Abstract
Background:
Genetic changes have been widely reported in association with cholangiocarcinoma (CCA), while epigenetic changes are poorly characterised. We aimed to further evaluate CpG-island hypermethylation in CCA at candidate loci, which may have potential as diagnostic or prognostic biomarkers.
Methods:
We analysed methylation of 26 CpG-islands in 102 liver fluke related-CCA and 29 adjacent normal samples using methylation-specific PCR (MSP). Methylation of interest loci was confirmed using pyrosequencing and/or combined bisulfite restriction analysis, and protein expression by immunohistochemistry.
Results:
A number of CpG-islands (OPCML, SFRP1, HIC1, PTEN and DcR1) showed frequency of hypermethylation in >28% of CCA, but not adjacent normal tissues. The results showed that 91% of CCA were methylated in at least one CpG-island. The OPCML was the most frequently methylated locus (72.5%) and was more frequently methylated in less differentiated CCA. Patients with methylated DcR1 had significantly longer overall survival (Median; 41.7 vs 21.7 weeks, P=0.027). Low-protein expression was found in >70% of CCA with methylation of OPCML or DcR1.
Conclusion:
Aberrant hypermethylation of certain loci is a common event in liver fluke-related CCA and may potentially contribute to cholangiocarcinogenesis. The OPCML and DcR1 might serve as methylation biomarkers in CCA that can be readily examined by MSP.
Keywords: DNA methylation, cholangiocarcinoma, liver fluke, OPCML, DcR1, biomarkers
Cholangiocarcinoma (CCA), a malignancy originating from biliary epithelium, has been considered as a rare disease, which globally accounts for 10–25% of primary liver cancers (Kamangar et al, 2006). As the incidence and mortality rates for intrahepatic CCA (ICC) have markedly increased worldwide (Patel, 2002; Khan et al, 2005), there has been growing interest in this cancer. Cholangiocarcinoma represents a major public health problem in Thailand, particularly in the northeast region where Opisthorchis viverrini (OV) infection remains highly endemic. The prevalence of OV infection in this region is 15.7%, while the average in Khon Kaen (the centre for CCA collection in the present study) is 24.5% (Sripa et al, 2007). Here, CCA accounts for the highest incidence worldwide with truncated age-standardised incidence of ages >35 years up to 317.6 per 100 000 population (Sriamporn et al, 2004). Infection with OV occurs when raw or undercooked cyprinoid fish with the infective stage of metacercariae is ingested (Sripa et al, 2007). However, OV infection alone does not cause CCA. Animal experiments suggest that OV infection in combination with nitrosamine administration is the causation of CCA in a dose-dependent manner (Thamavit et al, 1978, 1987). Honjo et al (2005) conducted a population-based case–control study on genetic and environmental risk factors for OV-related CCA. They showed that anti-OV antibody ⩾0.200 was the strongest risk indicator. Furthermore, dietary habits during the past 10 years, raw fish intake (OV infection) and fermented fish or pork intake (exogenous nitrosamine) were significantly associated with an increased risk. Alcohol drinking, but not smoking showed a higher risk in which alcohol may affect metabolic pathways of endogenous and exogenous nitrosamines. The prognosis of CCA is generally poor with <5% of patients surviving >5 years and has not significantly improved over the past three decades (Shaib and El-Serag, 2004). This can partly be attributed to the fact that CCA is difficult to diagnose and often presents at a late clinical stage. Biomarkers, which allow earlier detection of CCA could potentially impact on patient survival by allowing earlier treatment. Although surgical resection can be potentially curative, only a few patients are suitable for surgery because of spread of the disease when late presenting, and effective non-surgical therapies are limited (Patel, 2006).
Epidemiological and experimental evidence implicate the carcinogenic liver fluke as a major risk factor of CCA. It is possible that chronic inflammation driven by the infection makes the biliary epithelium more susceptible to neoplastic transformation. (Thamavit et al, 1978; IARC, 1994). However, the exact molecular events involved in cholangiocarcinogenesis are not well understood. Like other tumours, CCA is a result of a multistep process in which genetic and epigenetic aberrations of regulatory genes are accumulated. For instance, genetic alterations of K-ras (Ohashi et al, 1995) and p53 (Limpaiboon et al, 2002; Liu et al, 2006b) have frequently been reported in CCA. Aberrant DNA methylation at CpG-islands associated with transcriptional silencing is widely observed in cancers (Esteller, 2006). DNA methylation is a potential rich source of diagnostic, prognostic or predictive biomarkers of clinical outcome. Hypermethylation in CCA has been reported in tumour suppressor genes related to cell cycle (p16, 14–3–3σ, p73, p14, p15), apoptosis (DAPK, SEMA3B) and cell adhesion (APC, E-cadherin, TIMP3, THBS1) (Lee et al, 2002; Tannapfel et al, 2002; Tischoff et al, 2005; Yang et al, 2005; Chinnasri et al, 2009). Furthermore, several reports demonstrated hypermethylation of genes involved in DNA repair and carcinogen metabolism, such as hMLH1, MGMT and GSTP1 (Lee et al, 2002; Limpaiboon et al, 2005; Yang et al, 2005). However, while these previous studies have reported aberrant methylation at several loci in CCA, their evaluation as diagnostic or prognostic biomarkers is still limited.
To further elucidate the role of methylation in liver fluke-related CCA, we here analysed the methylation status by methylation-specific PCR (MSP) of a defined set of 26 CpG islands at genes, which function in cell cycle, apoptosis, DNA repair and cell–cell interaction, which have been previously reported in CCA and other types of cancer as frequently methylated. Methylation-specific PCR was performed in 102 primary CCA and where possible adjacent normal tissues. Methylation-specific PCR provides a relatively simple and cost-effective assay that can be used to assess methylation in clinical samples, however, for loci of interest, we have confirmed methylation status by bisulfite pyrosequencing or combined bisulfite restriction analysis (COBRA). We have identified differential hypermethylation of a number of CpG-islands and reported the associations to clinicopathological parameters.
Materials and methods
Samples and clinicopathological data
Frozen tissues of 102 primary CCA and 29 tumour-adjacent normal samples were used. Moreover, those CCA samples, which had paraffin-embedded sections were also used for immunohistochemistry. Clinicopathological data of CCA patients, including age at initial diagnosis, gender, gross types, histological grades and the overall survival time (excluding patients who died within 4 weeks after surgery) were also collected. All of the samples and data were kindly supplied by the Liver Fluke and Cholangiocarcinoma Research Centre, Khon Kaen University (Khon Kaen, Thailand). Among CCA samples, mean age at initial diagnosis was 55 years (45.2–64.8 years), and 68 males and 34 females were accounted. Cholangiocarcinoma samples were classified into three gross types; mass forming (MF, n=65), periductal infiltrating (PI, n=26) and intraductal growth (ID, n=11) type, and two histological grades; well differentiated (n=74) and less differentiated (included moderate and poor differentiation, n=25). Median overall survival time of CCA patients was 29 weeks. The study was reviewed and approved by the Khon Kaen University Ethics Committee for Human Research (HE510651).
DNA extraction and bisulfite modification
Genomic DNA was extracted from frozen liver tissues using DNeasy Tissue Kit (Qiagen, Hercules, CA, USA) according to the manufacturer's protocols. Genomic DNA (1 μg) from each sample was modified by sodium bisulfite as described by Herman et al (1996) with slight modification as previously reported (Sriraksa et al, 2010). The quality of bisulfite-converted DNA was verified by PCR amplification using Calponin-specific primer sets as previously described (Teodoridis et al, 2005; Sriraksa et al, 2010).
Methylation-specific PCR (MSP)
Methylated-specific primers and PCR conditions of 22 CpG-islands for MSP were obtained from methprimerDB (http://medgen.ugent.be/methprimerdb/index.php). In addition, the methylated-specific primers and PCR conditions for 14–3–3σ, DcR2, DR4 and DR5 were obtained from previous studies (Lee et al, 2002; van Noesel et al, 2002; Liu et al, 2008). Methylation-specific PCR (25 μl per reaction) was performed on a PTC-100 Thermal cycle (MJ Research INC., Ramsey, MN, USA) as the appropriate PCR conditions for each locus. Methylation-positive sample was defined by comparing the MSP product to those of diluted in vitro methyltransferase-treated placental DNA (positive control) and the weak signal was disregarded.
Pyrosequencing
Pyrosequencing of bisulfite modified DNA was used to validate methylation status of candidate loci, OPCML and DcR1, previously identified by MSP. The primers for OPCML amplification and sequencing were; 5′-GGGAGTGTGAGATGTATGTGAGTG-3′ (forward), 5′-biotin-TACCCCCAAAACCACAACTAATT-3′ (reverse) and 5′-AGAGGTAGGTTTGTTGTGT-3′ (sequencing) and for DcR1; 5′-TTGGTAGTGTAGTTGTGGGAATTTTT-3′ (forward), 5′-biotin-TCTATCCCCAAAATTCCCTAA-3′ (reverse) and 5′-GGTAGTGTAGTTGTGGGAA-3′ (sequencing). The schematics of these specific primers for pyrosequencing are shown in Supplementary Figure 1, upper and lower panels. DNA used as a template of PCR was bisulfite modified using EpiTect Bisulfite Kit (Qiagen, West Sussex, UK), and the annealing temperature and MgCl2 concentration for OPCML and DcR1 were 63 °C, 2 mM and 61 °C, 3.6 mM, respectively. Pyrosequencing of PCR products was performed using PyroGold Reagent kit (Biotage, Uppsala, Sweden) according to the manufacturer's instruction. The methylation percentage at individual CpG sites was then analysed using the Pyro Q-CpG software (Biotage).
Combined bisulfite restriction analysis (COBRA)
COBRA was performed to validate methylation status of OPCML, for which specific primers, amplification condition and restriction enzyme used were obtained from methprimerDB (http://medgen.ugent.be/methprimerdb/index.php). The positions of OPCML-specific primers for COBRA were located in the same region as for MSP (Supplementary Figure 1, upper panel). Digestion was performed by incubation at 37 °C for 2 h, before separation on a 2% agarose gel in which band intensity and methylation level were evaluated.
Immunohistochemistry
The expression of OPCML and DcR1 was examined in formalin fixed paraffin-embedded tissue sections of primary CCA using immunohistochemistry as previously described (Hussain et al, 2010). Antigen retrieval was performed by microwave heating in EDTA (pH 9.0) for 20 min. To block non-specific antibody binding, sections were incubated with 2.5% normal horse serum (Vector Laboratories, Peterborough, UK) for 10 min. Here, sections were incubated overnight at 4 °C with OPCML monoclonal mouse antibody (1 : 1000 dilution; Anti-human OBCAM: MAB27771, R&D Systems, Abingdon, UK) or DcR1 monoclonal mouse antibody (1 : 100 dilution; DcR1 (L19): sc-73890, Santa Cruz Biotechnology, Heidelberg, Germany). Positive (breast carcinoma) and negative (PBS instead of primary antibody) controls were also conducted. After washing with phosphate-buffered saline (PBS) containing 0.1% Tween-20, sections were incubated with secondary antibody for 30 min at room temperature (ImmPRESS Universal Antibody Polymer Detection Kit, Vector Laboratories). Antigen–antibody reaction was visualised using diaminobenzidine chromogen and sections were counterstained with haematoxylin. All slides were examined and scored by two independent observers. The staining was scored as negative (=0), or positive with weak (=1), moderate (=2) and strong (=3) intensity.
Statistical analysis
The statistical analysis was performed using SPSS software (SPSS version 16.0, Chicago, IL, USA). The correlation between methylation of each CpG-island and clinicopathological features of CCA patients including gender, gross types and histological grades, was analysed using χ2-test or Fisher's exact test. The correlation between methylation status and age at initial diagnosis was analysed using Mann–Whitney U-test. The association of methylation and overall survival time was determined using Kaplan–Meier method by log-rank test or univariate analysis by Cox regression. The association between MSP and pyrosequencing, COBRA or protein expression was analysed using χ2-test. Two sided P<0.05 was considered as statistically significant.
Results
Frequency of CpG-island methylation and concurrent methylation in cholangiocarcinoma
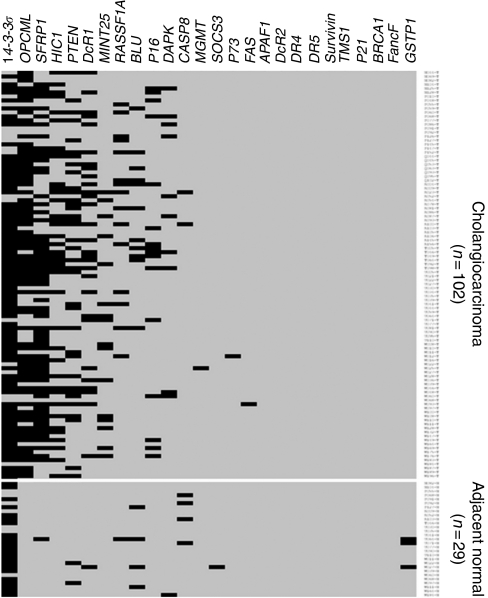
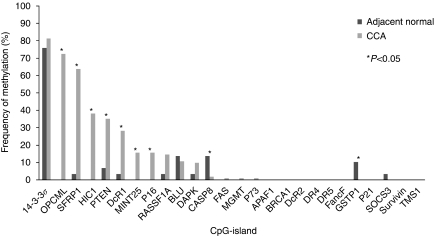
The CpG-island hypermethylation of 26 loci was investigated in 102 primary CCA and 29 matched adjacent normal tissues using MSP (Figures 1 and 2). All bisulfite-treated DNA samples were tested with Calponin-specific primer sets to demonstrate successful bisulfite modification before MSP analysis (Teodoridis et al, 2005; Sriraksa et al, 2010). The CpG-islands with high frequency of methylation in CCA included 14–3–3σ, OPCML, SFRP1, HIC1, PTEN and DcR1 (81.4, 72.5, 63.7, 38.2, 35.3 and 28.4%, respectively). Moderately frequent methylation was found for MINT25, p16, RASSF1A and BLU (15.7, 15.7, 14.7 and 10.8%, respectively) and low frequency was observed for DAPK, CASP8, FAS, MGMT and p73 in 1–10% of CCA. No methylation was detected for APAF1, BRCA1, DcR2, DR4, DR5, FancF, GSTP1, p21, SOCS3, Survivin and TMS1. Adjacent normal tissues showed no or very low levels of methylation at all loci, with the exception of 14–3–3σ for which a high frequency of methylation (75.9%) was found in normal adjacent tissue, and therefore methylation data for 14–3–3σ has been excluded from subsequent analysis. After excluding 14–3–3σ, 91% (93 out of 102) of CCA samples were methylated in at least one locus out of the 14 loci (OPCML, SFRP1, HIC1, PTEN, DcR1, MINT25, p16, RASSF1A, BLU, DAPK, CASP8, FAS, MGMT and p73).
Figure 1.
Methylation profile of 26 CpG-islands in cholangiocarcinoma and adjacent normal samples. Black and grey boxes represent positive and negative methylation by MSP, respectively.
Figure 2.
Histogram represented frequency of methylation of 26 CpG-islands in adjacent normal samples (n=29) and cholangiocarcinoma (n=102).
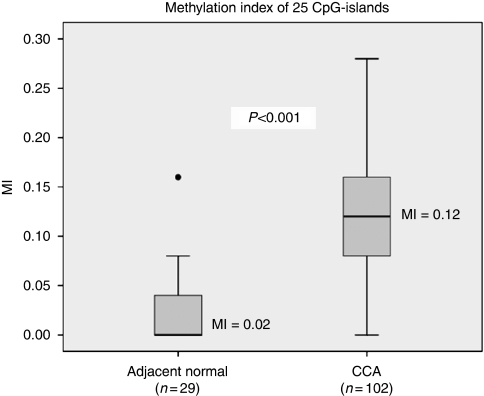
We analysed the tumours for a methylation index (MI), which was calculated from the ratio between the number of methylated CpG-islands and the number of analysed CpG-islands (Gutierrez et al, 2004). The average methylation index of CCA was significantly higher than that in adjacent normal tissues (average MI; 0.12 vs 0.02, P<0.001) as shown in Figure 3. Concordant methylation in CCA was found between pairs of CpG-island including OPCML and SFRP1 (P=0.007), OPCML and HIC1 (<0.001), and HIC1 and PTEN (P=0.002). No significant association between other pairs of CpG-island was observed. In addition, we also analysed the association of MI and clinicopathological data of CCA patients. However, no significant correlation of clinical data and MI was observed.
Figure 3.
Methylation index (MI) between adjacent normal samples and CCA in which CCA samples showed significantly higher MI than adjacent normal tissues.
OPCML
The highest methylation frequency was observed in 72.5% of CCA for OPCML, whereas no methylation of this locus was revealed in adjacent normal samples. In addition, methylation of OPCML was found more frequently in less differentiated type (88% 22 out of 25) than in the well-differentiated types of CCA (67.6% 50 out of 74) (P=0.047). To confirm the methylation detected by MSP, an independent method of methylation analysis, COBRA, was used. The correlation between MSP and COBRA results was analysed using χ2-test and significant association with MSP was found at OPCML by COBRA (80.0% of tested samples, P=0.025). To address whether methylation has effect on gene expression, immunohistochemistry was performed to determine OPCML protein expression in CCA samples. Low or no OPCML protein expression was detected in CCA, 63% (58 out of 92) weakly expressed OPCML and 26% (24 out of 92) exhibited no detectable expression. Low-protein expression was found in 88% (59 out of 67) of OPCML-hypermethylated CCA, however, as the vast majority of tumours (89%) had no or very low OPCML expression, it was not feasible to statistically analyse the association between expression and methylation.
DcR1
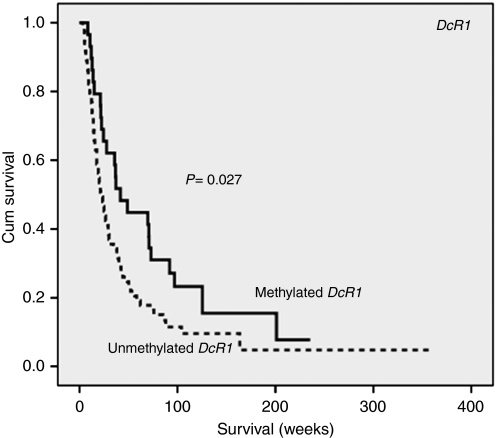
In the present study, DcR1 CpG island methylation was observed in 28.4% of CCA. Interestingly, patients with methylated DcR1 showed longer overall survival than those without (Median; 41.7 vs 21.7 weeks, P=0.027), Figure 4. The significance of methylated DcR1 on survival raised the question on the role of methylation status of other TRAIL-related genes in CCA. To address this question, we next analysed methylation of CpG islands at DcR2, DR4 and DR5 in CCA samples, but no methylation was detected at these loci. To confirm the methylation detected by MSP, we showed a significant correlation between MSP and pyrosequencing in each individual CpG site of DcR1 and also in the average of the four CpG sites examined. Moreover, a trend of correlation of methylation in CpG1 with patient survival was observed (P=0.08, data not shown), similar to that of MSP. Low or no DcR1 protein expression was detected in CCA, in which 54% (50 out of 92) showed low-DcR1 expression and 23% (22 out of 92) displayed no expression. Increased frequency of low-protein expression was also observed in 77% (20 out of 26) of DcR1-hypermethylated samples. In addition, low-DcR1 expression was observed in ID type (100% 11 out of 11), followed by MF (82.5% 47 out of 57) and PI types (58.3% 14 out of 24) (P=0.01).
Figure 4.
The association between methylation of DcR1 and overall survival time of cholangiocarcinoma patients. Patients with methylated DcR1 had longer overall survival time than those without (Kaplan–Meier analysis using log-rank test, P=0.027).
Discussion
In the present study, the majority of CCA (91%) shows aberrant methylation in at least one locus, suggesting that DNA methylation is a common event in CCA, which is in agreement with previous reports (Lee et al, 2002; Yang et al, 2005). The high-methylation frequency of 14–3–3σ found in CCA and adjacent normal tissues is similar to that observed in normal breast tissue and breast cancer (Umbricht et al, 2001) suggesting that methylation of 14–3–3σ is tissue specific regardless of pathological status. The high-methylation frequency presented for CpG islands at OPCML, SFRP1, HIC1, PTEN and DcR1 suggests that many signalling pathways could be involved in the development and progression of CCA, although does not demonstrate that all are drivers of carcinogenesis. However, association of methylated OPCML and DcR1 with patient clinicopathological data supports their role in tumour progression. The tumour suppressor gene OPCML (Opioid binding protein/cell adhesion molecule-like gene) belongs to the IgLON family of immunoglobulin (Ig) domain containing glycosylphosphatidylinositol (GPI)-anchored cell adhesion molecules that are involved in cell adhesion and cell–cell recognition (McNamee et al, 2002; Sellar et al, 2003). In addition, OPCML has been proposed as a stress- and p53-responsive gene, with the response impaired when the promoter becomes hypermethylated. Moreover, exogenous expression of OPCML in carcinoma cells lacking its expression leads to dramatic growth inhibition suggesting OPCML as a tumour suppressor (Sellar et al, 2003; Cui et al, 2008). The OPCML hypermethylation has been reported in several cancers for example, 33.3% of late stage ovarian cancer (Teodoridis et al, 2005), 70% of hepatocellular carcinoma (Liu et al, 2006a), 63.9% of invasive cervical cancer (Ye et al, 2008) and 57–100% of multiple carcinomas and lymphomas (Cui et al, 2008). We are the first to report OPCML methylation in CCA with high frequency (72.5%) with no methylation in normal adjacent tissue. The previous studies have demonstrated that CCA with less differentiation shows a poorer outcome and high incidence of metastases than well differentiated tumours (Nakajima et al, 1988; Nakanuma et al, 2000). The high incidence of OPCML methylation in CCA and its association with less differentiation together with its absence in normal tissue indicate that OPCML methylation could be used as an epigenetic biomarker for molecular prognosis and diagnosis of CCA.
The highly frequent methylation of SFRP1, HIC1 and PTEN could lead to an increase in proliferation, inhibition of apoptosis and promotion of survival advantage in CCA through increased Wnt/β-catenin signal transduction through the impaired p53-responsive pathway and activated PI3K and its downstream effectors as previously reported in other cancers (Wales et al, 1995; Finch et al, 1997; Melkonyan et al, 1997; Besson et al, 1999; Nicoll et al, 2001; Sulis and Parsons, 2003; Chen et al, 2005; Manning and Cantley, 2007; Takagi et al, 2008; Fleuriel et al, 2009).
The defect of programmed cell death through p53-dependent pathway has been observed in CCA with mutational inactivation (41.6%) and LOH of p53 (32%) as shown in our previous studies together with p53 deacetylation because of HIC1 methylation (38.2%) in this study. These phenomena impede p53 function, which allows cells to bypass apoptosis and survive DNA damage (Limpaiboon et al, 2002, 2004). It is surprising that death receptor-induced apoptosis was rarely methylated. These findings indicate the advantage for selective cancer treatment and good outcome in CCA patients. The decoy receptors have been postulated to account for TRAIL resistance as overexpression of DcR1 and/or DcR2 prevent cancer cells from TRAIL-induced apoptosis. Hypermethylation of DcR1 (28.4%), but not death receptors (DR4 and DR5) and other TRAIL signalling-related genes (APAF1, CASP8 and DcR2) in our study indicates the potential use of recombinant TRAIL or TRAIL receptor agonistic monoclonal antibodies as selective anti-tumour therapy in CCA. To overcome TRAIL resistance that always happens, combined treatment with standard chemotherapeutics can enhance TRAIL-induced apoptosis. Previous studies showed correlation of methylated DcR1 with improved prognosis in malignant mesothelioma and prostate cancer (Shivapurkar et al, 2004; Suzuki et al, 2006). As longer overall survival was observed in methylated DcR1 patients, DcR1 methylation may be useful as a prognostic marker of CCA.
In conclusion, promoter hypermethylation has been observed in many genes, which have important roles in carcinogenesis and progression of liver fluke-related CCA. Further study is warranted to validate the use of high-frequency methylated genes as potential biomarkers for diagnosis, prognosis and prediction of CCA patients. Moreover, tumour cells are significantly more sensitive to TRAIL-induced apoptosis than normal cells, thus the use of recombinant TRAIL or TRAIL receptor agonistic monoclonal antibodies for selective treatment of CCA in combination with chemotherapeutic drugs may improve patient survival.
Acknowledgments
This work was supported by the Thailand Research Fund through the Royal Golden Jubilee PhD Program (Grant no. PHD/0038/2548 code 5LKK/48/B1 to R Srisaksa and T Limpaiboon), Imperial College London Experimental Cancer Research Centre, the Centre for Research and Development of Medical Diagnostic Laboratories, Faculty of Associated Medical Sciences, Khon Kaen University, Thailand; and Epigenetics Unit, Department of Surgery and Oncology, Imperial College, London, UK. We would like to greatly thank the Liver Fluke and Cholangiocarcinoma Research Center, Khon Kaen University for all samples and clinical data. We thank Dr Chanvit Leelayuwat and Dr Jens M Teodoridis for very great suggestions. We also thank Dr Prasong Khaenam, Dr Janet Graham, Louisa Luk and Surasee Kamlua for some laboratory experiments.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Besson A, Robbins SM, Yong VW (1999) PTEN/MMAC1/TEP1 in signal transduction and tumorigenesis. Eur J Biochem 263: 605–611 [DOI] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB (2005) Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 123: 437–448 [DOI] [PubMed] [Google Scholar]
- Chinnasri P, Pairojkul C, Jearanaikoon P, Sripa B, Bhudhisawasdi V, Tantimavanich S, Limpaiboon T (2009) Preferentially different mechanisms of inactivation of 9p21 gene cluster in liver fluke-related cholangiocarcinoma. Hum Pathol 40: 817–826 [DOI] [PubMed] [Google Scholar]
- Cui Y, Ying Y, van Hasselt A, Ng KM, Yu J, Zhang Q, Jin J, Liu D, Rhim JS, Rha SY, Loyo M, Chan AT, Srivastava G, Tsao GS, Sellar GC, Sung JJ, Sidransky D, Tao Q (2008) OPCML is a broad tumor suppressor for multiple carcinomas and lymphomas with frequently epigenetic inactivation. PLoS One 3: e2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M (2006) Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer 94: 179–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch PW, He X, Kelley MJ, Uren A, Schaudies RP, Popescu NC, Rudikoff S, Aaronson SA, Varmus HE, Rubin JS (1997) Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc Natl Acad Sci USA 94: 6770–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleuriel C, Touka M, Boulay G, Guerardel C, Rood BR, Leprince D (2009) HIC1 (hypermethylated in cancer 1) epigenetic silencing in tumors. Int J Biochem Cell Biol 41: 26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MI, Siraj AK, Khaled H, Koon N, El-Rifai W, Bhatia K (2004) CpG island methylation in Schistosoma- and non-Schistosoma-associated bladder cancer. Mod Pathol 17: 1268–1274 [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 93: 9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo S, Srivatanakul P, Sriplung H, Kikukawa H, Hanai S, Uchida K, Todoroki T, Jedpiyawongse A, Kittiwatanachot P, Sripa B, Deerasamee S, Miwa M (2005) Genetic and environmental determinants of risk for cholangiocarcinoma via Opisthorchis viverrini in a densely infested area in Nakhon Phanom, northeast Thailand. Int J Cancer 117: 854–860 [DOI] [PubMed] [Google Scholar]
- Hussain F, Wang J, Ahmed R, Guest SK, Lam EW, Stamp G, El-Bahrawy M (2010) The expression of IL-8 and IL-8 receptors in pancreatic adenocarcinomas and pancreatic neuroendocrine tumours. Cytokine 49: 134–140 [DOI] [PubMed] [Google Scholar]
- IARC (1994) Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum 61: 1–241 [PMC free article] [PubMed] [Google Scholar]
- Kamangar F, Dores GM, Anderson WF (2006) Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 24: 2137–2150 [DOI] [PubMed] [Google Scholar]
- Khan SA, Thomas HC, Davidson BR, Taylor-Robinson SD (2005) Cholangiocarcinoma. Lancet 366: 1303–1314 [DOI] [PubMed] [Google Scholar]
- Lee S, Kim WH, Jung HY, Yang MH, Kang GH (2002) Aberrant CpG island methylation of multiple genes in intrahepatic cholangiocarcinoma. Am J Pathol 161: 1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpaiboon T, Khaenam P, Chinnasri P, Soonklang M, Jearanaikoon P, Sripa B, Pairojkul C, Bhudhisawasdi V (2005) Promoter hypermethylation is a major event of hMLH1 gene inactivation in liver fluke related cholangiocarcinoma. Cancer Lett 217: 213–219 [DOI] [PubMed] [Google Scholar]
- Limpaiboon T, Krissadarak K, Sripa B, Jearanaikoon P, Bhuhisawasdi V, Chau-in S, Romphruk A, Pairojkul C (2002) Microsatellite alterations in liver fluke related cholangiocarcinoma are associated with poor prognosis. Cancer Lett 181: 215–222 [DOI] [PubMed] [Google Scholar]
- Limpaiboon T, Sripa B, Wongkham S, Bhudhisawasdi V, Chau-in S, Teerajetgul Y (2004) Anti-p53 antibodies and p53 protein expression in cholangiocarcinoma. Hepatogastroenterology 51: 25–28 [PubMed] [Google Scholar]
- Liu S, Ren S, Howell P, Fodstad O, Riker AI (2008) Identification of novel epigenetically modified genes in human melanoma via promoter methylation gene profiling. Pigment Cell Melanoma Res 21: 545–558 [DOI] [PubMed] [Google Scholar]
- Liu WJ, Wang L, Wang JP, Li JQ, Zhang CQ, Zheng L, Yuan YF (2006a) Correlations of CpG island methylator phenotype and OPCML gene methylation to carcinogenesis of hepatocellular carcinoma. Ai Zheng 25: 696–700 [PubMed] [Google Scholar]
- Liu XF, Zhang H, Zhu SG, Zhou XT, Su HL, Xu Z, Li SJ (2006b) Correlation of p53 gene mutation and expression of P53 protein in cholangiocarcinoma. World J Gastroenterol 12: 4706–4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Cantley LC (2007) AKT/PKB signaling: navigating downstream. Cell 129: 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamee CJ, Reed JE, Howard MR, Lodge AP, Moss DJ (2002) Promotion of neuronal cell adhesion by members of the IgLON family occurs in the absence of either support or modification of neurite outgrowth. J Neurochem 80: 941–948 [DOI] [PubMed] [Google Scholar]
- Melkonyan HS, Chang WC, Shapiro JP, Mahadevappa M, Fitzpatrick PA, Kiefer MC, Tomei LD, Umansky SR (1997) SARPs: a family of secreted apoptosis-related proteins. Proc Natl Acad Sci USA 94: 13636–13641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Kondo Y, Miyazaki M, Okui K (1988) A histopathologic study of 102 cases of intrahepatic cholangiocarcinoma: histologic classification and modes of spreading. Hum Pathol 19: 1228–1234 [DOI] [PubMed] [Google Scholar]
- Nakanuma Y, Sripa B, Vatanasapt V, Leong ASY, Ponchon T, Ishak KG (2000) Intrahepatic cholangiocarcinoma. In: Hamilton SR, Aaltonen LA. (eds). World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of the Digestive system. pp 173–180. IARC Press: Lyon [Google Scholar]
- Nicoll G, Crichton DN, McDowell HE, Kernohan N, Hupp TR, Thompson AM (2001) Expression of the hypermethylated in cancer gene (HIC-1) is associated with good outcome in human breast cancer. Br J Cancer 85: 1878–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, Nakajima Y, Kanehiro H, Tsutsumi M, Taki J, Aomatsu Y, Yoshimura A, Ko S, Kin T, Yagura K, Konishi Y, Nakano H (1995) Ki-ras mutations and p53 protein expressions in intrahepatic cholangiocarcinomas: relation to gross tumor morphology. Gastroenterology 109: 1612–1617 [DOI] [PubMed] [Google Scholar]
- Patel T (2002) Worldwide trends in mortality from biliary tract malignancies. BMC Cancer 2: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T (2006) Cholangiocarcinoma. Nat Clin Pract Gastroenterol Hepatol 3: 33–42 [DOI] [PubMed] [Google Scholar]
- Sellar GC, Watt KP, Rabiasz GJ, Stronach EA, Li L, Miller EP, Massie CE, Miller J, Contreras-Moreira B, Scott D, Brown I, Williams AR, Bates PA, Smyth JF, Gabra H (2003) OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat Genet 34: 337–343 [DOI] [PubMed] [Google Scholar]
- Shaib Y, El-Serag HB (2004) The epidemiology of cholangiocarcinoma. Semin Liver Dis 24: 115–125 [DOI] [PubMed] [Google Scholar]
- Shivapurkar N, Toyooka S, Toyooka KO, Reddy J, Miyajima K, Suzuki M, Shigematsu H, Takahashi T, Parikh G, Pass HI, Chaudhary PM, Gazdar AF (2004) Aberrant methylation of trail decoy receptor genes is frequent in multiple tumor types. Int J Cancer 109: 786–792 [DOI] [PubMed] [Google Scholar]
- Sriamporn S, Pisani P, Pipitgool V, Suwanrungruang K, Kamsa-ard S, Parkin DM (2004) Prevalence of Opisthorchis viverrini infection and incidence of cholangiocarcinoma in Khon Kaen, Northeast Thailand. Trop Med Int Health 9: 588–594 [DOI] [PubMed] [Google Scholar]
- Sripa B, Kaewkes S, Sithithaworn P, Mairiang E, Laha T, Smout M, Pairojkul C, Bhudhisawasdi V, Tesana S, Thinkamrop B, Bethony JM, Loukas A, Brindley PJ (2007) Liver fluke induces cholangiocarcinoma. PLoS Med 4: e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraksa R, Chaopatchayakul P, Jearanaikoon P, Leelayuwat C, Limpaiboon T (2010) Verification of complete bisulfite modification using calponin-specific primer sets. Clin Biochem 43: 528–530 [DOI] [PubMed] [Google Scholar]
- Sulis ML, Parsons R (2003) PTEN: from pathology to biology. Trends Cell Biol 13: 478–483 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Shigematsu H, Shivapurkar N, Reddy J, Miyajima K, Takahashi T, Gazdar AF, Frenkel EP (2006) Methylation of apoptosis related genes in the pathogenesis and prognosis of prostate cancer. Cancer Lett 242: 222–230 [DOI] [PubMed] [Google Scholar]
- Takagi H, Sasaki S, Suzuki H, Toyota M, Maruyama R, Nojima M, Yamamoto H, Omata M, Tokino T, Imai K, Shinomura Y (2008) Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol 43: 378–389 [DOI] [PubMed] [Google Scholar]
- Tannapfel A, Sommerer F, Benicke M, Weinans L, Katalinic A, Geissler F, Uhlmann D, Hauss J, Wittekind C (2002) Genetic and epigenetic alterations of the INK4a-ARF pathway in cholangiocarcinoma. J Pathol 197: 624–631 [DOI] [PubMed] [Google Scholar]
- Teodoridis JM, Hall J, Marsh S, Kannall HD, Smyth C, Curto J, Siddiqui N, Gabra H, McLeod HL, Strathdee G, Brown R (2005) CpG island methylation of DNA damage response genes in advanced ovarian cancer. Cancer Res 65: 8961–8967 [DOI] [PubMed] [Google Scholar]
- Thamavit W, Bhamarapravati N, Sahaphong S, Vajrasthira S, Angsubhakorn S (1978) Effects of dimethylnitrosamine on induction of cholangiocarcinoma in Opisthorchis viverrini-infected Syrian golden hamsters. Cancer Res 38: 4634–4639 [PubMed] [Google Scholar]
- Thamavit W, Kongkanuntn R, Tiwawech D, Moore MA (1987) Level of Opisthorchis infestation and carcinogen dose-dependence of cholangiocarcinoma induction in Syrian golden hamsters. Virchows Arch B Cell Pathol Incl Mol Pathol 54: 52–58 [DOI] [PubMed] [Google Scholar]
- Tischoff I, Markwarth A, Witzigmann H, Uhlmann D, Hauss J, Mirmohammadsadegh A, Wittekind C, Hengge UR, Tannapfel A (2005) Allele loss and epigenetic inactivation of 3p21.3 in malignant liver tumors. Int J Cancer 115: 684–689 [DOI] [PubMed] [Google Scholar]
- Umbricht CB, Evron E, Gabrielson E, Ferguson A, Marks J, Sukumar S (2001) Hypermethylation of 14-3-3 sigma (stratifin) is an early event in breast cancer. Oncogene 20: 3348–3353 [DOI] [PubMed] [Google Scholar]
- van Noesel MM, van Bezouw S, Salomons GS, Voute PA, Pieters R, Baylin SB, Herman JG, Versteeg R (2002) Tumor-specific down-regulation of the tumor necrosis factor-related apoptosis-inducing ligand decoy receptors DcR1 and DcR2 is associated with dense promoter hypermethylation. Cancer Res 62: 2157–2161 [PubMed] [Google Scholar]
- Wales MM, Biel MA, el Deiry W, Nelkin BD, Issa JP, Cavenee WK, Kuerbitz SJ, Baylin SB (1995) p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med 1: 570–577 [DOI] [PubMed] [Google Scholar]
- Yang B, House MG, Guo M, Herman JG, Clark DP (2005) Promoter methylation profiles of tumor suppressor genes in intrahepatic and extrahepatic cholangiocarcinoma. Mod Pathol 18: 412–420 [DOI] [PubMed] [Google Scholar]
- Ye F, Zhang SF, Xie X, Lu WG (2008) OPCML gene promoter methylation and gene expression in tumor and stroma cells of invasive cervical carcinoma. Cancer Invest 26: 569–574 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.