Abstract
The Casitas-B-lineage Lymphoma (Cbl) proteins play an important role in regulating signal transduction pathways by functioning as E3-ubiquitin ligases. The Cbl proteins contain a conserved tyrosine kinase binding (TKB) domain that bind over a dozen proteins, including protein tyrosine kinases (PTKs) in a phosphorylation dependent manner. The cell surface expression levels of the PTKs are regulated by Cbl-mediated ubiquitination, internalization, and degradation. Dysfunction in this signaling cascade has resulted in prolonged activation of the PTKs and therefore implicated in inflammatory diseases and various cancers. Due to this negative regulatory function, Cbl has been largely ignored as a therapeutic target. However recent studies such as the identification of (a) gain of function c-Cbl mutations in subsets of myeloid cancer and (b) c-Cbl as a prostate basal cell marker that correlates with poor clinical outcome, suggests otherwise. Here we report the development of a competitive high throughput fluorescence polarization assay in a 384-well format to identify inhibitors of Cbl(TKB). The high throughput screen (HTS) readiness of the assay was demonstrated by screening the Prestwick chemical library®.
Keywords: High-throughput screening, Fluorescence polarization assay, Cbl-PTK inhibitors
Introduction
Extracellular peptide growth factors recognize and bind protein tyrosine kinases (PTKs) and transfer information to regulate key cellular functions such as growth, immune response etc.[1,2] This pathway is down regulated through ubiquitination followed by endosome directed degradation of the PTKs.[3] Cbl proteins are major components of this regulation by functioning as both a docking protein that recognizes phosphorylated PTKs and as an E3-ubiquitin ligase to tag the PTKs for degradation.[4,5] It has been well documented that dysfunction in this signal transduction pathway has been implicated in inflammatory diseases and cancers.[1–6]
Recently, several groups have identified clinically relevant, oncogenic c-Cbl mutations.[7,8] These mutations have been most frequently identified in patients with myleodysplastic myleoproliferative tumors.[9] These include chronic myelomonocytic leukemia (CMML), juvenile myelomonocytic leukeumia (JMML), and atypical chronic myeloid leukemia (aCML). The frequency of c-Cbl mutations occurs in 5% of aCML cases and in 10–15% of JMML and CMML cases.[10] Genetically, these mutations are not accompanied by CBL gene deletions, pointing to a gain-of function nature rather than a loss-of-function.[8] Biochemically, the exact mechanism of the gain-of-function Cbl mutations is unclear; however the end result translates to excessive PTK signaling. Current interpretation proposes that wild type Cbl acts as a tumor suppressor, while mutated Cbl acts as an oncoprotein.[9,10] Clinically relevant mutations, therefore, are the result of the loss of the negative E3 function and maintenance of the oncogenic adaptor function.[10] Taken together, these observations point to the possibility that small molecule inhibitors could be of therapeutic value to patients with cancers driven by Cbl mutations. Further, in an attempt to identify biomarkers, a recent prostate cancer clinical study showed elevated Cbl levels correlated with poor clinical outcome (i.e., decreased patient survival).[11] Although the molecular basis for this observation is unclear at the present time, it does suggest that Cbl could be a potential prostate cancer therapeutic target.
The mammalian Cbl family of proteins is comprised of three gene products: c-Cbl, Cbl-b and Cbl-c/Cbl-3.[12] The N-terminus of the Cbl family of proteins has a conserved tyrosine kinase binding (TKB) domain comprised of a SH2-like domain flanked by a four-helix bundle and a calcium-binding EF hand.[13,14] The TKB domains recognize and bind phosphorylated PTKs that are subsequently ubiquitinated and degraded.[15] The TKB domains also recognize proteins such as the Adaptor protein containing Pleckstrin homology and SH3 domain (APS) that is important for insulin signaling.[16] These illustrative examples highlight the importance of Cbl(TKB)-protein interactions in facilitating critical cellular functions.
The consensus recognition sequence and the binding modes of phosphopeptides recognized by the Cbl(TKB) domain has undergone several iterations of refinement starting with D(N/D)XpY, followed by (N/D)XpY(S/T)XXP found in several PTKs such as 70kDa ζ-associate protein kinase (ZAP70), epidermal growth factor receptor (EGFR), Src etc.[13–15,17,18] With the identification of additional Cbl binding partners such as APS and c-Met the binding motif’s RA(V/I)XNQpY(S/T) and DpYR respectively were proposed.[19–21] A recent comprehensive structural study showed that phosphopeptides with diverse sequences bind TKB at the same site albeit in two different orientations.[22] Their study also describes a unified model for the TKB-phosphopeptide binding anchored by a unique intrapeptidyl hydrogen bond (H-bond). The H-bond is formed between the phosphotyrosine (pY) and conserved asparagine (N) at the P-2 position or an arginine (R) at the P-1 position.[22]
The disease relevance and the functional diversity of Cbl, coupled with varied binding motifs recognized by the TKB domain prompted us to develop a high throughput screen to identify small molecule inhibitors for the phosphotyrosine binding site on Cbl(TKB). These small molecule inhibitors will serve as (a) potential drug leads against myeloproliferative disorders and (b) chemical probes to manipulate the signaling specifically mediated by the TKB-phosphoprotein interaction. Previously we have successfully used the fluorescence polarization (FP) technique to develop a BRCA1(BRCT) assay and used it to carry out a quantitative high throughput campaign which led to the identification of small molecule inhibitors.[23,24]
Here we describe the development and optimization of a high throughput Cbl FP assay using the TKB domain expressed as a His-tagged fusion protein and a fluorescently labeled ZAP70 phosphotyrosine peptide. The FP assay was used to demonstrate the phosphospecific nature of the Cbl(TKB)-ZAP70 interaction. To demonstrate high throughput feasiblity we used the optimized FP assay to screen the Prestwick chemical library® for inhibitors in a 384-well format. During the screen we used the unlabeled ZAP70 peptide as a positive control and demonstrate that the Cbl-FP assay is qHTS ready.
Materials and Methods
Reagents
The labeled peptide Flu-βA-TLNSDGpYTPEPA-CONH2 (1), the unlabeled, phosphorylated peptide Ac-TLNSDGpYTPEPA-CONH2 (2), and the unlabeled, non-phosphorylated peptide Ac-TLNSDGYTPEPA-CONH2 (3) (Table 1) were synthesized and HPLC purified either by us or by the Tufts University Core Facility, Boston MA. The peptide concentrations were determined by amino acid analysis at the protein chemistry core, UTMB or protein structure core facility, UNMC. The GST-Cbl(TKB) (kind gift from Dr. Hamid Band) was expressed and purified as previously described.[13] The His-Cbl(TKB) was generated by inserting the BamHI-EcoRI digest of the GST-Cbl(TKB) clone into a pET30a vector. The His-Cbl(TKB) was expressed in E. Coli and purified on a Ni-agarose column following the manufacturer’s (QIAGEN) directions. Protein concentrations were determined by the nanodrop method and using the Bicinchoninic acid protein assay (Pierce). The GST-Cbl(TKB) protein stock solution starts to precipitate after about a week even at 4°C, no such precipitation was observed for the His-Cbl(TKB) even after 6-weeks at 4°C.
Table 1.
ZAP70 peptides used in this study; FITC = Fluorescein Isothiocyanate
| Peptide | Sequence | Fluorescent Label |
|---|---|---|
| 1 | Flu-βA-TLNSDGpYTPEPA | FITC |
| 2 | Ac-TLNSDGpYTPEPA | None |
| 3 | Ac-TLNSDGYTPEPA | None |
Compound library
The Prestwick chemical library® contains 1120 small molecules, 90% being off-patent marketed drugs and 10% bioactive alkaloids or related substances, presenting a high degree of drug-likeliness. The active compounds were selected for their high chemical and pharmacological diversity as well as for their known bioavailability and safety in humans. Compounds were present at 2 mg/mL in DMSO (5.8 mM median concentration). In every 96-well drug plate, eighty compounds were added from A2 to H11 as 1 μL aliquots on a Biomek FX platform. From a separate source plate a concentration range (0.625 – 80 μM) of the unlabeled ZAP70 peptide (2) was added to the 1st and last columns of each plate as positive controls. The chemical library was a kind gift from John S. Dunn GCC Chemical Genomic Research Consortium.
General methods
All measurements were made on 384-well, low-volume, black round-bottom polystyrene NBS microplate (Corning) using a Spectramax M5 plate reader (MDS). The polarization values are reported in millipolarization units (mP) and were measured at an excitation wavelength of 485 nm and an emission wavelength of 538 nm. The data was fitted and the ΔmP, Kd and IC50 values were estimated using non-linear least square fit to a single site binding model (SigmaPlot 11.0).
Kd value estimation for fluorescein-labeled ZAP70 peptide to Cbl(TKB) domains
To each well, 1 μL of 2 μM labeled ZAP70 peptide 1 and 19 μL of increasing concentrations of His-Cbl(TKB) protein (9 nM – 20 μM) were added to the assay buffer (10 mM PBS, 137 mM NaCl, and 2.7 mM KCl). Fluorescence measurements were taken after 0-, 30-, 60-, and 180-min incubations. The experiment was repeated after incubating the assay plate for 30 min at variable temperatures (25 – 50°C in 5°C increments). The Kd of the probe determined using eq. 6 and eq. 39 (Q = 0.9) described by Roehrl et. al. was 0.7 ± 0.03 μM which is comparable to one obtained by non-linear least square fit.[25]
Estimation of IC50 and Ki values for unlabeled ZAP70 peptide
To each well, 10 μL of increasing concentrations of unlabeled ZAP70 peptide 2 (335 nM – 685 μM) was added. Then 5 μL of His-Cbl(TKB) (for final concentrations of 9, 5, 2.5, 1.25, or 0.3 μM) and 5 μL of labeled ZAP70 peptide 1 (for final concentration of 100 nM) were added to the reaction mixture, for a total volume of 20 μL. Fluorescence measurements were taken after a 30 min incubation period at 25°C. The Ki value was determined using the Coleska-Wang equation (http://sw16.im.med.umich.edu/software/calc_ki/).[26]
Independence of FP and total fluorescence
To each well, 15 μL of increasing concentrations of labeled ZAP70 peptide 1 (1.5 – 375 nM) and 5 μL of 20 μM His-Cbl(TKB) were added. The reaction mixture was incubated at 25°C for 30 min, followed by FP and total fluorescence measurements.
Dimethyl sulfoxide tolerance
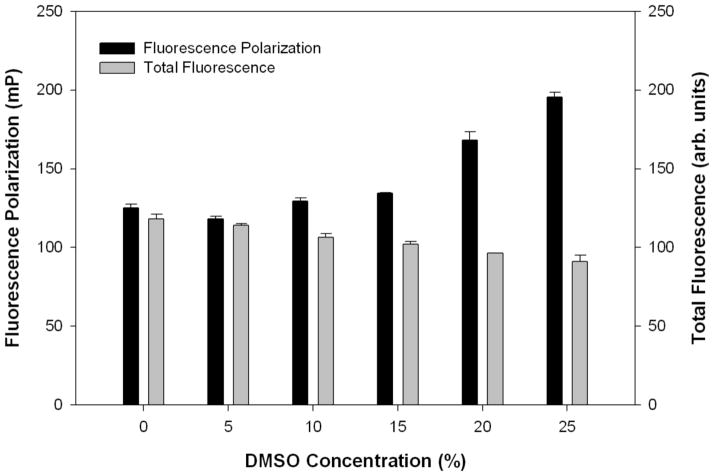
Increasing concentrations of dimethyl sulfoxide (DMSO) at 5 – 25% of the assay volume (20 μL) were added to make a final concentration of 100 nM labeled ZAP70 peptide 1 and 5 μM His-Cbl(TKB). FP and total fluorescence measurements were taken at 25°C after 30 min incubation.
Results and Discussion
Binding and inhibition studies to demonstrate phosphospecific peptide recognition by Cbl(TKB)
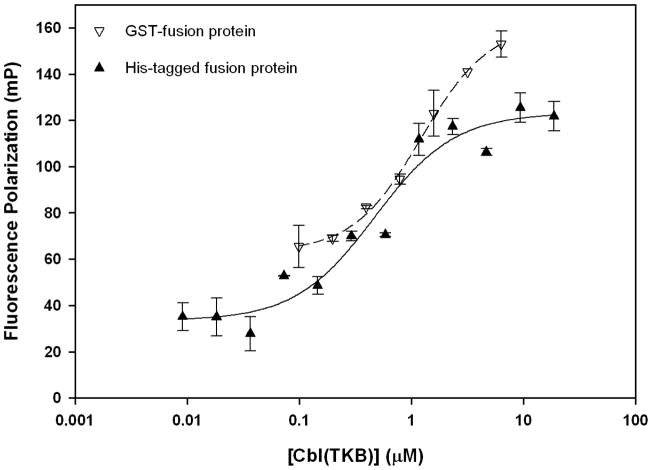
In separate experiments, two Cbl-fusion proteins (GST-Cbl(TKB) and His-Cbl(TKB)) were titrated into fluorescently labeled phosphotyrosine ZAP70 peptide (1). In both cases we observed a dose dependent increase in the fluorescence polarization indicating that the phosphorylated ZAP70 peptide binds to Cbl(TKB) (Figure 1) consistent with previously reported structure and biochemical studies. The binding affinity (Kd) and screening window (ΔmP) for each protein-peptide pair was determined by curve fitting the data. The labeled ZAP70 peptide (1) binds His-Cbl(TKB) protein with a higher affinity (0.47 ± 0.01 μM) compared to the GST-Cbl (1.2 ± 0.2 μM). We observe a slightly larger screening window with GST-Cbl(TKB) (~100 mP) compared to His-Cbl(TKB) (~90 mP) possibly due to the molecular weight difference between GST-Cbl(TKB) protein (62 kDa) and His-Cbl(TKB) protein (46 kDa). However additional time resolved data is needed to confirm this possibility. Additionally, our stability studies indicate that the His-Cbl(TKB) is stable for over 6-weeks at 4°C while we see precipitation of the GST-Cbl(TKB) protein after a week under identical conditions. Therefore for all subsequent studies we used the His-Cbl(TKB) protein.
Figure 1.
Cbl(TKB)-ZAP70 peptide binding isotherms. 100 nM ZAP70 peptide 1; Kd (His-Cbl) = 0.47 ± 0.01 μM, Kd (GST-Cbl) = 1.2 ± 0.2 μM, ΔmP(His-Cbl) = 90 units and ΔmP(GST-Cbl) = 99 units
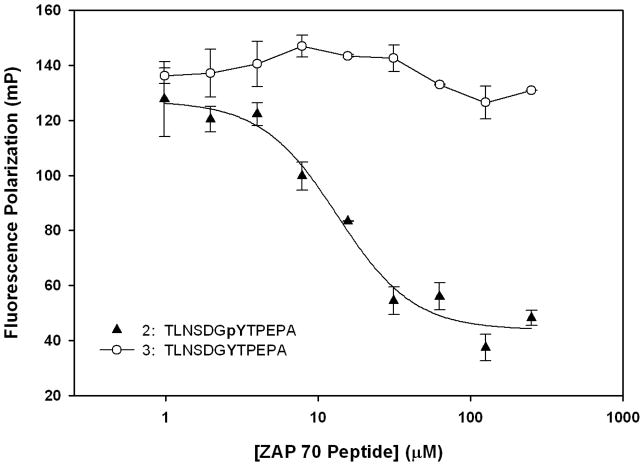
To demonstrate competitive inhibition (Figure 2) of the Cbl(TKB)-phosphopeptide interaction, we titrated the unlabeled ZAP70 peptide 2 into a mixture of His-Cbl(TKB) and fluorescently labeled ZAP70 peptide 1. The IC50 value for 2 obtained through curve fitting was 13.0 ± 1.9 μM. The inhibition constant (Ki) for 2 was estimated to be 0.4 ± 0.02 μM.[26] To determine the phosphospecific nature of the Cbl-phosphopeptide interaction we titrated a non-phosphorylated ZAP70 peptide 3 into the His-Cbl(TKB) and labeled ZAP70 peptide (1) mixture (Figure 2). We observed no decrease in the FP values with increasing concentrations of the ZAP70 peptide 3, thus confirming the phosphospecific nature of this binding interaction.
Figure 2.
Cbl(TKB)-ZAP70 peptide inhibition data. Unlabeled ZAP70 peptides 2 (Ac-TLNSDGpYTPEPA-CONH2) and 3 (Ac-TLNSDGYTPEPA-CONH2) were titrated into a mixture of His-Cbl(TKB)(9 μM) + 100nM ZAP70 peptide 1; IC50(His-Cbl) = 13.4 ± 1.9 μM and Ki = 0.4 ± 0.02 μM.
Assay development and optimization for high-throughput screening
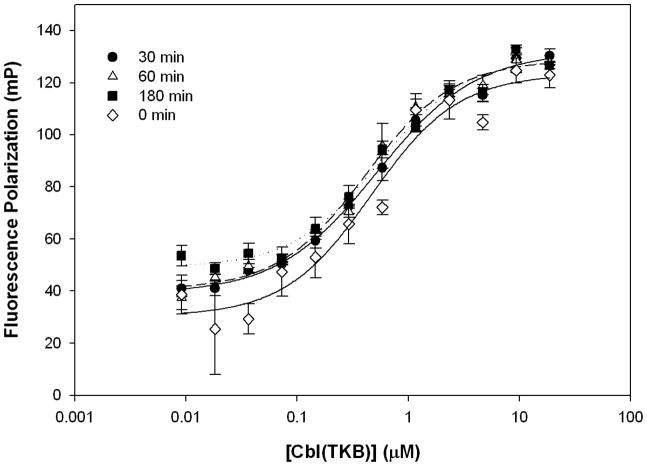
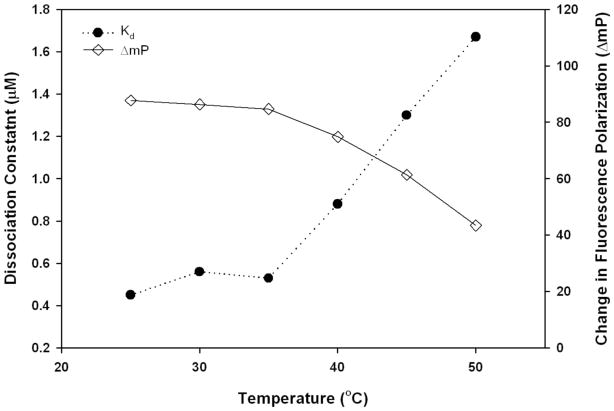
To develop an FP assay amenable to a high-throughput screening (HTS) format, the stability of the binding experiments to common HTS variables such as incubation time, temperature, total fluorescence, and DMSO concentrations were evaluated prior to the screen. To determine the binding stability over time, Kd and ΔmP values were measured at incubation periods of 0-, 30-, 60-, and 180-min. There was no significant change in the Kd or ΔmP values over the observed period (Figure 3). To facilitate rapid screening, a 30 min incubation period was chosen for further experiments. To identify the optimum temperature for the binding reaction Kd and ΔmP values were determined over a series of increasing temperatures. The Kd and ΔmP values remain constant throughout the 25°C – 35°C range, but are dramatically altered at higher temperatures (Figure 4). For practical considerations, we chose to incubate the reaction at room temperature (25°C) for subsequent screens. To demonstrate the independence of FP from total fluorescence, increasing concentrations of labeled ZAP70 peptide 1 were titrated (1.5 – 375 nM) against a constant His-Cbl(TKB) concentration (5 μM). With increasing concentrations of 1, the total fluorescence increases, while the FP remains constant (data not shown). Small molecules that are fluorescent will, therefore, not influence the HTS outcome. The effect of DMSO, a common solvent, on the FP and total fluorescence measurements was determined (Figure 5). Increasing amounts of DMSO were added to a constant amount of labeled ZAP70 peptide 1 and His-Cbl(TKB) (5 μM) mixture. FP and total fluorescence measurements are stable in the presence of up to 5% DMSO. Higher DMSO concentrations did affect FP and total fluorescence values (Figure 5).
Figure 3.
Stability of binding experiments over time. Cbl(TKB) (9 nM – 20 μM) was titrated against 100 nM ZAP70 peptide 1. FP measurements were taken at 25°C for each indicated time point (0 min, 30 min, 1h, 3h). (Kd (0min) = 0.46 ± 0.09 μM, Kd (30min) = 0.52 ± 0.04 μM, Kd (60min) = 0.41 ± 0.04 μM, Kd (180 min) = 0.54 ± 0.06 μM).
Figure 4.
Temperature dependence of Kd and ΔmP values. Cbl(TKB) (9 nM – 20 μM) was titrated against 100 nM ZAP70 peptide 1. The Kd and ΔmP values were obtained through a nonlinear least squares fit to a single-site binding model. For each temperature, values were measured after 30 min incubations.
Figure 5.
Stability of binding experiments with increasing DMSO concentrations. To increasing amounts of DMSO, a constant amount of His-Cbl(TKB) (5 μM) and ZAP70 peptide 1 (100 nM) were added. The total reaction volume was maintained constant in all wells. The measurements were made at 25°C after 30 min incubation.
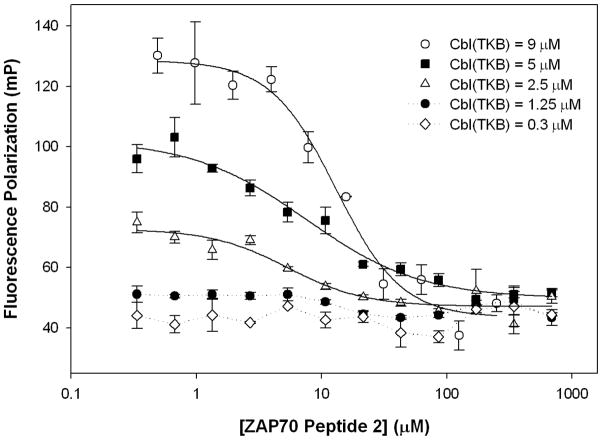
To determine the optimum amount of protein to use in the screen, a competition assay was performed using the unlabeled ZAP70 peptide 2, labeled ZAP70 peptide 1, and varying concentrations of His-Cbl(TKB). ZAP70 peptide 2 (335 nM – 685 μM), was titrated against a mixture of His-Cbl(TKB) (9, 5, 2.5, 1.25, or 0.31 μM) and labeled ZAP70 peptide 1 (100 nM) (Figure 6). A concentration of 9 μM His-Cbl(TKB) gives an IC50 value of 13.0 ± 1.7 μM and a ΔmP = 85.2 units. A concentration of 5 μM His-Cbl(TKB) gives an IC50 value of 7.5 ± 1.6 μM and ΔmP = 52.5, while a 2.5 μM His-Cbl(TKB) gives an IC50 value of 5.4 ± 1.6 μM and ΔmP = 25.7. His-Cbl(TKB) concentrations of 1.25 and 0.3 μM do not give measureable IC50 or ΔmP values. The average Ki calculated from the above experiments was 0.5 ± 0.2 μM. The ΔmP at a 5 μM concentration of His-Cbl(TKB) in the competition assay gives a reasonable screening window while minimizing the amount of necessary reagents, i.e., suitable for the HTS campaign. In summary, the optimized conditions for a high-throughput screen to identify small molecule inhibitors of Cbl(TKB) were defined as follows: (i) 5 μM Cbl(TKB), (ii) 100 nM fluorescently-labeled ZAP70 peptide 1, (iii) up to 5% DMSO concentration, and (iv) incubation for 30 min at 25°C.
Figure 6.
Effect of Cbl(TKB) concentration on assay parameters. Displacement of labeled ZAP70 peptide 1 from His-Cbl(TKB) by unlabeled ZAP70 peptide 2 in varying concentrations of His-Cbl(TKB). (IC50 @9 μM = 13.0 ± 1.7 μM, IC50 @5 μM = 7.5 ± 1.6 μM, IC50 @2.5 μM = 5.4 ± 1.6 μM, Ki(average) = 0.5 ± 0.2 μM).
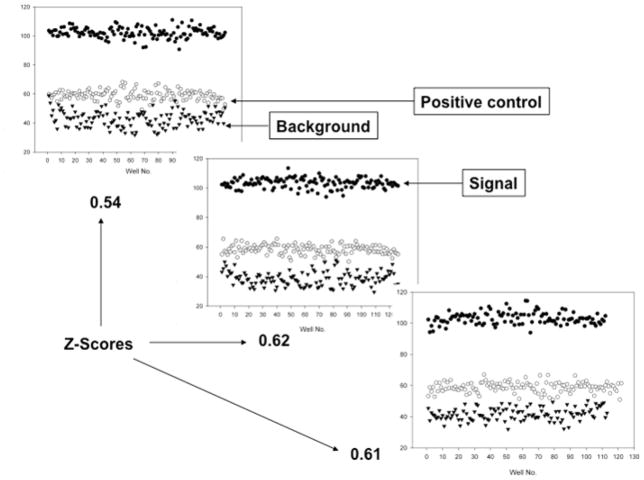
The stability of the reagents used in the screen has to be tested before an assay is deemed HTS ready. The statistical parameter Z-score is generally used to determine if an assay is HTS ready.[27] An assay is usually considered HTS ready if the Z-score for the assay is > 0.5 with 1.0 being the perfect score. We evaluated the stability of the reagents, inter-plate and day-to-day variations using Z-scores as the read out. Several 384-well plates with a third of the wells treated with 100 nM labeled ZAP70 peptide 1 alone (▾), another third of the plate with 5 μM His-Cbl(TKB) + 100 nM labeled ZAP70 peptide 1 (●) and the final third of the plate with 5 μM His-Cbl(TKB) + 100 nM labeled peptide 1 + 15.0 μM unlabeled ZAP70 peptide 2 (⚪) were generated. The plates were incubated for different times to a maximum of 24h. We found that the reagents were stable up 24h at 4°C as evidenced by the stable FP read outs for the three subsets of reagents. The Z scores variation was < 5% for the same day plate-to-plate reads (data not shown) while it was larger with a maximum of 20% for the inter-day plates (Figure 7). This suggests the need to normalize the screening data to a positive control if conducted over several days. Based on the optimized Kd, ΔmP, IC50, values and the stability of the proteins, we decided to use test our Cbl(TKB) assay in an HTS format by screening the Prestwick chemical library®.
Figure 7.
Representative plates from separate experiments conducted on three different days and their corresponding Z-scores.
Pilot screen to assess the feasibility of the optimized assay
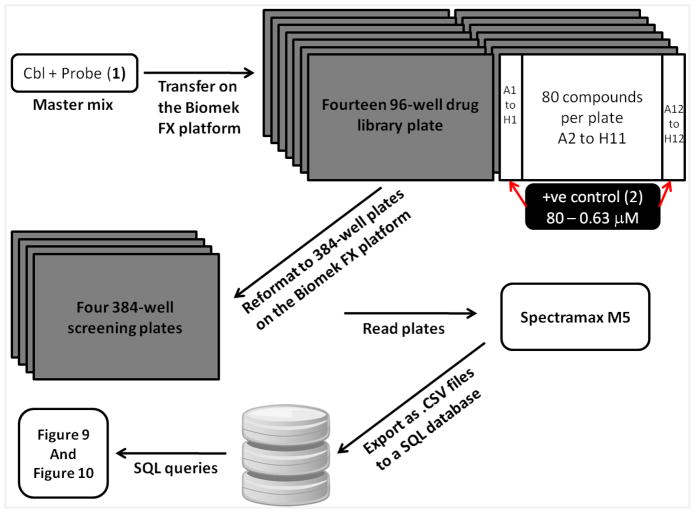
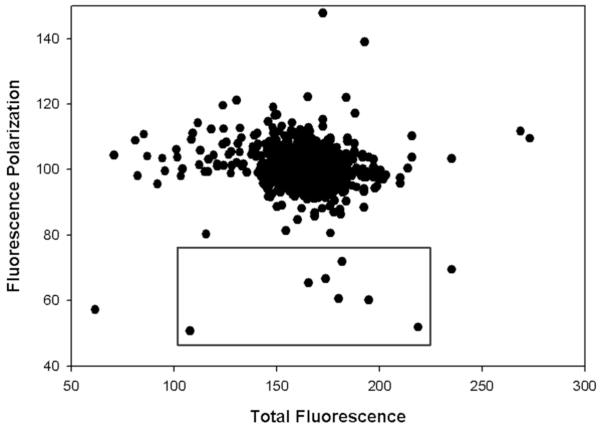
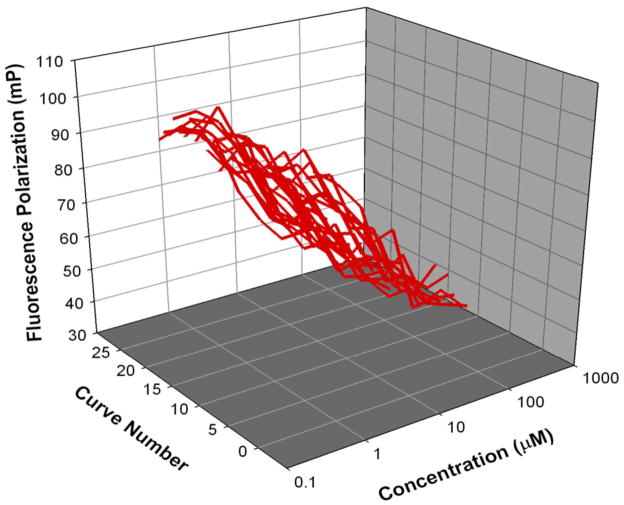
A schematic describing the screen and the relevant data processing is summarized in Figure 8. All the experiments were carried out on a Biomek FX platform that has an inline barcode reader and the Spectramax M5 plate reader. The Prestwick drug library was aliquoted as 1 μL DMSO solutions into bar coded 96-well drug plates. For a separate source plate a mixture of His-Cbl(TKB) and the labeled ZAP70 peptide 1 was transferred to the bar coded 96-well drug plates. The reagents were mixed and transferred to bar coded 384-well screening plates and incubated for 15 minutes on the deck. Fluorescence polarization and total fluorescence were measured for the 384-well plates on the plate reader and the data was captured as .CSV files onto a SQL server. The data were queried to generate the FP vs. TF plot. Hits were defined as compounds with > 50% inhibition on the FP axis and ± 150% of the positive control on the TF axis to eliminate fluorescence interference. This resulted in the identification of 7 compounds as hits (highlighted in the box in Figure 9). Dose-dependent validation of the hits identified showed that they were false positives. However, the validity of the (a) assay and (b) optimized conditions for the screen were confirmed by analyzing the results of the positive control (ZAP70 peptide 2) used (Figure 10). The average IC50 value (5.9 ± 0.4 μM) for the positive control from the screen was in agreement with our individually determined value (Figure 6, IC50 @ 5 μM = 7.5 ± 1.6 μM).
Figure 8.
Schematic describing the HTS.
Figure 9.
HTS of the Prestwick chemical library® using the fluorescence polarization Cbl assay (His-Cbl(TKB) = 5 μM and 100 nM of ZAP70 peptide 1); hits were defined as compounds with > 50% inhibition on the FP axis and ± 150% of the positive control on the TF axis to eliminate fluorescence interference.
Figure 10.
Inhibition curves of the positive control (ZAP70 peptide 2) from the HTS, the average IC50 value from the screen was 5.9 ± 0.4 μM.
In conclusion, we have developed and optimized an FP assay to identify small molecule inhibitors of the Cbl(TKB)-PTK protein-protein interaction. The assay was used to confirm the phosphospecific nature of the interaction. The FP assay was miniaturized to a 384-well format and variables that would affect the outcome of the screen were optimized. A small chemical library was screened to demonstrate HTS feasibility. The dose dependent response of the positive control suggests qHTS achievability. In short we have developed and optimized a HT-Cbl(TKB) assay that is ready for large scale HTS campaigns.
Acknowledgments
Funding in part from NIH (R01CA127239 to A.N and T32CA009476 to E.A.K) is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duan L, Reddi AL, Ghosh A, Dimri M, Band H. The Cbl family and other ubiquitin ligases: destructive forces in control of antigen receptor signaling. Immunity. 2004;21:7–17. doi: 10.1016/j.immuni.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer. 2008;8:835–50. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 3.Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Exp Cell Res. 2009;315:1610–8. doi: 10.1016/j.yexcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–12. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 5.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–40. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 6.Peschard P, Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 2003;3:519–23. doi: 10.1016/s1535-6108(03)00136-3. [DOI] [PubMed] [Google Scholar]
- 7.Caligiuri MA, Briesewitz R, Yu J, Wang L, Wei M, Arnoczky KJ, Marburger TB, Wen J, Perrotti D, Bloomfield CD, Whitman SP. Novel c-CBL and CBL-b ubiquitin ligase mutations in human acute myeloid leukemia. Blood. 2007;110:1022–4. doi: 10.1182/blood-2006-12-061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S, Tamura A, Honda H, Sakata-Yanagimoto M, Kumano K, Oda H, Yamagata T, Takita J, Gotoh N, Nakazaki K, Kawamata N, Onodera M, Nobuyoshi M, Hayashi Y, Harada H, Kurokawa M, Chiba S, Mori H, Ozawa K, Omine M, Hirai H, Nakauchi H, Koeffler HP, Ogawa S. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460:904–8. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 9.Ogawa S, Shih LY, Suzuki T, Otsu M, Nakauchi H, Koeffler HP, Sanada M. Deregulated intracellular signaling by mutated c-CBL in myeloid neoplasms. Clin Cancer Res. 2010;16:3825–31. doi: 10.1158/1078-0432.CCR-09-2341. [DOI] [PubMed] [Google Scholar]
- 10.Kales SC, Ryan PE, Nau MM, Lipkowitz S. Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Res. 2010;70:4789–94. doi: 10.1158/0008-5472.CAN-10-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight JF, Shepherd CJ, Rizzo S, Brewer D, Jhavar S, Dodson AR, Cooper CS, Eeles R, Falconer A, Kovacs G, Garrett MD, Norman AR, Shipley J, Hudson DL. TEAD1 and c-Cbl are novel prostate basal cell markers that correlate with poor clinical outcome in prostate cancer. Br J Cancer. 2008;99:1849–58. doi: 10.1038/sj.bjc.6604774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swaminathan G, Tsygankov AY. The Cbl family proteins: ring leaders in regulation of cell signaling. J Cell Physiol. 2006;209:21–43. doi: 10.1002/jcp.20694. [DOI] [PubMed] [Google Scholar]
- 13.Lupher ML, Jr, Reedquist KA, Miyake S, Langdon WY, Band H. A novel phosphotyrosine-binding domain in the N-terminal transforming region of Cbl interacts directly and selectively with ZAP-70 in T cells. J Biol Chem. 1996;271:24063–8. doi: 10.1074/jbc.271.39.24063. [DOI] [PubMed] [Google Scholar]
- 14.Meng W, Sawasdikosol S, Burakoff SJ, Eck MJ. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature. 1999;398:84–90. doi: 10.1038/18050. [DOI] [PubMed] [Google Scholar]
- 15.Lupher ML, Jr, Songyang Z, Shoelson SE, Cantley LC, Band H. The Cbl phosphotyrosine-binding domain selects a D(N/D)XpY motif and binds to the Tyr292 negative regulatory phosphorylation site of ZAP-70. J Biol Chem. 1997;272:33140–4. doi: 10.1074/jbc.272.52.33140. [DOI] [PubMed] [Google Scholar]
- 16.Yokouchi M, Wakioka T, Sakamoto H, Yasukawa H, Ohtsuka S, Sasaki A, Ohtsubo M, Valius M, Inoue A, Komiya S, Yoshimura A. APS, an adaptor protein containing PH and SH2 domains, is associated with the PDGF receptor and c-Cbl and inhibits PDGF-induced mitogenesis. Oncogene. 1999;18:759–67. doi: 10.1038/sj.onc.1202326. [DOI] [PubMed] [Google Scholar]
- 17.Kurakin A, Hoffman NG, Kay BK. Molecular recognition properties of the C-terminal Sh3 domain of the Cbl associated protein, Cap. J Pept Res. 1998;52:331–7. doi: 10.1111/j.1399-3011.1998.tb00657.x. [DOI] [PubMed] [Google Scholar]
- 18.Wong ES, Fong CW, Lim J, Yusoff P, Low BC, Langdon WY, Guy GR. Sprouty2 attenuates epidermal growth factor receptor ubiquitylation and endocytosis, and consequently enhances Ras/ERK signalling. Embo J. 2002;21:4796–808. doi: 10.1093/emboj/cdf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu J, Hubbard SR. Structural characterization of a novel Cbl phosphotyrosine recognition motif in the APS family of adapter proteins. J Biol Chem. 2005;280:18943–9. doi: 10.1074/jbc.M414157200. [DOI] [PubMed] [Google Scholar]
- 20.Katsanakis KD, Pillay TS. Cross-talk between the two divergent insulin signaling pathways is revealed by the protein kinase B (Akt)-mediated phosphorylation of adapter protein APS on serine 588. J Biol Chem. 2005;280:37827–32. doi: 10.1074/jbc.M505959200. [DOI] [PubMed] [Google Scholar]
- 21.Peschard P, Ishiyama N, Lin T, Lipkowitz S, Park M. A conserved DpYR motif in the juxtamembrane domain of the Met receptor family forms an atypical c-Cbl/Cbl-b tyrosine kinase binding domain binding site required for suppression of oncogenic activation. J Biol Chem. 2004;279:29565–71. doi: 10.1074/jbc.M403954200. [DOI] [PubMed] [Google Scholar]
- 22.Ng C, Jackson RA, Buschdorf JP, Sun Q, Guy GR, Sivaraman J. Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates. Embo J. 2008;27:804–16. doi: 10.1038/emboj.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lokesh GL, Rachamallu A, Kumar GD, Natarajan A. High-throughput fluorescence polarization assay to identify small molecule inhibitors of BRCT domains of breast cancer gene 1. Anal Biochem. 2006;352:135–41. doi: 10.1016/j.ab.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Simeonov A, Yasgar A, Jadhav A, Lokesh GL, Klumpp C, Michael S, Austin CP, Natarajan A, Inglese J. Dual-fluorophore quantitative high-throughput screen for inhibitors of BRCT–phosphoprotein interaction. Anal Biochem. 2008;375:60–70. doi: 10.1016/j.ab.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roehrl MH, Wang JY, Wagner G. A general framework for development and data analysis of competitive high-throughput screens for small-molecule inhibitors of protein-protein interactions by fluorescence polarization. Biochemistry. 2004;43:16056–66. doi: 10.1021/bi048233g. [DOI] [PubMed] [Google Scholar]
- 26.Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wang S. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332:261–73. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 27.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]