Abstract
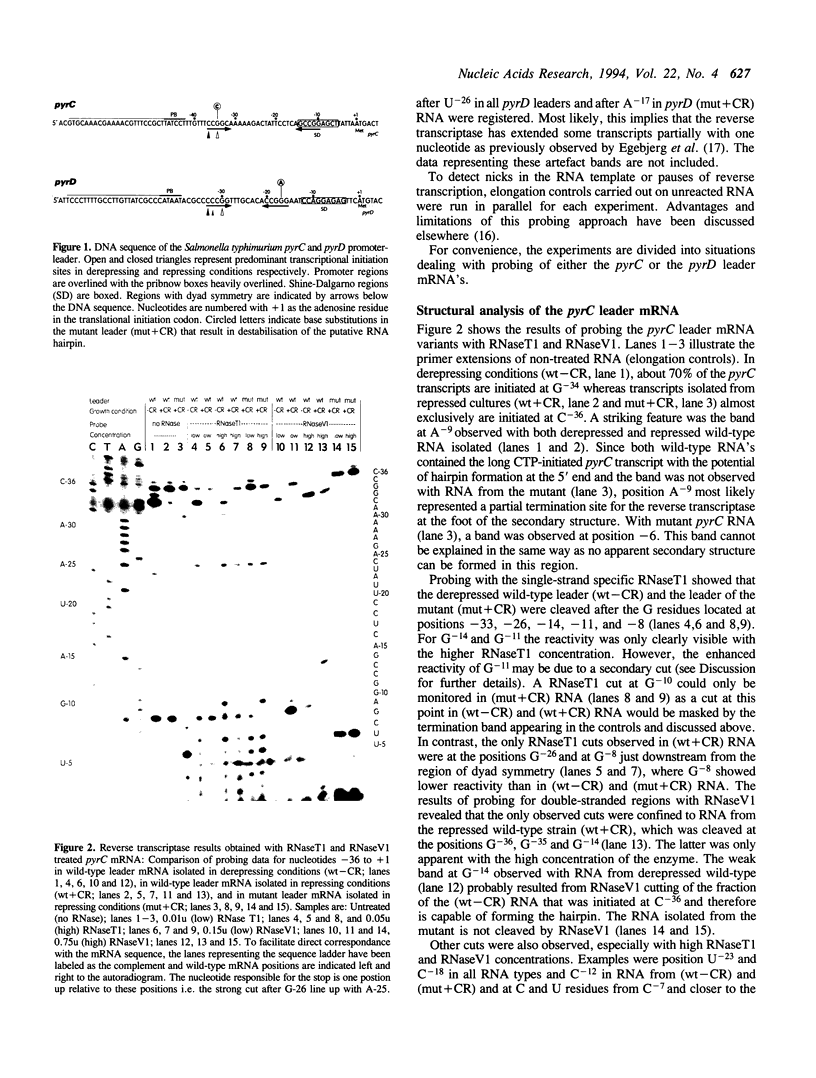
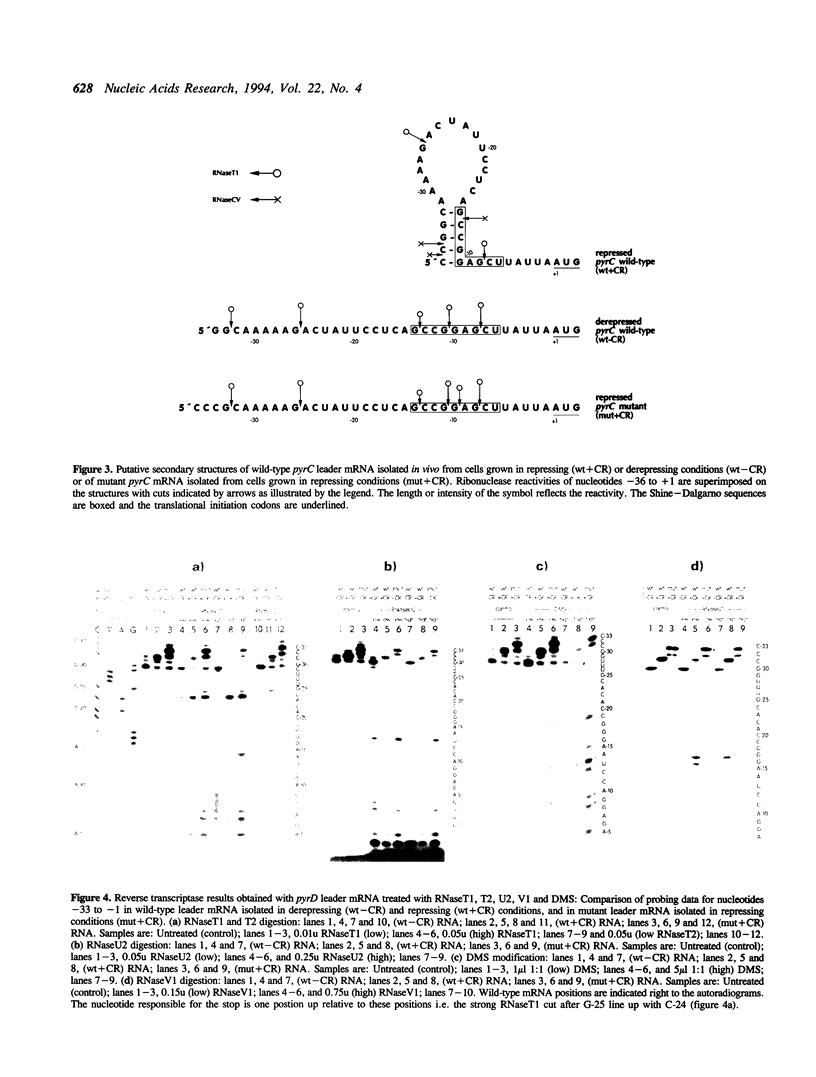
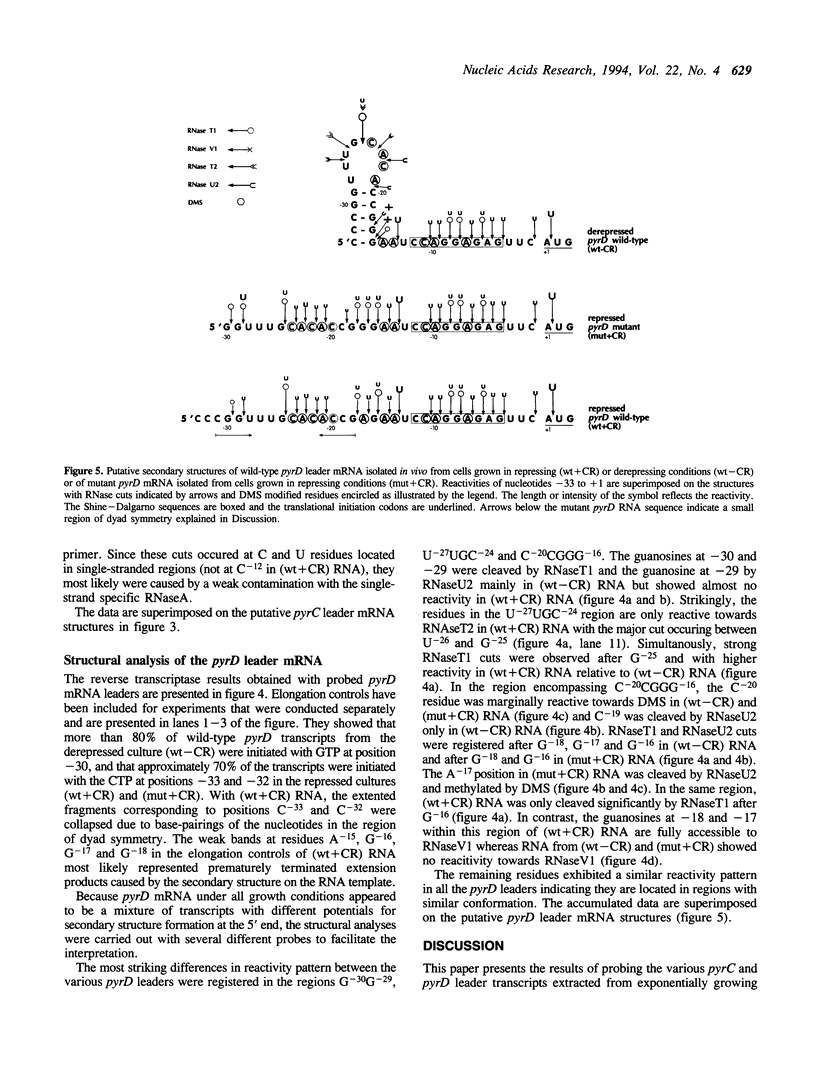
In Salmonella typhimurium, different conformations of the pyrC and pyrD leader transcripts are produced as a result of nucleotide sensitive selection of the transcriptional start site. The CTP-initiated transcripts, synthesized at high intracellular CTP/GTP pool ratios (repressing conditions), have the potential of forming a stable secondary structure at the 5' end, thereby sequestering the site for translational initiation. At low CTP/GTP pool ratios (derepressing conditions), transcription starts 2-3 bp further downstream, resulting in transcripts with limited potential for stem-loop formation and therefore open for translational initiation. The conformation of the leader regions of wild type pyrC and pyrD mRNA has been investigated by chemical and enzymatic probing of RNA isolated from cultures grown in repressing and derepressing conditions. As controls and to obtain further information on the relation between the leader RNA conformation and the regulatory mechanism, the probing experiments also included pyrC and pyrD mRNA from mutants that contain a base substitution at a position that destabilizes the putative hairpin. In accordance with predictions based on the nucleotide sequence, the results showed that the 5' end of pyrC and pyrD leader mRNA isolated from repressed cultures is folded into a secondary structure, whereas it is largely unstructured in mRNA isolated from derepressed cultures.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Davison J., Heusterspreute M., Merchez M., Brunel F. Vectors with restriction-site banks. I. pJRD158, a 3903-bp plasmid containing 28 unique cloning sites. Gene. 1984 Jun;28(3):311–318. doi: 10.1016/0378-1119(84)90148-3. [DOI] [PubMed] [Google Scholar]
- Egebjerg J., Leffers H., Christensen A., Andersen H., Garrett R. A. Structure and accessibility of domain I of Escherichia coli 23 S RNA in free RNA, in the L24-RNA complex and in 50 S subunits. Implications for ribosomal assembly. J Mol Biol. 1987 Jul 5;196(1):125–136. doi: 10.1016/0022-2836(87)90515-8. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick M. M., Neuhard J., Kelln R. A. Cloning, nucleotide sequence and regulation of the Salmonella typhimurium pyrD gene encoding dihydroorotate dehydrogenase. Eur J Biochem. 1990 Dec 12;194(2):573–578. doi: 10.1111/j.1432-1033.1990.tb15654.x. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Saenger W. Specific protein-nucleic acid recognition in ribonuclease T1-2'-guanylic acid complex: an X-ray study. Nature. 1982 Sep 2;299(5878):27–31. doi: 10.1038/299027a0. [DOI] [PubMed] [Google Scholar]
- Kelln R. A., Neuhard J. Regulation of pyrC expression in Salmonella typhimurium: identification of a regulatory region. Mol Gen Genet. 1988 May;212(2):287–294. doi: 10.1007/BF00334698. [DOI] [PubMed] [Google Scholar]
- Lu C. D., Kilstrup M., Neuhard J., Abdelal A. Pyrimidine regulation of tandem promoters for carAB in Salmonella typhimurium. J Bacteriol. 1989 Oct;171(10):5436–5442. doi: 10.1128/jb.171.10.5436-5442.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton N. P. Improved plasmid vectors for the isolation of translational lac gene fusions. Gene. 1984 Nov;31(1-3):269–273. doi: 10.1016/0378-1119(84)90220-8. [DOI] [PubMed] [Google Scholar]
- Neuhard J., Stauning E., Kelln R. A. Cloning and characterization of the pyrE gene and of PyrE::Mud1 (Ap lac) fusions from Salmonella typhimurium. Eur J Biochem. 1985 Feb 1;146(3):597–603. doi: 10.1111/j.1432-1033.1985.tb08693.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen K. I., Neuhard J. Dual transcriptional initiation sites from the pyrC promoter control expression of the gene in Salmonella typhimurium. Mol Gen Genet. 1991 Feb;225(2):249–256. doi: 10.1007/BF00269856. [DOI] [PubMed] [Google Scholar]
- Uchida T., Arima T., Egami F. Specificity of RNase U2. J Biochem. 1970 Jan;67(1):91–102. doi: 10.1093/oxfordjournals.jbchem.a129239. [DOI] [PubMed] [Google Scholar]
- Warrington R. C. Ribonuclease T1 catalyzed degradation of transfer RNA: an unusual alteration induced by urea. Biochim Biophys Acta. 1974 Jun 14;353(1):63–68. doi: 10.1016/0005-2787(74)90097-5. [DOI] [PubMed] [Google Scholar]
- Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]