Abstract
A fundamental function of autophagy conserved from yeast to mammals is mobilization of macromolecules during times of limited nutrient availability, permitting organisms to survive under starvation conditions. In yeast, autophagy is initiated following nitrogen or carbon deprivation, and autophagy mutants die rapidly under these conditions. Similarly, in mammals, autophagy is upregulated in most organs following initiation of starvation, and is critical for survival in the perinatal period following abrupt termination of the placental nutrient supply. The nutrient-sensing kinase, mammalian target of rapamycin, coordinates cellular proliferation and growth with nutrient availability, at least in part by regulating protein synthesis and autophagy-mediated degradation. This review focusses on the regulation of autophagy by Tor, a mammalian target of rapamycin, and Ulk1, a mammalian homolog of Atg1, in response to changes in nutrient availability. Given the importance of mitochondria in maintaining bioenergetic homestasis, and potentially as a source of membrane for autophagosomes during starvation, possible roles for mitochondria in this process are also discussed. Antioxid. Redox Signal. 14, 1953–1958.
Introduction
Autophagy is a catabolic process through which cellular components are sequestered within vesicles and delivered to lysosomes for degradation and recycling of basic metabolic units, including amino acids. This process is initiated by the elongation of phagophores into double-membrane bound, neutral pH vesicles known as autophagosomes that sequester cargo (10). The outer membranes of autophagosomes fuse with late endosomes and lysosomes, promoting the degradation of contents into basic bioenergetic units, such as amino acids and fatty acids that can be reutilized by the cell. Flux through the autophagy pathway is increased in response to various cellular stresses and is generally thought to promote cell survival, although there are instances where autophagy contributes to cell death (39).
Metabolic stress induces a robust autophagy response that is mediated by energy-sensing kinase cascades, including those involving mammalian target of rapamycin (mTOR), AMP regulated kinase, and cAMP-PKA (2, 11, 34, 37, 49). Although starvation-induced autophagy is considered a form of nonselective autophagy in budding yeast (32), the organism in which autophagy has been best characterized at the molecular/genetic level, given the importance of mitochondria in regulating cell survival and death in higher eukaryotes, it seems unlikely that degradation of mitochondria would be left to chance, especially during starvation. Indeed, a recent proteomics-based study suggested that there is ordered degradation of proteins even during amino acid starvation (26). Specifically, cytosolic proteins are degraded rapidly, while proteins annotated to various complexes and organelles, including mitochondria, are degraded at later time points.
In yeast, the serine–threonine kinase Atg1 plays a central role in switching between “selective” or cargo-initiated autophagy and starvation-induced autophagy (35). Atg1 forms different complexes, interacting with factors such as Atg17 that promote formation of large autophagosomes, or with cargo co-adapters such as Atg11 in a TOR-dependent manner (5, 6, 20, 49, 52). Recent evidence indicates that mammalian homologs of Atg1, Ulk1, and Ulk2 regulate autophagy in an mTOR-dependent manner (13, 18, 19), although it remains to be determined whether these Atg1 homologs regulate the specificity of autophagy in a manner similar to their yeast counterpart.
Ulk1 is Required for Efficient Amino Acid Starvation-Induced Autophagy and Mitochondrial Clearance
RNAi-mediated knock down of Ulk1 in cells results in a defect in amino acid starvation-induced autophagy in various cultured cell lines (3); however, unlike animals lacking nonredundant autophagy genes, such as Atg3, Atg5, and Atg7 (25, 28, 48), ulk1−/− animals do not suffer perinatal lethality, and are phenotypically unremarkable except for a defect in clearance of mitochondria and ribosomes specifically at terminal stages of erythroid maturation (29). Consistent with this observation, erythroid maturation involves a spike in Ulk1 but not Ulk2 expression (29). Although early ultrastructural studies demonstrated the presence of mitochondria in lysosomes and autophagic structures of maturing erythroid cells (15, 17, 23), the defect in the Ulk1-deficient mice provided the first genetic evidence that autophagy is involved in the clearance of organelles during red blood cell maturation. Whether this process represents a form of selective autophagy or shares features with starvation-induced autophagy remains to be determined.
Since Atg1 has a conserved essential role in autophagy (4, 35), and autophagy-deficient mice exhibit perinatal mortality that is associated with severe nutrient and energy insufficiency (25, 28, 48), the ability of Ulk1-deficient animals to survive the metabolic shock associated with birth suggests that another Atg1 homolog, such as Ulk2, plays an equally important, if not more important, role in the induction of autophagy at birth. Although RNAi-mediated knockdown of Ulk2 does not significantly affect starvation-induced autophagy in HEK293 cells (whereas knockdown of Ulk1 does), ectopic expression of Ulk2 (but not Ulk1) enhances the process (3, 4), suggesting that the relative expression of Ulk1 and Ulk2 in different cells may contribute to the dependence of any given cell on one or the other of the Atg1 homologs.
mTOR Negatively Regulates Ulk1
The mTOR complex 1 (mTORC1) branch of the mTOR signaling pathway coordinates upstream signals from growth factors, intracellular energy levels and amino acid availability with cell growth by modulating rates of protein synthesis and degradation (via autophagy). The raptor component of mTORC1 interacts with mTOR substrates and facilitates efficient phosphorylation by mTOR. Downstream targets of active mTORC1 include eukaryotic initiation factor 4E-binding proteins 1, 2, and 3, and the p70 S6 kinases (S6K1 and S6K2), which are involved in the regulation of protein translation, as well as cell growth and proliferation (8). Under nutrient replete conditions, mTORC1 also inhibits autophagy by phosphorylating and negatively regulating Ulk1 and its interacting partner, Atg13 (13, 18, 19).
Unlike yeast Atg1, whose interaction with Atg13 and Atg17 depends on TOR activity (21, 22), Ulk1 and Ulk2 form stable complexes with mAtg13 and FIP200 (which shares regions of homology with yeast Atg17), and both are required for Ulk1 association with phagophores under starvation conditions (13, 18, 19). Although the formation of the Ulk1–Atg13–FIP200 complex occurs in an mTOR-independent manner, association of this complex with the raptor component of mTORC1 occurs only in the presence of amino acids (18). This interaction promotes phosphorylation of both Ulk1 and Atg13 by mTOR and inhibition of Ulk1 kinase activity (13, 18, 19). Although, the functional significance of mTOR-mediated phosphorylation has not been formally established by identifying and disrupting the mTOR phosphorylation sites on Ulk1 and Atg13 in mammalian cells, yeast TORC1 directly phosphorylates Atg13 at multiple Ser residues, and expression of an unphosphorylatable Atg13 mutant bypasses the TORC1-mediated inhibition of autophagy under nutrient-rich conditions (22).
mTOR Activity is Regulated by Amino Acid Levels
Changes in growth factor availability or changes in ATP level are signaled to mTORC1 via tuberous sclerosis complex 1 and 2 (TSC1/2), a GTPase-activating protein complex that regulates the GTP-loading state of Rheb. Rheb is a Ras-related GTP-binding protein which, when bound to GTP, interacts with mTORC1 and is required for activation of mTORC1 by all signals (30). The mitogenically regulated PtdIns3K/Akt and Ras/MAPK cell signaling pathways activate mTOR via impairment of TSC2 function, whereas the energy sensing AMP-dependent protein kinase inhibits mTOR signaling by phosphorylating and activating TSC2 (8). In cells lacking TSC2, the mTORC1 pathway is resistant to growth factor withdrawal, but remains sensitive to amino acid withdrawal, suggesting that the TSC1/2 is not required for amino acid-mediated regulation of mTORC1 activity (47). Instead, activation of mTORC1 by amino acids involves its recruitment to the lysosomal surface, where it has been proposed that mTORC1 may find its co-activator, Rheb, which localizes to late endosomes/lysosomes (42, 43).
Amino acids promote the interaction of Rag GTPases (specifically Rag heterodimers containing GTP-bound RagB) with the raptor component of mTORC1 (44). The amino acid-induced localization of Rag proteins to the lysosomal compartment and activation of mTORC1 depends on a complex (termed the “Ragulator”) encoded by MAPKSP1, ROBLD2, and c11orf59 genes that interact with the Rag GTPases (43). Lysosomal localization of mTORC1 does not occur in the absence of amino acids, even with serum stimulation or constitutive activation of Rheb by TSC2 inactivation (43). Conversion of early to late endosomes is also required for amino acid-stimulated mTORC1 activation; and mTORC1 localizes to hybrid early/late endosomes when the conversion is blocked (by constitutively active Rab5 or impaired Rab 7 function) (12, 31).
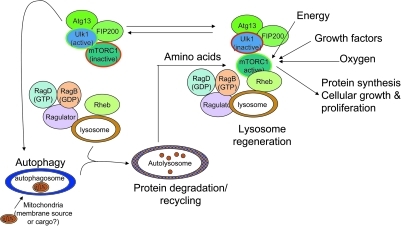
Although mTOR activity is inhibited following amino acid withdrawal, increased flux through the autophagy pathway leads to restoration of mTOR activity, which promotes lysosomal regeneration from autolysosomes (54). Since autophagy is involved in recycling amino acids, the amino acid-dependent lysosomal localization of mTORC1 may underlie this reactivation of mTOR under starvation conditions, and may also serve to limit the autophagy response by inhibiting Ulk1 kinase activity (Fig. 1).
FIG. 1.
Amino acid-dependent regulation of autophagy by mTOR and Ulk1. Amino acid-dependent regulation of autophagy by mTOR and Ulk1. The mTOR mediated repression of the Ulk1–Atg13–FIP200 complex is inhibited when growth factors, oxygen, amino acids and/or intracellular ATP levels becoming limiting. The active Ulk1–Atg13–FIP200 complex then promotes autophagy, which recycles cellular contents and restores amino acid pools. Increased amino acid levels are sensed by the “Ragulator” complex, which promotes re-activation of mTOR and limits the autophagy response. mTOR, mammalian target of rapamycin.
Role of Mitochondria in Starvation-Induced Autophagy
The liver is integrally involved in regulating nutrient availability in mammals. Therefore, it is not surprising that the earliest evidence of starvation-induced mitophagy came from ultrastructural examination of intact and degraded mitochondria in lysosomes of rat hepatocytes treated with glucagon (1). Starvation-induced autophagy is considered a form of nonselective autophagy in budding yeast, and while it is possible that mitochondria are randomly sequestered in autophagosomes with other cytoplasmic components, a recent proteomics-based study suggested that there is ordered degradation of proteins even during amino acid starvation, with mitochondrial proteins being among the last to be degraded (26).
More recent studies have highlighted an interesting relationship between starvation-induced expansion of the autophagosome/lysosome compartment, mitophagy and mitochondrial depolarization in starved rat hepatocytes (9, 41). When grown in complete media, GFP-LC3 fluorescence in hepatocytes (derived from GFP-LC3 transgenic mice) is distributed diffusely except for small dotted structures distributed in an apparently random fashion throughout the cytoplasm (24). Following nutrient deprivation, these LC3-positive structures become closely associated with individual mitochondria, some of which form crescent-shaped structures that eventually surround entire mitochondria. Mitochondrial depolarization follows autophagosome completion and is accompanied by acidification of the autophagosome and loss of GFP-LC3 fluorescence. Mitochondrial depolarization is not the signal for degradation, but occurs following sequestration in LC3+ vesicles. Interestingly, cyclosporine, an inhibitor of the mitochondrial permeability transition, does not block sequestration and formation of GFP-LC3-labeled autophagosomes, but rather suppresses mitochondrial depolarization, the acidification of autophagosomes and the proliferation of autophagosomes and autolysosomes during nutrient deprivation (9, 24). These studies suggest that sequestration and depolarization of mitochondria may contribute to autolysosome proliferation, a concept that is especially intriguing in light of recent evidence that mitochondria contribute membrane for autophagosome biogenesis during starvation (16). While the detection of mitophagy during starvation may seem to be at odds with the notion of ordered degradation of cellular contents during starvation observed in cultured cells (26), it may simply reflect the level of mitophagy required to preserve mitochondrial function under conditions of metabolic stress.
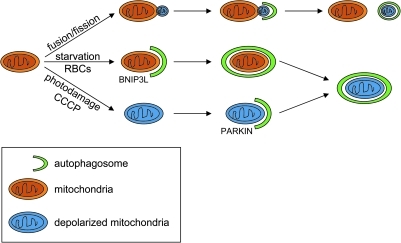
The observation that mitochondrial depolarization occurs after sequestration of mitochondria within phagophores also raises an important question regarding the signal targeting mitochondria for degradation (Fig. 2). There are many examples in which mitochondrial depolarization triggers clearance by autophagy. For example, when hepatocytes are subjected to irreversible laser-induced photodamage, GFP-LC3 fluorescence envelops and completely encircles depolarized mitochondria (24). In this case, since depolarization precedes sequestration, it likely signals recruitment of components of the autophagy machinery. Further evidence that the elimination of depolarized mitochondria by autophagy represents an important aspect of mitochondrial quality control comes from studies linking mitochondrial dynamics with autophagy. Specifically, mitochondria in pancreatic beta cells undergo frequent cycles of fusion and fission that often result in uneven daughter units: one with increased membrane potential and a high probability of subsequent fusion, and the other with decreased membrane potential, and reduced probability for a fusion event, which are degraded by autophagy after several hours (51). Interestingly, mitofusin, a gene involved in mitochondrial fusion, has recently been shown to be a target of the E3 ubiquitin ligase, Parkin, which localizes to mitochondria following loss of membrane potential and promotes recruitment of LC3 and degradation by autophagy (14, 36, 40). The Parkin-dependent degradation of mitofusin may provide a mechanism, at least in Parkin-expressing cells, to prevent fusion and recovery of depolarized mitochondria, promoting their degradation by autophagy (56). Since loss of mitochondrial membrane potential can clearly trigger a pathway resulting in degradation of depolarized mitochondria by autophagy, the observation that mitochondria are not depolarized in hepatocytes prior to their degradation suggests that additional signals remain to be identified.
FIG. 2.
Relationship between mitochondrial depolarization and clearance. The signal(s) responsible for degradation of mitochondria by autophagy may depend on the trigger. Mitochondrial dynamics (fission and fusion) play an important role in the maintenance of mitochondrial integrity under normal growth conditions and in response to cellular stress. Cycles of fusion and fission result in unequal division with segregation of damaged components into the depolarized daughter unit, followed by autophagy-mediated degradation of the depolarized mitochondria. Similarly, in response to photodamage or the uncoupling agent, carbonylcyanide-3-chlorophenyl hydrazone (CCCP), mitochondrial depolarization appears to be a trigger for autophagy-mediated degradation, at least in part through recruitment of the E3 ligase, Parkin. In starved hepatocytes and in maturing erythroid cells, mitochondrial clearance is not preceded by depolarization. Instead, mitochondrial dysfunction may be sensed by BH3 only proteins, such as BNIP3 or BCL2/adenovirus E1B 19 kDa interacting protein (BNIP3L).
The BH3-only protein, BNIP3L (Nix), is essential for mitochondrial clearance in erythroid cells (45, 46). Moreover, its mitochondrial localization and ability to directly interaction with Atg8 homologs suggest that it is involved in targeting the organelle for degradation by autophagy (38). The observation that mitochondria retained within autophagy-deficient (ulk1−/− and atg7−/−) red blood cells maintain their membrane potential (based on tetramethylrhodamine methyl ester staining) (29, 55) suggests that mitochondrial depolarization also follows sequestration within autophagosomes during terminal differentiation of red blood cells. Thus, it would be interesting to determine if BNIP3L is required for starvation-induced mitophagy in hepatocytes. Intriguingly, the treatment of Bnip3l- or Ulk1-deficient red blood cells with the mitochondrial uncoupler, carbonylcyanide-3-chlorophenyl hydrazone (CCCP), triggers clearance of mitochondria suggesting that loss of membrane potential may provide an alternative signal for targeting mitochondria to autophagosomes in red cells (29, 45). An unresolved twist to the story that needs to be further explored comes from the unexpected observation that mitochondria in Bnip3l-deficient murine embryonic fibroblasts show some resistance to depolarization in response to CCCP treatment (compared to wild-type MEFS), which is associated with impaired inhibition of mTOR (7). Perhaps BNIP3L, similar to BNIP3 (27), functions as a sensor of mitochondrial dysfunction (i.e., during starvation or differentiating red blood cells that are in the process of converting to a purely glycolytic pathway for ATP production), linking mitochondrial function with local mTOR activity and autophagy.
Clearly, additional studies are required to further elucidate the role of mitochondria as autophagy cargo or membrane source (or both) during starvation and other situations associated with increased flux through the autophagy pathway. Identification of the molecular signals responsible for targeting mitochondria to autophagsosomes will also no doubt provide tools necessary to dissect the complex relationship between mitochondria and autophagy. Finally, given the importance of Ulk1 in both starvation-induced autophagy and mitochondrial clearance, and as a downstream target of mTOR, understanding the mechanism by which this serine–threonine kinase regulates autophagy may unlock additional insights into the relationship between mitochondria and autophagy. Interestingly, Ulk1 regulates the trafficking of Atg9 (53), a transmembrane protein that in yeast is concentrated in a cluster of vesicle and tubules adjacent to mitochondria and is involved in the de novo formation of the phagophore assembly site and in phagophore expansion (33, 50, 53, 54).
Abbreviations Used
- mTOR
mammalian target of rapamycin
- mTORC1
mTOR complex 1
- TSC
tuberous sclerosis complex
Acknowledgments
M.K. is supported by funding from the National Institutes of Health (HL084199), the Burroughs Welcome Fund, and the American Lebanese Syrian Associated Charities (ALSAC).
References
- 1.Ashford TP. Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cardenas C. Miller RA. Smith I. Bui T. Molgo J. Muller M. Vais H. Cheung KH. Yang J. Parker I. Thompson CB. Birnbaum MJ. Hallows KR. Foskett JK. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell. 2010;142:270–283. doi: 10.1016/j.cell.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan EY. Kir S. Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 4.Chan EY. Longatti A. McKnight NC. Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheong H. Nair U. Geng J. Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheong H. Yorimitsu T. Reggiori F. Legakis JE. Wang CW. Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding WX. Ni HM. Li M. Liao Y. Chen X. Stolz DB. Dorn GW., 2nd Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunlop EA. Tee AR. Mammalian target of rapamycin complex 1: signalling inputs, substrates and feedback mechanisms. Cell Signal. 2009;21:827–835. doi: 10.1016/j.cellsig.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Elmore SP. Qian T. Grissom SF. Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 10.Eskelinen EL. New insights into the mechanisms of macroautophagy in mammalian cells. Int Rev Cell Mol Biol. 2008;266:207–247. doi: 10.1016/S1937-6448(07)66005-5. [DOI] [PubMed] [Google Scholar]
- 11.Filomeni G. Desideri E. Cardaci S. Graziani I. Piccirillo S. Rotilio G. Ciriolo MR. Carcinoma cells activate AMP-activated protein kinase-dependent autophagy as survival response to kaempferol-mediated energetic impairment. Autophagy. 2010;6:202–216. doi: 10.4161/auto.6.2.10971. [DOI] [PubMed] [Google Scholar]
- 12.Flinn RJ. Yan Y. Goswami S. Parker PJ. Backer JM. The late endosome is essential for mTORC1 signaling. Mol Biol Cell. 2010;21:833–841. doi: 10.1091/mbc.E09-09-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganley IG. Lam du H. Wang J. Ding X. Chen S. Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gegg ME. Cooper JM. Chau KY. Rojo M. Schapira AH. Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/Parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gronowicz G. Swift H. Steck TL. Maturation of the reticulocyte in vitro. J Cell Sci. 1984;71:177–197. doi: 10.1242/jcs.71.1.177. [DOI] [PubMed] [Google Scholar]
- 16.Hailey DW. Rambold AS. Satpute-Krishnan P. Mitra K. Sougrat R. Kim PK. Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heynen MJ. Tricot G. Verwilghen RL. Autophagy of mitochondria in rat bone marrow erythroid cells. Relation to nuclear extrusion. Cell Tissue Res. 1985;239:235–239. doi: 10.1007/BF00214924. [DOI] [PubMed] [Google Scholar]
- 18.Hosokawa N. Hara T. Kaizuka T. Kishi C. Takamura A. Miura Y. Iemura S. Natsume T. Takehana K. Yamada N. Guan JL. Oshiro N. Mizushima N. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung CH. Jun CB. Ro SH. Kim YM. Otto NM. Cao J. Kundu M. Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabeya Y. Kamada Y. Baba M. Takikawa H. Sasaki M. Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamada Y. Funakoshi T. Shintani T. Nagano K. Ohsumi M. Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamada Y. Yoshino K. Kondo C. Kawamata T. Oshiro N. Yonezawa K. Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent G. Minick OT. Volini FI. Orfei E. Autophagic vacuoles in human red cells. Am J Pathol. 1966;48:831–857. [PMC free article] [PubMed] [Google Scholar]
- 24.Kim I. Rodriguez-Enriquez S. Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komatsu M. Waguri S. Ueno T. Iwata J. Murata S. Tanida I. Ezaki J. Mizushima N. Ohsumi Y. Uchiyama Y. Kominami E. Tanaka K. Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kristensen AR. Schandorff S. Hoyer-Hansen M. Nielsen MO. Jaattela M. Dengjel J. Andersen JS. Ordered organelle degradation during starvation-induced autophagy. Mol Cell Proteomics. 2008;7:2419–2428. doi: 10.1074/mcp.M800184-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Kubli DA. Quinsay MN. Huang C. Lee Y. Gustafsson AB. Bnip3 functions as a mitochondrial sensor of oxidative stress during myocardial ischemia and reperfusion. Am J Physiol Heart Circ Physiol. 2008;295:H2025–H2031. doi: 10.1152/ajpheart.00552.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuma A. Hatano M. Matsui M. Yamamoto A. Nakaya H. Yoshimori T. Ohsumi Y. Tokuhisa T. Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 29.Kundu M. Lindsten T. Yang CY. Wu J. Zhao F. Zhang J. Selak MA. Ney PA. Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laplante M. Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L. Kim E. Yuan H. Inoki K. Goraksha-Hicks P. Schiesher RL. Neufeld TP. Guan KL. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem. 2010;285:19705–19709. doi: 10.1074/jbc.C110.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch-Day MA. Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mari M. Griffith J. Rieter E. Krishnappa L. Klionsky DJ. Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meley D. Bauvy C. Houben-Weerts JH. Dubbelhuis PF. Helmond MT. Codogno P. Meijer AJ. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- 35.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 36.Narendra D. Tanaka A. Suen DF. Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–168. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Novak I. Kirkin V. McEwan DG. Zhang J. Wild P. Rozenknop A. Rogov V. Lohr F. Popovic D. Occhipinti A. Reichert AS. Terzic J. Dotsch V. Ney PA. Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platini F. Perez-Tomas R. Ambrosio S. Tessitore L. Understanding autophagy in cell death control. Curr Pharm Des. 2010;16:101–113. doi: 10.2174/138161210789941810. [DOI] [PubMed] [Google Scholar]
- 40.Poole AC. Thomas RE. Yu S. Vincow ES. Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/Parkin pathway. PLoS ONE. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez-Enriquez S. Kim I. Currin RT. Lemasters JJ. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito K. Araki Y. Kontani K. Nishina H. Katada T. Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. J Biochem. 2005;137:423–430. doi: 10.1093/jb/mvi046. [DOI] [PubMed] [Google Scholar]
- 43.Sancak Y. Bar-Peled L. Zoncu R. Markhard AL. Nada S. Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sancak Y. Peterson TR. Shaul YD. Lindquist RA. Thoreen CC. Bar-Peled L. Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandoval H. Thiagarajan P. Dasgupta SK. Schumacher A. Prchal JT. Chen M. Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweers RL. Zhang J. Randall MS. Loyd MR. Li W. Dorsey FC. Kundu M. Opferman JT. Cleveland JL. Miller JL. Ney PA. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith EM. Finn SG. Tee AR. Browne GJ. Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 48.Sou YS. Waguri S. Iwata J. Ueno T. Fujimura T. Hara T. Sawada N. Yamada A. Mizushima N. Uchiyama Y. Kominami E. Tanaka K. Komatsu M. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stephan JS. Yeh YY. Ramachandran V. Deminoff SJ. Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tooze SA. The role of membrane proteins in mammalian autophagy. Semin Cell Dev Biol. 2010;21:677–682. doi: 10.1016/j.semcdb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 51.Twig G. Elorza A. Molina AJ. Mohamed H. Wikstrom JD. Walzer G. Stiles L. Haigh SE. Katz S. Las G. Alroy J. Wu M. Py BF. Yuan J. Deeney JT. Corkey BE. Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yorimitsu T. Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young AR. Chan EY. Hu XW. Kochl R. Crawshaw SG. High S. Hailey DW. Lippincott-Schwartz J. Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 54.Yu L. McPhee CK. Zheng L. Mardones GA. Rong Y. Peng J. Mi N. Zhao Y. Liu Z. Wan F. Hailey DW. Oorschot V. Klumperman J. Baehrecke EH. Lenardo MJ. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J. Randall MS. Loyd MR. Dorsey FC. Kundu M. Cleveland JL. Ney PA. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ziviani E. Whitworth AJ. How could Parkin-mediated ubiquitination of mitofusin promote mitophagy? Autophagy. 2010;6:660–662. doi: 10.4161/auto.6.5.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]