Abstract
Background:
Mitochondrial DNA (mtDNA) mutations occur in head and neck squamous cell carcinoma (HNSCC) and are most frequently detected in the displacement-loop (D-loop) region. The D-loop is considered to be important because it controls mitochondrial gene expression and mtDNA replication. There is currently no evidence that mtDNA mutations can be used as prognostic or predictive biomarkers in HNSCC.
Methods:
We used denaturing high performance liquid chromatography to screen the entire mitochondrial genome of six oral squamous cell carcinoma-derived cell lines and then focused on detecting D-loop abnormalities in 34 HNSCC tissue samples.
Results:
Mitochondrial DNA mutations are not ubiquitous in HNSCC because only half of the cell lines had detectable mtDNA abnormalities following screening of the entire mitochondrial genome and only 18% (6 of 34) of tissue samples had D-loop mutations. There was no correlation between D-loop mutations and determinates of clinical outcome; specifically, tumour stage and the expression of hypoxia-inducible genes included in a highly prognostic hypoxia metagene.
Conclusions:
Taken together, these data suggest that mtDNA D-loop mutations are stochastic events that may not significantly influence the biology of HNSCC and supports the hypothesis that mtDNA mutations in cancer represent bystander genotoxic damage as a consequence of tumour development and progression.
Keywords: head and neck squamous cell carcinoma, mitochondrial DNA, mutations, hypoxia
Head and neck squamous cell carcinoma (HNSCC) is a major world health problem; estimates indicate that there are over 400 000 new cases diagnosed each year (Cancer Research UK, 2010). Typically, most patients present with advanced disease which has a poor prognosis and more than a third of the patients die within 5 years of diagnosis (Cancer Research UK, 2010). The search for prognostic and predictive biomarkers is a key strategy that could be used to improve the management of patients with HNSCC.
Somatic mutations in mitochondrial DNA (mtDNA) have been increasingly observed in human cancers and have been proposed as important oncological biomarkers (Jakupciak et al, 2005; Chatterjee et al, 2006). Mitochondrial DNA mutations have been detected in HNSCCs and their premalignant counterparts (Ha et al, 2002; Chatterjee et al, 2006; Mithani et al, 2007). Mutations occur throughout the mitochondrial genome, but are most frequently detected in the displacement-loop (D-loop) region; around a third of HNSCC samples harbour D-loop mutations (range 2–67% Table 1; Fliss et al, 2000; Sanchez-Cespedes et al, 2001; Tan et al, 2003; Lièvre et al, 2006; Pai et al, 2006; Prior et al, 2006; Zhou et al, 2006, 2007; Dasgupta et al, 2010). The D-loop region is considered to be important because it is the major control site for mtDNA expression and it is also involved in mtDNA replication (Taanman, 1999).
Table 1. Prevalence of D-loop mutations in head and neck squamous cell carcinoma.
| Study | No. of patients | No. with D-loop mutations | Percentage % |
|---|---|---|---|
| Fliss et al (2000) | 13 | 3 | 23 |
| Sanchez-Cespedes et al (2001) | 51 | 19 | 37 |
| Tan et al (2003) | 18 | 12 | 67 |
| Lièvre et al (2006) | 109 | 23 | 21 |
| Pai et al (2006) | 67 | 41 | 61 |
| Prior et al (2006) | 30 | 16 | 53 |
| Zhou et al (2006) | 7 | 3 | 43 |
| Zhou et al (2007) | 83 | 24 | 29 |
| Dasgupta et al (2010) | 50 | 1 | 2 |
| Total | 428 | 142 | 33 |
The biological significance of mtDNA mutations in cancer remains unclear (Chatterjee et al, 2006). Nevertheless, evidence is emerging that mtDNA damage has direct effects on the malignant phenotype and that these effects are mediated by reactive oxygen species (ROS; Petros et al, 2005; Ishikawa et al, 2008). Interestingly, mitochondrial ROS contributes to stabilisation of HIF-1α protein in hypoxia, leading to the proposal that ROS act as ‘oxygen sensors’ to trigger the hypoxia response (Klimova and Chandel, 2008). Recently, Sun et al (2009) showed expression of mutant mitochondrial NADH dehydrogenase 2 (ND2) in a HNSCC-derived cell line increased ROS production, which resulted in HIF-1α stabilisation and a shift to aerobic glycolysis. Furthermore, persistent increased levels of ROS, in the context of mutated mtDNA, may represent one of the mechanisms that drive genomic instability. Reactive oxygen species are DNA damaging agents that could perpetuate mtDNA damage and also cause genotoxic damage to the nucleosome (Lee and Wei, 2009). The link between mtDNA abnormalities, increased ROS and hypoxia may be particularly significant in HNSCC because of the association between hypoxia and an adverse prognosis in the disease (Nordsmark et al, 2005; Winter et al, 2007; Buffa et al, 2010).
In the present study, we screened the entire mitochondrial genome of six oral squamous cell carcinoma-derived cell lines for mtDNA deletions and mutations using denaturing high performance liquid chromatography (DHPLC) and then focused on identifying D-loop abnormalities in a set of HNSCC tissue samples. For the first time, we examined the relationship between mtDNA mutations and the expression of hypoxia-inducible genes in HNSCC tissue samples.
Materials and methods
Cell lines
Six human oral squamous cell carcinoma-derived cell lines were used (Table 2). Four of the cell lines have been described previously (Prime et al, 1990). The two new cell lines (H764A and H764B) were a kind gift of Professor S Prime (University of Bristol). HaCaT cells (Boukamp et al, 1990) and primary cultures of normal oral keratinocytes grown from explanted biopsy material were used as controls for high-throughput screening of cell line mtDNA. Cell line matched normal DNA was used to precisely characterise ‘screen-detected’ mtDNA abnormalities. Genomic DNA extracted from contiguous fibroblast cultures was available for four of the cell lines (H314, H357, H400, and H413; Drugan et al, 1997) and DNA extracted from formalin-fixed paraffin-embedded tissue (striated muscle) was used for H764A and H764B.
Table 2. Derivation of the human oral squamous cell carcinoma-derived cell lines (Prime et al, 1990).
| Cell line | Age | Sex | Site | Grade | Stage |
|---|---|---|---|---|---|
| H314 | 82 | M | FOM | MD | II |
| H357 | 74 | M | T | WD | I |
| H400 | 55 | F | AP | MD | II |
| H413 | 53 | F | BM | MD | II |
| H764A | 70 | M | FOM | MD | IV |
| H764B | 70 | M | LN | MD | IV |
Abbreviations: AP=alveolar process; BM=buccal mucosa; FOM=floor of mouth; LN=lymph node; T=tongue; MD=moderately differentiated; WD=well-differentiated squamous cell carcinoma.
Tissue samples
The tissue samples (tumour and normal mucosa) were derived from patients with previously untreated primary HNSCC. The research was given a favourable opinion from the Newcastle and North Tyneside 2 Research Ethics Committee (Reference 09/H0907/58). The clinical features of the samples are summarised in Table 3. The tissues were harvested at the time of surgical resection and were immediately placed in RNAlater (Applied Biosystems, Warrington, UK) before storage in liquid nitrogen.
Table 3. The clinical features of squamous cell carcinoma tissue samples.
| Sample no. | Age | Sex | Site | Grade | Stage |
|---|---|---|---|---|---|
| 34 | 73 | M | Oral cavity | MD | IVA |
| 35 | 65 | M | Hypopharynx | MD | IVA |
| 38 | 60 | F | Oral cavity | MD | IVA |
| 41 | 38 | M | Larynx | MD | IVA |
| 42 | 71 | M | Hypopharynx | MD | IVA |
| 43 | 58 | M | Hypopharynx | Basaloid | IVA |
| 44 | 68 | F | Oral cavity | MD | II |
| 45 | 68 | M | Oral cavity | MD | II |
| 46 | 44 | M | Oropharynx | Basaloid | IVA |
| 47 | 69 | M | Oral cavity | PD | IVA |
| 51 | 59 | M | Oral cavity | MD | I |
| 54 | 58 | M | Larynx | PD | IVA |
| 59 | 38 | F | Oral cavity | MD | III |
| 60 | 64 | M | Oral cavity | MD | I |
| 61 | 44 | M | Oral cavity | MD | II |
| 62 | 70 | M | Larynx | NR | IVA |
| 63 | 59 | M | Oropharynx | Basaloid | IVA |
| 64 | 54 | M | Oropharynx | Basaloid | IVA |
| 65 | 70 | M | Larynx | MD | II |
| 67 | 56 | M | Oral cavity | MD | IVA |
| 68 | 64 | M | Larynx | MD | IVA |
| 70 | 66 | M | Oropharynx | Basaloid | IVA |
| 71 | 53 | M | Oral cavity | MD | III |
| 73 | 55 | M | Hypopharynx | MD | III |
| 75 | 76 | F | Larynx | MD | IVA |
| 76 | 43 | M | Oral cavity | MD | II |
| 78 | 53 | M | Larynx | MD | IVA |
| 80 | 63 | F | Larynx | MD | IVA |
| 82 | 63 | M | Larynx | PD | IVA |
| 83 | 56 | M | Larynx | PD | IVA |
| 85 | 57 | F | Oral cavity | MD | IVA |
| 87 | 54 | M | Oral cavity | MD | II |
| 88 | 61 | F | Oral cavity | MD | IVA |
| 89 | 69 | M | Hypopharynx | MD | IVA |
Abbreviations: MD=moderately differentiated; PD=poorly differentiated; NR=not recorded.
Preparation of DNA
Genomic DNA was extracted from cell pellets using a standard laboratory kit (QIAGEN Ltd, Crawley, UK). DNA was extracted from homogenised tissue samples using Tri Reagent (Sigma-Aldrich, Gillingham, UK) and cleaned using a standard laboratory kit (QIAGEN). Extracted DNA samples were diluted to 200 ng μl−1 for use in downstream analysis.
Long template PCR
Genomic DNA from the cell lines were screened for large-scale mtDNA deletions using the Expand Long Template PCR System as previously described (Roche Diagnostics Ltd, Burgess Hill, UK; Ray et al, 2000). DNA from a patient with mitochondrial myopathy, characterised by a 4976-bp mtDNA deletion, was used as a positive control (kind gift of Professor R Taylor, Newcastle University).
Mitoscreen assay and DHPLC analysis
Genomic DNA samples were screened for small deletions, insertions, and point mutations using the Mitoscreen Assay System (Transgenomic Ltd, Glasgow, UK). The system comprises 19 overlapping primer sets that amplify the entire mitochondrial genome. Reactions were carried out according to the manufacturer's standard protocol. Briefly, PCRs were performed in 20–40 μl reactions containing 100 ng of genomic DNA, 0.15 μM of each primer and 2.5 units of Optimase polymerase (Transgenomic Ltd). In all, 15 of the 19 primer sets generated large amplicons that required digestion with restriction enzymes before heteroduplex formation. Heteroduplexed DNA was analysed by DHPLC on a Transgenomic Wave DNA Fragment Analysis System 3500 using Navigator software (Transgenomic Ltd).
Automated sequencing
Denaturing high performance liquid chromatography fractions harbouring putative mtDNA abnormalities were eluted and dehydrated using a DNA concentrator (Gyrovap, Howe and Co., London, UK). DNA was resuspended in 5 μl of water and reamplified using appropriate primer sets. The PCR products were resolved on a 2% low melting point agarose gel (NuSieve 3.1 Agarose, Lonza Rockland Inc., Rockland, ME, USA) containing ethidium bromide. The DNA band representing the amplicon was excised from the gel and the DNA was extracted using the QIAquick Gel Extraction Kit (QIAGEN). The purified DNA was eluted in 30 μl of Tris/EDTA buffer. The sequencing reaction was carried out using the GenomeLab Dye Terminator Cycle Sequencing Kit (Beckman Coulter UK Ltd, High Wycombe, UK) according to the manufacturer's protocol. The products were run on a Beckman CEQ Genetic Analysis System 8000 (Beckman Coulter). Sequences were visualised using MacVector 7.0 software (Oxford Molecular Ltd, Oxford, UK) and compared with the mtDNA revised Cambridge reference sequence (MITOMAP, 2010).
Hypoxia-inducible gene expression
Tissue samples were prepared for quantitative real-time PCR (qRT–PCR) as described previously (Winter et al, 2007). Quantitative real-time PCR for CA9, PGAM1, SLC2A1, and VEGFA was performed using Taqman assays (Applied Biosystems) on a low-density array format with normalisation to appropriate endogenous reference genes. Cycle threshold (Ct) values were converted to a measure of gene expression relative to the endogenous reference gene expression using the 2−ΔCt formula before statistical analysis as previously described (Livak and Schmittgen, 2001).
Results
Screening of cell lines for mtDNA abnormalities
None of the cell lines contained any large-scale mtDNA deletions (data not shown). Using DNA extracted from normal oral keratinocytes and HaCaT cells as a comparator, DHPLC profiles from the cell line DNA had a number of abnormalities. The abnormalities included broader wave profiles, peak shifts, and a change in the number of fragments in those samples subject to restriction enzyme digestion. Whole mitochondrial genome screening of the six cell lines generated 114 amplicons representing around 100 kb of DNA. A total of 17 mtDNA abnormalities were detected using this method and abnormalities were identified in all of the cell lines.
Characterisation of cell line mtDNA abnormalities by sequence analysis
Sequencing of ‘screen-detected’ mtDNA abnormalities in the cell lines alongside matched normal DNA demonstrated that around half of the abnormalities (8 of 17) were common to both samples and represented mtDNA polymorphisms. H357, H400, and H413 had polymorphic mtDNA, but no somatic mtDNA mutations. By contrast, H314 had a complex mtDNA genotype with nine homoplasmic point mutations located in two regions of the mitochondrial genome, namely cytochrome oxidase I and the D-loop (Table 4). There were three base pair changes in the cytochrome oxidase I gene of H314 that were not present in the matched fibroblast DNA. The base changes were all synonymous and there was no predicted change in the amino-acid sequence of the codon. Six point mutations were detected in the D-loop; the matched fibroblasts followed the revised Cambridge reference sequence (MITOMAP, 2010). The base pair changes were all located in hypervariable segment 1 (HVS1) of the D-loop.
Table 4. Mitochondrial DNA mutations in H314 cells.
| Mitochondrial gene/region | D-loop region | Nucleotide position | mtDNA rCRS | Control H314 fibroblasts | Tumour H314 | Mutation status | Reported previouslya |
|---|---|---|---|---|---|---|---|
| COI | NA | 6152 | T | T | C | HM | No |
| COI | NA | 6179 | G | G | A | HM | No |
| COI | NA | 6180 | G | G | A | HM | No |
| D-loop | HVS1 | 16 064 | T | T | C | HM | No |
| D-loop | HVS1 | 16 093 | T | T | C | HM | No |
| D-loop | HVS1 | 16 094 | T | T | C | HM | No |
| D-loop | HVS1 | 16 102 | T | T | A | HM | No |
| D-loop | HVS1 | 16 124 | T | T | G | HM | No |
| D-loop | HVS1 | 16 145 | G | G | A | HM | No |
Abbreviations: COI=cytochrome oxidase I (np 5904–7445); D-loop=displacement-loop (np 16024–576); HVS1=hypervariable segment 1; NA=not applicable; mtDNA rCRS=mitochondrial DNA revised Cambridge reference sequence (MITOMAP, 2010); HM=homoplasmic.
Mutation reported on a publically available database (MITOMAP, 2010).
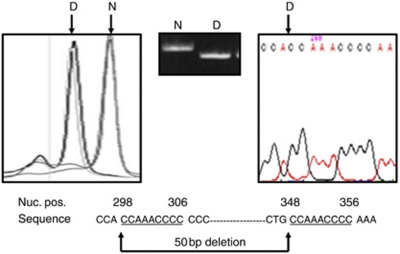
H764A and H764B, cell lines derived from the same patient representing a primary oral squamous cell carcinoma and a corresponding lymph node metastasis, respectively, had two sequence abnormalities not detected in the matched normal DNA. The first was a homoplasmic 50 bp deletion in the D-loop between nucleotide positions 298/306 and 348/356; the breakpoints were flanked by a 9-bp repeat sequence 5′-CCAAACCCC-3′ (Figure 1). The deletion was located in a non-coding, control region of the D-loop covering the mtTF1-binding site, replication primer, and conserved sequence blocks 2 and 3. An identical 50 bp deletion has previously been reported in four gastric cancers and a hepatocellular carcinoma (Burgart et al, 1995; Lee et al, 2004). The second sequence abnormality was a homoplasmic point mutation in the D-loop; G-A at nucleotide 16 129 in HVS1.
Figure 1.
H764A and H764B cell lines harboured a homoplasmic 50 bp deletion in the D-loop region. The deletion (D) was identified by DHPLC, DNA electrophoresis, and DNA sequencing (from left to right, respectively; N=matched normal DNA sample). The deletion was located between nucleotide positions (nuc. pos.) 298/306 and 348/356; the breakpoints were flanked by a 9-bp repeat sequence 5′-CCAAACCCC-3′.
Screening tissue samples for mtDNA abnormalities in the D-loop region
Twenty-two abnormalities were detected following screening of 102 amplicons in 34 HNSCC with matched normal mucosal samples. Sequencing of the variant amplicons demonstrated that 15 of the samples contained known sequence variants (polymorphisms) that were identical in the normal and tumour DNA (data not shown). Eight somatic mutations were detected in seven samples; sample 85 contained two mutations (Table 5). The majority (6 of 8) of the mutations were located in the D-loop; two were located in 12S rRNA gene. Eighteen percent (6 of 34) of the samples harboured a D-loop mutation and there was a mixture of homoplasmic and heteroplasmic mutations (Table 5; Figure 2). One of the D-loop mutations (146 T-C) has previously been reported in ovarian and prostate cancer (Figure 2A; MITOMAP, 2010).
Table 5. Mitochondrial mutations detected in head and neck squamous cell carcinoma tissue samples.
| Case | Mitochondrial gene/region | D-loop region | Nucleotide position | mtDNA rCRS | Control tissue | Tumour | Mutation status | Reported previouslya |
|---|---|---|---|---|---|---|---|---|
| 43 | D-loop | HVS2 | 153 | A | A | G | HM | No |
| 47 | D-loop | H-strand origin | 374 | A | A | A/G | HT | No |
| 64 | 12S rRNA | NA | 796 | G | G | A | HM | No |
| 83 | D-loop | HVS2 | 265 | T | T | T/A | HT | No |
| 85 | 12S rRNA | NA | 779 | T | T | T/G | HT | No |
| 85 | D-loop | HVS1 | 16 129 | G | A | G/A | HT | No |
| 87 | D-loop | HVS2 | 146 | T | T | C | HM | Ovarian carcinoma Prostate tumour |
| 88 | D-loop | HVS1 | 16 145 | G | G | G/A | HT | No |
Abbreviations: HVS1/2=hypervariable segment 1/2; NA=not applicable; mtDNA rCRS=mitochondrial DNA revised Cambridge reference sequence (MITOMAP, 2010); HM=homoplasmic; HT=heteroplasmic.
Mutation reported on a publically available database (MITOMAP, 2010).
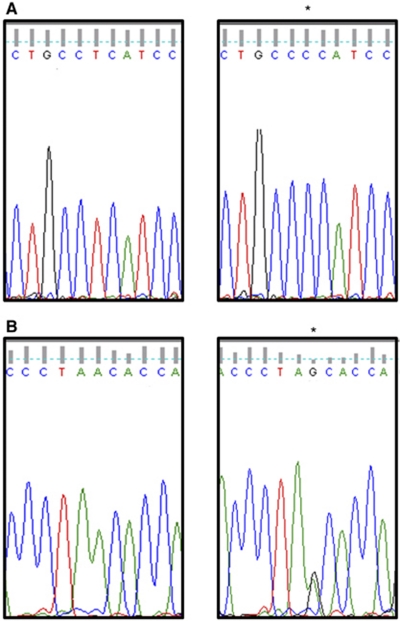
Figure 2.
D-loop mutations in head and neck squamous cell carcinoma tissue samples. (A) Homoplasmic mutation at 146 T-C (*) in sample 87. Sequence shown from nucleotide position 141–151 for normal sample (left panel) and tumour sample (right panel). (B) Heteroplasmic mutation at 374 A-G (*) in sample 47. Sequence shown from nucleotide position 369–379 for normal sample (left panel) and tumour sample (right panel). In the tumour, there are two peaks at 374; there is a mixture of A and G nucleotides.
Relationship of D-loop mutations to clinical parameters and hypoxia-inducible genes
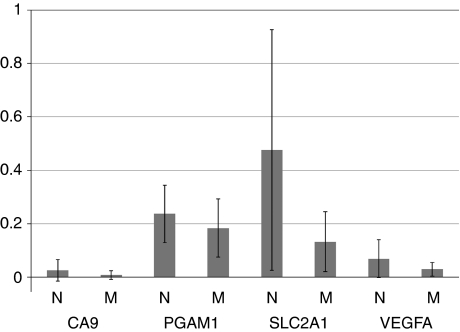
The samples with D-loop mutations did not have any distinct clinico-pathological characteristics; there was no clear association with patient age, gender, tumour location, grade, and stage. Analysis of overall survival showed no difference for patients with D-loop mutations compared with those patients with normal D-loop sequences. We investigated the relationship between mtDNA D-loop mutations and the expression of four hypoxia-inducible genes included in a highly prognostic hypoxia metagene (CA9, PGAM1, SLC2A1, and VEGFA; Winter et al, 2007; Buffa et al, 2010). For each of the four genes, the samples with D-loop mutations (n=6) had lower mean gene expression levels when compared with those samples that had normal D-loop sequences (n=23), however, there was no statistically significant difference in hypoxia gene expression between the two groups (CA9 0.03 vs 0.01, P=0.30; PGAM1 0.24 vs 0.18, P=0.29; SLC2A1 0.48 vs 0.13, P=0.08; VEGFA 0.07 vs 0.03, P=0.19; parametric t-test, SPSS Statistics 17.0, SPSS Inc., Chicago, IL, USA; Figure 3).
Figure 3.
The mean expression of four hypoxia-inducible genes using qRT–PCR for samples with normal (N) and mutated (M) D-loop sequences.
Discussion
In the current study, we screened oral squamous cell carcinoma-derived cell lines and HNSCC tissue samples for mtDNA mutations. Large-scale deletions of the mitochondrial genome have been reported in oral SCC (Tan et al, 2003; Shieh et al, 2004), however, our study and others (Zhou et al, 2007) have not been able to detect such abnormalities and therefore the significance of large-scale mtDNA deletions in HNSCC is unclear. We used DHPLC to screen samples for small-scale deletions, insertions, and point mutations. The majority of studies investigating mtDNA abnormalities in HNSCC have focused on small regions of the mitochondrial genome using PCR and sequencing (Fliss et al, 2000; Sanchez-Cespedes et al, 2001; Lièvre et al, 2006; Pai et al, 2006; Prior et al, 2006). Sequencing alone lacks sensitivity, because the technique only reliably identifies mutant species when they represent at least 25% of the amplicons in the sequencing reaction. By contrast, DHPLC is highly sensitive and detects mutations at very low thresholds; <5% (Birket and Birch-Machin, 2007). Denaturing high performance liquid chromatography also benefits from high specificity; identical DHPLC profiles between matched tumour and normal DNA effectively excludes mutation (Van den Bosch et al, 2000). The combination of DHPLC analysis and sequencing of enriched amplicon fractions is likely to have optimised mutation detection in our study.
We found that mtDNA mutations are not ubiquitous in HNSCC because half of the cell lines had no detectable mtDNA abnormalities following screening of the entire mitochondrial genome and only a small proportion (18%) of the HNSCC tissue samples had D-loop mutations. Pooled data from nine studies that looked for D-loop mutations in over 400 HNSCCs indicate that around a third of HNSCCs harbour D-loop mutations; however, there is wide variation between different studies (range 2–67% Table 1). The latter may reflect differences in the methods of mutation detection or the intrinsic genetic heterogeneity of HNSCCs. Significantly, our data are comparable with the largest series examined to date (n=109; Lièvre et al, 2006), which detected D-loop mutations in only 21% of the samples.
The D-loop is considered to be particularly important because it controls mitochondrial gene expression and it is involved in mtDNA replication (Taanman, 1999). More recently, there is evidence that the D-loop has a functional role in the formation of mitochondrial nucleoids and the organisation of mtDNA during segregation and replication (He et al, 2007; Holt et al, 2007). Of the mutations detected in the current study, the majority were T-C, G-A base transitions, which are associated with oxidative damage (Fliss et al, 2000; Zhou et al, 2007) and raise the possibility that ROS have a pivotal role in perpetuating mtDNA damage. While, elevated levels of ROS are known to stabilise HIF-1α and trigger the hypoxia response (Klimova and Chandel, 2008), we were unable to demonstrate a correlation between mtDNA mutations and hypoxia-inducible gene expression.
The 50-bp deletion identified in the D-loop of both H764A and its corresponding lymph node metastasis, H764B, has previously been reported in gastric adenocarcinomas and hepatocellular carcinomas (Burgart et al, 1995; Lee et al, 2004). Furthermore, this small-scale deletion was also present in the metastases of two of the gastric cancers (Burgart et al, 1995). These findings are consistent with the concept of clonal expansion and conservation of mtDNA mutations during tumour progression. The functional significance of the deletion is unknown; however, one possible mechanism is that the deletion affects nucleoid formation, by interfering with ATAD3 binding, leading to altered mtDNA copy number (Holt et al, 2007). Increased mtDNA copy number has been described in HNSCCs (Kim et al, 2004) and is also associated with increased oxidative stress (Lee and Wei, 2009). These factors may account for the accumulation of the deletion to homoplasmic levels and its persistence during tumour progression.
There are a number of studies which indicate that D-loop mutations have prognostic significance. Studies in non-small cell lung cancer, colorectal cancer, and breast cancer showed D-loop mutations are associated with poor prognosis (Matsuyama et al, 2003; Lièvre et al, 2005; Tseng et al, 2006). The only study in HNSCC found no correlation between D-loop mutations and prognosis or response to chemotherapy in 109 patients (Lièvre et al, 2006). Similarly, in the present study, there was no obvious correlation between D-loop mutations and prognostic indicators, specifically, tumour stage and the expression of hypoxia-inducible genes included in a highly prognostic hypoxia metagene (Winter et al, 2007; Buffa et al, 2010). These data are consistent with other studies in a variety of other cancers (oesophagus, gastric, lung, and ovarian), showing no correlation between mtDNA abnormalities with clinico-pathological features, which limits their utility as prognostic biomarkers in these diseases (Lee and Wei, 2009).
In conclusion, our study indicates that mtDNA mutations are infrequent in HNSCC, but most commonly located in the D-loop region. Displacement-loop mutations are likely to be stochastic events (i.e., subject to the variation of chance) that may not significantly influence the biology of HNSCC. Our data support the hypothesis that mtDNA mutations in cancer represent bystander genotoxic damage as a consequence of tumour development and progression (Taylor and Turnbull, 2005).
Acknowledgments
This work was supported by The Newcastle upon Tyne Hospitals NHS Charity and the JGW Paterson Foundation. We thank Professor John Lunec (Northern Institute for Cancer Research, Newcastle University) for the use of his DNA sequencing facilities. The statistical analysis was carried out in collaboration with Dr Simon Kometa (Research Computing Specialist, Statistics, Newcastle University).
References
- Birket M, Birch-Machin MA (2007) Ultraviolet radiation exposure accelerates the accumulation of aging-dependent T414G mitochondrial DNA mutation in human skin. Aging Cell 6: 557–564 [DOI] [PubMed] [Google Scholar]
- Boukamp P, Stanbridge EJ, Foo DY, Cerutti PA, Fusenig NE (1990) c-Ha-ras oncogene expression in immortalised human keratinocytes (HaCaT) alters growth potential in vivo, but lacks correlation with malignancy. Cancer Res 50: 2840–2847 [PubMed] [Google Scholar]
- Buffa FM, Harris AL, West CM, Miller CJ (2010) Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br J Cancer 102: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgart LJ, Zheng J, Shu Q, Strickler JG, Shibata D (1995) Somatic mitochondrial mutation in gastric carcinoma. Am J Pathol 147: 1105–1111 [PMC free article] [PubMed] [Google Scholar]
- Cancer Research UK (2010) http://info.cancerresearchuk.org/cancerstats/
- Chatterjee A, Mambo E, Sidransky D (2006) Mitochondrial DNA mutations in human cancer. Oncogene 25: 4663–4674 [DOI] [PubMed] [Google Scholar]
- Dasgupta S, Koch R, Westra WH, Califano JA, Ha PK, Sidransky D, Koch WM (2010) Mitochondrial DNA mutation in normal margins and tumors of recurrent head and neck squamous cell carcinoma patients. Cancer Prev Res (Phila) 3: 1205–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drugan CS, Stone A, Game SM, Prime SS (1997) The mitogenic effect of KGF and the expression of its cell surface receptor on cultured normal and malignant human oral keratinocytes and on contiguous fibroblasts. J Oral Pathol Med 26: 327–333 [DOI] [PubMed] [Google Scholar]
- Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D (2000) Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science 287: 2017–2019 [DOI] [PubMed] [Google Scholar]
- Ha PK, Tong BC, Westra WH, Sanchez-Cespedes M, Parrella P, Zahurak M, Sidransky D, Califano JA (2002) Mitochondrial C-tract alteration in premalignant lesions of the head and neck: a marker for progression and clonal proliferation. Clin Cancer Res 8: 2260–2265 [PubMed] [Google Scholar]
- He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, Reichelt S, Spelbrink JN, Walker JE, Holt IJ (2007) The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organisation. J Cell Biochem 178: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt IJ, He J, Mao CC, Boyd-Kirkup JD, Martinsson P, Sembongi H, Reyes A, Spelbrink JN (2007) Mammalian mitochondrial nucleoids: organising an independently minded genome. Mitochondrion 7: 311–321 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J (2008) ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science 320: 661–664 [DOI] [PubMed] [Google Scholar]
- Jakupciak JP, Wang W, Markowitz ME, Ally D, Coble M, Srivastava S, Maitra A, Barker PE, Sidransky D, O’Connell CD (2005) Mitochondrial DNA as a cancer biomarker. J Mol Diagn 7: 258–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MM, Clinger JD, Masayesva BG, Ha PK, Zahurak ML, Westra WH, Califano JA (2004) Mitochondrial DNA quantity increases with histopathologic grade in premalignant and malignant head and neck lesions. Clin Cancer Res 10: 8512–8515 [DOI] [PubMed] [Google Scholar]
- Klimova T, Chandel NS (2008) Mitochondrial complex III regulates hypoxic activation of HIF. Cell Death Differ 15: 660–666 [DOI] [PubMed] [Google Scholar]
- Lee HC, Li SH, Lin JC, Wu CC, Yeh DC, Wei YH (2004) Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutat Res 547: 71–78 [DOI] [PubMed] [Google Scholar]
- Lee HC, Wei YH (2009) Mitochondrial DNA instability and metabolic shift in human cancers. Int J Mol Sci 10: 674–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lièvre A, Blons H, Houllier AM, Laccourreye O, Brasnu D, Beaune P, Laurent-Puig P (2006) Clinicopathological significance of mitochondrial D loop mutations in head and neck carcinoma. Br J Cancer 94: 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lièvre A, Chapusot C, Bouvier AM, Zinzindohoué F, Piard F, Roignot P, Arnould L, Beaune P, Faivre J, Laurent-Puig P (2005) Clinical value of mitochondrial mutations in colorectal cancer. J Clin Oncol 23: 3517–3525 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Matsuyama W, Nakagawa M, Wakimoto J, Hirotsu Y, Kawabata M, Osame M (2003) Mitochondrial DNA mutation correlates with stage progression and prognosis in non-small cell lung cancer. Hum Mutat 21: 441–443 [DOI] [PubMed] [Google Scholar]
- Mithani SK, Taube JM, Zhou S, Smith IM, Koch WM, Westra WH, Califano JA (2007) Mitochondrial mutations are a late event in the progression of head and neck squamous cell cancer. Clin Cancer Res 13: 4331–4335 [DOI] [PubMed] [Google Scholar]
- MITOMAP (2010) A Human Mitochondrial Genome Database, http://www.mitomap.org
- Nordsmark M, Bentzen SM, Rudat V, Brizel D, Lartigau E, Stadler P, Becker A, Adam M, Molls M, Dunst J, Terris DJ, Overgaard J (2005) Prognostic value of tumor oxygenation in 397 head and neck tumors after primary radiation therapy. An international multi-center study. Radiother Oncol 77: 18–24 [DOI] [PubMed] [Google Scholar]
- Pai CY, Hsieh LL, Lee TC, Yang SB, Linville J, Chou SL, Yang CH (2006) Mitochondrial DNA sequence alterations observed between blood and buccal cells within the same individuals having betel quid (BQ)-chewing habit. Forensic Sci Int 156: 124–130 [DOI] [PubMed] [Google Scholar]
- Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC (2005) mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA 102: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prime SS, Nixon SV, Crane IJ, Stone A, Matthews JB, Maitland NJ, Remnant L, Powell SK, Game SM, Scully C (1990) The behaviour of human oral squamous cell carcinoma in cell culture. J Pathol 160: 259–269 [DOI] [PubMed] [Google Scholar]
- Prior SL, Griffiths AP, Baxter JM, Baxter PW, Hodder SC, Silvester KC, Lewis PD (2006) Mitochondrial DNA mutations in oral squamous cell carcinoma. Carcinogenesis 27: 945–950 [DOI] [PubMed] [Google Scholar]
- Ray AJ, Turner R, Nikaido O, Rees JL, Birch-Machin MA (2000) The spectrum of mitochondrial DNA deletions is a ubiquitous marker of ultraviolet radiation exposure in human skin. J Invest Dermatol 115: 674–679 [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Parrella P, Nomoto S, Cohen D, Xiao Y, Esteller M, Jeronimo C, Jordan RC, Nicol T, Koch WM, Schoenberg M, Mazzarelli P, Fazio VM, Sidransky D (2001) Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors. Cancer Res 61: 7015–7019 [PubMed] [Google Scholar]
- Shieh DB, Chou WP, Wei YH, Wong TY, Jin YT (2004) Mitochondrial DNA 4977-bp deletion in paired oral cancer and precancerous lesions revealed by laser microdissection and real-time quantitative PCR. Ann NY Acad Sci 1011: 154–167 [DOI] [PubMed] [Google Scholar]
- Sun W, Zhou S, Chang SS, McFate T, Verma A, Califano J (2009) Mitochondrial mutations contribute to HIF1α accumulation via increased reactive oxygen species and up-regulated pyruvate dehydrogenase kinase 2 in head and neck cancer. Clin Cancer Res 15: 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taanman JW (1999) The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta 1410: 103–123 [DOI] [PubMed] [Google Scholar]
- Tan DJ, Chang J, Chen WL, Agress LJ, Yeh KT, Wang B, Wong LJ (2003) Novel heteroplasmic frameshift and missense somatic mitochondrial DNA mutations in oral cancer of betel quid chewers. Genes Chromosomes Cancer 37: 186–194 [DOI] [PubMed] [Google Scholar]
- Taylor RW, Turnbull DM (2005) Mitochondrial DNA mutations in human disease. Nat Rev Genet 6: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, Wei YH, Lee HC (2006) Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer 45: 629–638 [DOI] [PubMed] [Google Scholar]
- Van den Bosch BJC, de Coo RFM, Scholte HR, Nijland JG, van den Bogaard R, de Visser M, de Die-Smulders CEM, Smeets HJM (2000) Mutation analysis of the entire mitochondrial genome using denaturing high performance liquid chromatography. Nucleic Acids Res 28: e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SC, Buffa FM, Silva P, Miller C, Valentine HR, Turley H, Shah KA, Cox GJ, Corbridge RJ, Homer JJ, Musgrove B, Slevin N, Sloan P, Price P, West CM, Harris AL (2007) Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res 67: 3441–3449 [DOI] [PubMed] [Google Scholar]
- Zhou S, Kachhap S, Sun W, Wu G, Chuang A, Poeta L, Grumbine L, Mithani SK, Chatterjee A, Koch W, Westra WH, Maitra A, Glazer C, Carducci M, Sidransky D, McFate T, Verma A, Califano JA (2007) Frequency and phenotypic implications of mitochondrial DNA mutations in human squamous cell cancers of the head and neck. Proc Natl Acad Sci USA 104: 7540–7545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Kassauei K, Cutler DJ, Kennedy GC, Sidransky D, Maitra A, Califano J (2006) An oligonucleotide microarray for high-throughput sequencing of the mitochondrial genome. J Mol Diagn 8: 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]