Abstract
Mitochondria, which convert energy for the cell, accumulate damage with age, and the resulting mitochondrial dysfunction has been linked to the development of degenerative diseases and aging. To curb the accumulation of damaged mitochondria, the cell has elaborated a number of mitochondrial quality control processes. We describe recent work suggesting that Parkin and PTEN-induced putative kinase 1 (PINK1), two gene products linked to familial forms of parkinsonism, may constitute one of the cell's mitochondrial quality control pathways—identifying impaired mitochondria and selectively trimming them from the mitochondrial network by mitophagy. In particular, we discuss the regulation of PINK1 protein expression and Parkin localization by the bioenergetic status of individual mitochondria; the mechanism by which PINK1 recruits Parkin to the outer mitochondrial membrane; and Parkin's promotion of mitophagy through its ubiquitination of outer mitochondrial membrane proteins. This recent work suggests that Parkin and PINK1 may be among the first mammalian proteins identified with a direct role in regulating mitophagy, and implicate a failure of mitophagy in the pathogenesis of Parkinson's disease. Antioxid. Redox Signal. 14, 1929–1938.
Introduction
The mitochondrial electron transport chain couples reduction of oxygen to proton transport across the inner mitochondrial membrane. The electrochemical potential this transport produces drives ATP production, calcium buffering, and mitochondrial protein import, among other mitochondrial processes that are critical for eukaryotic life. Side reactions in this conversion, however, produce harmful reactive oxygen (ROS) and nitrogen species that damage mitochondria and other cellular components. In animal models, both increased oxidative damage and the decreased energy production from dysfunctional mitochondria may exacerbate the aging phenotype (20, 33, 46). Additionally, these appear to contribute to specific diseases of aging, such as the neurodegenerative disorders Alzheimer's disease, amyotrophic lateral sclerosis, and Parkinson's disease (49).
To slow the accumulation of mitochondrial damage that occurs with aging, the cell has elaborated a number of quality control (QC) processes. Mitochondrial proteins damaged by local superoxide production can be broken down on site by mitochondrial proteases such as the mAAA protease complex (1). If the accumulated damage has not led to severe loss of function, mitochondria harboring oxidatively damaged mitochondrial DNA (mtDNA) can fuse with more bioenergetically active mitochondria to borrow missing components of their electron transport chain and/or mitochondrial transfer RNAs, a process known as functional complementation (3). Mitochondria that have accumulated greater damage and are no longer able to maintain a membrane potential, however, appear to be targeted to lysosomes and degraded in a process known as quality control autophagy (hereafter QC mitophagy), after which they may be replaced by the progeny of more bioenergetically active mitochondria. We and others have proposed that PTEN-induced putative kinase 1 (PINK1) and Parkin (both of which are linked to recessive parkinsonism) constitute one of the cell's QC mitophagy pathways. The PINK1/Parkin QC pathway appears to be bioenergetically regulated at the level of the individual mitochondrion, which allows for the selective recognition and elimination of dysfunctional mitochondria by targeted mitophagy. Recent advances in our understanding of PINK1/Parkin QC mitophagy are the focus of this review.
PINK1/Parkin Quality Control Pathway in Drosophila
Parkin, a largely cytosolic E3 ubiquitin ligase, was shown in 1998 to cause a recessive form of parkinsonism (18). Studying a Drosophila model of Parkin disease, a few years later Leo Pallanck and colleagues provided the first strong evidence linking Parkin to a mitochondrial quality control pathway (12, 50). Drosophila lacking Parkin appear grossly normal after ecclosure. As the flies age, however, they lose their ability to fly due to degeneration of their flight muscles, the males are sterile, and a cluster of their dopaminergic neurons degenerates. Interestingly, mitochondria in the affected tissues appear very dysmorphic by electron and light microscopy and are dysfunctional with less efficient oxidative phosphorylation and increased ROS production. These mitochondrial defects precede visible derangement of the muscle fibers, suggesting that the mitochondrial dysfunction is an early event in the degeneration. Additionally, the mitochondrial dysfunction and muscle degeneration is progressive and can be suppressed by decreasing basal levels of oxidative stress (through upregulation of the glutathione detoxifying pathway), pointing to loss of a mitochondrial quality control pathway as a possible cause of the Parkin null phenotype.
Shortly after the mitochondrial kinase PINK1 was also linked to families with recessive parkinsonism (47), three groups independently observed that Drosophila null for Pink1 have the same unusual phenotype as Parkin knockout flies (5, 38, 53). This suggested that Parkin and Pink1 may function in the same genetic pathway, a notion that was supported by the observation that knockout of both Parkin and Pink1 appeared no worse than knockout of either Parkin or Pink1 on its own. Additionally, these groups observed that Parkin overexpression partially rescued the phenotype of Pink1 null flies, whereas Pink1 overexpression failed to rescue the phenotype of Parkin null flies. This observation suggested not only that Pink1 and Parkin operate in a common pathway but also that Pink1 acts genetically upstream of Parkin in the pathway. Finally, because Pink1 had a strong mitochondrial localization signal and exogenously expressed Pink1 is clearly targeted to mitochondria in Drosophila and human cells, by linking Parkin to Pink1 (a bone fide mitochondrial protein), these studies supported the contention that Parkin may play a primary role in a mitochondrial quality control pathway.
Conservation of a PINK1/Parkin Mitochondria Quality Control Pathway in Mammals
Studies in mice, most notably by Jie Shen's group (10, 34), supported the notion that PINK1 and Parkin are important for preserving mitochondrial function in the brain. Loss of Pink1 or Parkin in mice results in a less dramatic phenotype than is observed in Drosophila, and unlike humans, loss of Pink1 and Parkin function in mice does not result in degeneration of dopaminergic neurons in the substantia nigra, or parkinsonism. Nonetheless, mice null for Pink1 or Parkin exhibit synaptic dysfunction in neurons projecting to the striatum and this synaptic dysfunction correlates with progressive loss of mitochondrial function and increased oxidative stress in the striatum with age. The phenotype of the Pink1 and Parkin double knockout is similar to the single knockouts (19), which is at least consistent with the notion that Pink1 and Parkin may function in a common pathway in mice.
Loss of function mutations in PINK1 or Parkin have also been associated with mitochondrial dysfunction in cells from patients with familial forms of parkinsonism (9, 26, 27), suggesting that PINK1 and Parkin may form an evolutionarily conserved quality control pathway that protects against mitochondrial dysfunction.
Regulation of Parkin Mitochondrial Localization by Mitochondrial Membrane Potential
While Parkin and PINK1 seemed to play a role in mitochondrial quality control in vivo, the molecular basis for this protection was obscure. In particular, because Parkin appeared to be predominately a cytosolic protein under basal conditions, it was unclear whether Parkin suppressed the mitochondrial dysfunction observed in vivo from a distance or through specific interactions with mitochondrial proteins.
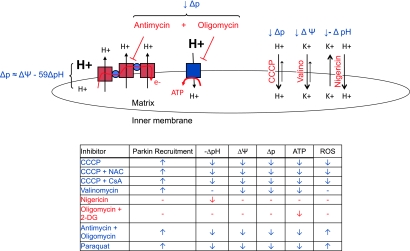
Screening for conditions under which Parkin might be on mitochondria, we found that Parkin's subcellular localization can be regulated by the bioenergetic status of individual mitochondria in the cell (29) (Fig. 1). While Parkin normally resides in the cytosol, it is robustly recruited to mitochondria treated with carbonyl cyanide m-chlorophenylhydrazone (CCCP), a drug that increases the inner membrane permeability to protons, thereby dissipating both the voltage (ΔΨ) and chemical component (ΔpH) of the protonmotive force. In HeLa cells, the recruitment begins ∼25 min after treatment with CCCP, although the dynamics of Parkin recruitment differ between cell types (31). Upon further examination, it appeared that Parkin recruitment is predominately responsive to the voltage component (ΔΨ) of the membrane potential, as it could also be recruited to mitochondria by valinomycin, which increases inner membrane permeability to potassium (thereby dissipating the electrical but not the chemical component of the protonmotive force), and not by nigericin, which reduces ΔpH but not ΔΨ, by promoting K+/H+ exchange across the inner membrane (30). As was later shown by Przedborski and colleagues, Parkin can also be recruited by antimycin in combination with oligomycin, which leads to collapse of the protonmotive force by blocking proton translocation by the respiratory chain and the F1Fo-ATPase (48). Additionally, in Rho0 cells, which lack mtDNA and a functional ETC, Parkin is recruited after inhibition of the F1-ATPase by azide, leading to collapse of the residual protonmotive force in these cells (45). The induction of Parkin recruitment by CCCP is unlikely to be due to increased mitochondrial ROS production, as CCCP treatment leads to a decrease in ROS production in HeLa cells (23), and the CCCP-induced recruitment of Parkin is not blocked by antioxidants, such as N-acetylcysteine (8, 29), or inhibitors of permeability transition, such as cyclosporine (48).
FIG. 1.
Schematic depicting ion transport across the inner mitochondrial membrane. Parkin is recruited to mitochondria with low voltage (ΔΨ) across the inner mitochondrial membrane. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
We find that in addition to chemical uncouplers and oxidative phosphorylation inhibitors, Parkin can be recruited to mitochondria by relatively long exposure to oxidative stress (after treatment with paraquat), chronic loss of mitochondrial fusion, or loss of mitochondrial DNA integrity (29, 45). However, in each of these cases the recruitment of Parkin correlates with loss of ΔΨ at the level of individual mitochondria, suggesting that collapse of ΔΨ is likely the signal for Parkin recruitment in these cases as well. Although it was recently suggested that treatment with H2O2 induces Parkin recruitment in the absence of ΔΨ collapse, the interpretation of these data is difficult as membrane potential was assessed at the population level while Parkin recruitment was assessed in individual cells by microscopy (41). This discrepancy may be resolved by examination of membrane potential and Parkin recruitment at the level of individual mitochondria after treatment with H2O2. Collapse of membrane potential has now been shown to recruit both endogenous and tagged Parkin to mitochondria by several independent groups and in a variety of cell lines (8, 11, 15, 22, 25, 29, 45, 48, 57). These include HEK293, HeLa, and 143B osteosarcoma cells; neuronal-like M17 and SH-SY5Y cells; Drosophila S1 cells; primary and transformed mouse embryonic fibroblasts; primary human fibroblasts; and primary cortical neurons. Together, these findings suggest that the regulation of Parkin localization by collapse of ΔΨ is robust and may be evolutionarily conserved in metazoans.
Study of cells with bioenergetically heterogenous populations of mitochondria (due to paraquat-induced damage, a chronic fusion defect, or an mtDNA mutation) provides additional clues as to how Parkin is regulated by membrane potential (29, 45). In a cell in which some mitochondria are bioenergetically active and some are depolarized, Parkin is preferentially recruited to the mitochondria that have lost membrane potential. This suggests that the recruitment of Parkin to depolarized mitochondria is regulated in a mitochondria-autonomous fashion: that is, the molecular events between loss of membrane potential and Parkin recruitment are confined to the individual mitochondrion that has suffered ΔΨ collapse.
PINK1 Signals ΔΨ Collapse to Parkin
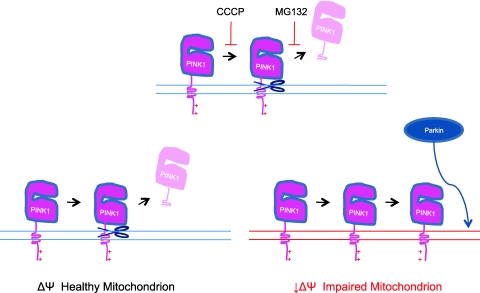
PINK1 appears to play a pivotal role in communicating ΔΨ collapse to Parkin. PINK1, which is most likely anchored to the outer mitochondrial membrane by a transmembrane domain (at least under depolarizing conditions), is normally expressed at low levels on mitochondria, due to its constitutive two-step processing on bioenergetically active mitochondria (24, 25, 31). In the presence of a membrane potential, newly imported PINK1 is rapidly cleaved from mitochondria by an unidentified protease and is subsequently degraded in a proteasome-dependent manner. Interestingly, the first proteolytic step is voltage sensitive, and, like most outer membrane proteins, the import of PINK1 is voltage insensitive. Thus, while PINK1 is normally kept at low levels on healthy mitochondria, it rapidly and selectively accumulates on mitochondria that have lost membrane potential.
As was originally reported by Kim and colleagues, increasing PINK1 levels in the cell is sufficient to recruit Parkin to mitochondria (17), and as we and others demonstrated, the expression of PINK1 is necessary for the recruitment of Parkin to depolarized mitochondria (25, 31, 48, 57).
Together, these findings suggested to us a partial model for the communication of ΔΨ collapse to Parkin (31) (Fig. 2). In this model, the selective inhibition of PINK1 cleavage after ΔΨ collapse leads to its rapid accumulation on mitochondria, and the accumulated PINK1 selectively recruits Parkin to the depolarized mitochondria. This model predicts that stable expression of PINK1 on the outer mitochondrial membrane should robustly recruit Parkin to mitochondria in the absence of ΔΨ collapse. To test this hypothesis, we generated a PINK1 fusion protein with increased stability on the outer mitochondrial membrane. The stabilized PINK1 fusion protein recruited Parkin to mitochondria more efficiently than full-length PINK1, consistent with the proposed model in which mitochondrial dysfunction induces Parkin recruitment by stabilizing PINK1.
FIG. 2.
Cartoon depicting regulation of PINK1 processing by voltage potential across the inner mitochondrial membrane. PTEN-induced putative kinase 1 (PINK1) is proteolytically cleaved and degraded in healthy mitochondria but stabilized on mitochondria with low inner membrane voltage (ΔΨ). (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
In addition to the questions it resolves, the model also highlights what is still poorly understood about how Parkin is recruited and activated by ΔΨ collapse: namely, (i) how ΔΨ collapse inhibits PINK1 cleavage, that is, the identity of the ΔΨ sensor in the system; and (ii) how accumulated PINK1 recruits and activates Parkin.
In answer to the first question, it is tempting to speculate that PINK1's import and cleavage may be coupled and that these coupled processes may constitute the ΔΨ sensor of the PINK1/Parkin QC pathway. PINK1 appears to be targeted to mitochondria differently depending on whether or not the mitochondria have a membrane potential. In the presence of a membrane potential, a canonical mitochondrial targeting signal in PINK1's N terminus appears capable of engaging the translocase of the inner membrane subunit Tim23 (Tim23) import pathway, which is ΔΨ dependent. Fusion of PINK1's mitochondrial targeting signal (MTS) to cyan fluorescent protein targets cyan fluorescent protein to the matrix, demonstrating that the MTS is sufficient for targeting along the canonical Tim23 pathway (44). Similarly, removal of the putative transmembrane (TM) sequence directly after PINK1's MTS results in targeting of PINK1 to the matrix (56). This suggests that in the absence of the TM, which acts as a stop transfer, PINK1 is likely imported by the Tim23 pathway to the matrix. However, PINK1 continues to be targeted to mitochondria in the presence of uncouplers like CCCP and valinomycin (which inhibit the Tim23 pathway) and does not require most of its canonical MTS for targeting to the outer membrane (11, 24, 25, 31, 48, 56). We suggest that these two modes of mitochondrial targeting, one in the presence of a membrane potential and one in the absence of a membrane potential, may act as the ΔΨ sensor. In this scenario, PINK1's MTS would engage the Tim23 complex in the presence of a membrane potential. This interaction would position PINK1 for cleavage by the protease, maintaining low levels of PINK1 on mitochondria and suppressing Parkin activation. In the absence of a membrane potential, however, PINK1 would not engage the Tim23 complex and would not be cleaved. Instead, it would be targeted to the outer membrane where it can accumulate and activate Parkin. Whether this model is correct and exactly how this might work will require additional study into the mechanism of PINK1 mitochondrial import in the presence and absence of a membrane potential and the mechanism of PINK1 cleavage (e.g., the identification of PINK1's cleavage site and the protease(s) responsible for its cleavage).
How accumulated PINK1 on the outer mitochondrial membrane recruits Parkin to mitochondria and activates Parkin is more mysterious. PINK1 appears able to direct Parkin to mitochondria with collapsed ΔΨ, and thus it seems PINK1 is able to selectively increase the affinity of some mitochondria for Parkin but not others. In addition, because PINK1 with its catalytic lysine mutated fails to recruit Parkin, likely PINK1's kinase activity is required for Parkin recruitment and activation (11, 31, 48).
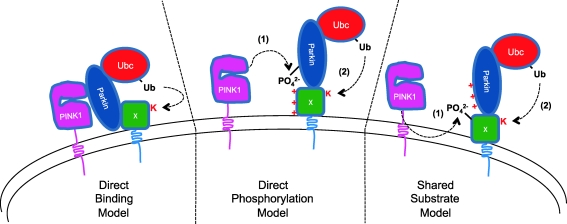
Several models for the recruitment and activation of Parkin by PINK1 have been proposed and each has strengths and weaknesses (Fig. 3). In what might be called the “binding” model, PINK1 and Parkin are postulated to form a complex. In this model, under basal conditions most of Parkin is in the cytosol because little PINK1 is present on the outer membrane to bind Parkin. As PINK1 accumulates in response to mitochondrial damage, it binds Parkin, thereby anchoring it to the surface of the impaired mitochondria. The binding model explains why Parkin has increased affinity for some mitochondria and not others (some mitochondria display more PINK1 on their surface and therefore some mitochondria have a higher Parkin binding capacity than others), and in support of the model several groups have reported that Parkin and PINK1 coimmunoprecipitate (42, 51). In addition, both Parkin and PINK1's kinase domain are on the cytosolic face of the outer mitochondrial membrane after depolarization, making a direct interaction at least plausible (31, 48, 56). However, the model does not explain why PINK1's catalytic lysine is required for Parkin recruitment to mitochondria. While it is possible that mutating the PINK1's catalytic lysine disrupts Parkin binding, PINK1 K219M appears to coimmunoprecipitate Parkin as well as wild-type PINK1 (42), suggesting that this is unlikely to be the reason PINK1's kinase activity is required for Parkin recruitment.
FIG. 3.
Several models for the recruitment of Parkin to mitochondria by the kinase PINK1. In the direct binding model, PINK1 recruits Parkin through direct binding. In the direct phosphorylation model, phosphorylation of Parkin by PINK1 increases Parkin's affinity for depolarized mitochondria. In the shared substrate model, PINK1's phosphorylation of mitochondrial proteins increases their affinity for the E3 ubiquitin ligase Parkin. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
In what might be called the direct phosphorylation model, PINK1 renders Parkin active and competent for recruitment through direct phosphorylation. While this model explains why PINK1's kinase activity is required for Parkin recruitment, it has more difficulty explaining how PINK1 directs Parkin to a subset of mitochondria. In this model it is also not clear why PINK1 lacking its mitochondrial-targeting N-terminus fails to induce Parkin recruitment until after its association with mitochondria is forced, for example, through induced heterodimerization (31). The evidence for direct phosphorylation of Parkin by PINK1 is mixed, with some groups reporting direct phosphorylation of purified Parkin or Parkin peptides by PINK1 (17, 42), and one group failing to see phosphorylation of Parkin by PINK1 in vitro (48). Assessing whether Parkin is inducibly phosphorylated after depolarization in cell culture would help determine whether Parkin phosphorylation is necessary to increase its affinity for mitochondria.
Alternatively, in what might be called the shared substrate model, PINK1 may locally phosphorylate mitochondrial proteins, thereby increasing their affinity for Parkin. Parkin, in turn, would promote their ubiquitination. This would explain both the requirement for PINK1's kinase activity as well as PINK1's ability to direct Parkin to a subset of mitochondria. No PINK1 substrates have been described on the outer mitochondrial membrane, but recently mitochondrial assembly regulatory factor (Marf), a Drosophila ortholog of the mitofusins, was identified as a Parkin substrate in Drosophila, and voltage-dependent anion-selective channel protein 1 (VDAC1) was identified as a Parkin substrate in mammalian cells (11, 57). It will be interesting to learn whether the mitofusins or VDAC1 are also substrates of PINK1 and whether their phosphorylation is required for their ubiquitination by Parkin.
These models, of course, are not mutually exclusive. For instance, it may be that PINK1 directly or indirectly induces a conformational change in Parkin and increases the affinity of a subset of mitochondria for Parkin.
Recruited Parkin Tags Impaired Mitochondria for Degradation
Shortly after recruitment of Parkin to impaired mitochondria, a subset of the mitochondria are engulfed by autophagosomes (29). Amazingly, in cells overexpressing Parkin and treated with a depolarizing agent or overexpressed PINK1, this induced mitophagy can go to completion (29, 31). All mitochondria within the cell can be degraded within 24–96 h, and cells lacking mitochondria can survive for up to 2 weeks under typical cell culture conditions. Although this phenomenon (complete degradation of mitochondria) has been reported previously in the context of cells undergoing apoptosis in the presence of caspase inhibitors and in the maturation of specialized cell type such as reticulocytes (13, 52), to our knowledge PINK1 and Parkin are the first factors described that are minimally sufficient to induce complete mitophagy in the metazoan cell.
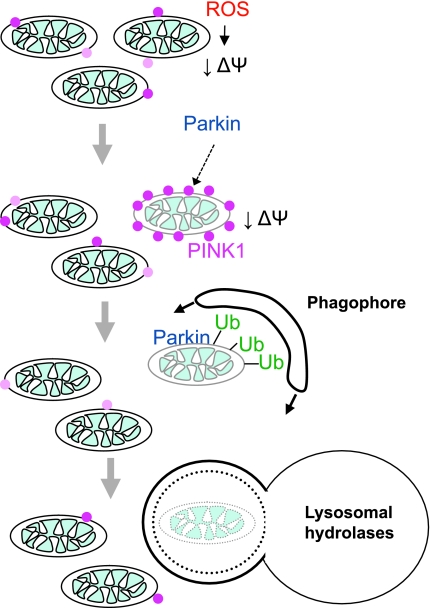
Together, our findings also suggested a model in which PINK1 and Parkin are regulated by the bioenergetic status of individual mitochondria, enabling them to survey the mitochondrial network and selectively trim dysfunctional mitochondria from it (Fig. 4). To test this hypothesis more rigorously, we examined the effect of altering Parkin expression in cells containing a stable proportion of wild-type mtDNA and mtDNA with a deleterious mutation in the CoxIV subunit, which leads to mitochondrial dysfunction (45). We hypothesized that if Parkin was able to target and selectively remove dysfunctional mitochondria containing the mutant DNA, then over time we should see a shift in the ratio of wild-type and mutant mtDNA. In cells with 2–6-fold higher levels of Parkin expression than is observed in endogenous tissues, almost all of the mutant mtDNA was removed, allowing the cells to repopulate with wild-type mtDNA. This reversion to wild-type mtDNA was accompanied by a restoration in cytochrome c oxidase enzymatic activity. These findings support the hypothesis that the PINK1-Parkin pathway is able to selectively identify and remove dysfunctional mitochondria from the cell. It will be interesting to learn whether manipulating Parkin expression levels can similarly alter the rate of deleterious mtDNA mutations in vivo.
FIG. 4.
PINK1 and Parkin target depolarized mitochondria for autophagic degradation. PINK1 selectively accumulates on mitochondria with low voltage (ΔΨ) across their inner membranes. The accumulated PINK1 directs Parkin to mitochondria with low ΔΨ. Parkin tags the dysfunctional mitochondria with ubiquitin, which likely serves as the signal for their subsequent degradation in lysosomes. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Parkin's E3 Ubiquitin Ligase Activity May Be Required for Mitophagy
How Parkin induces mitophagy after its recruitment and activation by PINK1 is not fully resolved, but a number of advances have been made recently (Fig. 5). Parkin promotes the robust polyubiquitination of mitochondrial proteins after its recruitment to mitochondria (11, 25). In addition, mutations that disrupt Parkin's RING domains or lead to loss of the RING2 domain, which is required for Parkin's ubiquitin ligase activity in vitro (4, 43), disrupt recruitment of Parkin to mitochondria (11, 25, 31). These results suggest that Parkin's mitochondrial recruitment may be mediated by its binding to substrates. A mutation located in a loop within RING1, R275W, appears to disrupt Parkin's ubiquitination of mitochondrial proteins as well as its ability to induce mitophagy without affecting its recruitment to mitochondria (11, 31). This correlation between mitochondria ubiquitination and mitophagy supports the notion that Parkin's induction of mitophagy requires the ubiquitination of mitochondrial proteins.
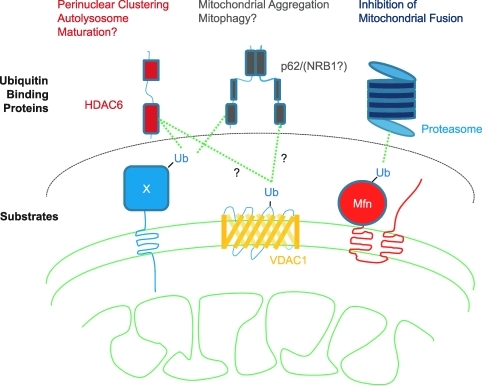
FIG. 5.
Cartoon depicting substrates ubiquitinated by Parkin and ubiquitin-binding adapter proteins recruited by Parkin. Ubiquitination of mitochondrial membrane proteins by Parkin results in mitochondrial quality control likely mediated by different ubiquitin-binding adaptor proteins. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
To date, two potential substrates of Parkin have been proposed on the outer mitochondrial membrane, one in a mammalian system, VDAC1 (11), and one in the Drosophila system, Marf (40, 57), which is an ortholog of the mammalian mitofusins.
VDAC1 is ubiquitinated by overexpressed Parkin in mammalian cells, after mitochondrial depolarization (11). Ubiquitination of VDAC1 did not appear to affect its stability, suggesting that VDAC1 ubiquitination may function in a nonproteasomal pathway, possibly serving as a positive signal for the autophagy of mitochondria. Consistent with this view, depletion of VDAC1 by RNA interference was reported to cause a modest reduction in Parkin recruitment and Parkin-induced mitophagy after depolarization.
To explain how ubiquitination of VDAC1 may lead to mitophagy, it was suggested that an adaptor protein, p62, which can bind both ubiquitin and a protein integral to autophagosomes, light chain 3-II, may recruit autophagosomes to ubiquitinated VDAC1. A model of this sort has been proposed for other forms of selective autophagy, including the autophagy of ubiquitinated proteins in the face of ubiquitin proteasome system insufficiency (2, 37), peroxisomes (16), and some intracellular bacteria (55). In support of this model, it was reported that p62 is recruited to mitochondria in a Parkin- and depolarization-dependent manner and that knockdown of p62 substantially inhibits mitophagy (8, 11, 22). While a model of this sort may be correct (Parkin tags an outer membrane protein with ubiquitin, which selectively recruits autophagosomes via an adapator protein), whether VDAC1 and p62 are necessary or sufficient for Parkin-induced mitophagy is in need of additional confirmation. At least in mouse embryonic fibroblasts lacking VDAC1 or p62, VDAC1 (49) and p62 (28, 32) appear to be dispensible for Parkin-induced mitophagy, although p62 appears to be required for mitochondrial clustering (28, 32). Additionally, we have been unable to confirm the findings that depletion of p62 by RNAi leads to a substantial reduction of mitophagy in HeLa cells (28). Finally, p62 can be recruited to mitochondria by a mitochondrion-anchored ubiquitin fusion protein, but it fails to induce substantial mitophagy, suggesting that p62 recruitment to mitochondria may also be insufficient for mitophagy (28). Therefore, VDAC1 and p62 may not be responsible for Parkin-induced mitophagy and/or there may be redundancy both in outer membrane proteins carrying the ubiquitin tag and in the adaptor proteins linking ubiquitinated mitochondria to autophagosomes. NBR1 (neighbor of BRCA1 gene 1), for instance, appears to be at least partially functionally redundant with p62 and may be a good candidate for additional investigation.
In addition, it was recently suggested that the Drosophila mitofusin ortholog Marf may be a substrate of Parkin in Drosophila cells (40, 57). The mitofusins are GTPases on the outer mitochondrial membrane and mediate outer membrane fusion. Thus, their elimination or inhibition by ubiquitination would be predicted to promote fission of the mitochondrial network. Consistent with this prediction, ubiquitination of Marf by Parkin appears to promote mitochondrial fission. Parkin-induced mitochondrial fission, in turn, appears to be necessary for mitophagy to proceed (40, 57). This finding is consistent with previous reports that mitochondrial fission is necessary for efficient mitophagy and suggests that Marf ubiquitination may be a permissive event for Parkin-induced mitophagy, at least in Drosophila. These findings are also consistent with genetic data in Drosophila suggesting that genetic alterations that promote net fission of the mitochondrial network (e.g., knockdown of Drosophila mitofusins or overexpression of the mitochondrial fission protein Fis1) can partially compensate for loss of Parkin or PINK1 (7, 39, 54). It will be important to determine whether the ubiquitination of mitofusins by Parkin is conserved in mammalian cells.
Together, these data support the notion that Parkin is acting as an E3 ubiquitin ligase on the outer mitochondrial membrane after its recruitment resulting from treatment with CCCP. As new Parkin substrates are identified on mitochondria, it will be important to distinguish between proteins that are incidentally ubiquitinated by overexpressed Parkin and substrates that play a substantial role in Parkin-induced mitophagy. Making this distinction may be challenging if Parkin has multiple mitochondrial substrates, each of which contributes something to mitophagy and none of which is strictly necessary for mitophagy. If Parkin has multiple substrates that serve as a positive signal for mitophagy, it also will be important to determine the rules that govern which substrate(s) contributes to the pro-mitophagy signal and which does not.
In addition to promoting mitochondrial fission and mitophagy, Parkin appears to promote the minus-end transport of depolarized mitochondria along microtubules. Shortly after Parkin recruitment, mitochondria clump and gather in a perinuclear location (29, 48). This is not observed to the same extent in cells lacking Parkin, suggesting that it is a Parkin-dependent phenomenon. The perinuclear gathering of mitochondria requires a microtubular network and is likely driven by increased retrograde transport of mitochondria by the dynein motor (22, 48). Consistent with this notion, overexpression of dynactin (which inhibits dynein-driven retrograde transport) suppresses the retrograde transport of mitochondria after Parkin recruitment (22, 48). Recently, it was suggested that this perinuclear gathering of mitochondria is a prerequisite for Parkin-induced mitophagy, as in fibroblasts mitochondria are not cleared by Parkin if the microtubule network is disrupted by nocodazole treatment (22). In HeLa cells, however, we find that nocodazole does not inhibit mitophagy (28). Additionally, in mouse embryonic fibroblasts lacking p62, mitophagy appears to proceed without mitochondrial clumping as an intermediate (28, 32). Together, these findings suggest that additional work may be required to sort out the role the microtubular network plays in Parkin-induced mitophagy.
In some cell types such as the follicular cells of Drosophila, Parkin's primary role may be to promote the transport of mitochondria to the minus end of microtubules (6). In this system, because of the unusual radiation of microtubules from the plus end in the center of the cell to the minus end in the periphery, Parkin-dependent minus-end movement leads to the dispersion rather than the concentration of mitochondria in the cell. In the absence of Parkin, mitochondria become aggregated in the center of follicular cells.
The molecular mechanism governing Parkin regulation of minus-end transport of mitochondria along microtubules is not known. It was suggested that histone deacetylase 6 (HDAC6), which Yao and colleagues had previously found mediates the minus-end movement of ubiquitinated protein aggregates to the microtubule organizing center (14), may be responsible for this minus-end transport of mitochondria (22). They find that like p62, HDAC6, which has a ubiquitin binding domain, is recruited to mitochondria that have been ubiquitinated by Parkin. Additionally, they report that HDAC6 as well as cortactin promotes Parkin-induced mitophagy. While the authors attribute the promotion of mitophagy to minus-end transport, it is also possible that HDAC6 and cortactin are promoting mitophagy by increasing the rate of autophagosome-lysosomal fusion through actin remodeling, as Lee, Yao, and colleagues suggest occurs in the context of quality control mitophagy (21). HDAC6's involvement in Parkin-induced minus-end microtubular transport might be tested in follicular cells of Drosophila, where the situation is likely not confounded by a potential role for HDAC6 in the later stages of mitophagy. Whatever the mechanism, as HDAC6 overexpression was shown to promote the autophagic clearance of protein aggregates in Drosophila models of Huntington's disease and Alzheimer's disease previously (35, 36), it will be interesting to learn whether HDAC6 overexpression is similarly able to clear dysfunctional mitochondria from Parkin- and/or Pink1-deficient flies.
Summary
Recent studies have identified Parkin and PINK1 as mediators of a mitochondrial quality control pathway. PINK1 expression levels are regulated by the bioenergetic status of individual mitochondria and drive recruitment of Parkin from the cytosol to impaired mitochondria. On the outer mitochondrial membrane, Parkin promotes the ubiquitination of the mitochondrial fusion protein, mitofusin, thereby isolating the damaged mitochondrion from the mitochondrial network. Parkin also ubiquitinates VDAC1 and likely other proteins, and ultimately promotes the selective degradation of the damaged mitochondria by autophagy. Teasing out how PINK1 expression is regulated by membrane potential, how PINK1 directs Parkin to impaired mitochondria, and how Parkin induces mitophagy remain the central questions for future work on the PINK1/Parkin mitophagy pathway. Also important will be determining whether this pathway is of physiological importance in vivo, and how therapeutic regulation of this pathway might be used to treat diseases resulting from mitochondrial dysfunction.
Abbreviations Used
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
- HDAC6
histone deacetylase 6
- Marf
mitochondrial assembly regulatory factor
- mtDNA
mitochondrial DNA
- MTS
mitochondrial targeting signal
- NBR1
neighbor of BRCA1 gene 1
- PINK1
PTEN-induced putative kinase 1
- QC
quality control
- ROS
reactive oxygen species
- Tim23
translocase of the inner membrane subunit Tim23
- TM
transmembrane
- VDAC1
voltage-dependent anion-selective channel protein 1
Acknowledgments
R.J.Y. and D.P.N. are funded by the National Institutes of Health intramural program. D.P.N is a member of the U.S. National Institutes of Health-Oxford-Cambridge Scholars Program.
References
- 1.Arnold I. Langer T. Membrane protein degradation by AAA proteases in mitochondria. Biochim Biophys Acta. 2002;1592:89–96. doi: 10.1016/s0167-4889(02)00267-7. [DOI] [PubMed] [Google Scholar]
- 2.Bjorkoy G. Lamark T. Brech A. Outzen H. Perander M. Overvatn A. Stenmark H. Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H. Chan DC. Physiological functions of mitochondrial fusion. Ann N Y Acad Sci. 2010;1201:21–25. doi: 10.1111/j.1749-6632.2010.05615.x. [DOI] [PubMed] [Google Scholar]
- 4.Chung KK. Zhang Y. Lim KL. Tanaka Y. Huang H. Gao J. Ross CA. Dawson VL. Dawson TM. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat Med. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- 5.Clark IE. Dodson MW. Jiang C. Cao JH. Huh JR. Seol JH. Yoo SJ. Hay BA. Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 6.Cox RT. Spradling AC. Clueless, a conserved Drosophila gene required for mitochondrial subcellular localization, interacts genetically with parkin. Dis Model Mech. 2009;2:490–499. doi: 10.1242/dmm.002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng H. Dodson MW. Huang H. Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding WX. Ni HM. Li M. Liao Y. Chen X. Stolz DB. Dorn Ii GW. Yin XM. Nix is critical to two distinct phases of mitophagy: reactive oxygen species (ROS)-mediated autophagy induction and Parkin-ubiqutin-p62-mediated mitochondria priming. J Biol Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Exner N. Treske B. Paquet D. Holmstrom K. Schiesling C. Gispert S. Carballo-Carbajal I. Berg D. Hoepken HH. Gasser T. Kruger R. Winklhofer KF. Vogel F. Reichert AS. Auburger G. Kahle PJ. Schmid B. Haass C. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautier CA. Kitada T. Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geisler S. Holmstrom KM. Skujat D. Fiesel FC. Rothfuss OC. Kahle PJ. Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 12.Greene JC. Whitworth AJ. Kuo I. Andrews LA. Feany MB. Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm TM. Braun A. Trigatti BL. Brugnara C. Sakamoto M. Krieger M. Andrews NC. Failure of red blood cell maturation in mice with defects in the high-density lipoprotein receptor SR-BI. Blood. 2002;99:1817–1824. doi: 10.1182/blood.v99.5.1817. [DOI] [PubMed] [Google Scholar]
- 14.Kawaguchi Y. Kovacs JJ. McLaurin A. Vance JM. Ito A. Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 15.Kawajiri S. Saiki S. Sato S. Sato F. Hatano T. Eguchi H. Hattori N. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010;584:1073–1079. doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Kim PK. Hailey DW. Mullen RT. Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y. Park J. Kim S. Song S. Kwon SK. Lee SH. Kitada T. Kim JM. Chung J. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 18.Kitada T. Asakawa S. Hattori N. Matsumine H. Yamamura Y. Minoshima S. Yokochi M. Mizuno Y. Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 19.Kitada T. Tong Y. Gautier CA. Shen J. Absence of nigral degeneration in aged parkin/DJ-1/PINK1 triple knockout mice. J Neurochem. 2009;111:696–702. doi: 10.1111/j.1471-4159.2009.06350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kujoth GC. Hiona A. Pugh TD. Someya S. Panzer K. Wohlgemuth SE. Hofer T. Seo AY. Sullivan R. Jobling WA. Morrow JD. Van Remmen H. Sedivy JM. Yamasoba T. Tanokura M. Weindruch R. Leeuwenburgh C. Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 21.Lee JY. Koga H. Kawaguchi Y. Tang W. Wong E. Gao YS. Pandey UB. Kaushik S. Tresse E. Lu J. Taylor JP. Cuervo AM. Yao TP. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JY. Nagano Y. Taylor JP. Lim KL. Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J. Gao X. Qian M. Eaton JW. Mitochondrial metabolism underlies hyperoxic cell damage. Free Radic Biol Med. 2004;36:1460–1470. doi: 10.1016/j.freeradbiomed.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Lin W. Kang UJ. Characterization of PINK1 processing, stability, and subcellular localization. J Neurochem. 2008;106:464–474. doi: 10.1111/j.1471-4159.2008.05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuda N. Sato S. Shiba K. Okatsu K. Saisho K. Gautier CA. Sou YS. Saiki S. Kawajiri S. Sato F. Kimura M. Komatsu M. Hattori N. Tanaka K. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortiboys H. Thomas KJ. Koopman WJ. Klaffke S. Abou-Sleiman P. Olpin S. Wood NW. Willems PH. Smeitink JA. Cookson MR. Bandmann O. Mitochondrial function and morphology are impaired in parkin-mutant fibroblasts. Ann Neurol. 2008;64:555–565. doi: 10.1002/ana.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muftuoglu M. Elibol B. Dalmizrak O. Ercan A. Kulaksiz G. Ogus H. Dalkara T. Ozer N. Mitochondrial complex I and IV activities in leukocytes from patients with parkin mutations. Mov Disord. 2004;19:544–548. doi: 10.1002/mds.10695. [DOI] [PubMed] [Google Scholar]
- 28.Narendra D. Kane LA. Hauser DN. Fearnley IM. Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narendra D. Tanaka A. Suen DF. Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narendra D. Tanaka A. Suen DF. Youle RJ. Parkin-induced mitophagy in the pathogenesis of Parkinson disease. Autophagy. 2009;5:706–708. doi: 10.4161/auto.5.5.8505. [DOI] [PubMed] [Google Scholar]
- 31.Narendra DP. Jin SM. Tanaka A. Suen DF. Gautier CA. Shen J. Cookson MR. Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okatsu K. Saisho K. Shimanuki M. Nakada K. Shitara H. Sou YS. Kimura M. Sato S. Hattori N. Komatsu M. Tanaka K. Matsuda N. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orr WC. Sohal RS. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263:1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- 34.Palacino JJ. Sagi D. Goldberg MS. Krauss S. Motz C. Wacker M. Klose J. Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 35.Pandey UB. Batlevi Y. Baehrecke EH. Taylor JP. HDAC6 at the intersection of autophagy, the ubiquitin-proteasome system and neurodegeneration. Autophagy. 2007;3:643–645. doi: 10.4161/auto.5050. [DOI] [PubMed] [Google Scholar]
- 36.Pandey UB. Nie Z. Batlevi Y. McCray BA. Ritson GP. Nedelsky NB. Schwartz SL. DiProspero NA. Knight MA. Schuldiner O. Padmanabhan R. Hild M. Berry DL. Garza D. Hubbert CC. Yao TP. Baehrecke EH. Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 37.Pankiv S. Clausen TH. Lamark T. Brech A. Bruun JA. Outzen H. Overvatn A. Bjorkoy G. Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 38.Park J. Lee SB. Lee S. Kim Y. Song S. Kim S. Bae E. Kim J. Shong M. Kim JM. Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 39.Poole AC. Thomas RE. Andrews LA. McBride HM. Whitworth AJ. Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poole AC. Thomas RE. Yu S. Vincow ES. Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rakovic A. Grunewald A. Seibler P. Ramirez A. Kock N. Orolicki S. Lohmann K. Klein C. Effect of endogenous mutant and wild-type PINK1 on Parkin in fibroblasts from Parkinson disease patients. Hum Mol Genet. 2010;19:3124–3137. doi: 10.1093/hmg/ddq215. [DOI] [PubMed] [Google Scholar]
- 42.Sha D. Chin LS. Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-kappaB signaling. Hum Mol Genet. 2010;19:352–363. doi: 10.1093/hmg/ddp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimura H. Hattori N. Kubo S. Mizuno Y. Asakawa S. Minoshima S. Shimizu N. Iwai K. Chiba T. Tanaka K. Suzuki T. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 44.Silvestri L. Caputo V. Bellacchio E. Atorino L. Dallapiccola B. Valente EM. Casari G. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 45.Suen DF. Narendra DP. Tanaka A. Manfredi G. Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci U S A. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trifunovic A. Wredenberg A. Falkenberg M. Spelbrink JN. Rovio AT. Bruder CE. Bohlooly YM. Gidlof S. Oldfors A. Wibom R. Tornell J. Jacobs HT. Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 47.Valente EM. Abou-Sleiman PM. Caputo V. Muqit MM. Harvey K. Gispert S. Ali Z. Del Turco D. Bentivoglio AR. Healy DG. Albanese A. Nussbaum R. Gonzalez-Maldonado R. Deller T. Salvi S. Cortelli P. Gilks WP. Latchman DS. Harvey RJ. Dallapiccola B. Auburger G. Wood NW. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 48.Vives-Bauza C. Zhou C. Huang Y. Cui M. de Vries RL. Kim J. May J. Tocilescu MA. Liu W. Ko HS. Magrane J. Moore DJ. Dawson VL. Grailhe R. Dawson TM. Li C. Tieu K. Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitworth AJ. Theodore DA. Greene JC. Benes H. Wes PD. Pallanck LJ. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson's disease. Proc Natl Acad Sci U S A. 2005;102:8024–8029. doi: 10.1073/pnas.0501078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong H. Wang D. Chen L. Choo YS. Ma H. Tang C. Xia K. Jiang W. Ronai Z. Zhuang X. Zhang Z. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest. 2009;119:650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue L. Fletcher GC. Tolkovsky AM. Mitochondria are selectively eliminated from eukaryotic cells after blockade of caspases during apoptosis. Curr Biol. 2001;11:361–365. doi: 10.1016/s0960-9822(01)00100-2. [DOI] [PubMed] [Google Scholar]
- 53.Yang Y. Gehrke S. Imai Y. Huang Z. Ouyang Y. Wang JW. Yang L. Beal MF. Vogel H. Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y. Ouyang Y. Yang L. Beal MF. McQuibban A. Vogel H. Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng YT. Shahnazari S. Brech A. Lamark T. Johansen T. Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 56.Zhou C. Huang Y. Shao Y. May J. Prou D. Perier C. Dauer W. Schon EA. Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ziviani E. Tao RN. Whitworth AJ. Drosophila Parkin requires PINK1 for mitochondrial translocation and ubiquitinates Mitofusin. Proc Natl Acad Sci U S A. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]