from these mice (Figures 6C and 6D, respectively). These results suggested that the reduced fibrotic response of knockout mice to bleomycin injury might be due, at least in part, to inhibition of myofibroblast differentiation, as a result of MeCP2 deficiency.
Figure 6.
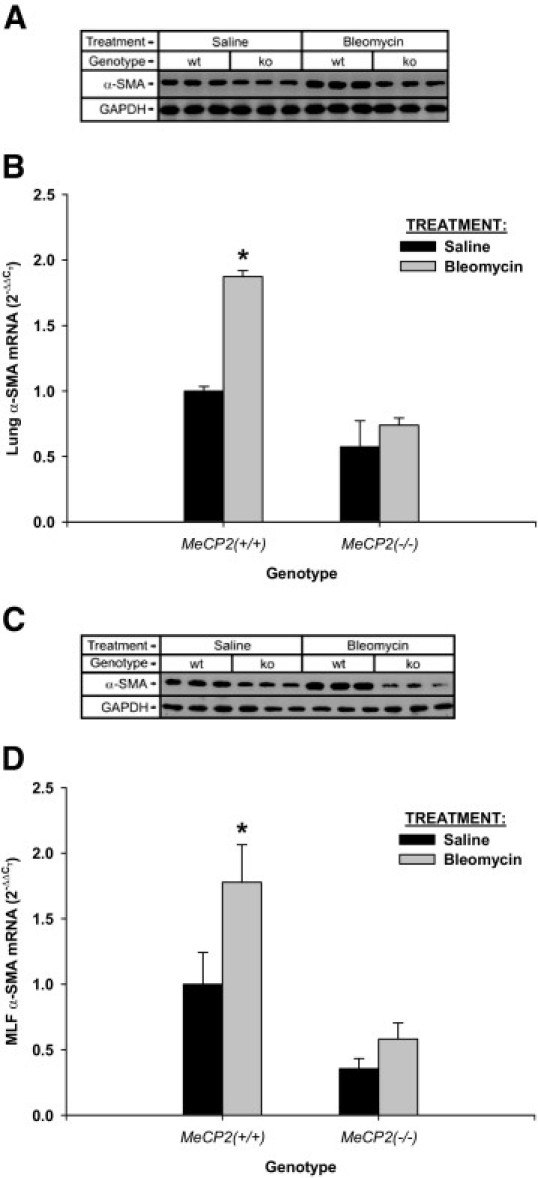
Effects of MeCP2 deficiency on myofibroblast differentiation in pulmonary fibrosis. MeCP2 null (“ko”) mice (MeCP2−/−) and its wild-type (“wt”) littermates (MeCP2+/+) were treated with bleomycin or saline as indicated. After harvesting the lungs were analyzed for α-SMA protein by Western blotting (A) and mRNA by real time PCR (B). A: GAPDH was used as a loading control. B: The results were expressed as 2−ΔΔCT, with 18S rRNA used as the reference and the level in saline-treated wild type mice used as calibrator. C and D: Similar analyses of α-SMA protein and mRNA, respectively, were undertaken for lung fibroblasts isolated from lungs of mice treated as in A and B. Data were shown as mean ± SE from triplicate samples. Statistically significant difference from respective saline-treated controls (*P < 0.05).
Although TGFβ could induce α-SMA, despite suppressed MeCP2 expression (Figures 2B and 2D), it is unclear if this were the case in the total absence of MeCP2. To address this issue, lung fibroblasts from wild type and MeCP2 knockout mice were analyzed for α-SMA expression in response to TGFβ. The results showed that TGFβ was able to induce α-SMA expression at both protein and mRNA levels in the total absence of MeCP2 (Figure 7A and B, respectively). The fold induction was comparable in both wild-type and MeCP2-deficient cells, thus indicating that TGFβ induced α-SMA did not depend on or was not mediated by MeCP2 and that the reduced fibrosis in MeCP2 knockout mice was not due to impairment of TGFβ-induced myofibroblast differentiation.
Figure 7.
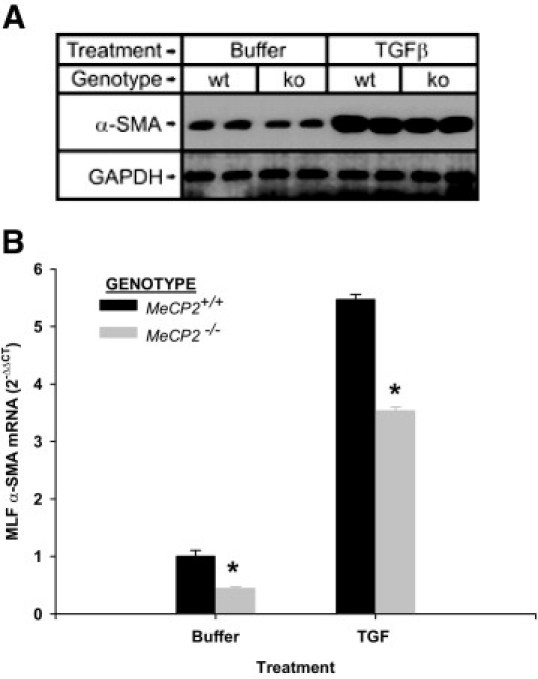
Effect of MeCP2 deficiency on TGFβ response. Mouse lung fibroblasts were isolated from MeCP2 null mice (MeCP2−/−) and its wild-type littermates (MeCP2+/+). They were either treated with TGFβ at 4 ng/mL or buffer only for 48 hours (A) or 6 hours (B), respectively. The total protein and total RNA were then analyzed for α-SMA gene expression by Western blot (A) and real time PCR analysis (B). Mean ± SE was shown for the bar graph with N = 3. Statistically significant difference from respective wild-type control (*P < 0.05). The effect of TGFβ treatment was statistically significant in both wild-type and MeCP2-deficient cells.
Discussion
DNA methylation is a key epigenetic regulatory mechanism that plays an essential role in mammalian development.36,37 Previously, three CpG islands in the α-SMA gene are shown to play a critical role in the regulation of myofibroblast differentiation, which is related to their methylation status.19 In this study, preferential binding of MeCP2 to the methylated DNA fragment in the α-SMA gene could be demonstrated using two approaches. First, using electrophoretic mobility shift assay, the intensity of a shifted band in samples with methylated probe was higher than that in samples with unmethylated probes. Second, consistent with the previous finding that only the promoter region of α-SMA is highly methylated in rat lung fibroblasts,19 a ChIP assay using anti-MeCP2 antibody successfully pulled down a complex containing the CpG islands of the α-SMA gene promoter region. However, it failed to pull down a DNA fragment corresponding to the first intronic region, which was previously shown to be poorly methylated.19 Thus, these two methods confirmed that methylated DNA fragments had a greater affinity for MeCP2 relative to unmethylated sequences. The gel shift assay also suggested that MeCP2 binding to the methylated promoter sequences might be important in regulation of α-SMA gene expression in intact cells.
MeCP2 is best known for its role in the development of the nervous system and mutations in the MeCP2 gene are associated with Rett syndrome and other forms of X-linked mental retardation.32,33 Male MeCP2 mutant mice exhibit tremors, progressive motor dysfunction, oily disheveled fur, hypoactivity, myoclonic seizures, and kyphosis. Approximately 10% of male mutants die between 10 and 12 months of age. Heterozygous female mice exhibit a milder phenotype. All mutant male mice and 62% of female heterozygotes exhibit a repetitive clasping movement of their forelimbs and exhibit tremors.32 Although MeCP2 was found in all cells in the body,33 its role outside of nervous system is less clear. Initially, MeCP2 is considered to be a repressor of gene expression through binding to the methylated CpG islands present in the target gene sequence.25 However, subsequent studies reveal that only 6% of the MeCP2 binding sites are in CpG islands, and that 63% of MeCP2-bound promoters belong to actively expressed genes, of which only 6% are highly methylated.26,27 In contrast to the earlier findings, these latter results indicate that the main function of MeCP2 is something other than silencing methylated promoters. The results of the current study favor the latter interpretation based on the following: i) Ectopically enhanced MeCP2 gene expression stimulated α-SMA gene expression, which appears to be additive to induction by TGFβ; ii) suppression of MeCP2 gene expression by siRNA or its deficiency in cells from MeCP2 knockout mice led to a reduced α-SMA gene expression, but did not affect TGFβ inducibility; and iii) MeCP2-deficient mice had much less pulmonary fibrosis, as assessed by lung histopathology, hydroxyproline content, collagen I expression, as well as by impaired de novo myofibroblast differentiation assessed by α-SMA expression. The totality of these results suggested a significant modulatory role of MeCP2 in fibroblast α-SMA expression and myofibroblast differentiation. It was not essential for inducibility by TGFβ, but augmented its effect on α-SMA gene expression. The effects of MeCP2 deficiency in the bleomycin model indicated that this MeCP2-mediated augmentation might be essential for pulmonary fibrosis. However, MeCP2 regulation of α-SMA gene expression appears not to be exclusively via direct binding to and regulation of the α-SMA gene. A recent study using hepatic stellate cells describes an indirect mechanism by which diminished miRNA targeting of MeCP2 results in elevated cellular levels of this protein, resulting in suppression of PPARγ (a known repressor of α-SMA expression) expression and thus heightened α-SMA expression.23 In this case, however, MeCP2 is shown to be acting as a repressor with PPARγ as the target gene. Thus, MeCP2, while preferentially binding to methylated DNA, has the potential of acting in a complex fashion, both as a repressor and activator of gene expression, although the mechanistic basis for this dichotomy is unclear. Previous investigations on the MeCP2-DNA interaction discovered that MeCP2 might assemble novel secondary chromatin structures independent of DNA modification.24 However, it is unclear if this dichotomy depends on binding to methylated versus unmethylated sequences, despite abundant studies showing that MeCP2 preferentially binds to methylated DNA.31 Nevertheless, the findings in this study favored the interpretation of a direct activator role for MeCP2 vis-à-vis the α-SMA gene in lung fibroblasts, which might be of importance in pathogenesis of pulmonary fibrosis.
Acknowledgments
We thank Matthew Ullenbruch for his assistance in the animal model studies.
Footnotes
Supported by grants HL28737, HL31963, HL52285, HL77297, and HL91775 from the National Institutes of Health and used the Cell and Molecular Biology Core of the Michigan Diabetes Research and Training Center funded by DK020572 from the National Institute of Diabetes and Digestive and Kidney Diseases.
References
- 1.Phan S.H. The myofibroblast in pulmonary fibrosis. Chest. 2002;122:286S–289S. doi: 10.1378/chest.122.6_suppl.286s. [DOI] [PubMed] [Google Scholar]
- 2.Scotton C.J., Chambers R.C. Molecular targets in pulmonary fibrosis: the myofibroblast in focus. Chest. 2007;132 doi: 10.1378/chest.06-2568. 1311-1121. [DOI] [PubMed] [Google Scholar]
- 3.Darby I.A., Hewitson T.D. Fibroblast differentiation in wound healing and fibrosis. Int Rev Cytol. 2007;257:143–179. doi: 10.1016/S0074-7696(07)57004-X. [DOI] [PubMed] [Google Scholar]
- 4.Thannickal V.J., Toews G.B., White E.S., Lynch J.P., 3rd, Martinez F.J. Mechanisms of pulmonary fibrosis. Annu Rev Med. 2004;55:395–417. doi: 10.1146/annurev.med.55.091902.103810. [DOI] [PubMed] [Google Scholar]
- 5.Gharaee-Kermani M., Hu B., Phan S.H., Gyetko M.R. Recent advances in molecular targets and treatment of idiopathic pulmonary fibrosis: focus on TGFbeta signaling and the myofibroblast. Curr Med Chem. 2009;16:1400–1417. doi: 10.2174/092986709787846497. [DOI] [PubMed] [Google Scholar]
- 6.Pardo A., Selman M. Idiopathic pulmonary fibrosis: new insights in its pathogenesis. Int J Biochem Cell Biol. 2002;34:1534–1538. doi: 10.1016/s1357-2725(02)00091-2. [DOI] [PubMed] [Google Scholar]
- 7.Boström H., Willetts K., Pekny M., Levéen P., Lindahl P., Hedstrand H., Pekna M., Hellström M., Gebre-Medhin S., Schalling M., Nilsson M., Kurland S., Törnell J., Heath b.J.K., Betsholtz C. PDGF-A signaling is a critical event in lung alveolar myofibroblast development and alveogenesis. Cell. 1996;85:863–873. doi: 10.1016/s0092-8674(00)81270-2. [DOI] [PubMed] [Google Scholar]
- 8.Hu B., Wu Z., Phan S.H. Smad3 mediates transforming growth factor-beta-induced alpha-SMA expression. Am J Respir Cell Mol Biol. 2003;29:397–404. doi: 10.1165/rcmb.2003-0063OC. [DOI] [PubMed] [Google Scholar]
- 9.Hu B., Wu Z., Liu T., Ullenbruch M.R., Jin H., Phan S.H. Gut-enriched Krüppel-like factor interaction with Smad3 inhibits myofibroblast differentiation. Am J Respir Cell Mol Biol. 2007;36:78–84. doi: 10.1165/rcmb.2006-0043OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu B., Wu Z., Jin H., Hashimoto N., Liu T., Phan S.H. CCAAT/enhancer-binding protein beta isoforms and the regulation of alpha-SMA gene expression by IL-1 beta. J Immunol. 2004;173:4661–4668. doi: 10.4049/jimmunol.173.7.4661. [DOI] [PubMed] [Google Scholar]
- 11.Hu B., Ullenbruch M.R., Jin H., Gharaee-Kermani M., Phan S.H. An essential role for CCAAT/enhancer binding protein beta in bleomycin-induced pulmonary fibrosis. J Pathol. 2007;211:455–462. doi: 10.1002/path.2119. [DOI] [PubMed] [Google Scholar]
- 12.Cogan J.G., Subramanian S.V., Polikandriotis J.A., Kelm R.J., Jr., Strauch A.R. Vascular smooth muscle alpha-actin gene transcription during myofibroblast differentiation requires Sp1/3 protein binding proximal to the MCAT enhancer. J Biol Chem. 2002;277:36433–36442. doi: 10.1074/jbc.M203232200. [DOI] [PubMed] [Google Scholar]
- 13.Qiu P., Feng X.H., Li L. Interaction of Smad3 and SRF-associated complex mediates TGF-beta1 signals to regulate SM22 transcription during myofibroblast differentiation. J Mol Cell Cardiol. 2003;35:1407–1420. doi: 10.1016/j.yjmcc.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Buck M., Kim D.J., Houglum K., Hassanein T., Chojkier M. c-Myb modulates transcription of the alpha-SMA gene in activated hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol. 2000;278:G321–G328. doi: 10.1152/ajpgi.2000.278.2.G321. [DOI] [PubMed] [Google Scholar]
- 15.Liu T., Hu B., Choi Y.Y., Chung M., Ullenbruch M., Yu H., Lowe J.B., Phan S.H. Notch1 signaling in FIZZ1 induction of myofibroblast differentiation. Am J Pathol. 2009;174:1745–1755. doi: 10.2353/ajpath.2009.080618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noseda M., Fu Y., Niessen K., Wong F., Chang L., McLean G., Karsan A. Smooth muscle α-actin is a direct target of Notch/CSL. Circ Res. 2006;98:1468–1470. doi: 10.1161/01.RES.0000229683.81357.26. [DOI] [PubMed] [Google Scholar]
- 17.Hinz B., Phan S.H., Thannickal V.J., Galli A., Bochaton-Piallat M.L., Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 19.Hu B., Gharaee-Kermani M., Wu Z., Phan S.H. Epigenetic regulation of myofibroblast differentiation by DNA methylation. Am J Pathol. 2010;177:21–28. doi: 10.2353/ajpath.2010.090999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niki T., Rombouts K., De Bleser P., De Smet K., Rogiers V., Schuppan D., Yoshida M., Gabbiani G., Geerts A. A histone deacetylase inhibitor, trichostatin A, suppresses myofibroblastic differentiation of rat hepatic stellate cells in primary culture. Hepatology. 1999;29:858–867. doi: 10.1002/hep.510290328. [DOI] [PubMed] [Google Scholar]
- 21.Glenisson W., Castronovo V., Waltregny D. Histone deacetylase 4 is required for TGFbeta1-induced myofibroblastic differentiation. Biochim Biophys Acta. 2007;1773:1572–1582. doi: 10.1016/j.bbamcr.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Guo W., Shan B., Klingsberg R.C., Qin X., Lasky J.A. Abrogation of TGF-beta1-induced fibroblast-myofibroblast differentiation by histone deacetylase inhibition. Am J Physiol Lung Cell Mol Physiol. 2009;297:L864–L870. doi: 10.1152/ajplung.00128.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann J., Chu D.C., Maxwell A., Oakley F., Zhu N.L., Tsukamoto H., Mann D.A. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology. 2010;138:705–714. doi: 10.1053/j.gastro.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mann J., Oakley F., Akiboye F., Elsharkawy A., Thorne A.W., Mann D.A. Regulation of myofibroblast transdifferentiation by DNA methylation and MeCP2: implications for wound healing and fibrogenesis. Cell Death Differ. 2007;14:275–285. doi: 10.1038/sj.cdd.4401979. [DOI] [PubMed] [Google Scholar]
- 25.Lewis J.D., Meehan R.R., Henzel W.J., Maurer-Fogy I., Jeppesen P., Klein F., Bird Al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 26.Yasui D.H., Peddada S., Bieda M.C., Vallero R.O., Hogart A., Nagarajan R.P., Thatcher K.N., Farnham P.J., Lasalle J.M. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci USA. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chahrour M., Jung S.Y., Shaw C., Zhou X., Wong S.T., Qin J., Zoghbi H.Y. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikitina T., Shi X., Ghosh R.P., Horowitz-Scherer R.A., Hansen J.C., Woodcock C.L. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol Cell Biol. 2007;27:864–877. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Georgel P.T., Horowitz-Scherer R.A., Adkins N., Woodcock C.L., Wade P.A., Hansen J.C. Chromatin compaction by human MeCP2: Assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 30.Gregory R.I., Randall T.E., Johnson C.A., Khosla S., Hatada I., O'Neill L.P., Turner B.M., Feil R. DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1. Mol Cell Biol. 2001;21:5426–5436. doi: 10.1128/MCB.21.16.5426-5436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nan X., Tate P., Li E., Bird A. DNA methylation specifies chromosomal localization of MeCP. Mol Cell Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shahbazian M., Young J., Yuva-Paylor L., Spencer C., Antalffy B., Noebels J., Armstrong D., Paylor R., Zoghbi H. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 33.Hite K.C., Adams V.H., Hansen J.C. Recent advances in MeCP2 structure and function. Biochem Cell Biol. 2009;87:219–227. doi: 10.1139/o08-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen S., Zhou Z., Greenberg M.E. Medicine: activating a repressor. Science. 2008;320:1172–1173. doi: 10.1126/science.1159146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crooke S.T., Bradner W.T. Bleomycin, a review. J Med. 1976;7:333–428. [PubMed] [Google Scholar]
- 36.Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 37.Miranda T.B., Jones P.A. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]


