Abstract
The Single Nucleotide Polymorphism database (dbSNP) is a variation database at the National Center for Biotechnology Information (NCBI). It is a public repository of submitted nucleotide variations and is part of NCBI’s search and retrieval system Entrez. This chapter describes two basic protocols to search dbSNP effectively, one to perform a text-based search and another to perform a sequence-based search. The unit also describes one of the result display formats called GeneView to obtain information about all submitted SNPs in a particular gene.
Keywords: single nucleotide polymorphism (SNP), variation, NCBI, dbSNP
INTRODUCTION
The Single Nucleotide Polymorphism database (dbSNP) is a variation database at the National Center for Biotechnology Information (NCBI; (Sherry et al., 2001; Sayers et al., 2010). It is a public repository of submitted nucleotide variations. As the name suggests, it includes single nucleotide polymorphisms (SNPs) but it also includes other variations such as small insertions/deletions and microsatellites or short tandem repeats. The database can be accessed from http://www.ncbi.nlm.nih.gov/guide/variation/. The page also lists other variation databases at NCBI such as the Database of Genomic Structural Variation (dbVar), Database of Genotypes and Phenotypes (dbGaP), and Database of Major Histocompatibility Complex (dbMHC). This unit gives a brief overview of dbSNP, describes how to search in dbSNP (Basic Protocols 1 and 2), and reviews one of the result display formats called GeneView.
Overview of dbSNP
dbSNP is a database that includes entries submitted by public laboratories and private organizations for a large number of organisms. Each submission includes information about the actual nucleotide variation and the 5’ and 3’ flanking sequences. It may also include other information such as genotype and frequency information. Each submitted entry is assigned a unique ID number that begins with letters “ss” (submitted SNP). If a number of submitted SNP entries align to the same position on the genome assembly, then they are also reported as a part of a group called the Reference SNP cluster (refSNP), which is assigned a new ID number that begins with letters “rs”. This procedure is performed periodically in the process that results in a new database “build”. The current build number and the build history can be obtained from http://www.ncbi.nlm.nih.gov/projects/SNP/buildhistory.cgi. More information about the build process and computed information from the submitted data can be obtained from http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=handbook&part=ch5 and is described in the Commentary section below.
Searching dbSNP
The database can be searched by various ways as described below.
Searching dbSNP in Entrez
The search box in dbSNP could be the first start to search the database using a text word or a phrase as a query. However, since dbSNP is part of NCBI’s search and retrieval system Entrez, similar to all databases in Entrez, this database can be searched effectively using the Limits page or the Preview/Index page. The Limits page in dbSNP lists options to restrict your query by a variety of criteria such as organism, chromosome, type of SNP, functional classification of SNP and success rate. The user can select multiple criteria which will be combined by the term “AND” in a majority of the cases. The Preview/Index page also can be used to build a query by using Boolean operators “AND”, “OR” and “NOT”. A help document for searching in Entrez is available at http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=helpentrez&part=EntrezHelp.
The list of the fields that can be used to restrict the query in dbSNP is described in the web page http://www.ncbi.nlm.nih.gov/snp.
BASIC PROTOCOL 1
SEARCHING dbSNP USING THE ENTREZ LIMITS SEARCH OPTION
This protocol describes how to use the Limits page to search for all human SNPs that cause a change in the amino acid, are associated with phenotype(s), are cited in publications, and have known 3-D protein structures for the wild type amino acid.
The nucleotide variations that lead to a change in the codon and would encode a different amino acid are labeled as non-synonymous in dbSNP. The synonymous SNPs are the nucleotide substitutions that do not lead to a change in the amino acid. Non-synonymous changes are further classified as missense, nonsense and frame-shift. Missense changes are the substitutions that lead to change in an amino acid. Nonsense variations result in a stop codon causing termination of the protein. Frame-shift variations are a result of an insertion/deletion leading to a change in a reading frame (Guttmacher and Collins, 2002; Feero et al., 2010). Refer to the Commentary section below for more details.
Some changes in the amino acids are tolerated and do not change the function of the protein. However, some changes are so drastic that they may cause a change in the observed phenotype. The changes in human genes associated with genetic diseases/phenotypes are reported in the Online Mendelian Inheritance in Man (OMIM) database at NCBI (Unit 1.2; Hamosh et al., 2002).
All databases in Entrez are interlinked and the Limits or Preview/Index search option can be used to obtain the associated information in other databases. For example, the SNPs in dbSNP with links to the OMIM database can be obtained from the SNP_OMIM filter menu in the Preview/Index search option or through the OMIM link under the Annotation box in the Limits search option (See Commentary section).
Necessary Resources
Hardware
Computer with Internet access
Software
An up-to-date Web browser, such as Firefox, Internet Explorer, or Safari
Searching dbSNP using the Entrez limits search option
-
1
Go to NCBI web page http://www.ncbi.nlm.nih.gov/.
-
2
Select the “SNP” database from the “All Databases” pull down menu at the top of the page (Figure 1.19.1). Alternatively, click on “Variation” link in the green box on the left of the web page. This gives a list of variation- related resources at NCBI. Click on the link “Database of Single Nucleotide Polymorphisms (dbSNP)”.
-
3
Click on the “Limits” tab (Figure 1.19.2).
-
4
In the Organism box, click on the box next to Homo sapiens (Figure 1.19.3).
-
5
Under the Function class, select the box next to coding non-synonymous (Figure 1.19.3).
-
6
In the Annotation box, click on the boxes next to Structure, OMIM and Cited in publication. Please note that the Annotation box is located on the same Web page as shown in Figure 1.19.3 and can be accessed by scrolling down the page.
-
7
Click on the Go button at the top of the page. Currently, the results of the above search include 19 entries (Figure 1.19.4 and Table 1.19.1).
Figure 1.19.1.
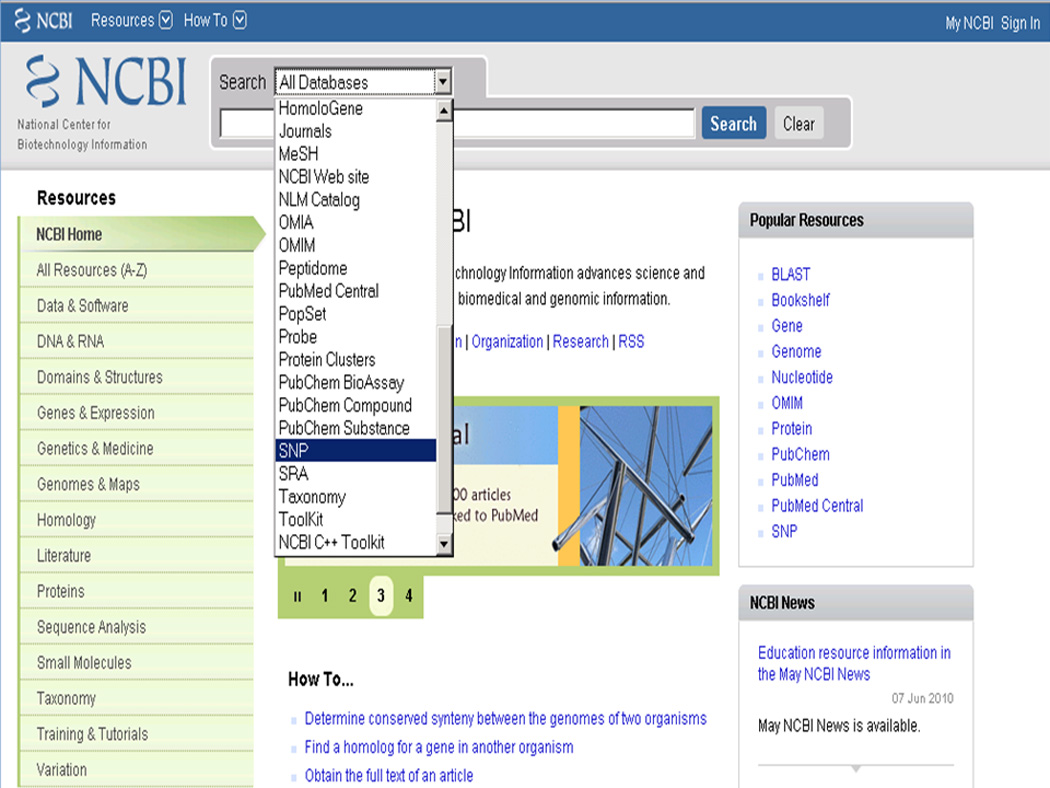
Selection of the SNP database from the NCBI Search menu. Note the Variation link in the green box on the left side of the page; this is another way to select the SNP database which is one of the variation databases at NCBI.
Figure 1.19.2.
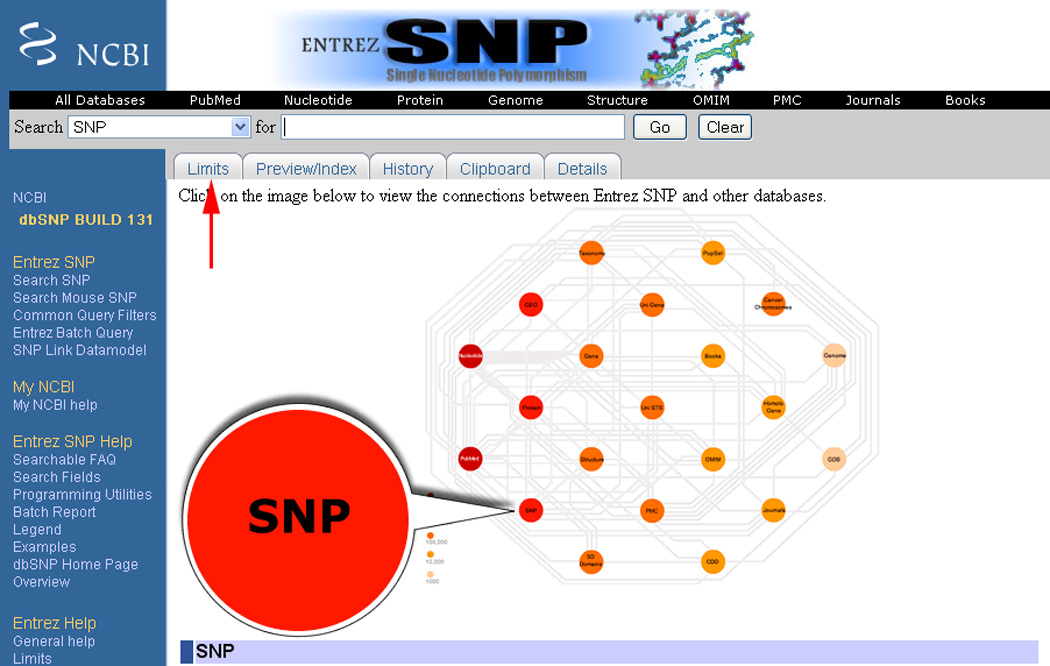
Location of the Limits tab on the Entrez SNP search page indicated by the red arrow.
Figure 1.19.3.
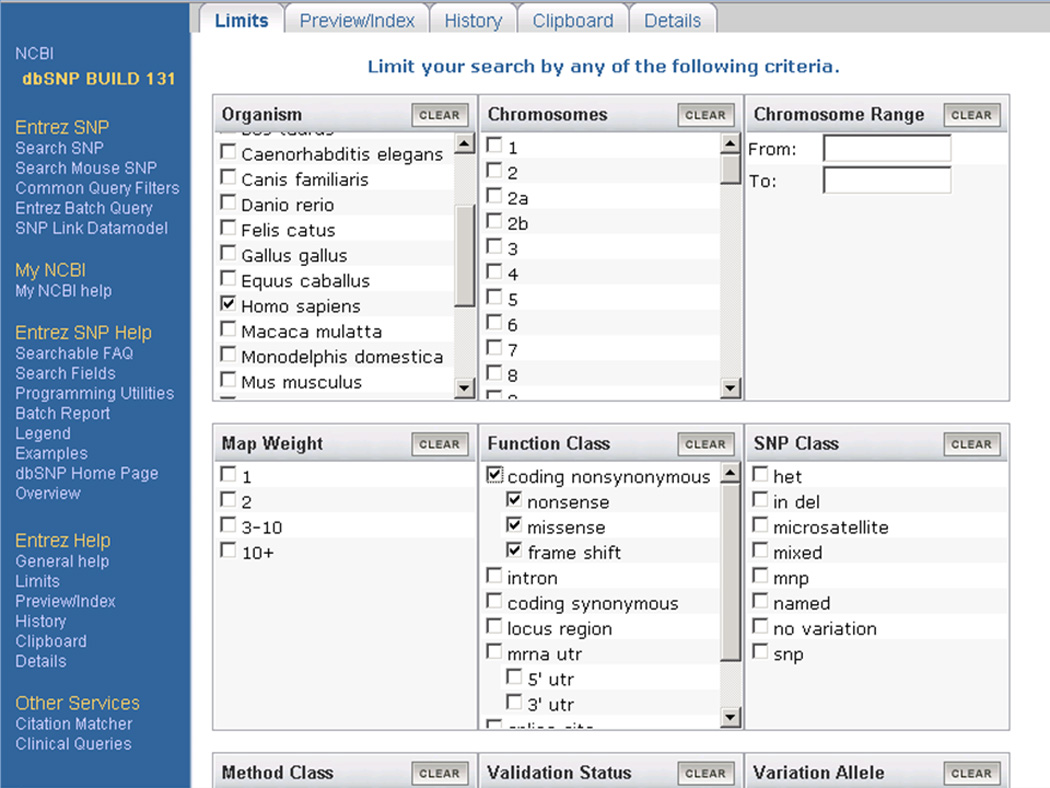
Selection of Homo sapiens as an organism and coding nonsynonymous as the Function Class in the Entrez dbSNP Limits page. Please note that the Annotation box is located on the same page as shown here and can be accessed by scrolling down the page.
Figure 1.19.4.
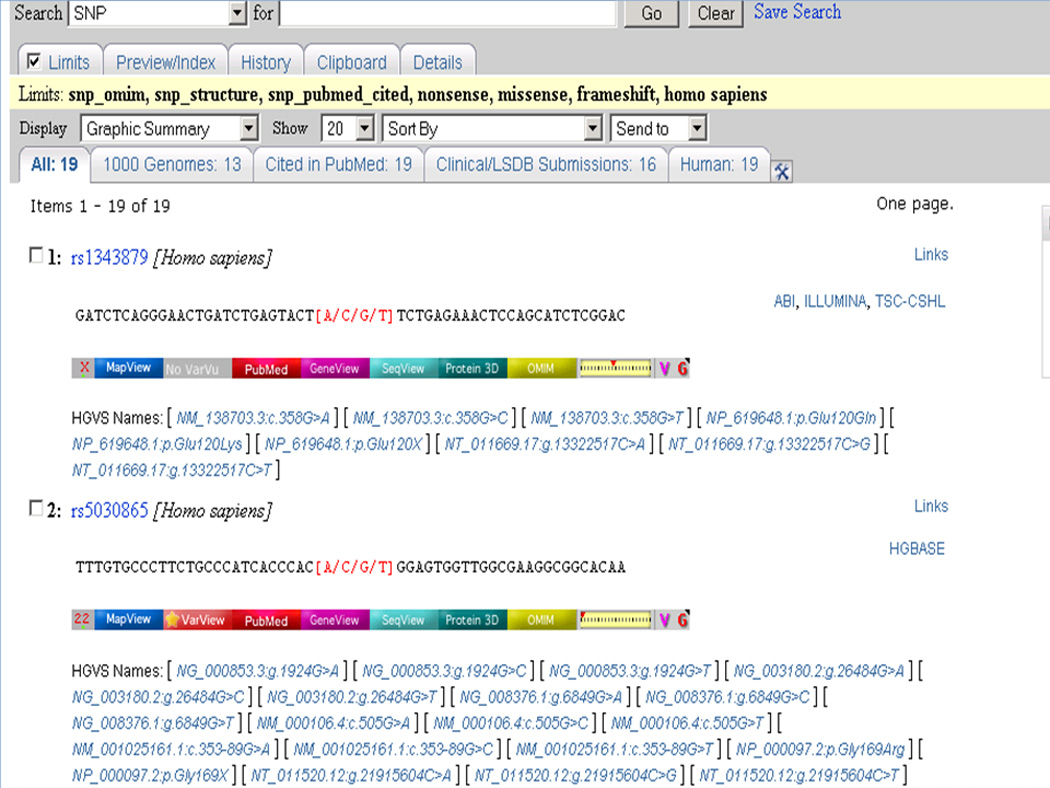
Results of the search in Basic Protocol 1. There are 19 human entries containing SNPs that cause a change in the amino acid, are associated with phenotype(s), are cited in publications, and have known 3-D protein structures for the wild type amino acid.
Table 1.19.1.
Descriptions of links from search performed in Basic Protocol 1.
| Link to NCBI’s MapViewer | |
| Link to Variation Viewer | |
| Link to publications | |
| Link to GeneView report of SNPs | |
| Link to Sequence Viewer | |
| Link to protein 3-D structure | |
| Link to OMIM database entry | |
| percentage of heterozygosity indicated by red arrow | |
| SNP is validated | |
| genotype information is available |
More information found at http://www.ncbi.nlm.nih.gov/corehtml/query/Snp/EntrezSNPlegend.html.
Viewing a single RefSNP
-
8In the second entry shown in Figure 1.19.4, the rs number (rs5030865) is hyperlinked. Click on the rs number to see the RefSNP record with detailed information about the SNP such as nucleotide variation, Human Gene Variation Society (HGVS) nomenclature as per the rules (Horaitis et al., 2007), locations on different assemblies, locations on the gene and mRNA, submitted SNPs (ss records) in this reference record, FASTA sequence adjacent to the SNP and, if submitted, information about population diversity, hetrozygosity and validation.In this example (Figure 1.19.5), the HGVS Names column reports the nucleotide variation (G to T, A or C) and its location on the Reference Sequence (RefSeq) gene records (NG_000853.3, NG_003180.2 and NG_008376.1), RefSeq mRNA records (NM_000106.4 and NM_001025161.1), genomic contig record (NT_011520.12), and the corresponding change in the protein record (NP_000097.2) (Pruitt et al., 2007). For more details about the RefSeq database refer to the Commentary section.More information about the clinical association of the SNP can be obtained from the Variation Viewer (VarView) and OMIM links on this page.
-
9Scroll down to see the GeneView section of the page reports actual variation, the codon(s) and their locations (Figure 1.19.6).Again, in this record, the nucleotide changes are from G to A, C or T as shown under the Allele change column in the mRNA panel. This view gives the position of the nucleotide on the chromosome (22, 42525035) in the GRCh37 assembly, accession number of the contig and location on the contig (NT_011520.12, 21915604), name of the gene (CYP2D6), RefSeq entry for the gene record and location on the record (NG_008376, 5849), RefSeq mRNA record accession number and position (NM_000106.4, 595), and the RefSeq protein record accession number and the amino acid position (NP_000097.2, 169). The gene is present on the minus strand as indicated by the arrows pointing towards the left in the Sequence Viewer region. The assembly GRCh37 panel provides information for the plus strand sequence of chromosome 22. Hence, the nucleotide in the Allele column of the Assembly panel is C, a complementary nucleotide G is present in the Allele column in the RefSeqGene panel and in the AlleleChange column in the mRNA panel. Also, a “-’ appears in the SNP to mRNA column. Thus, the G to A, C or T change on the NM_000106.4 at position 595 leads to a change in the 169th amino acid in NP_000097.2 from glycine to arginine, arginine or stop, respectively.
Figure 1.19.5.
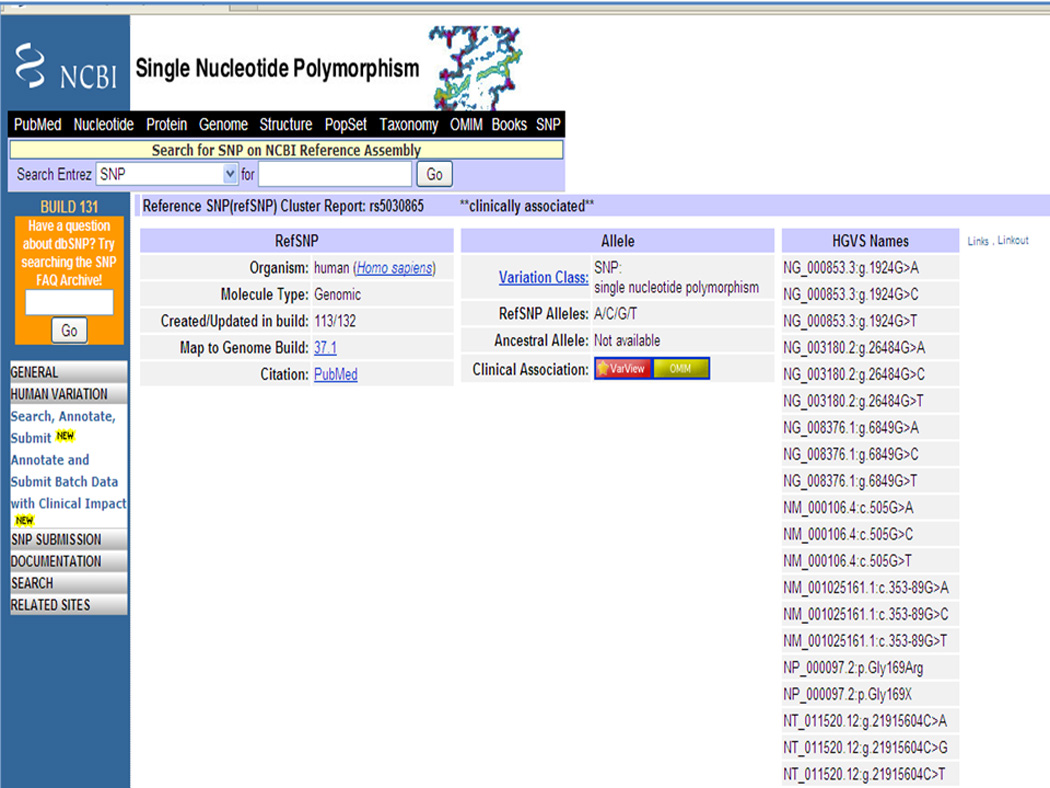
Top portion of the rs5030865 entry displaying the SNP build 131, allele A/C/G/T and HGVS names. More information about the clinical association of the SNP can be obtained from the Variation Viewer (VarView) and OMIM links on this page.
Figure 1.19.6.
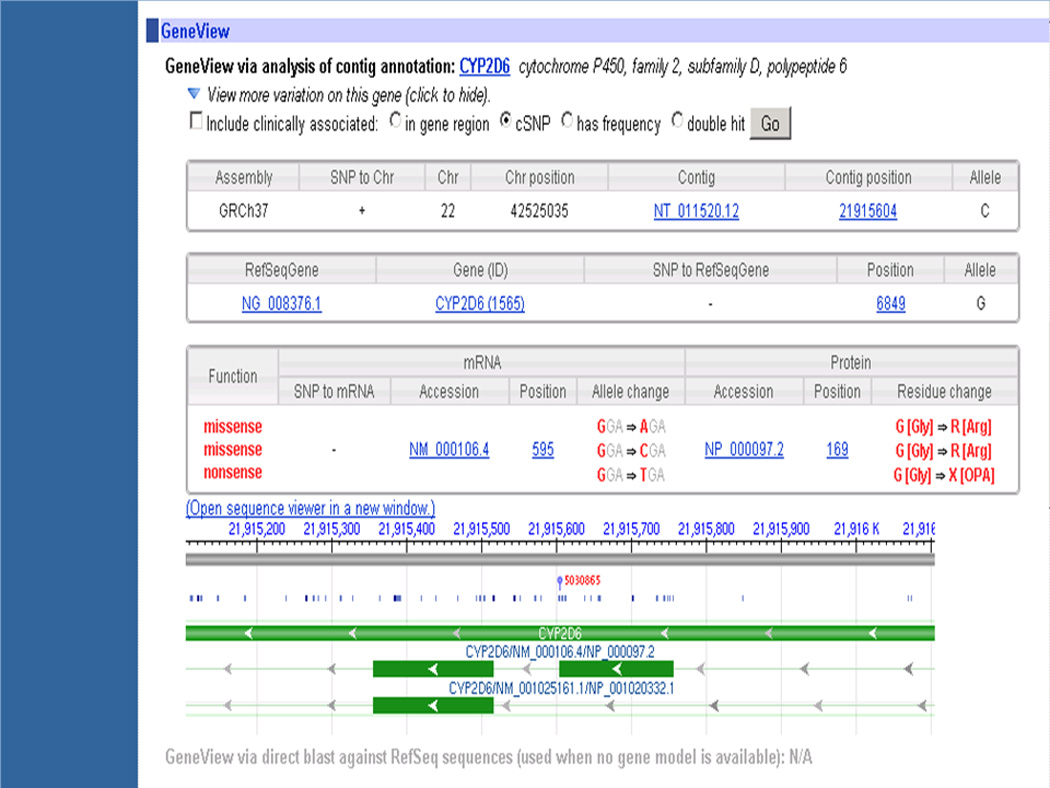
GeneView section of the rs5030865 entry. "See text for details."
Finding all SNPs in a gene
-
10Select the radio button next to “in gene region” under the title “View more variations on this gene” and then click on the Go button to get a list of all SNPs annotated on the gene CYP2D6.This page lists all reported SNPs in the gene region (2000 bases on the 5’-end and 300 bases on the 3’-end of the annotated gene). The default page lists their locations on one of the annotated mRNA variants and corresponding proteins with respect to one of the assemblies. In Figure 1.19.7, there are three assemblies listed in the column Contig Label: GRCh37 is the human assembly by the Genome Reference Consortium http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/index.shtml; the HuRef assembly represents a composite haploid version of the diploid genome sequence from a single individual (Levy et al., 2007); and Celera is the assembly submitted by the Celera Company (Venter et al., 2001). For more details about the human genome assembly refer to the Commentary section.
-
11Scroll down to the next section of the CYP2D6 SNPs annotations page (step 10), which shows the details about individual SNPs such as their location with respect to the chromosome, gene, mRNA and protein, type of the change (function class), validation and heterozygosity information (Figure 1.19.8).Each function class or location is color coded: near the 3’ or 5’ end (white), intron (yellow), non-synonymous nonsense and missense (red), synonomus (green) and frame-shift (blue). Validation status is indicated by different icons as described in http://www.ncbi.nlm.nih.gov/projects/SNP/snp_legend.cgi?legend=validation.In Figure 1.19.8, the first three red colored rows describe SNP rs61731577 (dbSNP rs# cluster id column). The nucleotide T (dbSNP allele column and Function contig reference listed in row 3) at position 42522600 (Chr. Position column) on chromosome 22 and 1560 on NM_000106 (mRNA pos column) is either reported to be G (dbSNP allele column and Function nonsense listed in row 2) or A (dbSNP allele column and Function nonsense listed in row 1). These variations lead to a change in the 3rd nucleotide (Codon pos column) of the codon for the 490th amino acid (Amino acid pos column). These variations cause the amino acid to change from Tyr[Y] to termination xxx[X] (Protein residue column) and are thus nonsense (Function column) variations. The next two green rows describe a synonymous SNP rs28371736 from nucleotide C at chromosome position 42522621 and mRNA position 1539 to T leading to a change in the 3rd nucleotide of codon 483. This nucleotide variation still encodes for the same amino acid Phe[F]. There is a link to the 3-D structure of the protein, for both variations, (“yes” link in the column 3D) for viewing in NCBI’s application Cn3D (Wang et al., 2000).
Figure 1.19.7.
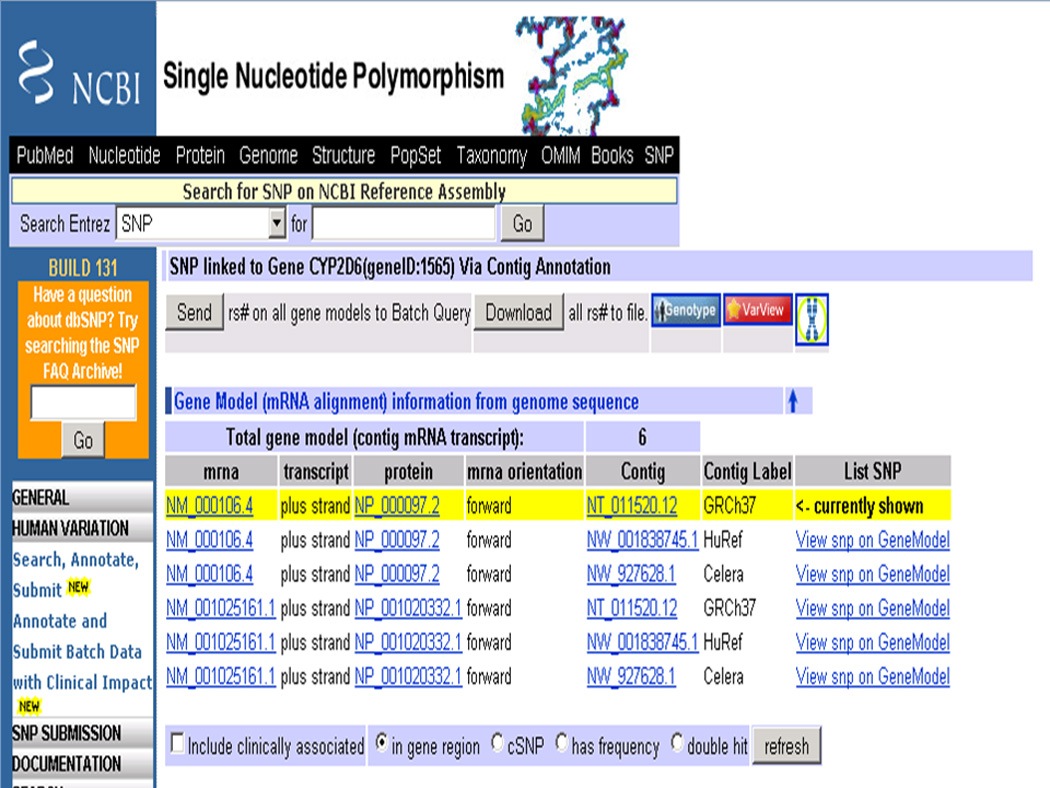
Top portion of the SNP GeneView report for the gene CYP2D6. Locations of SNPs as shown in Figure 1.19.8 are with respect to the transcript variant NM_000106 (and corresponding protein NP_000097) and the GRCh37 assembly. The user can change the assembly and/or the transcript by using the option “View snp on Gene model” in the List SNP column.
Figure 1.19.8.
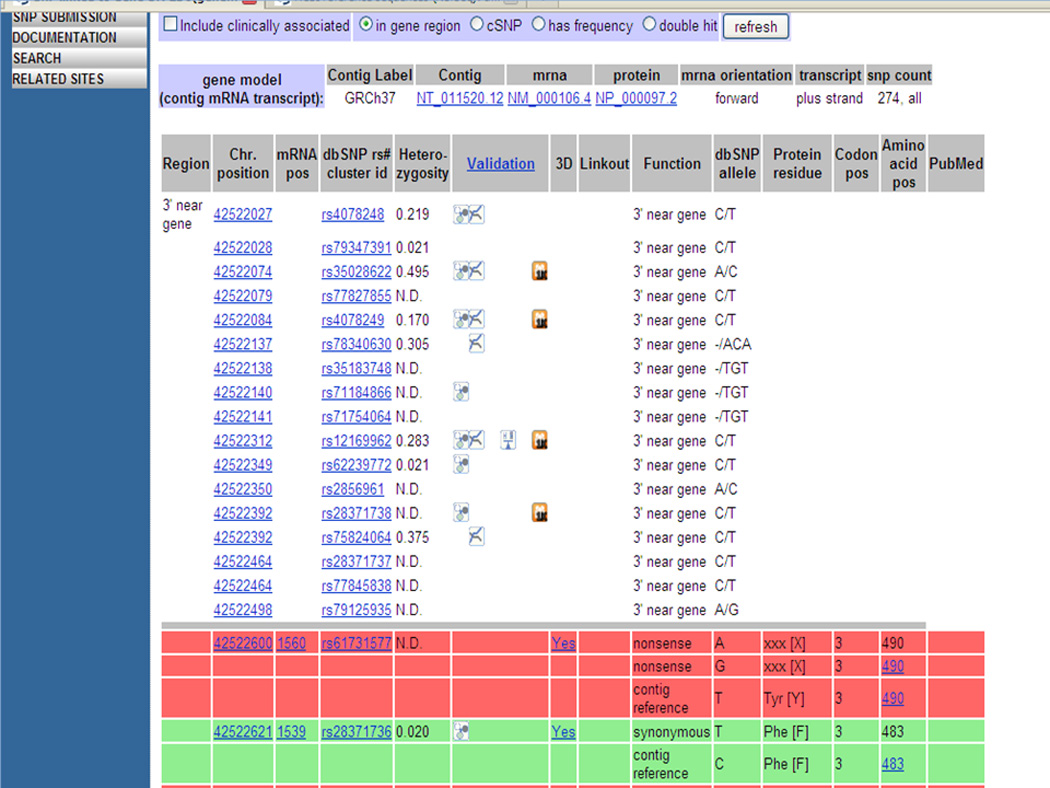
Portion of the SNP GeneView page displaying detailed information about each SNP in the gene CYP2D6. "See text for details."
Finding Clinically Associated SNPs
-
12In the same page, select the box next to “Include clinically associated” and click on the “refresh” button to obtain clinically associated SNPs.An extra column for “Clinically Associated” is added in the GeneView table and the SNPs which are clinically associated are indicated by the OMIM logo in that column (Figure 1.19.9). For example, in rs1135840, a variation leading to a change from threonine to serine at the 486th position of NP_000097.2 is reported to be clinically associated in the OMIM database. A click on the OMIM logo takes the user to the OMIM database for more information.
Figure 1.19.9.
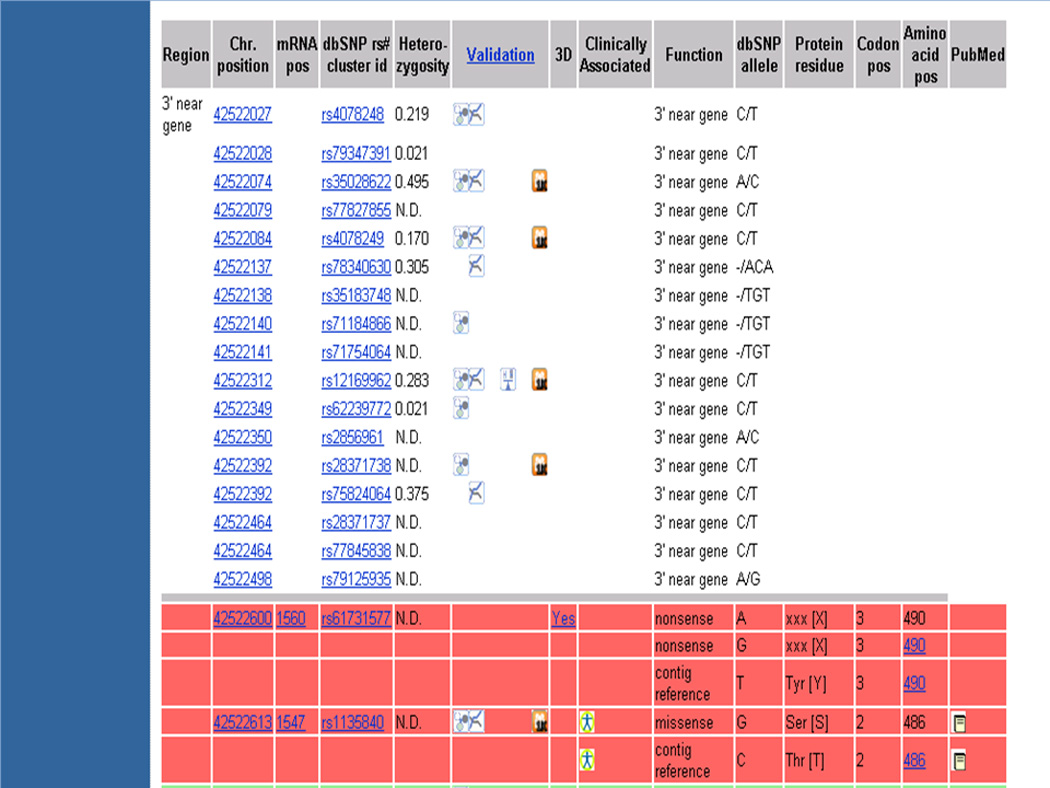
Results of the SNP GeneView page for the gene CYP2D6 including clinically associated SNPs. An extra column for “Clinically Associated” is added in the GeneView table compared to Figure 1.19.8 and the SNPs which are clinically associated are indicated by the OMIM logo in that column.
ALTERNATE PROTOCOL
SEARCHING dbSNP USING THE ENTREZ PREVIEW/INDEX SEARCH OPTION
This protocol describes how to use the Preview/Index page to search for all human non-synonymous SNPs cited in publications with known 3-D protein structures for the wild type amino acid, and SNPs associated with phenotype(s).
Searching dbSNP using the Preview/Index option
Go to NCBI web page http://www.ncbi.nlm.nih.gov/.
- Select the “SNP” database from the “All Databases” pull down menu (Figure 1.19.1).Alternatively, click on the “Variation” link in the green box on the left of the web page. This gives a list of variation- related resources at NCBI. Click on the link “Database of Single Nucleotide Polymorphisms (dbSNP)”.
Click on the “Preview/Index” tab.
From the All Fields pull down menu, select the Filter option (Figure 1.19.10).
Click on the Index button to obtain a list of searching criteria in the Filter menu (Figure 1.19.10).
Select the coding non-synonymous option and click on the AND button (Figure 1.19.11). This adds "coding nonsynonymous"[Filter] in the search box at the top of the page.
- In order to further restrict the search results to SNP entries that have links to the OMIM database, add the “snp omim” option from the Filter menu.The filter menu lists the possible criteria in an alphabetical manner. The “Down” button can be used to scroll down the list. An easy way to reach the criteria beginning with the words snp is to type “snp” in the box next to the Filter menu and click on the Index button.
-
Select the “snp omim” option and click on the AND button. The query in the search box at the top will change to "coding nonsynonymous"[Filter] AND "snp omim"[Filter].
To further restrict the search results to SNP entries that have links to the PubMed database, select the “snp pubmed cited” option and click on the AND button.
To further restrict the search results to SNP entries that have links to the structure database, select the “snp structure” option and click on the AND button.
To restrict search results to human as an organism, change the Filter menu to Organism, type human in the search box next to it and click on the AND button. The search box at the top of the page will have the following query built "coding nonsynonymous"[Filter] AND "snp omim"[Filter] AND "snp pubmed cited"[Filter] AND "snp structure"[Filter] AND human[Organism].
- Click on the Go button next to the query at the top of the page.The results page is similar to the one obtained in Basic Protocol 1. The advantage of the Preview/index page is that it can be used to build queries with Boolean operators NOT and OR as well. It can also be used to search in a range of parameters such as the publication dates. For example, "coding nonsynonymous"[Filter] NOT human[Organism] "2009 12 29":"2010 01 25" [Publication Date].More information about searching in dbSNP is available from the help document: http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=helpsnpfaq&part=Search
Figure 1.19.10.
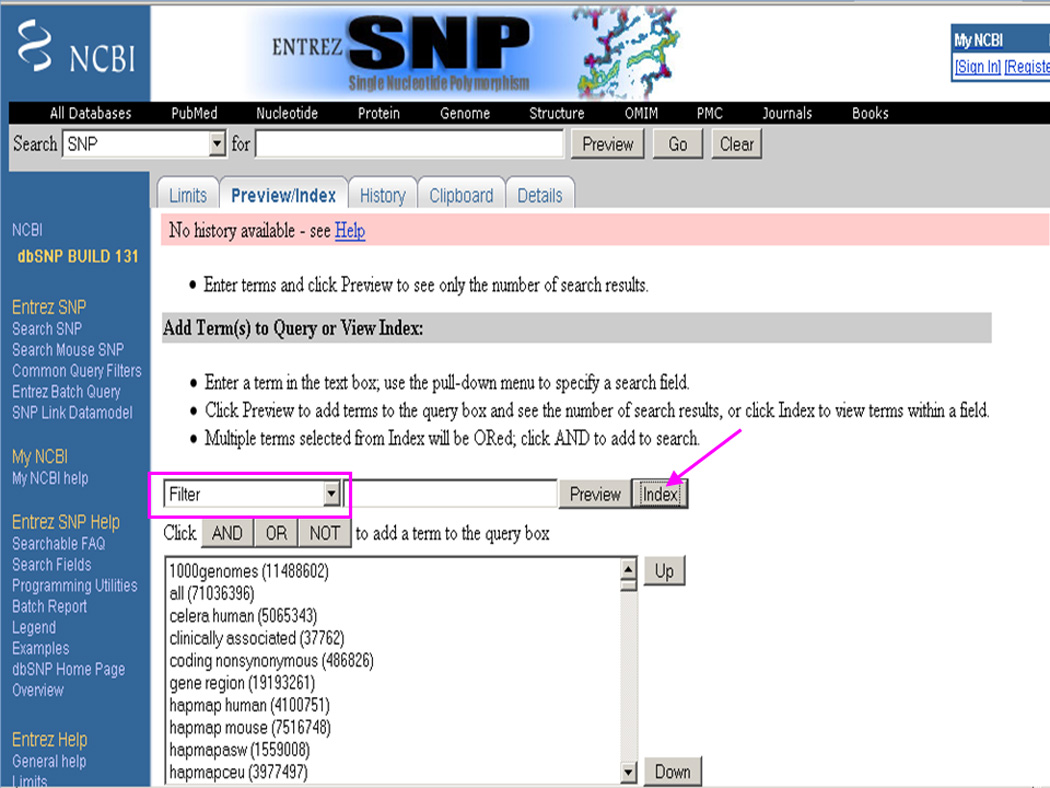
Accessing the list of criteria in the Filter menu in the Entrez dbSNP Preview/Index page. From the All Fields pull down menu, Filter option is selected and then the Index button is clicked to obtain a list of searching criteria in the Filter menu.
Figure 1.19.11.
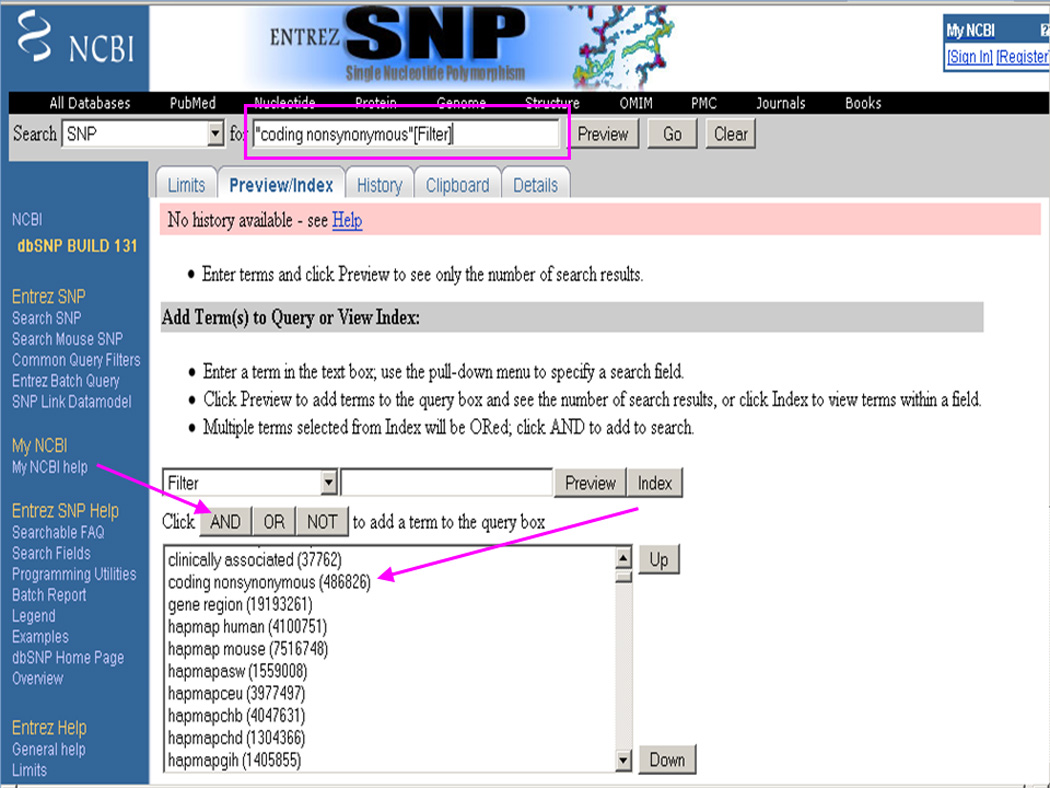
Building a query to search for the coding non-synonymous SNPs using the Filter menu in the Entrez dbSNP Preview/Index page and then clicking on the AND button. Note the query "coding nonsynonymous"[Filter] generated in the search box at the top of the page.
BASIC PROTOCOL 2
SEARCHING dbSNP USING A QUERY SEQUENCE
An alternative way to search SNPs is to start with a sequence instead of a term by using SNP BLAST. Basic Local Alignment Search Tool (BLAST) is a sequence comparison tool at NCBI (Altschul, Gish et al., 1990). SNP BLAST offers a way to compare a sequence with all sequences in dbSNP. Thus, this resource is useful in several ways, for example, to determine whether an SNP identified in a laboratory is already submitted to dbSNP or to identify all reported SNPs in a sequence.
Necessary Resources
Hardware
Computer with Internet access
Software
An up-to-date Web browser, such as Firefox, Internet Explorer, or Safari
We will take as an example an already submitted SNP rs1815739.
-
1
Go to NCBI web page http://www.ncbi.nlm.nih.gov/.
-
2
Select the “SNP” database from the “All Databases” pull down menu (Figure 1.19.1)
-
3Type rs1815739 in the query box and click on the Go button.CAUTION: if you have used the Limits page for searching earlier, uncheck the Limits box listed underneath the search bar.
-
4
Click on the rs1815739 link to view the full record (Figure 1.19.12).
-
5Click on the Fasta link as shown in Figure 1.19.12 to reach the sequence panel and copy the sequence (Figure 1.19.13).The SNP is C/T at position 251 as reported in the definition line as alleles=’C/T’ and allelePos=251, respectively.>gnl|dbSNP|rs1815739|allelePos=251|totalLen=501|taxid=9606|snpclass=1|alleles='C/T'|mol=Genomic|build=131SNP variations are encoded using IUPAC notation in the BLAST database (Units 3.3 and 3.4). IUPAC nomenclature is described at http://www.ncbi.nlm.nih.gov/SNP/iupac.html. Thus, this SNP is reported as “Y” in the FASTA sequence (see Unit 3.9).
-
6Go to the dbSNP home page http://www.ncbi.nlm.nih.gov/projects/SNP/Note that this page is different from the Entrez SNP page used in Basic Protocol 1. This page can also be accessed from the Entrez SNP page shown in Figure 1.19.2 by clicking on the “dbSNP Home Page” link in the side blue bar.
-
7
Click on the Search link in the dark blue column to the left.
-
8Click on the BLAST SNP link (Figure 1.19.14).Select the “No” radio button next to “Use megablast” to use the blastn program for a more relaxed/broader search (Figure 1.19.15). By default, this program uses megablast (see Unit 3.3; Morgulis et al., 2008; Baxevanis and Ouellette, 2001; Stover and Cavalcanti, 2009).
-
9
Select human as the organism from the list under “Choose a snp blast database” (Figure 1.19.15).
-
10
Paste the rs1815739 sequence in the FASTA format (see Unit 3.9), obtained as described above (steps 1–5), in the box under “Query Sequence” (Figure 1.19.15).
-
11
Click on the “Submit Query” button.
-
12
In the new page, click on the “View report” button.
Figure 1.19.12.
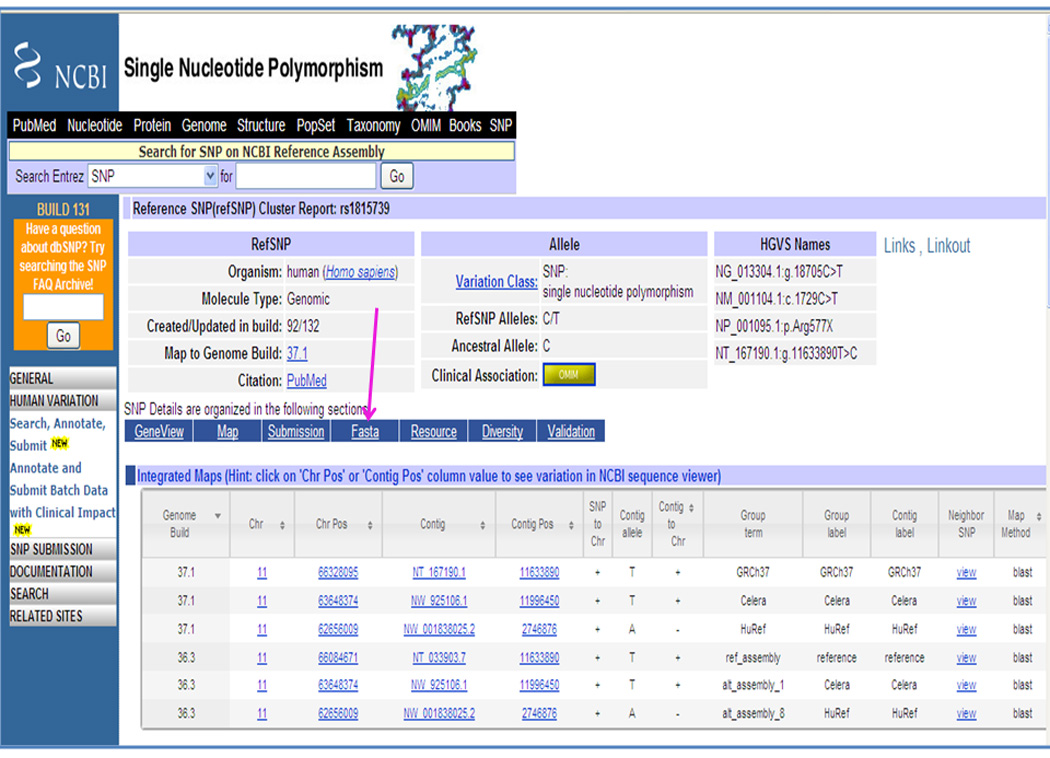
Top portion of the rs1815739 record in dbSNP. Click on the Fasta link as shown by an arrow to reach the portion of the page where sequence is listed as displayed in Figure 1.19.13.
Figure 1.19.13.

FASTA sequence of the entry rs1815739 obtained by clicking on the Fasta link shown in Figure 1.19.12.
Figure 1.19.14.
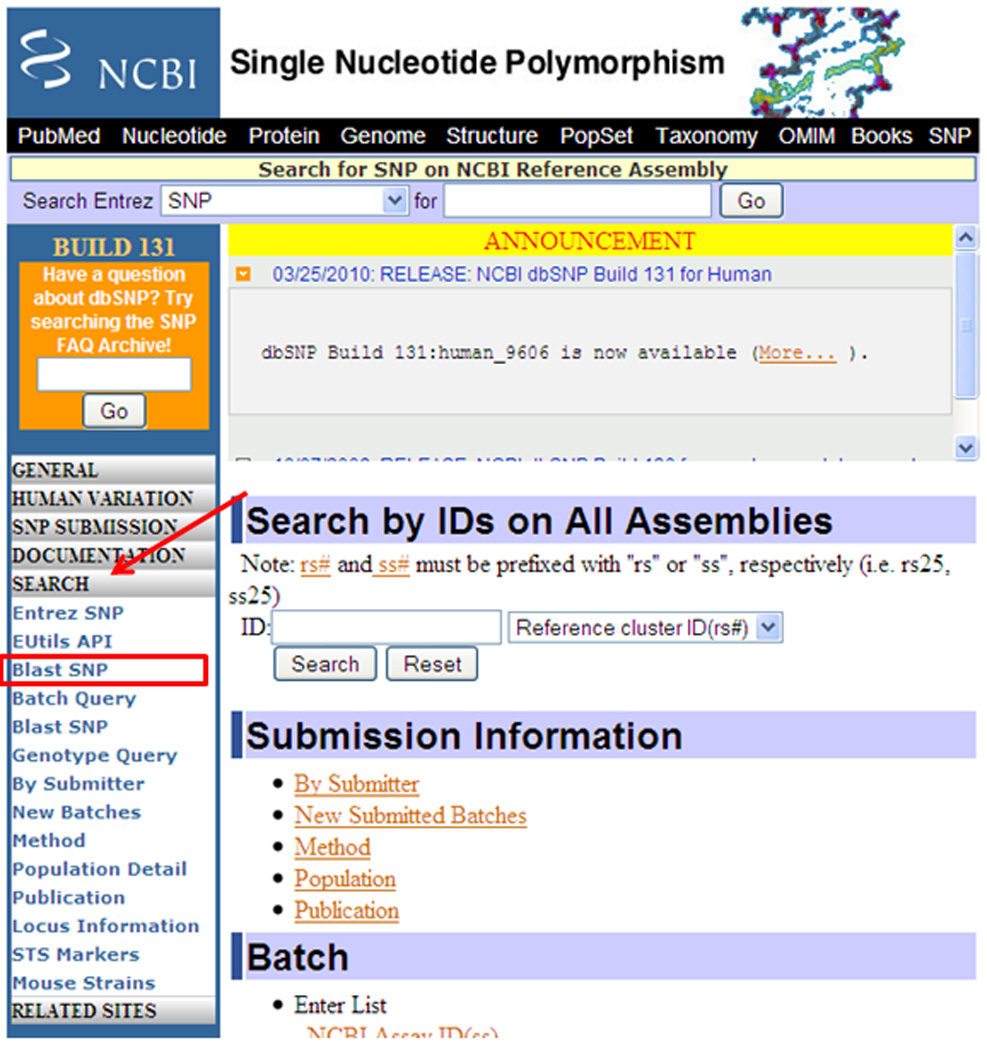
BLAST SNP link from the dbSNP home page. Click on the Search link in the blue bar on the dbSNP home page, shown by an arrow, to visualize the search options. One of them is BLAST SNP.
Figure 1.19.15.
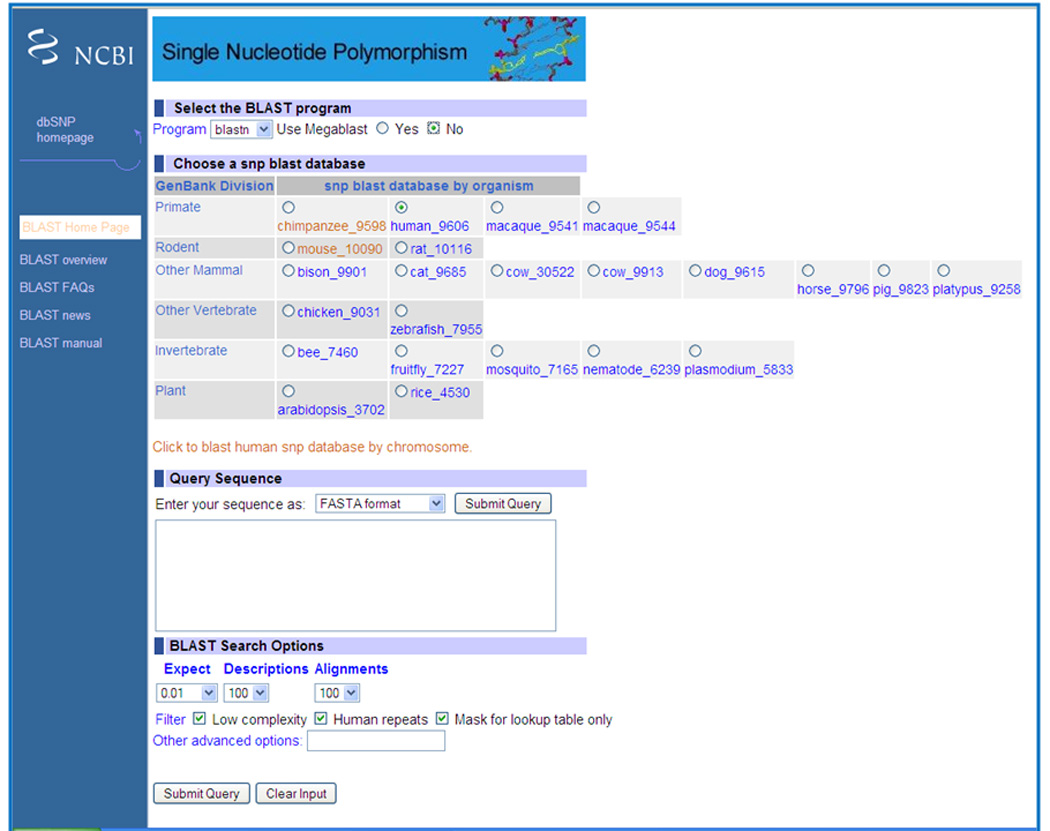
SNP BLAST query set up page. Select the “No” radio button next to “Use megablast” to use the blastn program.
Understanding the results
-
13Scroll down to the alignments panel. Figure 1.19.16 shows the alignment of the first hit from the results; it is the self hit of the query to the rs1815739 entry from the database labeled Sbjct in the alignment.The 100% match as shown next to “Identities” demonstrates that the SNP is already present in dbSNP. Note the nucleotide at position 251 in the alignment is Y indicating the nucleotide at this position is either a C or T. If the query sequence has any other nucleotide (A or G) at this position then the SNP is a novel SNP.
Figure 1.19.16.

First alignment in the results of the SNP BLAST search for query rs1815739. The 100% match as shown next to “Identities” demonstrates that the SNP is already present in dbSNP. Note the nucleotide at position 251 in the alignment is Y indicating the nucleotide at this position is either a C or T.
COMMENTARY
Background Information
Submitted information
dbSNP includes information provided by the submitters and also the information computed by NCBI based on the submitted information. The main component of the submitted information is of course the sequence itself. Minimum requirement for the total length of the submitted sequence is 100 nucleotides to ensure an adequate sequence for accurate mapping of the variation on the reference genome sequence. The sequence may include that obtained during variation assays and the one from the published sequence. The minimum requirement for the flanking sequence on the 5’end and 3’end of the variation is 25 nucleotides each. To meet the minimum requirement for the total length, the sequence can be obtained from the sequence databases. Each submitter provides information about the allele as G, A, T, or C and not in the IUPAC code and also provides information about techniques (method) used to assay variation. Other information such as population, frequency and genotype is optional.
Computed information
dbSNP computes additional information from the submitted data in the process called the “build”. The sequence data is aligned to the most recent genome assembly in order to compute additional information. Updates to the builds are released periodically, especially after the release of a new genome assembly. Computed information includes RefSNP clusters, location of the variation on the genome, mRNA, protein and 3-D structure, variation functional class, population diversity and links to other databases at NCBI.
RefSNP clustering and orientation
When a RefSNPs entry (rs) is generated, the submitted entries (ss) aligning to the same position on the genome are clustered. The ss entry with the longest flanking sequence is called the "RefSNP exemplar" and its flanking sequence and orientation is assigned to the rs record. If in the later builds, a new ss record with a longer flanking sequence has been added then it becomes the new exemplar sequence. However, the orientation of the RefSNP flanking sequence is not altered but the sequence of the new exemplar is reversed.
Types of SNPs
Since the genetic code is redundant different codons can code for the same amino acid. Thus, some nucleotide variations can still code for the same amino acid even if the codon is different, such variations are called synonymous SNPs in dbSNP. However, some variations encode a different amino acid, such variations are called non-synonymous SNPs in dbSNP. As mentioned above, non-synonymous changes are further classified as missense, nonsense and frame-shift. For example, by the standard genetic code (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=tgencodes#SG1), the codon TGT codes for amino acid cysteine. The substitution T/C at the third nucleotide position represents a synonymous change coding for the same amino acid cysteine. However, the substitution T/G at the same position represents a non-synonymous missense change coding for a different amino acid, tryptophan, and the substitution T/A at the same position represents a non-synonymous nonsense change resulting in a stop codon.
RefSeq Database
NCBI generates reference nucleotide sequences (RefSeq) from the submitted sequences. GenBank is a repository of submitted sequences and is a redundant database, accepting submissions from multiple sources, even if they are in the same region. RefSeq provides one sequence entry per molecule such as the genome, mRNA and protein and is, thus, a non-redundant database (Baxevanis and Ouellette, 2001). It has a characteristic accession number format that includes two letters, an underscore character and six or eight digits. The prefixes for RefSeq entries used in this chapter are:
NT_ genome contig sequence (contiguous sequence generated during the genome assembly)
NM_ mRNA
NP_ protein
More accession number prefixes can be found at the RefSeq web page: http://www.ncbi.nlm.nih.gov/refseq/key.html#accessions
Additional Searching Options
Additional dbSNP searching options such as submitter information, assay method, population, or the location between two markers can be found at http://www.ncbi.nlm.nih.gov/projects/SNP/. This page also lists options to search the dbSNP database using a large number of ss or rs accession numbers. SNPs with associated genotype information can be searched from http://www.ncbi.nlm.nih.gov/projects/SNP/snp_gf.cgi. SNPs between different mouse strains can be searched from http://www.ncbi.nlm.nih.gov/projects/SNP/MouseSNP.cgi. To automate a large number of Entrez dbSNP searching and retrieving tasks within software applications, NCBI’s Entrez Programming Utilities (eUtils) can be used. More information is found at http://www.ncbi.nlm.nih.gov/SNP/SNPeutils.htm. The entire dbSNP database is available at ftp://ftp.ncbi.nih.gov/snp/.
Other SNP-related databases
Below is a list of some additional databases/resources for obtaining information about SNPs. This list is not comprehensive and one of the ways to identify more of such resources is from the database issues published by Nucleic Acids Research.
GeneSNPs Database at http://www.genome.utah.edu/genesnps/ This is the database of SNPs identified in the genes that are considered to be susceptible to environmental exposure as a part of the Environmental Genome Project http://www.niehs.nih.gov/research/supported/programs/egp/ at the National Institutes of Environmental Health Sciences (NIEHS).
-
International HapMap Project at http://hapmap.ncbi.nlm.nih.gov/
The International HapMap Project is an international collaboration to generate a haplotype map (HapMap) of the human genome from different populations. The data can be visualized using a genome browser provided on the web page and also can be downloaded in bulk.
-
Human Genome Variation Genotype-to-Phenotype database, (HGVbaseG2P) at www.hgvbaseg2p.org
This database is a compilation of summary level findings from human genetic association studies such as obtained from Genome Wide Association Studies (GWAS) and sets submitted by individual laboratories. It also provides web-based tools for browsing, visualization and mining.
-
The Genome Variation Server (GVS): http://gvs.gs.washington.edu/GVS/
This database is hosted by the SeattleSNPs which is a part of the National Heart Lung and Blood Institute's (NHLBI) Programs for Genomic Applications (PGA). It includes data in dbSNP and HapMap database, and provides analysis tools such as linkage disequilibrium plots, tag SNPs, and merging populations.
-
Pharmacogenomics Knowledge Base (PharmGKB): http://www.pharmgkb.org/
This resource includes curated information and visualization tools for genes and variants associated with drug response.
Troubleshooting
Sometimes the variation nucleotide sequences between the ss and rs records may not match. In addition, these further may not match the genome or gene sequence. In such cases, check for the orientations between the following: the ss and rs flanking sequences, the rs flanking and contig sequences, the rs flanking and genome sequences and the gene/mRNA and genome sequences. For example, the flanking sequence of an ss record may be in the reverse orientation with respect to that of its rs record or the gene may be placed on the minus strand of the genome.
Footnotes
Internet Resources
http://www.ncbi.nlm.nih.gov/ NCBI home page
http://www.ncbi.nlm.nih.gov/sites/entrez?db=snp NCBI Entrez SNP page
http://www.ncbi.nlm.nih.gov/guide/variation/ NCBI Variation Databases
http://www.ncbi.nlm.nih.gov/projects/SNP/buildhistory.cgi dbSNP build history page
http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=handbook&part=ch5 Kitts, A. and Sherry S. 2009 The Single Nucleotide Polymorphism Database (dbSNP) of Nucleotide Sequence Variation. The NCBI Handbook, Chapter 5. National Center for Biotechnology Information, Bethesda, Md.
http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=helpentrez&part=EntrezHelp NCBI Entrez help document
http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=helpsnpfaq&part=Search dbSNP search help document
http://www.ncbi.nlm.nih.gov/snp dbSNP search fields
http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=tgencodes Genetic codes at the NCBI Taxonomy database
http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/index.shtml Genome Reference Consortium
http://www.ncbi.nlm.nih.gov/projects/SNP/snp_legend.cgi?legend=validation dbSNP validation legend
http://www.ncbi.nlm.nih.gov/corehtml/query/Snp/EntrezSNPlegend.html Entrez SNP figure legends
http://www.ncbi.nlm.nih.gov/SNP/iupac.html IUPAC nomenclature code at dbSNP
http://www.ncbi.nlm.nih.gov/projects/SNP/snp_blastByOrg.cgi SNP BLAST page
http://www.ncbi.nlm.nih.gov/projects/SNP/ Additional dbSNP searching options
http://www.ncbi.nlm.nih.gov/projects/SNP/snp_gf.cgi Genotype query page at dbSNP
http://www.genome.utah.edu/genesnps/ Gene SNP database
http://hapmap.ncbi.nlm.nih.gov/ International HapMap Project
www.hgvbaseg2p.org Human Genome Variation Genotype-to-Phenotype database, (HGVbaseG2P)
http://gvs.gs.washington.edu/GVS/ The Genome Variation Server
http://www.pharmgkb.org/ Pharmacogenomics Knowledge Base (PharmGKB)
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baxevanis AD, Ouellette BFF. Bioinformatics: a practical guide to the analysis of genes and proteins. 2nd ed. N.Y: John Wiley and Sons; 2001. [Google Scholar]
- Feero WG, Guttmacher AE, Collins FS. Genomic medicine--an updated primer. N Engl J Med. 2010;362(21):2001–2011. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- Guttmacher AE, Collins FS. Genomic medicine--a primer. N Engl J Med. 2002;347(19):1512–1520. doi: 10.1056/NEJMra012240. [DOI] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30(1):52–54. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horaitis O, Talbot CC, Jr, Phommarinh M, Phillips KM, Cotton RG. A database of locus-specific databases. Nature Genetics. 2007;39(4):425. doi: 10.1038/ng0407-425. [DOI] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, Lin Y, MacDonald JR, Pang AW, Shago M, Stockwell TB, Tsiamouri A, Bafna V, Bansal V, Kravitz SA, Busam DA, Beeson KY, McIntosh TC, Remington KA, Abril JF, Gill J, Borman J, Rogers YH, Frazier ME, Scherer SW, Strausberg RL, Venter JC. The diploid genome sequence of an individual human. PLoS Biol. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schaffer AA. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24(16):1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(Database issue):D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Mizrachi I, Ostell J, Panchenko A, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang Y, John Wilbur W, Yaschenko E, Ye J. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38(Database issue):D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover NA, Cavalcanti ARO. Using NCBI BLAST. Curr. Protoc. Essential. Biol. Tech. Unit 11.1 [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends in Biochemical Sciences. 2000;25(6):300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]


