Abstract
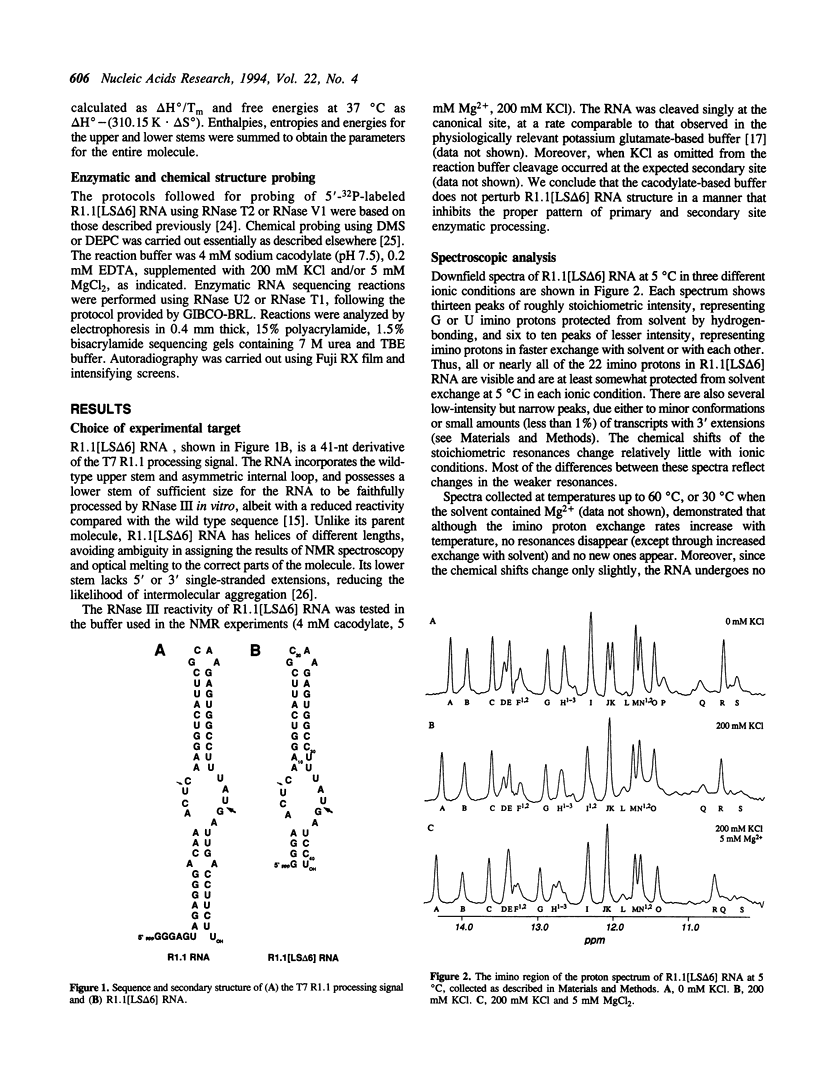
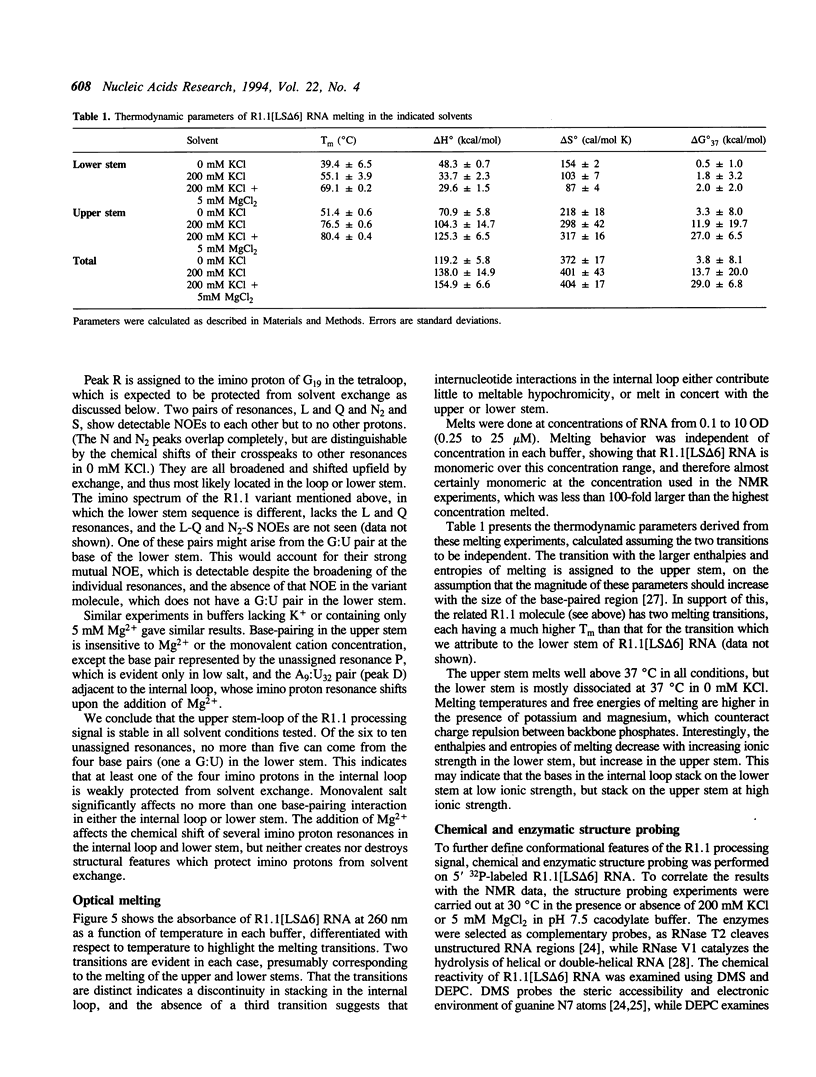
The structure of a ribonuclease III processing signal from bacteriophage T7 was examined by NMR spectroscopy, optical melting, and chemical and enzymatic modification. A 41 nucleotide variant of the T7 R1.1 processing signal has two Watson-Crick base-paired helices separated by an internal loop, consistent with its predicted secondary structure. The internal loop is neither rigidly structured nor completely exposed to solvent, and seems to be helical. The secondary structure of R1.1 RNA is largely insensitive to the monovalent cation concentration, which suggests that the monovalent cation sensitivity of secondary site cleavage by RNase III is not due to a low salt-induced RNA conformational change. However, spectroscopic data show that Mg2+ affects the conformation of the internal loop, suggesting a divalent cation binding site(s) within this region. The Mg(2+)-dependence of RNase III processing of some substrates may reflect not only a requirement for a divalent cation as a catalytic cofactor, but also a requirement for a local RNA conformation which is divalent cation-stabilized.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auron P. E., Weber L. D., Rich A. Comparison of transfer ribonucleic acid structures using cobra venom and S1 endonucleases. Biochemistry. 1982 Sep 14;21(19):4700–4706. doi: 10.1021/bi00262a028. [DOI] [PubMed] [Google Scholar]
- Branch A. D., Benenfeld B. J., Robertson H. D. Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6590–6594. doi: 10.1073/pnas.82.19.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelladurai B. S., Li H., Nicholson A. W. A conserved sequence element in ribonuclease III processing signals is not required for accurate in vitro enzymatic cleavage. Nucleic Acids Res. 1991 Apr 25;19(8):1759–1766. doi: 10.1093/nar/19.8.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelladurai B., Li H., Zhang K., Nicholson A. W. Mutational analysis of a ribonuclease III processing signal. Biochemistry. 1993 Jul 27;32(29):7549–7558. doi: 10.1021/bi00080a029. [DOI] [PubMed] [Google Scholar]
- Dunn J. J. RNase III cleavage of single-stranded RNA. Effect of ionic strength on the fideltiy of cleavage. J Biol Chem. 1976 Jun 25;251(12):3807–3814. [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodberg J., Dunn J. J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988 Mar;170(3):1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines D. S., Strauss K. I., Gillespie D. H. Cellular response to double-stranded RNA. J Cell Biochem. 1991 May;46(1):9–20. doi: 10.1002/jcb.240460104. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Kime M. J., Moore P. B. Nuclear Overhauser experiments at 500 MHz on the downfield proton spectrum of a ribonuclease-resistant fragment of 5S ribonucleic acid. Biochemistry. 1983 May 24;22(11):2615–2622. doi: 10.1021/bi00280a004. [DOI] [PubMed] [Google Scholar]
- Kime M. J., Moore P. B. Physical evidence for a domain structure in Escherichia coli 5 S RNA. FEBS Lett. 1983 Mar 7;153(1):199–203. doi: 10.1016/0014-5793(83)80147-1. [DOI] [PubMed] [Google Scholar]
- Li H. L., Chelladurai B. S., Zhang K., Nicholson A. W. Ribonuclease III cleavage of a bacteriophage T7 processing signal. Divalent cation specificity, and specific anion effects. Nucleic Acids Res. 1993 Apr 25;21(8):1919–1925. doi: 10.1093/nar/21.8.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libonati M., Carsana A., Furia A. Double-stranded RNA. Mol Cell Biochem. 1980 Aug 16;31(3):147–164. doi: 10.1007/BF00225848. [DOI] [PubMed] [Google Scholar]
- Linn T., St Pierre R. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol. 1990 Feb;172(2):1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard R. E., Kumar A. Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res. 1981 Oct 10;9(19):5125–5140. doi: 10.1093/nar/9.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowman H. B., Draper D. E. On the recognition of helical RNA by cobra venom V1 nuclease. J Biol Chem. 1986 Apr 25;261(12):5396–5403. [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson A. W., Niebling K. R., McOsker P. L., Robertson H. D. Accurate in vitro cleavage by RNase III of phosphorothioate-substituted RNA processing signals in bacteriophage T7 early mRNA. Nucleic Acids Res. 1988 Feb 25;16(4):1577–1591. doi: 10.1093/nar/16.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayotatos N., Truong K. Cleavage within an RNase III site can control mRNA stability and protein synthesis in vivo. Nucleic Acids Res. 1985 Apr 11;13(7):2227–2240. doi: 10.1093/nar/13.7.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Tan R., Calnan B. J., Frankel A. D., Williamson J. R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992 Jul 3;257(5066):76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- Robertson H. D. Escherichia coli ribonuclease III. Methods Enzymol. 1990;181:189–202. doi: 10.1016/0076-6879(90)81121-a. [DOI] [PubMed] [Google Scholar]
- Robertson H. D., Webster R. E., Zinder N. D. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem. 1968 Jan 10;243(1):82–91. [PubMed] [Google Scholar]
- Romaniuk P. J., de Stevenson I. L., Ehresmann C., Romby P., Ehresmann B. A comparison of the solution structures and conformational properties of the somatic and oocyte 5S rRNAs of Xenopus laevis. Nucleic Acids Res. 1988 Mar 25;16(5):2295–2312. doi: 10.1093/nar/16.5.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Richardson C. C. Processing of mRNA by ribonuclease III regulates expression of gene 1.2 of bacteriophage T7. Cell. 1981 Dec;27(3 Pt 2):533–542. doi: 10.1016/0092-8674(81)90395-0. [DOI] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Kierzek R., Turner D. H. Context dependence of hydrogen bond free energy revealed by substitutions in an RNA hairpin. Science. 1992 Apr 10;256(5054):217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- Searle M. S., Williams D. H. On the stability of nucleic acid structures in solution: enthalpy-entropy compensations, internal rotations and reversibility. Nucleic Acids Res. 1993 May 11;21(9):2051–2056. doi: 10.1093/nar/21.9.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., Brown N. H., Gall J. G., Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steege D. A., Cone K. C., Queen C., Rosenberg M. Bacteriophage lambda N gene leader RNA. RNA processing and translational initiation signals. J Biol Chem. 1987 Dec 25;262(36):17651–17658. [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Szewczak A. A., White S. A., Gewirth D. T., Moore P. B. On the use of T7 RNA polymerase transcripts for physical investigation. Nucleic Acids Res. 1990 Jul 25;18(14):4139–4142. doi: 10.1093/nar/18.14.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly B., Varani G., Tinoco I., Jr The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry. 1993 Feb 2;32(4):1078–1087. doi: 10.1021/bi00055a013. [DOI] [PubMed] [Google Scholar]
- Wyatt J. R., Chastain M., Puglisi J. D. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques. 1991 Dec;11(6):764–769. [PubMed] [Google Scholar]
- Ziemke P., McCarthy J. E. The control of mRNA stability in Escherichia coli: manipulation of the degradation pathway of the polycistronic atp mRNA. Biochim Biophys Acta. 1992 Apr 6;1130(3):297–306. doi: 10.1016/0167-4781(92)90442-3. [DOI] [PubMed] [Google Scholar]
- Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989 Apr 7;244(4900):48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]



