Abstract
The solely known function of Cu,Zn-superoxide dismutase (SOD1) is to catalyze the dismutation of superoxide anion into hydrogen peroxide. Our objective was to determine if SOD1 catalyzed murine liver protein nitration induced by acetaminophen (APAP) and lipopolysaccharide (LPS). Liver and plasma samples were collected from young adult SOD1 knockout mice (SOD1-/-) and wild-type (WT) mice at 5 or 6 h after an ip injection of saline, APAP, or LPS. Hepatic nitrotyrosine formation was induced by APAP and LPS only in the WT mice. The diminished hepatic protein nitration in the SOD1-/- mice was not directly related to plasma nitrite and nitrate concentrations. Similar genotype differences were seen in liver homogenates treated with a bolus of peroxynitrite. Adding only the holo, but not the apo-SOD1 enzyme into the liver homogenates enhanced the reaction in an activity-dependent fashion and nearly eliminated the genotype difference at the high doses. Mass Spectrometry showed 4 more nitrotyrosine residues in bovine serum albumin and 10 more nitrated protein candidates in the SOD1-/- liver homogenates by peroxynitrite with added SOD1. In conclusion, the diminished hepatic protein nitration mediated by APAP or LPS in the SOD1-/- mice was due to the lack of SOD1 activity per se.
Keywords: SOD1, Protein nitration, Acetaminophen, Peroxynitrite, Lipopolysaccharide, Mouse
Although copper,zinc-superoxide dismutase (SOD1) has been widely considered a major intracellular antioxidant enzyme in mammals, its solely known function is to catalyze the conversion of superoxide anion into less active hydrogen peroxide. Indeed, SOD1 is protective against oxidative stress associated with acute paraquat toxicity, myocardial ischemia/reperfusion injury, and N-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced dopaminergic neurodegeneration [1-3]. Mice deficient in SOD1 develop hepatocarcinogenesis [4] and retinal degeneration [5] in late life. However, overexpression of human SOD1 accelerated mitochondrial vacuolization, axonal degeneration, and premature motor neuron death in mice [6]. Knockout of SOD1 rendered mice resistant to acetaminophen (APAP)-mediated hepatotoxicity [7, 8]. These paradoxical findings strongly suggest that SOD1 may exert functions more than just superoxide dismutation. In fact, SOD1 was shown to promote peroxynitrite-mediated nitrotyrosine formation in vitro [9, 10].
Peroxynitrite is formed by a rapid diffusion-limited, radical-radical coupling reaction between nitric oxide (NO) and superoxide anion [11, 12]. Peroxynitrite can directly react with target molecules such as glutathione via one or two-electron oxidations [13], or yield radical NO2 via homolysis that nitrates tyrosine residuals of proteins into nitrotyrosine [14]. This particular chemical modification of tyrosine residues is called protein nitration [12, 15]. With a potential in inactivating or altering functions of the modified proteins [16-18], protein nitration has been suggested as an early biomarker for the degenerations of motor neurons in amyotrophic lateral sclerosis (ALS) and of brains in Alzheimer disease [19-22]. It has also been proposed as a mediator of APAP-induced hepatotoxicity [23-28]. Coincidentally, alterations of SOD1 expression are related to Down Syndrome [29], ALS [30], and body resistance to acetaminophen toxicity [7, 31].
Despite these pathological implications, the earlier observed in vitro catalytic role of SOD1 in nitrotyrosine formation [9, 10] has been largely dismissed or neglected. There has been no experiment to test a cause-effect relationship between SOD1 and protein nitration in vivo. The extremely short biological half-life of peroxynitrite [32] and the lack of specific in vivo models have made such a test very difficult, if not impossible. The development of SOD1 knockout (SOD1-/-) mice [1] and the demonstration of several drugs such as APAP to induce protein nitration in tissues [24, 25, 33, 34] provide us with an unprecedented opportunity to study the in vivo role of SOD1 in nitrotyrosine formation. Furthermore, the application of mass spectrometry allows identification of specific nitrated protein candidates in tissue homogenates and particular residues in selected proteins. Therefore, we used SOD1-/- and their wild-type (WT) mice and conducted a series of in vivo and in vitro experiments to determine: 1) if SOD1 knockout diminished hepatic protein nitration induced by different types and doses of stimuli; 2) if such impact of SOD1 was independent of NO production; 3) if the changes in protein nitration due to SOD1 knockout could be reversed by the holo- or apo-SOD1 enzyme; and 4) if SOD1 catalyzed nitrotyrosine formation selectively in target proteins of liver homogenates and(or) certain tyrosine residues in bovine serum albumin (BSA).
Materials and methods
Chemicals and Antibodies
All chemicals including bovine SOD1 protein were purchased from Sigma Chemical Co. (St. Louis, MO) unless otherwise indicated. Peroxynitrite was purchased from Calbiochem (San Diego, CA). Mouse anti-nitrotyrosine antibody was from Upstate Biotechnology (Lake Placid, NY), and anti-mouse IgG was from Pierce (Rockford, IL). Human holo- and apo-SOD1 protein (Cu, Zn-depleted) [35] were kindly provided by Dr. John Hart, University of Texas Health Science Center at San Antonio.
Animals and Treatments
Our experiments were approved by the Institutional Animal Care and Use Committee at Cornell University and conducted in accordance with the NIH guidelines for animal care. The SOD1-/- and WT mice were generated from 129SVJ × C57BL/6 mice [1]. All used mice were 8-10 weeks old and were fed a diet containing adequate levels of all required nutrients. To determine if SOD1 knockout exerted a general impact on hepatic protein nitration, we injected (intraperitoneal) mice with two different stimuli: APAP and lipopolysaccharide (LPS). The former is a widely used analgesic and antipyretic drug that raises plasma nitrate/nitrite concentrations and produces nitrotyrosine adducts in centrilobular areas of liver [24, 25, 27]. The latter induces the release of NO and superoxide in targeted tissues, leading to the formation of peroxynitrite and protein nitration [33, 36]. Based on preliminary experiments, we selected injections of APAP at 300 and 600 mg/kg of body weight and of LPS at 5 mg/kg of body weight. The two compounds were prepared in phosphate-buffered saline (PBS). The WT and SOD1-/- mice (n = 4-8 for each treatment by genotype) were euthanized for blood and liver sample collection at 5 and 6 h post the injection of APAP and LPS, respectively. For both stimuli, the same number of WT and SOD1-/- mice were injected with PBS and euthanized as the respective controls.
Hepatic Protein Nitration, Antibody Verification, and Plasma NO Production
Both immunohistochemistry and Western blot were used to characterize the impact of SOD1 knockout on liver nitrotyrosine formation. The Histomouse™-MAX kit (Zymed, South San Francisco, CA) was used for immunostaining of liver sections, and 12% gel was run to separate liver homogenates (50 μg protein/lane) for Western analysis. Specificity of the antibody against nitrotyrosine was verified by using 10 mM sodium dithionite to reduce nitrotyrosine on the blotting membrane and incubating the antibody with 20 μM 3-nitrotryosine overnight for a prior block [37]. Protein concentration was measured by the Lowry method [38]. As an indicator of NO production, plasma nitrate and nitrite concentrations were measured [39, 40] to test if the impact of SOD1 knockout on hepatic protein nitration was related to NO formation.
In Vitro Analysis of SOD1-catalyzed Protein Nitration
A series of experiments with mouse liver homogenates or BSA were conducted to determine: 1) if the observed genotype difference in the induced hepatic protein nitration was actually due to the SOD1 activity, but not other changes associated with the gene ablation; 2) if the hepatic protein nitration in the SOD1-/- mice was limited by formation of peroxynitrite; and 3) if the catalysis of SOD1 enzyme in protein nitration was dependent on its activity (the presence of metal Cu). Liver samples of WT and SOD1-/- mice treated with PBS or APAP were homogenized in 50 mM potassium phosphate buffer, pH 7.8, 0.1% Triton X-100, 1.34 mM diethylenetriaminepentaacetic acid. The homogenates were centrifuged at 14,000 g for 20 min at 4°C, and the resulting supernatants were adjusted to the same protein concentration (7.5 mg/ml). The same buffer was also used to prepare BSA solution (7.5 mg/ml). For each reaction, 16.4 nmol of peroxynitrite [41] was mixed with 150 μg of liver homogenate protein (20 μl) or 300 μg of BSA (40 μl) in the presence or absence of different amounts of SOD1 enzyme. The added SOD activities ranged from 1 to 4-times of the WT activity, and the mixture volumes were controlled to ensure identical peroxynitrite concentrations between different treatments. The mixture was incubated for 5 min at 37°C, and then put on ice. The SOD activities of bovine SOD1 and human holo-SOD1 were 3.4 × 104 and 9.6 × 104 units/mg protein, respectively. Little SOD activity was detected with human apo-SOD1 protein. Total SOD activity was measured using a water-soluble formazan dye kit (Dojindo Molecular Technologies, Gaithersburg, MD), following the manufacturer's instructions. After the peroxynitrite treatment, 50 μg of protein from the liver homogenate mixture or 5 μg of protein from the BSA mixture was used for analysis of nitrotyrosine formation by Western blot (as described above) and mass spectrometry (see below).
Nitrotyrosine Characterization by Mass Spectrometry
After the peroxynitrite-mediated nitrotyrosine formation was verified by Western blot, a portion of BSA solution (containing 76 pmol of protein) was reduced, alkylated, and digested with trypsin as described previously [42]. The peroxynitrite-treated WT and SOD1-/- liver homogenates (200 μg of protein) were pre-incubated with 2 μg of the anti-nitrotyrosine antibody for 1 h before adding 20 μl of protein G agarose beads for overnight incubation at 4°C with constant agitation. The mixture was then centrifuged at 3,000 g for 1 min and the precipitate was washed for 5 times with 1 ml of the homogenization buffer. After being boiled for 5 min in the sample buffer, the resulting supernatants were loaded onto two lanes (10 vs 90% of sample) of SDS-PAGE (12% gel). The lane with the 10% sample was used for Western blot analysis, while the other lane was stained with deep purple (Amersham, Piscataway, NJ). Bands corresponding to positive Western blot bands were excised, digested in situ, and extracted [42]. The in-gel extracted tryptic peptides were reconstituted in 15 μl of 0.5% formic acid containing 2% acetonitrile, and 6.4 μl of this solution were used for nanoLC-MS/MS analysis as described previously [43]. The solution-based BSA digests were treated in a similar manner. The MS/MS data generated from nanoLC/ESI-based information dependant acquisition (IDA) analysis were submitted to Mascot 1.9 for database searching using in-house licensed Mascot local server and the search was performed to query to NCBInr database (taxonomy: Mammals) with one missed cleavage site by trypsin allowed. The peptide tolerance was set to 2 Da and MS/MS tolerance was set to 0.8 Da. Only significant scores for the peptides defined by Mascot probability analysis (www.matrixscience.com/help/scoring_help.html#PBM) greater than “identity” were considered for the peptide identification.
Computer Modeling
A three-dimensional model for the sequence of BSA was generated from comparative protein modeling using the program MODELLER [44, 45]. The input data for MODELLER were: (a) the template structure, corresponding to the experimentally-determined structure of human serum albumin, monomer A, obtained from the Protein Data Bank code 1AO6, and (b) a sequence alignment of BSA and human serum albumin, obtained by using the program BLAST [46, 47].
Statistical Analysis
Numerical data were analyzed using the general linear model procedure in SAS (release 6.11, SAS Institute, Cary, NC). Statistical significance was defined as P < 0.05.
Results
Knockout of SOD1 Diminished both APAP and LPS-induced Liver Protein Nitration
Little hepatic SOD activity was detected in the SOD1-/- mice, and the treatment of 600 mg APAP/kg resulted in a decrease (P < 0.05) of the enzyme activity in the WT mice (Table 1). Western blot analysis showed strong bands of hepatic nitrotyrosine formation in the WT mice treated with 300 mg of APAP/kg (Fig. 1A), 600 mg of APAP/kg (Fig. 1B), and LPS (Fig. 1C). The same treatments produced little hepatic protein nitration in the SOD1-/- mice under the present condition. Immunostaining was performed to further confirm this genotype difference in mice treated with 300 mg APAP/kg (Fig. 1D). While the PBS-treated WT and SOD1-/- mice showed little background, there was intense staining (brown color) scattered in the hepatic lobules of the WT mice treated with APAP. The stained positive fine granules appeared in cytoplasm within the degenerated hepatocytes in the central lobular regions. The intravascular erythrocytes, fibroblast-like cells in the triads and endothelial cells were also stained positive. In contrast, very little hepatocellular nitrotyrosine staining was detected in the APAP-treated SOD1-/- mice.
Table 1.
Effect of APAP or LPS on total SOD activity in mouse liver
| WT | SOD1-/- | |
|---|---|---|
| PBS | 1194±131 | 16.5±5.3b |
| APAP (300 mg/kg) | 1180±40 | 20.6±5.5b |
|
| ||
| PBS | 1266±61 | 10.8±0.6b |
| APAP (600 mg/kg) | 1021±56a | 12.2±1.2b |
|
| ||
| PBS | 1315±97 | 12.9±0.8b |
| LPS (5 mg/kg) | 1360±73 | 14.5±3.3b |
Mice were treated with APAP for 5 h or for 6 h. Values are means ± S.E. (n=3-5).
indicates P < 0.05 compared to PBS treatment within genotypes;
indicates P < 0.05 compared to WT within the same treatment. PBS, phosphate-buffered saline; APAP, acetaminophen; LPS, lipopolysaccharide; WT, wild-type.
Fig. 1.
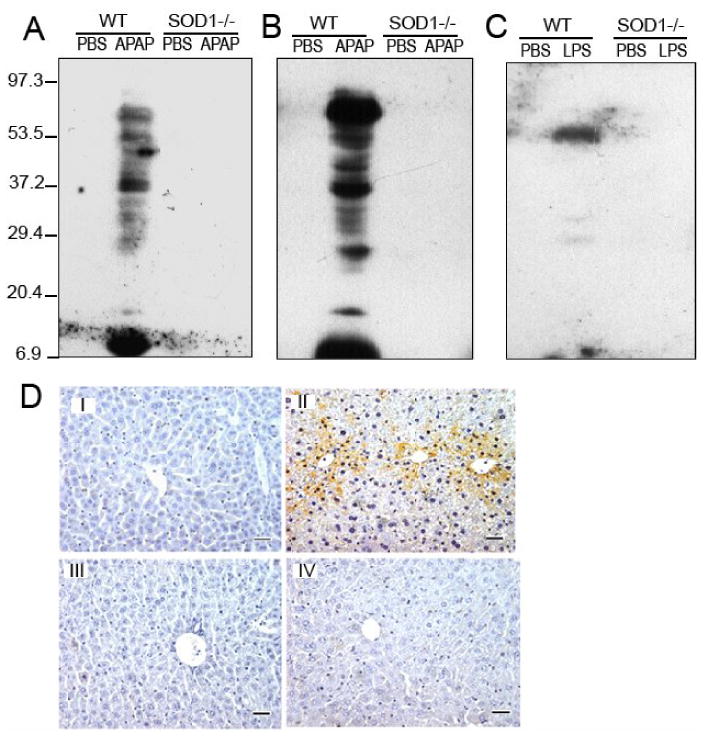
A, B, and C. Effect of SOD1 knockout on the APAP and LPS-induced liver nitrotyrosine protein adducts by Western blot analysis in mice treated with APAP (300 mg/kg, 5 h; A), APAP (600 mg/kg, 5 h; B), or LPS (5 mg/kg, 6 h; C). Each lane was loaded with 50 μg protein. D. Effect of SOD1 knockout on the APAP-induced liver nitrotyrosine adducts by immunostaining. Liver samples were collected from WT or SOD1-/- mice at 5 h after the injection of PBS (I and III) or APAP (II and IV; 300 mg/kg), and were formalin-fixed, paraffin-embedded, and sectioned. The nitrotyrosine formation was assessed with the Histomouse™-MAX kit (Zymed Inc.) using the mouse anti-nitrotyrosine antibody (Upstate Biotechnology, Lake Placid, NY). There was intense staining (brown color) scattered in the hepatic lobules of the WT mice treated with APAP, representing the induced nitrotyrosine formation (Original magnification 100×). In contrast, little hepatocellular nitrotyrosine staining was detected in the APAP-treated SOD1-/- mice or the PBS-treated mice. The images are representatives of four independent analyses.
Plasma NO Concentration Varied with Treatments and Genotypes
Compared with the PBS-injected controls, both APAP doses of 300 (Fig. 2A) and 600 mg/kg (Fig. 2B) led to increase (P < 0.05) in plasma NO concentrations in the WT mice. The increases in the SOD1-/-were significant (P < 0.05) only at the dose of 600 mg/kg. Plasma NO concentrations in the SOD1-/- were 79 and 49% lower (P < 0.05) than those in the WT mice treated with 300 and 600 mg APAP/kg, respectively. The LPS treatment caused similar increases (P < 0.05) in plasma NO concentrations in the two genotypes (Fig. 2C).
Fig. 2.
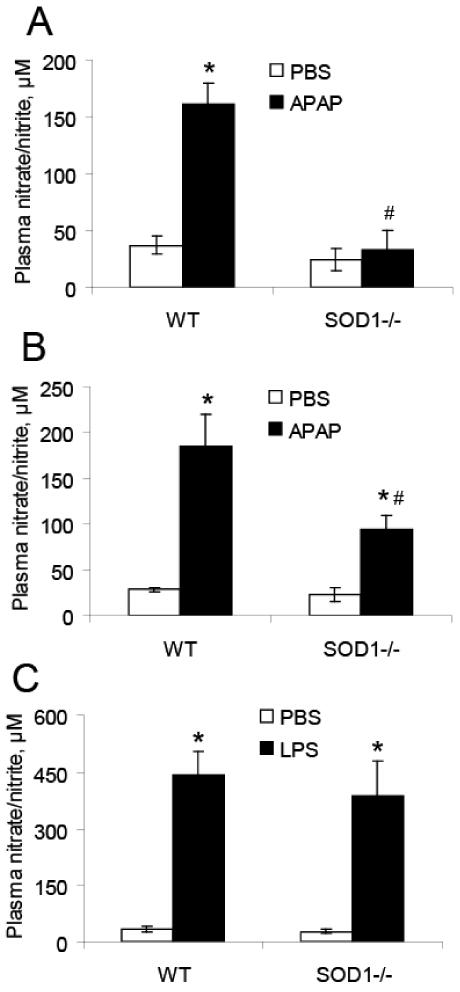
Effects of SOD1 knockout on the APAP or LPS-induced NO formation as measured by plasma nitrate and nitrite concentrations. A. Mice treated with PBS or APAP (300 mg/kg, 5 h, n = 4 - 6). B. Mice treated with PBS or APAP (600 mg/kg, 5 h, n = 4 - 6). C. Mice treated with PBS or LPS (5 mg/kg, 6 h, n = 6 - 8). * P < 0.05 vs. the PBS-treated mice within genotypes; # P < 0.05 vs. WT within the same treatment.
Bovine SOD1 Rescued the Peroxynitrite-mediated Protein Nitration in Liver Homogenates of SOD1-/- Mice
Compared with the respective controls, the peroxynitrite treatment produced much stronger formation of nitrotyrosine in the liver homogenates of WT mice than that in the SOD1-/- mice (Fig. 3 & 4A). Adding the SOD1 enzyme into the liver homogenates greatly enhanced the reactions in both genotypes, and the enhancements were SOD activity-dependent (Fig. 3). Accordingly, the dose of 4-times of WT activity was selected for further analyses. There was little total SOD activity in the liver homogenates of the SOD1-/- mice, and the activity was decreased by 27% (P < 0.05) in the WT mice after the peroxynitrite treatment (Fig. 4B, upper graph). When bovine SOD1 was added into the liver homogenates, total SOD activities in both genotypes were increased, and were not significantly decreased by the peroxynitrite treatment (Fig. 4B, lower graph). Likewise, the peroxynitrite-mediated nitration of BSA was greatly enhanced by the addition of bovine SOD1 enzyme (Fig. 4C). The added bovine SOD1 activity in the BSA reaction mixture was decreased (P < 0.05) after the peroxynitrite treatment (Fig. 4D).
Fig. 3.
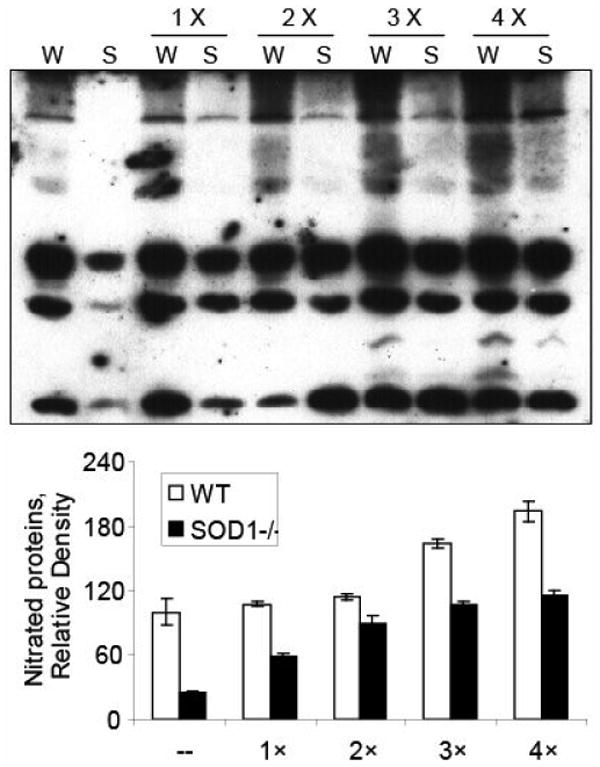
Effects of different doses of bovine SOD1 enzyme addition on the peroxynitrite-mediated protein nitration in liver homogenates. A total of 150 μg of protein was treated with 0.8 mM peroxynitrite in the absence or presence of added SOD1 protein. The upper panel shows representative Western blot of protein nitration (50 μg protein/lane). The lower panel (bar graphs) shows the relative density of the nitrated proteins (n = 2). W, WT; S, SOD1-/-; 1 ×, 2 ×, 3 × and 4 ×, times of SOD activity added as the WT level.
Fig. 4.
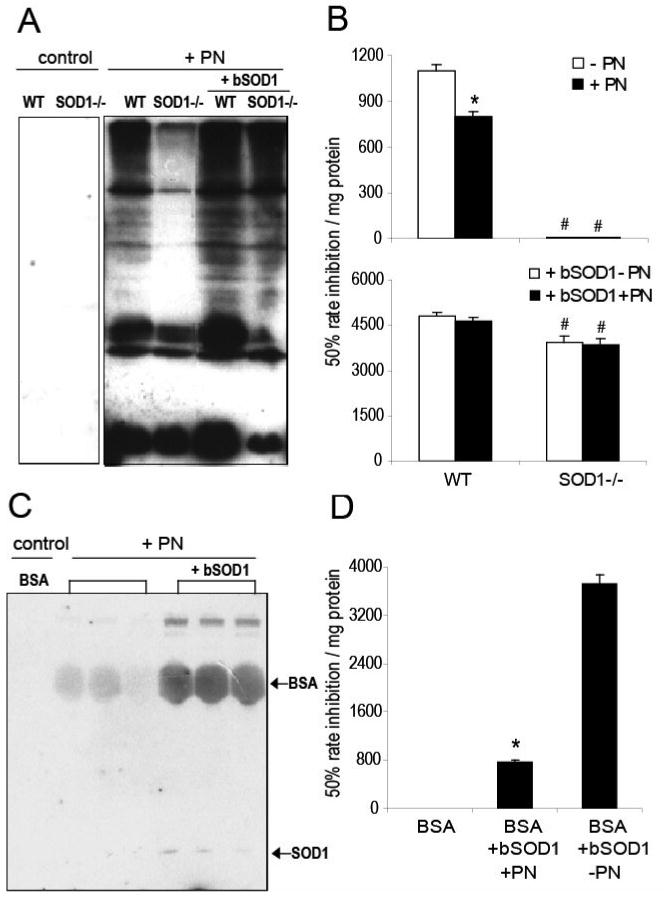
Effects of bovine SOD1 enzyme (bSOD1) addition on the peroxynitrite-mediated protein nitration in liver homogenates and BSA. For liver homogenates, a total of 150 μg of protein was treated with 0.8 mM peroxynitrite in the absence or presence of added SOD1 protein (20 μg). For BSA, a total of 300 μg of protein was treated with 0.4 mM peroxynitrite in the absence or presence of added SOD1 protein (40 μg). A. Western blot analysis of protein nitration of liver homogenates mixture (50 μg protein/lane). B. SOD activity of liver homogenates mixture measured before and after peroxynitrite treatment (n = 3 - 6). C. Western blot analysis of BSA nitration (5 μg protein/lane). D. SOD activity in the BSA reaction mixture measured before and after the peroxynitrite treatment in the presence of added SOD1 protein (n = 3). * P < 0.05 vs. before the peroxynitrite treatment. # P < 0.05 vs. WT.
Apo-SOD1 Was Unable to Catalyze the Peroxynitrite-mediated Nitrotyrosine Formation
Adding the apo-human SOD1 protein into the liver homogenates of WT and SOD1-/- mice or the BSA mixture failed to enhance the peroxynitrite-mediated nitrotyrosine formation (Fig. 5). In contrast, the addition of human holo-SOD1 enzyme greatly enhanced the formation in all three cases. The SOD activity in the mixture with the human holo-SOD1 protein was 4,860 ± 205 units/mg of protein and was decreased by 21% in the BSA mixture after the peroxynitrite treatment.
Fig. 5.
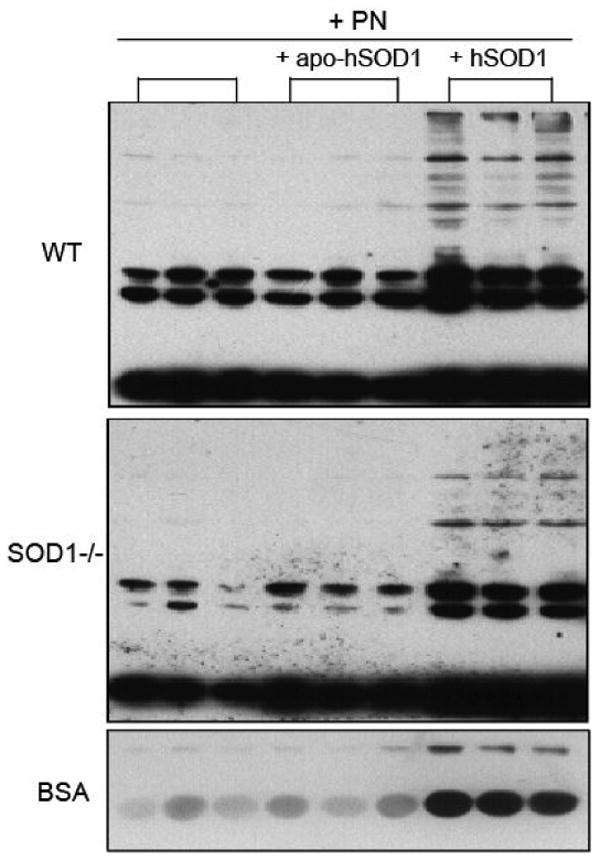
Effect of adding the human holo-SOD1 (hSOD1) and the metal-depleted apo-SOD1 (apo-hSOD1) enzymes on the peroxynitrite-mediated protein nitration in liver homogenates and BSA. For liver homogenates, a total of 150 μg of protein was treated with 0.8 mM peroxynitrite in the absence or presence of added holo-SOD1 or apo-SOD1 protein (6.5 μg). For BSA, a total of 300 μg of BSA was treated with 0.4 mM peroxynitrite in the absence or presence of added holo-SOD1 or apo-SOD1 protein (13 μg). After the reaction, 50 μg of liver homogenate protein or 5 μg of BSA was loaded per lane for Western blot analysis.
The SOD1-Catalyzed Nitrotyrosine Formation Was Residue and Protein Specific
After BSA was incubated with peroxynitrite alone, three tyrosine residues (Y161, Y357, and Y424) were found to be nitrated. Four additional nitrotyrosine residues: Y108, Y179, Y184, and Y475 were detected in BSA incubated with peroxynitrite with added bovine SOD1 enzyme. A representative MS/MS-based identification of nitrotyrosine in BSA (Supplemental Fig. 1) showed that Y475 was nitrated by peroxynitrite in the presence of bovine SOD1 enzyme, and all the ions in the y ion series starting at y8 and higher were shifted by 45.0 amu when compared to the corresponding native peptide of BSA treated with peroxynitrite alone. This shift confirmed nitration at tyrosine residue. No nitrotyrosine was found in the bovine SOD1 protein on the basis of 60% sequence coverage including both Y108 and Q131 that were previously suspected to be the nitrated targets [10]. When the liver homogenates of SOD1-/- mice were incubated with peroxynitrite alone, only 10 proteins were identified from the reaction mixture immuno-precipitated by the nitrotyrosine antibody. In contrast, there were 20 proteins identified from that of the liver precipitates treated with peroxynitrite and added bovine SOD1 enzyme. The 10 candidate proteins uniquely associated with bovine SOD1 catalysis are listed in Table 2.
Table 2.
Candidate nitrated proteins in liver homogenate associated with SOD1 catalysisa
| GI # | Protein name | Function | Location |
|---|---|---|---|
| 200246 | Pyruvate carboxylase | Catalyzes the initial reactions of glucose and lipid synthesis from pyruvate in a tissue specific manner | Mito
Cyto |
| 22477996 | Glutathione s-transferase, mu 2 | Conjugation of reduced glutathione to a wide number of exogenous and endogenous hydrophobic electrophiles | Cyto |
| 19354269 | Methyltransferase like 7b | Probable methyltransferase, highly expressed in liver and kidney | ER |
| 74141423 | Glyceraldehyde-3-phosphate dehydrogenase | Involved in carbohydrate degradation, glycolysis and pyruvate from D-glyceraldehyde 3-phosphate | Mito
Cyto |
| 13097441 | Sarcosine dehydrogenase | Involved in amine and polyamine degradation, sarcosine degradation, and formaldehyde and glycine from sarcosine | Mito
Cyto |
| 18044708 | Fibrinogen, gamma polypeptide | Yields monomers that polymerize into fibrin and acts as a cofactor in platelet aggregation | Sec |
| 1304157 | Heat shock 70kd protein 5 (glucose-regulated protein) | Probably plays a role in facilitating the assembly of multimeric protein complexes inside the endoplasmic reticulum | Cyto
ER |
| 18314540 | Dihydroxyacetone kinase 2 homolog (yeast) | Catalyzes the phosphorylation of dihydroxyacetone, involved in lipid metabolism | Cyto |
| 6753272 | Catalase | Protect cells from the toxic effects of hydrogen peroxide | Mito
Cyto |
| 82958328 | Ribosomal protein s14 | Structural constituent of ribosome | Cyto |
Liver homogenates of SOD1-/- mice were treated with peroxynitrite in the presence or absence of bovine SOD1 enzyme. The homogenates were then immuno-precipitated using the mouse antibody against nitrotyrosine, and processed for Mass Spectrometry. Listed are proteins uniquely resulted from the presence of SOD1 enzyme. Proteins are sorted based on http://david.abcc.ncifcrf.gov/tools.jsp and http://ca.expasy.org/. Cyto, cytoplasm; ER, endoplasmic reticulum; Mito, mitochondrion; Pero, peroxisome; and Sec, secreted.
Discussion
The most significant finding from this study is that knockout of SOD1 attenuated or diminished hepatic protein nitration in mice induced by two types/doses of stimuli. Although the two stimuli APAP and LPS display different chemical properties, metabolic fates, target organs, and abilities to induce NO or peroxynitrite formation [48, 49], impacts of SOD1 knockout on their induced hepatic protein nitration remained similar. The inability of either compound to induce strong hepatic protein nitration in the SOD1-/- mice as in the WT indicates a general need for SOD1 to catalyze the process in vivo. Because the catalytic role of SOD1 in protein nitration has been shown in vitro [9, 10, 40, 50], our results provide direct evidence for such a role of SOD1 in vivo.
The diminished hepatic protein nitration in the SOD1-/- mice was unlikely due to a limited NO formation or secondary changes associated with SOD1 knockout. Both treatments of APAP and LPS resulted in similar differences in hepatic protein nitration between the SOD1-/- and WT mice, despite a large variation (0 to 79%) of genotype differences in plasma NO concentrations. Although NO is needed for the formation of peroxynitrite and inhibition of NO synthesis attenuates protein nitration in some instances [28, 51], protein nitration may take place at baseline of NO production, and thereby is not strictly dependent on elevated NO concentrations [24, 26, 28]. In fact, expression of inducible nitric oxide synthase in liver was not different between WT and the SOD1-/- mice (data not shown). More convincingly, adding the same amount of peroxynitrite in the liver homogenates produced remarkably less nitrotyrosine in the SOD1-/- mice than that in the WT mice. This in vitro genotype difference resembled the in vivo case. Thus, the lack of SOD1 activity, instead of peroxynitrite supply, was the primary or limiting factor responsible for the attenuated hepatic protein nitration in the SOD1-/- mice. Furthermore, adding bovine SOD1 enzyme into the liver homogenates of both genotypes resulted in activity-dependent increases in the peroxynitrite-induced nitrotyrosine formation. The genotype difference was nearly eliminated at the high level of SOD1 enzyme addition. Only the holo-SOD1 enzyme, but not the Cu,Zn-depleted apo-SOD1 protein, was able to enhance the peroxynitrite-mediated nitrotyrosine formation in BSA and liver homogenates. These types of rescues strongly suggest that the lack of SOD1 activity per se, instead of compensatory changes or an accelerated nitrotyrosine degradation [52, 53] caused by the SOD1 knockout, was responsible for the greatly attenuated hepatic protein nitration in the APAP or LPS-treated SOD1-/- mice.
The catalytic or promoting role of SOD1 in in vivo hepatic protein nitration demonstrated in the present study is in contrast to observations from several previous experiments. Overexpression of SOD1 in mice prevented nitration of tyrosine hydroxylase [54] and APAP toxicity [31]. Those impacts by the elevated SOD1 activity represented pharmacological rather than the actual metabolic roles of SOD1. That protection was likely to decrease the stead-state levels of superoxide and consequently the formation of peroxynitrite. Meanwhile, copper deficient mouse embryos with low SOD1 activity showed an increased protein nitration [55], and a 3-wk ethanol feeding enhanced liver nitrotyrosine formation in SOD1-/- mice [56]. In these two cases, the elevated protein nitration was probably caused by secondary metabolic changes [57, 58] resulting from complex changes in body other than just SOD1 activity decrease by copper deficiency, and the interaction between long-term ethanol feeding and SOD1 ablation. Because the above-described impacts of SOD1 overexpression and knockout on protein nitration seem to be asymmetric, we compared the responses of SOD1 heterozygous mice (SOD1+/-) to the APAP-induced hepatic protein nitration and the related toxicity with those of the WT and SOD1-/- (Supplemental Fig. 2 & 3 and Supplemental Table 1). Our data show that liver protein nitration, mouse survival time, and mouse resistance to hepatotoxicity of APAP were SOD1 activity dose-dependent. All responses of the SOD1+/- mice were between the WT and SOD1-/- mice.
Our proteomic analysis provides several new leads to the catalytic mechanism by SOD1 in protein nitration. While the inability of the apo-SOD1 protein to enhance the peroxynitrite-mediated nitrotyrosine formation confirms the earlier view that Cu in SOD1 protein is involved in the reaction between peroxynitrite and the enzyme to form an intermediate with the reactivity of nitronium ion that nitrates tyrosine residues of proteins [10, 15], nitrotyrosine was not detected in the two tryptic peptides of the added bovine SOD1 protein covering two previously-suspected target residues, Y108 and Q131 [10]. Thus, other residues in the enzyme might be served as the intermediate transfer for passing the nitro-group to the target proteins. In addition, other types of modifications of the SOD1 protein by the peroxynitrite treatment might contribute to the post-reaction decrease in SOD1 activity in the mixture. Based on the modeled crystal structure of BSA (Fig. 6), locations of the three tyrosine residues nitrated by peroxynitrite alone were as follows: Y161 is buried, Y357 is situated in small cleft, and Y424 is on the surface. Two (Y108 and Y184) of the four additional tyrosine residues nitrated by peroxynitrite with added bovine SOD1 enzyme are buried in the inside of structure. Although six of the seven nitrated tyrosine residues by the latter treatment lie in regions with highly positive values of the electrostatic potential, surrounded by a number of positively charged residues (Supplemental Fig. 4), significant structural rearrangement of BSA may be required for the nitration of the buried tyrosine residues. Conceivably, the peroxynitrite treatment could cause BSA to undergo a local conformation change to render certain buried regions available, and SOD1 enzyme might facilitate the process. Based on the genome annotation database DAVID [59], the 10 unique proteins that were presumably nitrated by peroxynitrite under the catalysis by SOD1 enzyme bear the following functional annotation: 4 in mitochondrion, 8 in cytoplasm, 3 in endoplasmic reticulum or peroxisome, 8 related to metabolism, 7 with catalytic activity (4 with transferase activity and 3 with oxidoreductase activity), and 4 dealing with generation of precursor metabolites and energy. More interestingly, at least 4 out of the 10 proteins have been previously shown to be related to APAP toxicity [60, 61]. It is our future interest to find out if and how nitration of these proteins in mouse liver affects their resistance to APAP toxicity.
Fig. 6.
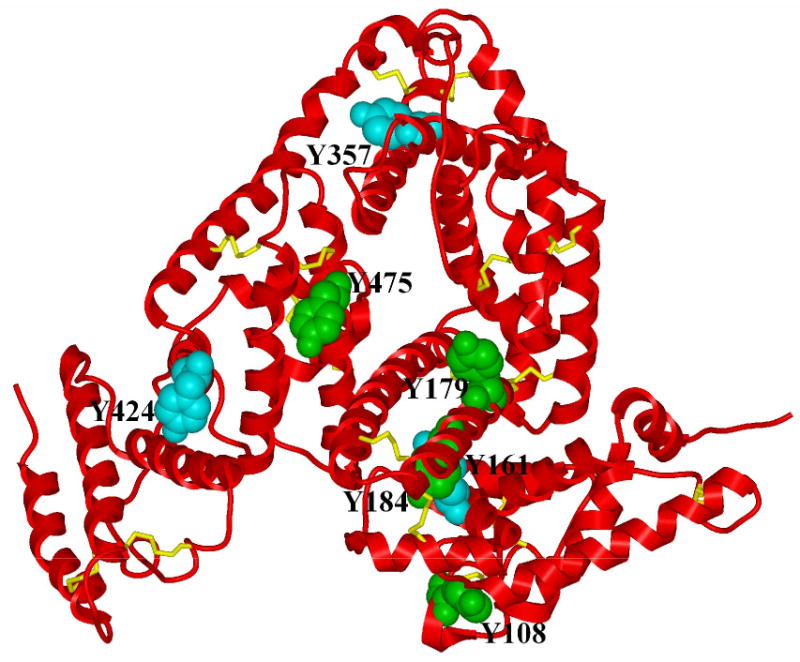
A simplified representation of a three-dimensional model of the bovine serum albumin (BSA) structure describing the position of nitrated tyrosine residues. The backbone of the structure is shown by using a ribbon representation colored red. Nitrated tyrosine residues are shown using a CPK representation. Residues in cyan correspond to those nitrated when BSA was treated with peroxynitrite alone. Residues in green were found in BSA after treatment with peroxynitrite in the presence of added bovine SOD1 enzyme. All other side chains in the structure were omitted except for cysteine residues involved in S-S bonds which are shown in yellow using a stick representation.
All together, our data show that the SOD1 activity was important for the catalysis of APAP and LPS-induced hepatic protein nitration in mice. The catalysis was not directly related to plasma NO concentrations, peroxynitrite availability, or secondary alternations associated with the SOD1 knockout. More than a decade after the demonstration of SOD1 in catalyzing nitrotyrosine formation in vitro [9, 10, 14], we have provided direct evidence for such a role of SOD1 in vivo. The application of mass spectrometry and proteomic analysis helps elucidate the structural mechanism for that catalytic function of SOD1 and the metabolic implications.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Health grant DK53108 to XGL and with the resources of the Computational Biology Service Unit from Cornell University which is partially funded by Microsoft Corporation.
Abbreviations used
- ALS
amyotrophic lateral sclerosis
- APAP
acetaminophen
- BSA
bovine serum albumin
- LPS
lipopolysaccharide
- NO
nitric oxide
- PBS
phosphate-buffered saline
- SOD1
copper,zinc-superoxide dismutase
- SOD1-/-
SOD1 knockout mice
- WT
wild-type mice
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ho YS, Gargano M, Cao J, Bronson RJ, Heimler I, Hutz RJ. Reduced fertility in female mice lacking copper-zinc superoxide dismutase. J Biol Chem. 1998;273:7765–7769. doi: 10.1074/jbc.273.13.7765. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida T, Maulik N, Engelman RM, Ho YS, Das DK. Targeted disruption of the mouse Sod1 gene makes the hearts vulnerable to ischemic reperfusion injury. Circ Res. 2000;86:264–269. doi: 10.1161/01.res.86.3.264. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Graham DG, Montine TJ, Ho YS. Enhanced N-methyl-4-phenyl-1,2,3,6-tetrahydropyridine toxicity in mice deficient in Cu,Zn-superoxide dismutase or glutathione peroxidase. J Neuropathol Exp Neurol. 2000;59:53–61. doi: 10.1093/jnen/59.1.53. [DOI] [PubMed] [Google Scholar]
- 4.Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson Roberts L, Van Remmen H, Epstein CJ, Huang TT. Cu,Zn-SOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- 5.Imamura Y, Noda S, Hashizume K, Shinoda K, Yamaguchi M, Uchiyama S, Shimizu T, Mizushima Y, Shirasawa T, Tsubota K. Drusen, choroidal neovascularization, and retinal pigment epithelium dysfunction in SOD1-deficient mice: A model of age-related macular degeneration. Proc Natl Acad Sci USA. 2006;103:11282–11287. doi: 10.1073/pnas.0602131103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaarsma D, Haasdijk ED, Grashorn JA, Hawkins R, van Duijn W, Verspaget HW, London J, Holstege JC. Human Cu,Zn- superoxide dismutase (SOD1) overexpression in mice causes mitochondrial vacuolization, axonal degeneration, and premature motoneuron death and accelerates motoneuron disease in mice expressing a familial amyotrophic lateral sclerosis mutant SOD1. Neurobiol Dis. 2000;7:623–643. doi: 10.1006/nbdi.2000.0299. [DOI] [PubMed] [Google Scholar]
- 7.Lei XG, Zhu JH, McClung JP, Aregullin M, Roneker CA. Mice deficient in Cu,Zn-superoxide dismutase are resistant to acetaminophen toxicity. Biochem J. 2006;399:455–461. doi: 10.1042/BJ20060784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu JH, Zhang X, McClung JP, Lei XG. Impact of Cu,Zn-superoxide dismutase and Se-dependent glutathione peroxidase-1 knockouts on acetaminophen-induced cell death and related signaling in murine liver. Exp Biol Med (Maywood) 2006;231:1726–1732. doi: 10.1177/153537020623101109. [DOI] [PubMed] [Google Scholar]
- 9.Crow JP, Sampson JB, Zhuang Y, Thompson JA, Beckman JS. Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J Neurochem. 1997;69:1936–1944. doi: 10.1046/j.1471-4159.1997.69051936.x. [DOI] [PubMed] [Google Scholar]
- 10.Ischiropoulos H, Zhu L, Chen J, Tsai M, Martin JC, Smith CD, Beckman JS. Peroxynitrite-mediated tyrosine nitration catalyzed by superoxide dismutase. Arch Biochem Biophys. 1992;298:431–437. doi: 10.1016/0003-9861(92)90431-u. [DOI] [PubMed] [Google Scholar]
- 11.Cudd A, Fridovich I. Electrostatic interactions in the reaction mechanism of bovine erythrocyte superoxide dismutase. J Biol Chem. 1982;257:11443–11447. [PubMed] [Google Scholar]
- 12.Beckman JS, Ye YZ, Anderson PG, Chen J, Accavitti MA, Tarpey MM, White CR. Extensive nitration of protein tyrosines in human atherosclerosis detected by immunohistochemistry. Biol Chem Hoppe Seyler. 1994;375:81–88. doi: 10.1515/bchm3.1994.375.2.81. [DOI] [PubMed] [Google Scholar]
- 13.Denicola A, Radi R. Peroxynitrite and drug-dependent toxicity. Toxicology. 2005;208:273–288. doi: 10.1016/j.tox.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 14.Beckman JS, Ischiropoulos H, Zhu L, van der Woerd M, Smith C, Chen J, Harrison J, Martin JC, Tsai M. Kinetics of superoxide dismutase- and iron-catalyzed nitration of phenolics by peroxynitrite. Arch Biochem Biophys. 1992;298:438–445. doi: 10.1016/0003-9861(92)90432-v. [DOI] [PubMed] [Google Scholar]
- 15.Beckman JS, Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am J Physiol. 1996;271:C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 16.Giasson BI, Duda JE, Murray IVJ, Chen Q, Souza JM, Hurtig HI, Ischiropoulos H, Trojanowski JQ, Lee VM. Oxidative damage linked to neurodegeneration by selective alpha -synuclein nitration in synucleinopathy lesions. Science. 2000;290:985–989. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- 17.Cassina AM, Hodara R, Souza JM, Thomson L, Castro L, Ischiropoulos H, Freeman BA, Radi R. Cytochrome c nitration by peroxynitrite. J Biol Chem. 2000;275:21409–21415. doi: 10.1074/jbc.M909978199. [DOI] [PubMed] [Google Scholar]
- 18.Gow AJ, Duran D, Malcolm S, Ischiropoulos H. Effects of peroxynitrite-induced protein modifications on tyrosine phosphorylation and degradation. FEBS Lett. 1996;385:63–66. doi: 10.1016/0014-5793(96)00347-x. [DOI] [PubMed] [Google Scholar]
- 19.Beal MF, Ferrante RJ, Browne SE, Matthews RT, Kowall NW, Brown RH., Jr Increased 3-nitrotyrosine in both sporadic and familial amyotrophic lateral sclerosis. Ann Neurol. 1997;42:644–654. doi: 10.1002/ana.410420416. [DOI] [PubMed] [Google Scholar]
- 20.Hensley K, Maidt ML, Yu Z, Sang H, Markesbery WR, Floyd RA. Electrochemical analysis of protein nitrotyrosine and dityrosine in the Alzheimer brain indicates region-specific accumulation. J Neurosci. 1998;18:8126–8132. doi: 10.1523/JNEUROSCI.18-20-08126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casoni F, Basso M, Massignan T, Gianazza E, Cheroni C, Salmona M, Bendotti C, Bonetto V. Protein nitration in a mouse model of familial amyotrophic lateral sclerosis: possible multifunctional role in the pathogenesis. J Biol Chem. 2005;280:16295–16304. doi: 10.1074/jbc.M413111200. [DOI] [PubMed] [Google Scholar]
- 22.Ischiropoulos H, Beckman JS. Oxidative stress and nitration in neurodegeneration: Cause, effect, or assoication? J Clin Invest. 2003;111:163–169. doi: 10.1172/JCI17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michael SL, Pumford NR, Mayeux PR, Niesman MR, Hinson JA. Pretreatment of mice with macrophage inactivators decreases acetaminophen hepatotoxicity and the formation of reactive oxygen and nitrogen species. Hepatology. 1999;30:186–195. doi: 10.1002/hep.510300104. [DOI] [PubMed] [Google Scholar]
- 24.Hinson JA, Pike SL, Pumford NR, Mayeux PR. Nitrotyrosine-protein adducts in hepatic centrilobular areas following toxic doses of acetaminophen in mice. Chem Res Toxicol. 1998;11:604–607. doi: 10.1021/tx9800349. [DOI] [PubMed] [Google Scholar]
- 25.Knight TR, Ho YS, Farhood A, Jaeschke H. Peroxynitrite is a critical mediator of acetaminophen hepatotoxicity in murine livers: protection by glutathione. J Pharmacol Exp Ther. 2002;303:468–475. doi: 10.1124/jpet.102.038968. [DOI] [PubMed] [Google Scholar]
- 26.Michael SL, Mayeux PR, Bucci TJ, Warbritton AR, Irwin LK, Pumford NR, Hinson JA. Acetaminophen-induced hepatotoxicity in mice lacking inducible nitric oxide synthase activity. Nitric Oxide. 2001;5:432–441. doi: 10.1006/niox.2001.0385. [DOI] [PubMed] [Google Scholar]
- 27.Gardner CR, Heck DE, Yang CS, Thomas PE, Zhang XJ, DeGeorge GL, Laskin JD, Laskin DL. Role of nitric oxide in acetaminophen-induced hepatotoxicity in the rat. Hepatology. 1998;27:748–754. doi: 10.1002/hep.510270316. [DOI] [PubMed] [Google Scholar]
- 28.Hinson JA, Bucci TJ, Irwin LK, Michael SL, Mayeux PR. Effect of inhibitors of nitric oxide synthase on acetaminophen-induced hepatotoxicity in mice. Nitric Oxide. 2002;6:160–167. doi: 10.1006/niox.2001.0404. [DOI] [PubMed] [Google Scholar]
- 29.Groner Y, Elroy-Stein O, Avraham KB, Schickler M, Knobler H, Minc-Golomb D, Bar-Peled O, Yarom R, Rotshenker S. Cell damage by excess CuZnSOD and Down's syndrome. Biomed Pharmacother. 1994;48:231–240. doi: 10.1016/0753-3322(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 30.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Mutations in Cu,Zn-superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 31.Mirochnitchenko O, Weisbrot-Lefkowitz M, Reuhl K, Chen L, Yang C, Inouye M. Acetaminophen toxicity. Opposite effects of two forms of glutathione peroxidase. J Biol Chem. 1999;274:10349–10355. doi: 10.1074/jbc.274.15.10349. [DOI] [PubMed] [Google Scholar]
- 32.Denicola A, Souza JM, Radi R. Diffusion of peroxynitrite across erythrocyte membranes. Proc Natl Acad Sci USA. 1998;95:3566–3571. doi: 10.1073/pnas.95.7.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elsasser TH, Kahl S, MacLeod C, Nicholson B, Sartin JL, Li C. Mechanisms underlying growth hormone effects in augmenting nitric oxide production and protein tyrosine nitration during endotoxin challenge. Endocrinology. 2004;145:3413–3423. doi: 10.1210/en.2004-0063. [DOI] [PubMed] [Google Scholar]
- 34.Lu Y, Wang X, Cederbaum AI. Lipopolysaccharide-induced liver injury in rats treated with the CYP2E1 inducer pyrazole. Am J Physiol Gastrointest Liver Physiol. 2005;289:G308–G319. doi: 10.1152/ajpgi.00054.2005. [DOI] [PubMed] [Google Scholar]
- 35.Doucette PA, Whitson LJ, Cao X, Schirf V, Demeler B, Valentine JS, Hansen JC, Hart PJ. Dissociation of human copper-zinc superoxide dismutase dimers using chaotrope and reductant. Insights into the molecular basis for dimer stability. J Biol Chem. 2004;279:54558–54566. doi: 10.1074/jbc.M409744200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C, Walker LM, Hinson JA, Mayeux PR. Oxidant stress in rat liver after lipopolysaccharide administration: effect of inducible nitric-oxide synthase inhibition. J Pharmacol Exp Ther. 2000;293:968–972. [PubMed] [Google Scholar]
- 37.Viera L, Ye YZ, Estevez AG, Beckman JS. Immunohistochemical methods to detect nitrotyrosine. Methods Enzymol. 1999;301:373–381. doi: 10.1016/s0076-6879(99)01101-5. [DOI] [PubMed] [Google Scholar]
- 38.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 39.Pollock JS, Forstermann U, Mitchell JA, Warner TD, Schmidt HH, Nakane M, Murad F. Purification and characterization of particulate endothelium-derived relaxing factor synthase from cultured and native bovine aortic endothelial cells. Proc Natl Acad Sci USA. 1991;88:10480–10484. doi: 10.1073/pnas.88.23.10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu JH, Lei XG. Double null of selenium-glutathione peroxidase-1 and copper, zinc-superoxide dismutase enhances resistance of mouse primary hepatocytes to acetaminophen toxicity. Exp Biol Med (Maywood) 2006;231:545–552. doi: 10.1177/153537020623100508. [DOI] [PubMed] [Google Scholar]
- 41.Fu Y, Sies H, Lei XG. Opposite roles of selenium-dependent glutathione peroxidase-1 in superoxide generator diquat- and peroxynitrite-induced apoptosis and signaling. J Biol Chem. 2001;276:43004–43009. doi: 10.1074/jbc.M106946200. [DOI] [PubMed] [Google Scholar]
- 42.Zhang S, Van Pelt CK, Henion JD. Automated chip-based nanoelectrospray-mass spectrometry for rapid identification of proteins separated by two-dimensional gel electrophoresis. Electrophoresis. 2003;24:3620–3632. doi: 10.1002/elps.200305585. [DOI] [PubMed] [Google Scholar]
- 43.Morris RM, Fung JM, Rahm BG, Zhang S, Freedman DL, Zinder SH, Richardson RE. Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Appl Environ Microbiol. 2007;73:320–326. doi: 10.1128/AEM.02129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez R, Sali A. Comparative protein structure modeling. Introduction and practical examples with modeller. Methods Mol Biol. 2000;143:97–129. doi: 10.1385/1-59259-368-2:97. [DOI] [PubMed] [Google Scholar]
- 46.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.James LP, Mayeux PR, Hinson JA. Acetaminophen-induced hepatotoxicity. Drug Metab Dispos. 2003;31:1499–1506. doi: 10.1124/dmd.31.12.1499. [DOI] [PubMed] [Google Scholar]
- 49.Su GL. Lipopolysaccharides in liver injury: molecular mechanisms of Kupffer cell activation. Am J Physiol Gastrointest Liver Physiol. 2002;283:G256–G265. doi: 10.1152/ajpgi.00550.2001. [DOI] [PubMed] [Google Scholar]
- 50.McBride AG, Borutaite V, Brown GC. Superoxide dismutase and hydrogen peroxide cause rapid nitric oxide breakdown, peroxynitrite production and subsequent cell death. Biochim Biophys Acta. 1999;1454:275–288. doi: 10.1016/s0925-4439(99)00046-0. [DOI] [PubMed] [Google Scholar]
- 51.Fries DM, Paxinou E, Themistocleous M, Swanberg E, Griendling KK, Salvemini D, Slot JW, Heijnen HF, Hazen SL, Ischiropoulos H. Expression of inducible nitric-oxide synthase and intracellular protein tyrosine nitration in vascular smooth muscle cells: role of reactive oxygen species. J Biol Chem. 2003;278:22901–22907. doi: 10.1074/jbc.M210806200. [DOI] [PubMed] [Google Scholar]
- 52.Kotamraju S, Tampo Y, Keszler A, Chitambar CR, Joseph J, Haas AL, Kalyanaraman B. Nitric oxide inhibits H2O2-induced transferrin receptor-dependent apoptosis in endothelial cells: Role of ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 2003;100:10653–10658. doi: 10.1073/pnas.1933581100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Souza JM, Choi I, Chen Q, Weisse M, Daikhin E, Yudkoff M, Obin M, Ara J, Horwitz J, Ischiropoulos H. Proteolytic degradation of tyrosine nitrated proteins. Arch Biochem Biophys. 2000;380:360–366. doi: 10.1006/abbi.2000.1940. [DOI] [PubMed] [Google Scholar]
- 54.Ara J, Przedborski S, Naini AB, Jackson-Lewis V, Trifiletti RR, Horwitz J, Ischiropoulos H. Inactivation of tyrosine hydroxylase by nitration following exposure to peroxynitrite and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) Proc Natl Acad Sci USA. 1998;95:7659–7663. doi: 10.1073/pnas.95.13.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beckers-Trapp ME, Lanoue L, Keen CL, Rucker RB, Uriu-Adams JY. Abnormal development and increased 3-nitrotyrosine in copper-deficient mouse embryos. Free Radic Biol Med. 2006;40:35–44. doi: 10.1016/j.freeradbiomed.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 56.Kessova IG, Ho YS, Thung S, Cederbaum AI. Alcohol-induced liver injury in mice lacking Cu, Zn-superoxide dismutase. Hepatology. 2003;38:1136–1145. doi: 10.1053/jhep.2003.50450. [DOI] [PubMed] [Google Scholar]
- 57.Patsouris D, Reddy JK, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha mediates the effects of high-fat diet on hepatic gene expression. Endocrinology. 2006;147:1508–1516. doi: 10.1210/en.2005-1132. [DOI] [PubMed] [Google Scholar]
- 58.Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 59.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 60.Thome-Kromer B, Bonk I, Klatt M, Nebrich G, Taufmann M, Bryant S, Wacker U, Kopke A. Toward the identification of liver toxicity markers: a proteome study in human cell culture and rats. Proteomics. 2003;3:1835–1862. doi: 10.1002/pmic.200300552. [DOI] [PubMed] [Google Scholar]
- 61.Welch KD, Wen B, Goodlett DR, Yi EC, Lee H, Reilly TP, Nelson SD, Pohl LR. Proteomic identification of potential susceptibility factors in drug-induced liver disease. Chem Res Toxicol. 2005;18:924–933. doi: 10.1021/tx050011b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


