Abstract
To advance knowledge on polyplex structure and composition, fluorescence resonance energy transfer (FRET) and anisotropy measurements were applied to polyplexes of rhodamine-labeled polyethylenimine (PEI) and fluorescein-labeled double-stranded oligodeoxynucleotide (ODN). About 25 kDa PEI was compared with low-molecular-weight PEI of 2.7 kDa. FRET reached maxima at amine to phosphate (N/P) ratios of 2 and 3 for 2.7 kDa and 25 kDa PEI, respectively, with similar average distances between donor and acceptor dye molecules in polyplexes. Anisotropy measurements allowed estimating the bound fractions of PEI and ODN. At N/P = 6, all ODN was bound, but only 58% of PEI 25 kDa and 45% of PEI 2.7 kDa. In conclusion, the higher molecular weight of PEI may conformationally restrict the availability of amino groups for charge interaction with phosphate groups in ODN. Moreover, significant fractions of both types of PEI remain free in solution at N/P ratios frequently used for transfection. FRET and anisotropy measurements provide effective tools for probing polyplex compositions and designing optimized delivery systems.
Introduction
Polyethylenimine (PEI) is a cationic polymer, which achieves high transfection efficiency as nonviral carriers for nucleic acid therapeutics such as plasmid DNA and as oligonucleotides (Boussif et al., 1995; Kircheis et al., 2001; Vinogradov et al., 2004; Akinc et al., 2005). Due to its high density of positive charges, it has the capability to protect nucleic acids from enzymatic degradation by formation of condensed complexes (polyplexes) and it mediates endosomal escape by a “proton sponge” mechanism (Boussif et al., 1995; Akinc et al., 2005). Complex formation between PEI or PEI-derivatives and nucleic acids has been investigated with various techniques, including dynamic light scattering (DLS) for size and zeta potential measurement, dye exclusion (Kunath et al., 2003), gel retardation (Fischer et al., 2005), and DNAse digestion with most of the studies focused on the fate of nucleic acids, mainly with regard to condensation and protection upon complex formation. However, the fate of the other partner in such complex formulations, PEI, has not been studied as extensively. PEI of different molecular weights and structure (branched vs. linear) may lead to different polyplex structure, resulting in differences in stability and release of the loaded nucleic acids (Fischer et al., 1999; Godbey et al., 1999). While it is obvious that adequate amounts of PEI are necessary for complete condensation of nucleic acids, it has been reported that polyplexes prepared at high amine group to phosphate group (N/P) ratios are particularly efficient transfection agents. However, a high N/P ratio can also cause toxicity as a consequence of free PEI in the formulation (Zou et al., 2000; Boeckle et al., 2004). Therefore, a good quantitative method for determination of the fractions of each component, which are either incorporated into complexes or remain free in solution, is desirable. Measurement of the fractional distribution by chromatographic separation of complex and free PEI (Boeckle et al., 2004) is feasible, but this multi-step process potentially introduces errors by re-equilibration and/or loss of free PEI due to absorption on surfaces. Techniques able to detect bound and free fractions in a homogenous phase, thus not requiring prior separation, should avoid such problems. The present studies utilized 2 techniques for characterization of polyplexes between PEI and a model double-stranded oligodeoxynucleotide (ODN), which is a 20-mer containing the NF-κB cis element (Morishita et al., 1997; Fischer et al., 2005). A commercially available branched PEI with MW 25 kDa (PEI25) and custom-synthesized low-molecular-weight PEI with MW 2.7 kDa (PEI2.7) (Fischer et al., 1999) were used for complex formation.
Fluorescence resonance energy transfer (FRET) is a distance-dependent excited-state interaction, in which emission of one fluorophore (donor) is coupled to the excitation of another (acceptor) in close proximity of several nanometers, leading to decreased donor emission and increased acceptor emission (SELVIN, 2000; Hillisch et al., 2001). It has been used as a tool for measuring distance data and monitoring conformational changes in biological macromolecules. In the present study, we applied FRET to obtain structural information on polyplexes between PEI and ODN and estimate the average distance between 2 fluorophores in double-labeled complexes of rhodamine-labeled PEI and fluorescein-labeled ODN (TMR-PEI/FL-ODN).
Fluorescence anisotropy provides information on molecular movement and has been widely used for studying interactions between macromolecules, and between macromolecules and small molecules. In this study, we applied the steady-state anisotropy technique to monitor interactions between PEI and ODN and to determine the fraction of free PEI after complex formulation.
Materials and Methods
Double-stranded ODNs
Single-stranded 20-mer ODNs containing the NF-κB cis element (Morishita et al., 1997) and corresponding 5′-fluorescein conjugated ODNs were purchased from MWG Biotech (Highpoint). ODNs were prepared by annealing the single-stranded ODN with equimolar amounts of complementary ODN at a final concentration of 1 μg/μL.
Polyethylenimine
Branched PEI with MW 25 kDa (PEI25) was obtained from BASF AG. Low-molecular weight PEI with MW 2.7 kDa (PEI2.7) was synthesized and characterized as described previously (von Harpe et al., 2000), and generously provided by Dr. T. Kissel (Marburg, Germany).
Labeling of PEIs with rhodamine
PEIs were fluorescently labeled using carboxytetramethylrhodamine N-hydroxysuccinimide (TMR-NHS) according to the supplier's instruction (Pierce). Briefly, 1 mg of PEI (2.7 or 25 kDa) in 1 mL HEPES buffer (10 mM HEPES, 5% glucose, pH 7.4) was mixed with 0.5 mg of TMR-NHS in 100 μL DMSO and incubated on ice for 2 hours. Unincorporated TMR-NHS was removed by column chromatography on Sephadex G25 (PD-10; Amersham) using HEPES buffer as eluent. It was determined that ∼1.9% of the amines (corresponding to 10.8 amines for 25 kDa PEI and 1.2 amine for 2.7 kDa) were conjugated to TMR in TMR-PEI.
FRET measurements of PEI-ODN complexes
Single-labeled complexes (PEI/FL-ODN) were prepared with FL-ODN and unlabeled PEI, and double-labeled complexes (TMR-PEI/FL-ODN) with TMR-PEI and FL-ODN. The desired amounts of ODN and 18 μg of PEI were diluted separately in HEPES buffer (10 mM HEPES, 5% glucose, pH 7.4) to a final volume of 500 μL each. After 10 minutes of incubation at room temperature, the PEI solutions were transferred to the ODN solution and vortexed immediately. After 10 minutes of incubation at room temperature, 1 mL of HEPES buffer was added. The amounts of PEI were calculated from the desired amine/phosphate (N/P) ratio, assuming 43.1 g/mol for each repeating unit of PEI containing one amine, and 330 g/mol for each repeating unit of ODN containing one phosphate. The amounts of fluorescence dyes were kept constant in all preparations, and the TMR to FL molar ratio in double-labeled complexes was adjusted to 1 by mixing TMR-PEI and unlabeled PEI accordingly. The fluorescence intensities were measured using a spectrofluorometer (F2500; Hitachi) at excitation wavelength 480 nm rather than 488 nm (= absorption maximum of FL) to reduce direct excitation of TMR, and emission scanning from 510 to 610 nm with a slit width of 10 nm. The decrease in FL emission intensity at 518 nm as a result of fluorescence quenching was expressed as percent quenching (%Q) according to
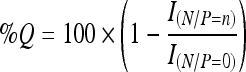 |
(1) |
where I(N/P=0) and I(N/P=n) are the FL emission intensity of free FL-ODN in the absence of PEI (N/P = 0), and FL emission intensity of complexes at N/P = n, respectively.
Fluorescence quenching may occur by several mechanisms such as static quenching by complex formation in ground state and dynamic quenching by collision, charge transfer, and FRET. As the decreased emission intensity of FL in double-labeled complexes represents the sum of fluorescence quenching by complex formation and FRET, the contribution of FRET on the fluorescence quenching can be obtained from the difference between the FL emission intensity of the double-labeled complexes and single-labeled complexes at corresponding N/P ratio. The Q value by energy transfer (Q energy transfer), also known as FRET efficiency (E), was calculated for each N/P ratio from the FL emission intensity of the single-labeled complexes (Isingle, N/P = n) and double-labeled complexes (Idouble, N/P = n) and was expressed in percent:
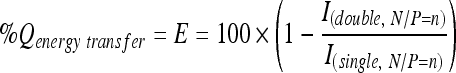 |
(2) |
From the calculated value of E, the average distance (R) between the 2 fluorophores in the double-labeled complexes was determined according to
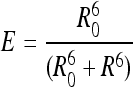 |
(3) |
where R0 is the Förster radius of the FL-TMR dye pair, that is, the distance at which energy transfer for the donor–acceptor pair is 50% of maximum.
Measurement of steady-state fluorescence anisotropy
For each PEI 2.7 and PEI25, a series of single-labeled complexes (TMR-PEI/ODN) was prepared with unlabeled ODN and TMR-PEI, as described above. The complexes were prepared with a constant amount of TMR-PEI (18 μg) and varying amounts of ODN to yield P/N ratios between 0.042 and 1. Fluorescence intensities were measured using a T-mode C61/2000 spectrofluorometer (Photon Technology International). The excitation wavelength was set to 550 nm, and emission intensities were scanned from 570 to 630 nm for TMR-PEI/ODN. The steady-state anisotropy (r) was then calculated as
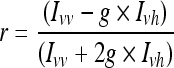 |
(4) |
where Ivv is emission intensity of vertically polarized light and Ivh is emission intensity of horizontally polarized light, when excitation light is vertically polarized (Shinitzky and Barenholz, 1978). The parameter “g” (g-factor) relates the relative sensitivity of the 2 emission channels and can be obtained as g = Ihv/Ihh, with the polarization of excitation set to horizontal (Parker et al., 2004).
Measurement of zeta potential
Zeta potential of the complexes was determined by DLS using the particle size analyzer Nicomp 380ZLS (Particle Sizing Systems). The scattered light was detected at 23°C at an angle of 18.9°. A viscosity value of 0.933 mPa·s and a refractive index of 1.333 were used for the data analysis. The instrument was routinely calibrated using latex microsphere suspensions (0.09 μm, 0.26 μm; Duke Scientific Corp).
Results and Discussion
FRET of PEI/ODN complexes
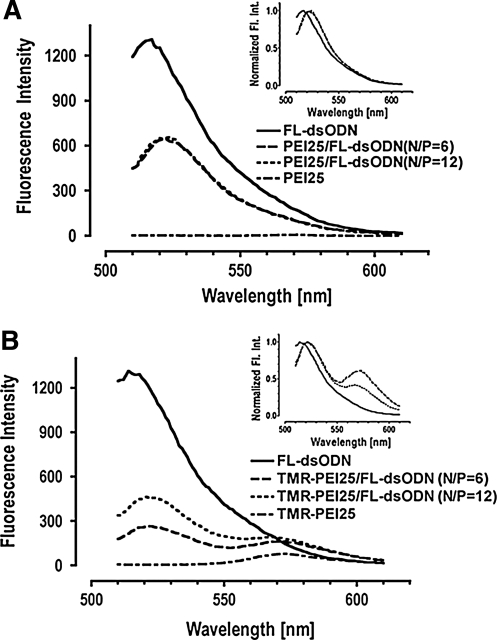
We measured the decrease in emission intensities of FL in single-labeled complex (PEI/FL-ODN) and double-labeled complex TMR-PEI/FL-ODN and calculated FRET from donor quenching. The alternative way of FRET estimation by monitoring the increase in acceptor emission intensity (acceptor sensitization) often requires complex formulation at acceptor to donor dye ratios on the order of 5:1 (Itaka et al., 2002), necessitating higher levels of dye substitution than applied here, thus introducing the risk of altering the physical properties of the molecules of interest. Figure 1 shows examples of emission spectra of FL-ODN and complexes with PEI25 (graphs for PEI2.7 look similar). The decrease in FL emission and slight red shift in the emission maximum is due to environmental changes for FL in complexes as compared to free solution. Several mechanisms cause fluorescence quenching upon complex formation in single-labeled complexes (Fig. 1A), including self-quenching between FL dye molecules in close proximity, and static quenching of FL by complex formation in ground state, thus suppressing excitation. Our data for single-labeled complexes are consistent with results reported by van Rompay et al. (2001) for complex formation between rhodamine-conjugated ODNs and several cationic polymers. Double-labeled complexes, TMR-PEI25/FL-ODN, had markedly decreased FL emission compared to single-labeled complexes at the same N/P ratio, and an increase in the TMR emission (Fig. 1B).
FIG. 1.
Representative fluorescence emission spectra and their normalized spectra (insets) of free FL-ODN and (A) single-labeled complexes, PEI25/FL-ODN, and (B) double-labeled complexes, TMR-PEI25/FL-ODN. The excitation wavelength was 480 nm and emission spectrum was scanned from 510 to 610 nm. The decrease in emission intensity and shift of emission maxima of FL are a consequence of complex formation between PEI and FL-ODN in ground state (A). Energy transfer between donor (FL) and acceptor (TMR) leads to further decrease in FL emission and increase in TMR emission (B). PEI, polyethylenimine; ODN, oligodeoxynucleotide; TMR, carboxytetramethylrhodamine.
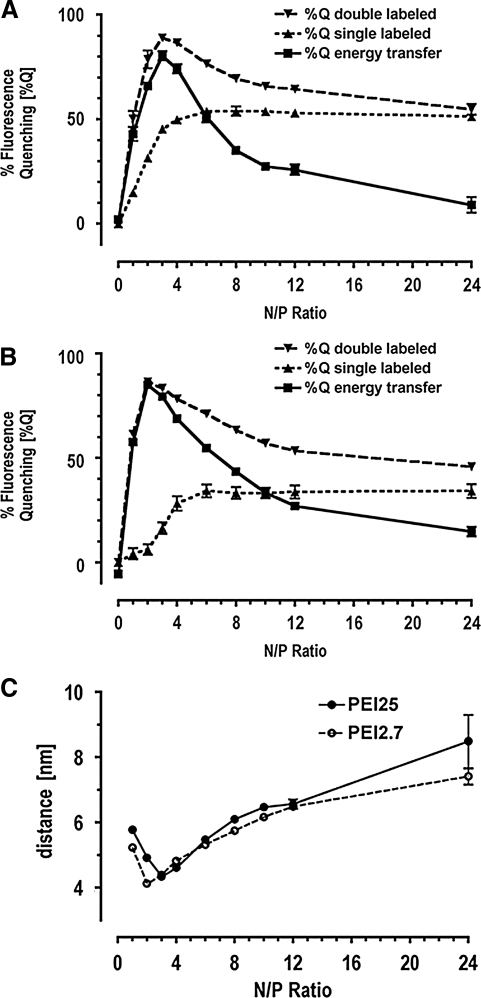
The % Quenching of FL emission intensities in single-labeled complexes, PEI25/FL-ODN and PEI2.7/ODN, increased steeply up to an N/P ratio of 4 and reached a plateau above an N/P ratio of 6 (Fig. 2A, B), suggesting saturation of the amount of PEI in complexes, leaving a significant fraction of free PEI at higher N/P ratios. PEI25/FL-ODN (Fig. 2A) and PEI2.7/FL-ODN (Fig. 2B) differed in the higher plateau for the PEI25 compared to PEI2.7 complexes (% Quenching ∼55% vs. ∼30%). Hence, the interaction of PEI25 with ODN is different from that of PEI2.7, indicating a different degree of condensation.
FIG. 2.
Quenching of fluorescence emission in PEI25/ODN complexes (A) and PEI2.7/ODN complexes (B) as a function of N/P ratio. %Qdouble labeled is the quenching curve for double-labeled complexes (TMR-PEI/FL-ODN) and %Qsingle labeled is the quenching curve for single-labeled complexes (PEI/FL-ODN). %Qenergy transfer is equivalent to energy transfer efficiencies (%E), providing the basis for calculation of distance between dyes within complexes. (C) Calculated average distances between the fluorescent dyes in FL-ODN and TMR-PEI plotted at different N/P ratios.
Single- and double-labeled complexes displayed the same degree of red shift of the FL signal. Moreover, comparing the single- and double-labeled complexes at a given N/P ratio accounts for any environmental change of the fluorophore. Quench maxima for double-labeled complexes were observed at N/P ratio 3 and 2 for PEI25 and PEI2.7, respectively (Fig. 2A, B). At least 2 factors contribute to the decline in quenching after the maximum and convergence of the quenching curves for single-labeled and double-labeled complexes. First, an increasing fraction of TMR-PEI does not participate in complex formation at higher N/P ratios, thus lessening the contribution of energy transfer to total quenching. Second, the net charge of the complexes goes from negative at low N/P ratio to neutral, then to positive at high N/P ratio. A necessary condition for FRET is alignment of the dipole moments of donor and acceptor during the lifetime of the donor's excited state. Strong local electric fields, either at low or high N/P ratio, can restrict the rotation of dipole moments and change the orientation factor resulting in low FRET efficiency. Highest rotational freedom and FRET efficiency occurs between these 2 extremes when the electric field inside the complex is neutralized. This explanation of electrostatic effect is supported by Zeta potential measurement of the complexes as a function of the N/P ratio, which approaches a value of zero around N/P ratio of 3 (data not shown). In addition, DLS size measurements of the complexes indicated aggregate formation around N/P equal 3 (for PEI2.7/ODN), and around N/P equal 4 (for PEI25/ODN), which is a consequence of charge neutralization and lack of electrostatic repulsion. The efficiency of energy transfer (%E) was used to estimate the distance (R) between donor and acceptor in the double-labeled complexes, TMR-PEI/FL-ODN, by Equation 2 and 3 (Materials and Methods section). The Förster radius, R0, was set as 5.5 nm, a value used by several other groups working with DNA complexes (Edelman et al., 2003; Wang et al., 2003). Therefore, the distances calculated here are estimates, which, nevertheless, provide useful information for comparison of complexes generated with different PEI. A plot of calculated average distances over increasing N/P ratio showed minima for both TMR-PEI25/FL-ODN and TMR-PEI2.7/FL-ODN (Fig. 2C), which correspond to the maxima of energy transfer (compare Fig. 2A and B). While the distance minima were not significantly different from each other (4.34 ± 0.11 nm vs. 4.12 ± 0.01 nm; mean ± SE, n = 3), they occurred at different N/P ratios, N/P equal 3 in the case of 25 kDa PEI, and N/P equal 2 for 2.7 kDa PEI. Therefore, maximum packing density in complexes containing 2.7 kDa PEI was reached at a lower amount of amine groups, which implies that the amine groups in 2.7 kDa PEI are, on average, better accessible for interaction with ODN than those in 25 kDa PEI.
Fluorescence anisotropy of PEI-ODN complexes
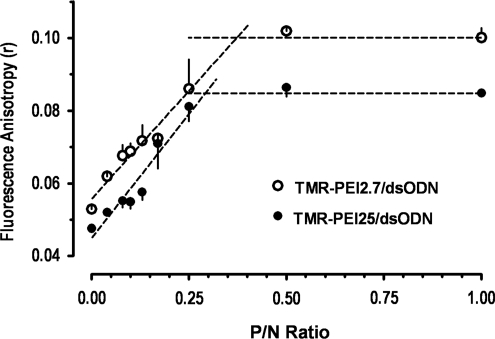
When small molecules participate in complex formation, for example, during incorporation of PEI and ODN into polyplexes, the rotational movement slows down, resulting in less depolarization of the emitted light (Kakehi et al., 2001). Hence, we measured steady-state fluorescence polarization anisotropy and used it to characterize single-labeled complexes, TMR-PEI/ODN. Steady-state anisotropy of the complexes was plotted as a function of P/N ratio TMR-PEI/ODN. The anisotropy of TMR-PEI/ODN complexes showed linear increase at lower P/N ratios (Fig. 3) and reached a plateau at a P/N ratio of about 0.29 (for TMR-PEI25) and 0.37 (for TMR-PEI2.7), demonstrating saturation. Confirming the FRET data, the saturation at higher P/N ratio (= lower N/P) indicates that PEI2.7 has more amine groups available for interaction with ODN phosphate groups than PEI25 (Fig. 4).
FIG. 3.
Fluorescence anisotropy (r) of the complexes between TMR-PEI and ODN as a function of P/N ratio. Increasing amounts of ODN were added to constant amounts of TMR-PEI.
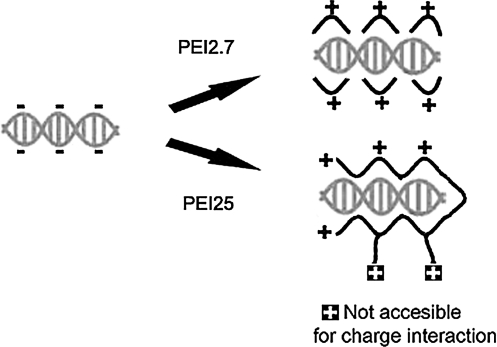
FIG. 4.
Schematic demonstrating complexation between PEIs and ODN. Positively charged PEIs complex negatively charged ODN by electrostatic interaction. Due to steric hindrance, positively charged amines of PEI25 are less accessible to the negative-charged phosphates of ODN than those of PEI2.7.
Assuming a linear proportion between anisotropy and bound fraction of TMR-PEI up to the saturation point, linear regression analysis of these data points resulted in highly significant correlation coefficients (r2 = 0.9576 and 0.9314 for PEI2.7 and PEI25, respectively). Based on this analysis, the fractions of bound PEI molecules in each preparation were calculated as 45% (for PEI2.7) and 58% (for PEI25) at P/N ratio of 0.17 (corresponding to N/P ratio 6). Applying a different technique, fluorescence correlation spectroscopy, Clamme et al. calculated that only 14% of PEI25 was bound in complexes with plasmid DNA at N/P ratios equal to 6 or 10 (Clamme et al., 2003). The discrepancy between Clamme's and our result, that is, 58% vs. 14% of PEI25 bound at N/P ratio 6, is probably due to the structural difference of the DNA molecules (20 bp ODN vs. 5.8 kbp plasmid DNA). The structure of plasmid DNA may sterically constrain interactions between phosphate groups and PEI amino groups in PEI, leaving a large fraction of DNA phosphate groups inaccessible to PEI (Clamme et al., 2003). In comparison, the small size and flexibility of the 20 bp ODN makes the phosphate groups more accessible to amino groups for charge interactions, leading to a higher fraction of bound PEI. It is, however, unexpected that the bound fraction of PEI, as measured with correlation spectroscopy, remained constant when the N/P ratio increased from 6 to 10 (Clamme et al., 2003), which is in contrast to the present data, where the bound fraction decreased from 62% (N/P = 6 or P/N = 0.17) to 35% (N/P = 10 or P/N = 0.1).
In conclusion, the presented data demonstrate the suitability of FRET and steady-state anisotropy measurements for characterization of multimolecular complexes between PEI and ODN. While a sufficiently high N/P ratio is necessary for complete condensation of ODN, the data show that significant amounts of both PEI2.7 and PEI25 preparations existed in free form at N/P ratios usually employed for transfection. Structural and compositional information gained by FRET and steady-state anisotropy on polymer/oligonucleotide complexes could be a valuable complement to other techniques available for characterization of such complexes (eg, fluorescence correlation spectroscopy). Beyond studying PEI polymers and ODNs, as in the present experiments, these methods should be applicable to the wide array of complexes between DNA or RNA and other cationic polymers, which are currently explored in transfection experiments.
Acknowledgments
This research was supported by grants from Sangji University Research Funds, Grant 5R01-NS045043-02 from National Institutes of Health (NIH), and grants from National Science Foundation and Petroleum Research Fund.
Author Disclosure Statement
No competing financial interests exist.
References
- AKINC A. THOMAS M. KLIBANOV A.M. LANGER R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- BOECKLE S. VON GERSDORFF K. VAN DER PIEPEN S. CULMSEE C. WAGNER E. OGRIS M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J. Gene Med. 2004;6:1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- BOUSSIF O. LEZOUALC'H F. ZANTA M.A. MERGNY M.D. SCHERMAN D. DEMENEIX B. BEHR J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLAMME J.P. AZOULAY J. MELY Y. Monitoring of the formation and dissociation of polyethylenimine/DNA complexes by two photon fluorescence correlation spectroscopy. Biophys. J. 2003;84:1960–1968. doi: 10.1016/S0006-3495(03)75004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EDELMAN L.M. CHEONG R. KAHN J.D. Fluorescence resonance energy transfer over approximately 130 basepairs in hyperstable lac repressor-DNA loops. Biophys. J. 2003;84(2 Pt 1):1131–1145. doi: 10.1016/S0006-3495(03)74929-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FISCHER D. BHATTACHARYA R. OSBURG B. BICKEL U. Inhibition of monocyte adhesion on brain-derived endothelial cells by NF-kappaB decoy/polyethylenimine complexes. J. Gene Med. 2005;7:1063–1076. doi: 10.1002/jgm.747. [DOI] [PubMed] [Google Scholar]
- FISCHER D. BIEBER T. LI Y. ELSASSER H.P. KISSEL T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm. Res. 1999;16:1273–1279. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- GODBEY W.T. WU K.K. MIKOS A.G. Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J. Biomed. Mater. Res. 1999;45:268–275. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- HILLISCH A. LORENZ M. DIEKMANN S. Recent advances in FRET: distance determination in protein-DNA complexes. Curr. Opin. Struct. Biol. 2001;11:201–207. doi: 10.1016/s0959-440x(00)00190-1. [DOI] [PubMed] [Google Scholar]
- ITAKA K. HARADA A. NAKAMURA K. KAWAGUCHI H. KATAOKA K. Evaluation by fluorescence resonance energy transfer of the stability of nonviral gene delivery vectors under physiological conditions. Biomacromolecules. 2002;3:841–845. doi: 10.1021/bm025527d. [DOI] [PubMed] [Google Scholar]
- KAKEHI K. ODA Y. KINOSHITA M. Fluorescence polarization: analysis of carbohydrate-protein interaction. Anal. Biochem. 2001;297:111–116. doi: 10.1006/abio.2001.5309. [DOI] [PubMed] [Google Scholar]
- KIRCHEIS R. WIGHTMAN L. WAGNER E. Design and gene delivery activity of modified polyethylenimines. Adv. Drug Deliv. Rev. 2001;53:341–358. doi: 10.1016/s0169-409x(01)00202-2. [DOI] [PubMed] [Google Scholar]
- KUNATH K. VON HARPE A. FISCHER D. PETERSEN H. BICKEL U. VOIGT K. KISSEL T. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J. Control. Release. 2003;89:113–125. doi: 10.1016/s0168-3659(03)00076-2. [DOI] [PubMed] [Google Scholar]
- MORISHITA R. SUGIMOTO T. AOKI M. KIDA I. TOMITA N. MORIGUCHI A. MAEDA K. SAWA Y. KANEDA Y. HIGAKI J. OGIHARA T. In vivo transfection of cis element “decoy” against nuclear factor-kappaB binding site prevents myocardial infarction. Nat. Med. 1997;3:894–899. doi: 10.1038/nm0897-894. [DOI] [PubMed] [Google Scholar]
- PARKER A. MILES K. CHENG K.H. HUANG J. Lateral distribution of cholesterol in dioleoylphosphatidylcholine lipid bilayers: cholesterol-phospholipid interactions at high cholesterol limit. Biophys. J. 2004;86:1532–1544. doi: 10.1016/S0006-3495(04)74221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SELVIN P.R. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 2000;7:730–734. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- SHINITZKY M. BARENHOLZ Y. Fluidity parameters of lipid regions determined by fluorescence polarization. Biochim. Biophys. Acta. 1978;515:367–394. doi: 10.1016/0304-4157(78)90010-2. [DOI] [PubMed] [Google Scholar]
- VAN ROMPAEY E. ENGELBORGHS Y. SANDERS N. DE SMEDT S.C. DEMEESTER J. Interaction between oligonucleotides and cationic polymers investigated by fluorescence correlation spectroscopy. Pharm. Res. 2001;18:928–936. doi: 10.1023/a:1010975908915. [DOI] [PubMed] [Google Scholar]
- VINOGRADOV S.V. BATRAKOVA E.V. KABANOV A.V. Nanogels for oligonucleotide delivery to the brain. Bioconjug. Chem. 2004;15:50–60. doi: 10.1021/bc034164r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VON HARPE A. PETERSEN H. LI Y. KISSEL T. Characterization of commercially available and synthesized polyethylenimines for gene delivery. J. Control. Release. 2000;69:309–322. doi: 10.1016/s0168-3659(00)00317-5. [DOI] [PubMed] [Google Scholar]
- WANG L. GAIGALAS A.K. BLASIC J. HOLDEN M.J. GALLAGHER D.T. PIRES R. Fluorescence resonance energy transfer between donor-acceptor pair on two oligonucleotides hybridized adjacently to DNA template. Biopolymers. 2003;72:401–412. doi: 10.1002/bip.10482. [DOI] [PubMed] [Google Scholar]
- ZOU S.M. ERBACHER P. REMY J.S. BEHR J.P. Systemic linear polyethylenimine (L-PEI)-mediated gene delivery in the mouse. J. Gene Med. 2000;2:128–134. doi: 10.1002/(SICI)1521-2254(200003/04)2:2<128::AID-JGM95>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]